Abstract
The African clawed frog Xenopus laevis has been instrumental to investigations of both development and cell biology, but the utility of this model organism for genetic and proteomic studies is limited by its long generation time and unsequenced pseudotetraploid genome. Xenopus tropicalis, which is a small, faster-breeding relative of X. laevis, has recently been adopted for research in developmental genetics and functional genomics, and has been chosen for genome sequencing. We show that X. tropicalis egg extracts reconstitute the fundamental cell cycle events of nuclear formation and bipolar spindle assembly around exogenously added sperm nuclei. Interestingly, X. tropicalis spindles were ∼30% shorter than X. laevis spindles, and mixing experiments revealed a dynamic, dose-dependent regulation of spindle size by cytoplasmic factors. Measurements of microtubule dynamics revealed that microtubules polymerized slower in X. tropicalis extracts compared to X. laevis, but that this difference is unlikely to account for differences in spindle size. Thus, in addition to expanding the range of developmental and cell biological experiments, the use of X. tropicalis provides novel insight into the complex mechanisms that govern spindle morphogenesis.
Introduction
The African clawed frog Xenopus laevis has been instrumental in studies of gene regulation and development (Brown, 2004), and in the past two decades, unfertilized eggs have been used to prepare cell-free extracts that have made important contributions to our understanding of cell cycle regulation (Murray, 1991), DNA replication (Dasso and Newport, 1990), and spindle assembly (Desai et al., 1999). However, X. laevis suffers from a long generation time that makes forward genetics prohibitively time consuming, as well as a pseudotetraploid genome that would obscure many phenotypes. The lack of genomic information has limited homology-based searches to existing EST libraries and complicated protein identification by mass spectrometry. Xenopus tropicalis, which is a small, faster breeding, diploid relative of X. laevis, has recently been adopted for research in developmental genetics and functional genomics, and it is undergoing genome sequencing (Hirsch et al., 2002; Klein et al., 2002). We investigated whether X. tropicalis egg extracts could be used for in vitro cell biology experiments and found that they could similarly reconstitute the fundamental cell cycle events of nuclear formation and bipolar spindle assembly around exogenously added sperm nuclei. X. laevis antibodies revealed similar staining patterns on X. tropicalis spindles and precipitated homologous proteins detected as single isoforms by Western blot, in contrast to the multiple X. laevis variants. X. tropicalis spindles were significantly smaller than X. laevis spindles formed around the same chromosome source. Extract-mixing experiments revealed the presence of cytoplasmic factors that regulate spindle size in a dynamic and dose-dependent fashion. Measurement of individual microtubule dynamics and spindle poleward flux rates did not reveal differences likely to account for the observed changes in spindle size. We propose that microtubule regulatory factors in X. tropicalis extracts respond differently to stabilizing agents, such as chromosomes, to generate smaller microtubule structures. Thus, X. tropicalis eggs can be used in the same types of assays previously established for X. laevis, benefit from more molecular and genetic tools, and be used to investigate interesting cellular scaling phenomena, such as spindle size regulation.
Results and discussion
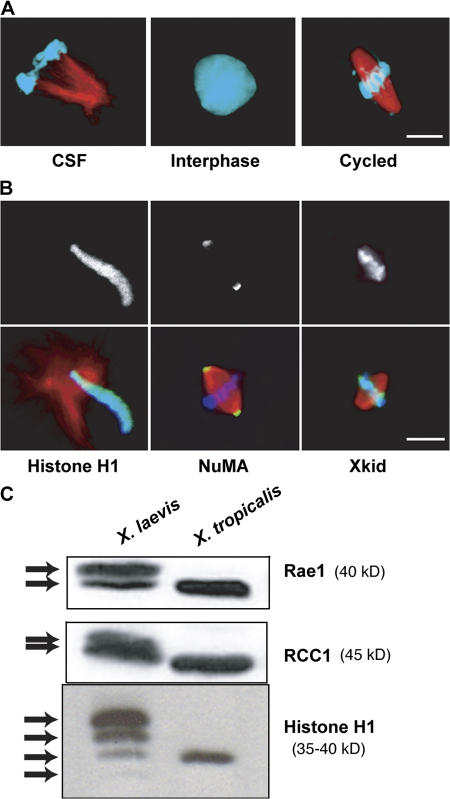
We set out to evaluate the utility of X. tropicalis for in vitro cell biological and biochemical investigations. X. tropicalis eggs (∼0.6 mm diam) are approximately one fifth the volume of those of X. laevis (1.2 mm diam). To test whether X. tropicalis eggs could be used to prepare functional cellular extracts, we collected, dejellied, and crushed unfertilized eggs, which, like those of X. laevis, are arrested in metaphase of meiosis II by cytostatic factor (CSF) activity (Murray, 1991). Metaphase-arrested X. tropicalis egg extracts assembled spindle structures around exogenously added sperm nuclei, entered interphase, and replicated DNA when released from the arrest, and then cycled back into mitosis (Fig. 1 A). Although yields of extract per frog were 10–20% that of X. laevis, X. tropicalis egg extracts effectively recapitulated cell cycle events in vitro.
Figure 1.
X. tropicalis egg extracts recapitulate major cell cycle events in vitro. (A) CSF-arrested egg extracts prepared from eggs of X. tropicalis and supplemented with X. laevis sperm nuclei and X-rhodamine–labeled tubulin formed “half spindles” in metaphase-arrested extracts, interphase nuclei after release from CSF arrest, and bipolar spindles when induced to reenter metaphase. Microtubules are red and DNA is blue. (B) X. tropicalis extract reactions were stained with fluorescently labeled antibody (α-H1 and α-NuMA), or by immunofluorescence (α-Xkid) using antibodies raised to the X. laevis proteins. Staining patterns recapitulated those in X. laevis reactions. In overlays, the stained protein is green. (C) CSF extracts pooled from eggs of multiple X. laevis or X. tropicalis frogs were blotted for Rae1, RCC1, and histone H1. In X. laevis extracts, all proteins gave multiple bands, presumably because of multiple genes for each protein, whereas X. tropicalis contained single isoforms. Bars, 10 μm.
The utility of X. tropicalis extracts would be maximized if reagents generated for X. laevis could be applied. Fluorescence microscopy revealed that antibodies against X. laevis histone H1 (chromatin component; Maresca et al., 2005a), nuclear mitotic apparatus protein (NuMA; spindle pole component; Merdes et al., 1996), and Xenopus kinesin-like DNA-binding protein (Xkid; Funabiki and Murray, 2000) gave identical staining patterns in X. tropicalis extracts, compared with X. laevis (Fig. 1 B and not depicted). Furthermore, the addition of an inhibitory antibody to Xkid resulted in chromosome congression defects in X. tropicalis spindle reactions that were very similar to those observed in X. laevis (unpublished data), suggesting that many reagents will be useful in both species. Because X. laevis has a pseudotetraploid genome, many genes are present in multiple copies, and without selective pressure, some may be expressed but may not be functional, like one of the Vg1 isoforms (Birsoy et al., 2005). Whether or not the isoforms are functional, there is frequently more than one. X. laevis Rae1 (mRNA export factor/spindle regulator; Blower et al., 2005), RCC1 (guanine exchange factor for Ran; Nishitani et al., 1990), and histone H1 were represented by multiple bands on a Western blot, whereas X. tropicalis contained a single band for each protein (Fig. 1 C). This suggests that the pseudotetraploid genotype of X. laevis contributes to the complexity of the proteome, and the use of X. tropicalis could simplify protein analysis.
X. laevis biochemistry is not underpinned by genomic information, making identification of proteins by mass spectrometry difficult. To test whether X. laevis proteins could be identified using the X. tropicalis database, we immunoprecipitated the microtubule-associated developmental regulator Xenopus nuclear factor 7 (Xnf7; Maresca et al., 2005b) from both X. laevis and X. tropicalis extracts (Fig. 2 A), and then used MALDI mass spectrometry to identify each protein using databases from both species. Xnf7 was identified from both immunoprecipitates using either database (Fig. 2 B), although the number of peptides identified was higher when queried against the database of the same species. Conceptual trypsin digestion of Xnf7 from both species and comparison of the peptides revealed that although the two proteins are highly conserved (Fig. 2 C), only 45% of the peptides have identical masses (not depicted). This analysis suggests that although the X. tropicalis database will greatly facilitate the identification of X. laevis proteins by mass spectrometry, it will be more efficient to identify homologous X. tropicalis proteins.
Figure 2.
Identification of X. tropicalis and X. laevis Xnf7 by mass spectrometry. (A) Xnf7 was immunoprecipitated from X. laevis or X. tropicalis extracts and subjected to SDS-PAGE. Excised bands were processed for MALDI-TOF mass spectrometry. (B) Xnf7-derived peptide masses from each band were compared against databases from both Xenopus species. The number of peptides that showed a perfect match against each database is shown. (C) Schematic representation of the Xnf7 domain structure showing the number of identical/conserved amino acids in each domain. Domains are in regular font and linker regions are in italics.
While validating the use of X. tropicalis extracts, we noticed that spindles assembled around X. laevis sperm nuclei in X. tropicalis extracts were considerably smaller (∼30% shorter) than those assembled in X. laevis extract (Fig. 3 A). Comparing the fluorescence area of the two types of spindles revealed an approximately threefold difference (unpublished data), indicating a substantially greater microtubule mass in X. laevis spindles compared with those of X. tropicalis. This prompted us to examine the poorly understood phenomenon of spindle scaling further.
Figure 3.
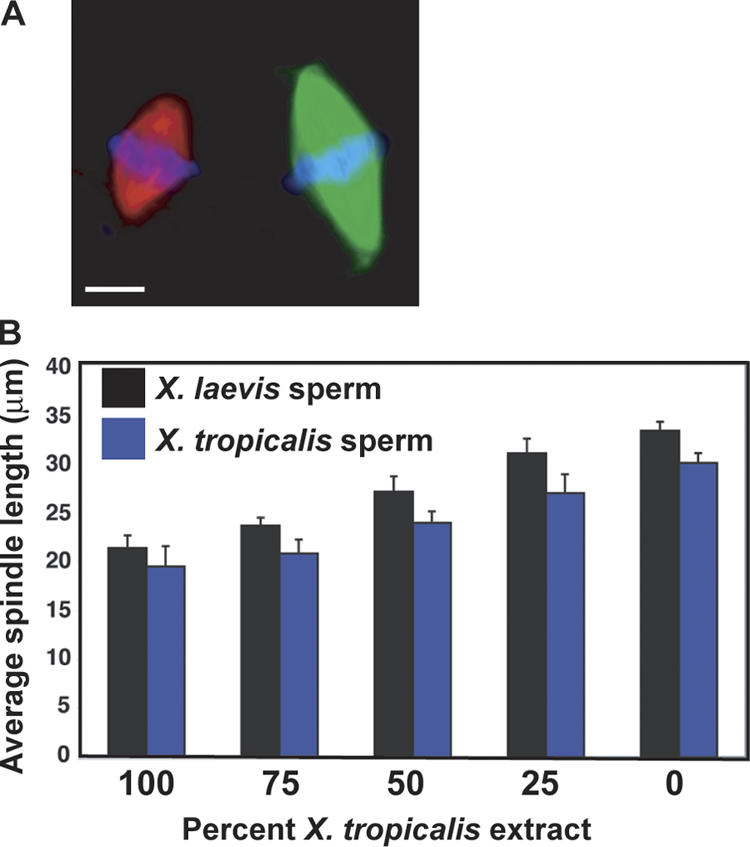
Comparison of spindle length between X. tropicalis and X. laevis. (A) Spindles assembled around X. laevis sperm nuclei in either X. laevis or X. tropicalis egg extracts were visualized using Hoechst dye (blue, DNA) and the incorporation of X-rhodamine tubulin (red microtubules, X. tropicalis), or Alexa Fluor 488 tubulin (green microtubules, X. laevis). Bar, 10 μm. (B) Mixed reactions with the indicated proportion of X. tropicalis extract were combined with X. laevis or X. tropicalis sperm nuclei. Spindle length was measured from pole to pole. A linear relationship was observed between the proportion of X. laevis extract present and spindle length. Error bars are the SD.
Spindle sizes were extract dependent, suggesting that they may be defined by diffusible cytoplasmic components. To determine whether these factors reflect a balance of activities present in both extracts, or a dominant activity in one of the extracts, we combined extracts in different proportions and examined spindle length in the mixed reactions. We found that spindle length had a direct and linear dependence on the proportion of X. laevis to X. tropicalis extract (Fig. 3 B), suggesting equilibrium behavior of cytoplasmic regulatory activities. Previous work has also demonstrated a role for chromatin mass in determining spindle size (Nicklas and Gordon, 1985). To investigate this phenomenon, we compared spindle length in X. laevis, X. tropicalis, and mixed extracts using sperm from X. tropicalis, whose diploid genome (1.7 × 109 bp) is ∼55% that of X. laevis (3 × 109 bp; Hirsch et al., 2002), and found that spindles assembled around X. tropicalis chromosomes were ∼10% shorter in all cases (Fig. 3 B). Therefore, we conclude that although chromatin mass does influence spindle length, soluble cytoplasmic factors are the major determinant in Xenopus egg extracts.
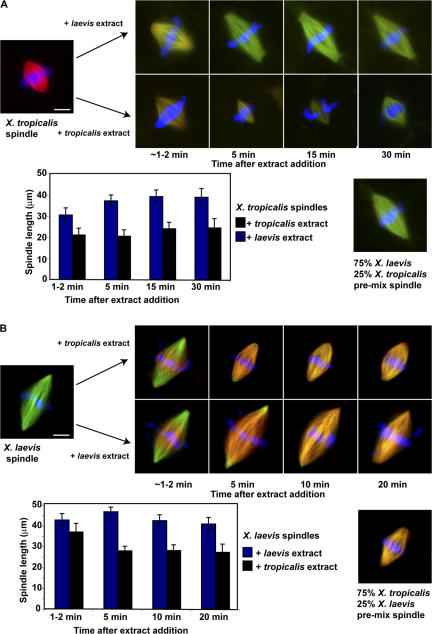
To examine whether spindle size regulation is a static or dynamic process, we added fresh extract to preassembled spindles. To X. tropicalis extracts containing spindles that had incorporated X-rhodamine–labeled tubulin, we added three volumes of either X. tropicalis or X. laevis extract containing Alexa Fluor 488 tubulin, and examined spindle length at various time points after mixing. Whereas X. tropicalis extract did not affect spindle length over the course of the experiment, addition of X. laevis extract caused rapid growth of X. tropicalis spindles, by ∼5 μm in length within 2 min, and to the size of premixed (75% X. laevis, 25% X. tropicalis) reactions within 5 min (Fig. 4 A). Reciprocally, the addition of X. tropicalis extract to preformed X. laevis spindles rapidly shrank them to the size of the premixed controls, whereas addition of X. laevis extract did not (Fig. 4 B). The added extract tubulin appeared to incorporate at the plus ends of growing microtubules in the central spindle (Fig. 4, A and B). These results demonstrate that soluble cytoplasmic factors dynamically govern spindle length in Xenopus extracts, in agreement with results obtained in Drosophila melanogaster cells (Goshima et al., 2005), and indicate that nonmicrotubule spindle matrix elements determining length, if they exist, cannot be purely static structures (Chang et al., 2004).
Figure 4.
Spindle length determination is highly dynamic in Xenopus extracts. (A) Spindles assembled in X. tropicalis extracts containing X-rhodamine tubulin were diluted with three volumes of X. laevis or X. tropicalis extract supplemented with Alexa Fluor 488 tubulin, and fixed for examination at various time points. Quantification of spindle length over time after mixing revealed steady-state spindle lengths were reached within ∼5 min, and they corresponded to those in the premixed control of 75% X. laevis 25% X. tropicalis extract. (B) The same experiment described in A, except that the spindles were assembled in X. laevis extract containing Alexa Fluor 488 tubulin, and diluted with X. tropicalis or X. laevis extract containing X-rhodamine tubulin. Quantification of spindle length over time after mixing is shown. Error bars are the SDs. Bars, 10 μm.
We found that chromatin bead spindles and asters induced by addition of DMSO or RanGTP were smaller in X. tropicalis extracts compared with X. laevis extracts (unpublished data). To determine if differences in global microtubule dynamics could account for the differences in spindle and aster size, we measured the parameters of microtubules nucleated from centrosomes in X. laevis and X. tropicalis extracts (Table I). Overall, although the microtubule growth rate was ∼20% slower in the X. tropicalis extracts (14.7 μm/min vs. 18.5 μm/min in X. laevis; P < 0.015 in t test), catastrophe and rescue frequencies were similar, and the calculated steady-state microtubule lengths were not significantly different (Table I; Verde et al., 1992). Another mode of microtubule turnover in the spindle is poleward microtubule flux, which is when microtubules coordinately polymerize at their plus ends and depolymerize at their minus ends as antiparallel microtubules slide apart (Khodjakov and Kapoor, 2005). We measured these rates using speckle microscopy to mark the spindle microtubule lattice, and found values in both extracts similar to those previously described for X. laevis (Table I; 1.79 ± 0.33 μm/min for X. laevis; 2.25 ± 0. 25 μm/min for X. tropicalis; Desai et al., 1998). Thus, our results indicate that the coordinated microtubule sliding with balanced plus end polymerization and minus end depolymerization are not significantly different in the two extracts.
Table I.
Dynamic parameters of individual microtubules nucleated by centrosomes in X. laevis and X. tropicalis extracts and rates of poleward flux
| X. laevis | X. tropicalis | t test | |
|---|---|---|---|
| n = 50 | n = 79 | ||
| Growth rate (μm/min) | 18.5; SD = 8.8 | 14.7; SD = 4.4 | 0.013 |
| Shrinkage rate (μm/min) | 21.8; SD = 9.3 | 17.6; SD = 5.0 | 0.19 |
| Catastrophe frequency (events/min) | 5.8; SD = 1.1 | 5.9; SD = 4.7 | 0.96 |
| Rescue frequency (events/min) | 3.0; SD = 2.6 | 4.3; SD = 4.9 | 0.44 |
| Average length (μm) | 5.7 | 6.3 | 0.42 |
| Flux rates (μm/min) | 1.79; SD = 0.33 | 2.25; SD = 0.25 | 0.31 |
Microtubule dynamics were measured from individual centrosomes in X. laevis and X. tropicalis CSF extracts. Dynamic parameters were computed using a custom algorithm. A t test was used to compute the significance of the differences. n denotes the number of individual microtubules measured for each extract system, taken from three different extracts. Average microtubule lengths were calculated according to Verde et al. (1992). Poleward flux rates were measured using time-lapse fluorescence speckle microscopy in 3 different extracts, with 5 spindles each (15 spindles total) for X. laevis and X. tropicalis.
What is the underlying cause of spindle size difference in the two extracts? One possibility is that we are unable to make precise enough measurements to distinguish potentially causal differences in the dynamics of microtubules in the two extracts. Alternatively, other microtubule dynamic parameters, such as the frequency of nucleation, pausing, or severing, may generate differences in spindle size (Srayko et al., 2006). Although different in morphology, centrosomal microtubules in the two extracts grew to similar lengths, whereas microtubule structures induced by taxol, DMSO, and RanGTP were significantly smaller in X. tropicalis extracts (unpublished data). An intriguing possibility is that extract factors in the two systems respond differently to microtubule stabilizing/destabilizing agents, including mitotic chromatin. We think that this is possible because we have identified extract factors, such as the microtubule plus-end binding protein Xorbit, whose depletion does not obviously affect centrosomally nucleated microtubules, but strongly destabilizes spindle microtubules (Hannak and Heald, 2006). Extract-dependent changes in the activity of spindle factors or their regulation caused by a different kinase/phosphatase balance or RanGTP signal surrounding chromosomes could locally alter microtubule stability and overall spindle size. The challenge will now be to identify molecules that can account for the observed differences in spindle size in the egg extracts and compare the activities of the orthologous factors between the two Xenopus species.
In conclusion, X. tropicalis provides molecular advantages over X. laevis as a genetically and proteomically tractable system that can be applied to address cell biological questions using in vitro approaches. Although it could be expected that the smaller X. tropicalis eggs would have smaller spindles, our results show that this is because of a difference in cytoplasmic factors, rather than the size of the cell itself. By comparing cytoplasmic activities that are intrinsic to X. laevis and X. tropicalis extracts, new insights can be gained into mechanisms regulating cellular morphogenesis.
Materials and methods
Preparation of CSF extracts from X. tropicalis
CSF-arrested X. tropicalis extracts were prepared essentially as previously described for X. laevis (Murray, 1991; Desai et al., 1999), with the following exceptions. To induce egg laying, frogs were primed with 10 U of human chorionic gonadotropin (HCG) ∼16 h before a hormone boost of 200 U HCG. Laying commenced ∼4–5 h after the second HCG injection, and eggs were collected into water at 27–28°C (http://tropicalis.berkeley.edu/home/). Frogs were also squeezed ∼6 h after the second HCG injection, and we found no substantial difference in the quality of the laid versus squeezed eggs. Pooled eggs were dejellied using 3% cysteine in water adjusted to pH 7.8 with NaOH. Incubation of frogs at temperatures below ∼23°C inhibited egg laying, and resulted in poor extracts. Once the extract was prepared it was stored at 16°C, as extended storage at 4°C resulted in a loss of CSF arrest. Typical extract yield was 200–400 μl per frog.
Spindle size determination in mixed extract
X. laevis and X. tropicalis extracts were mixed in different proportions and supplemented with either X. laevis or X. tropicalis sperm nuclei prepared by standard procedures (Murray, 1991) at a concentration of 500 sperm/μl, and X-rhodamine tubulin at 0.125 mg/ml. Cycling reactions were performed, and reactions were diluted into spindle fix (30% glycerol, 1× BRB80, and 0.5% Triton X-100), spun onto coverslips, fixed in –20°C methanol, and mounted in Vectashield according to standard procedures (Desai et al., 1999). Images were collected with a fluorescence microscope (BX51; Olympus) and a cooled charge-coupled device camera (Orca II; Hamamatsu), and spindle lengths were measured using MetaMorph software (Molecular Devices). Spindle area measurements were made using thresholded images in MetaMorph. Mixing experiments were performed three independent times by three different investigators, and the results were averaged.
Immunoprecipitation and mass spectrometry of Xnf7
The Xnf7 polyclonal antibody was coupled to protein A dynabeads, as previously described (Maresca et al., 2005b). For immunoprecipitating Xnf7, 110 μl of either X. tropicalis or X. laevis CSF extract was subjected to three successive 45-min incubations on ice with anti-Xnf7–coated dynabeads. The beads from each round were pooled and washed extensively with XB before eluting for 5 min at room temperature into SDS sample buffer and retrieving the beads on a magnet. Half of the supernatant was subjected to SDS-PAGE, the gel was stained with Gradipure colloidal G-250 Coomassie blue stain (NuSep), and the indicated bands were excised and subjected to mass spectrometry at the University of California Berkeley Mass Spectrometry Facility.
Dynamics of spindle size determination
CSF reactions containing X. laevis sperm nuclei and 25 μl X. tropicalis extract (with X-rhodamine tubulin) or X. laevis extract (with Alexa Fluor 488 tubulin) were cycled through interphase and back into metaphase by the addition of 25 μl of the same type of extract (no sperm). Once metaphase spindles had assembled, the extract was split into two tubes, each containing 25 μl of the reaction mixture. As a control, 75 μl of the same type of extract supplemented with the other labeled tubulin was added to one of the tubes, while 75 μl of the opposing extract was added to the other 25-μl reaction. Each of the 100-μl reactions were quickly split into 4 separate tubes, and spindles from each condition were diluted and spun down onto coverslips as described in Spindle size determination in mixed extract for imaging and length measurements. All spindle reactions were incubated at ∼23°C.
Microtubule dynamics and flux measurements
Centrosomes were prepared from KE37 cells as previously described (Chretien et al., 1997) and stored at −80°C. CSF extracts were prepared as described in the section Preparation of CSF extracts from X. tropicalis, and supplemented with either rhodamine tubulin (Cytoskeleton) or Alexa Fluor 488 tubulin (a gift from T. Whittman, University of California, San Francisco, San Francisco, CA) at 0.2 mg/ml. Centrosome reactions consisted of 8 μl of extract plus labeled tubulin, 1 μl of centrosomes, and 1 μl of Oxyrase. X. laevis extracts were incubated at ∼20°C, and X. tropicalis extracts were incubated at ∼23°C. To image centrosomes, 1 μl of extract was squashed under a 22-mm2 coverslip and imaged using a 100×/1.3 NA objective. All glassware was base cleaned and stored in 95% ethanol until the time of use. Images were acquired every 3 s for 1–3 min. Microtubule lengths were measured if the microtubule could be followed for at least five successive frames. Microtubule lengths were measured as the distance from the center of the centrosome to the tip of the microtubule. Dynamics measurements were calculated using a custom-made spreadsheet (a gift from R. Tournebize, Institute Pasteur, Paris, France). Calculation of Fcat and Fres were made by manual inspection of raw growth and shrinkage measurements.
MT flux measurements were made using fluorescence speckle microscopy by incubation of cycled spindles with rhodamine-labeled tubulin at a concentration of 1 μg/ml (Kapoor and Mitchison, 2001). 2 μl of extract was gently squashed under a 22 × 22-mm coverslip previously outlined using a PAP pen. Images of speckles were collected every 5 s for 2 min using a 60×/1.4 NA objective. Speckle movements were tracked on kymographs to calculate the rate of flux. Measurements were made from at least five separate spindles from three different extracts for both X. laevis and X. tropicalis.
Acknowledgments
We thank members of the Heald lab, especially J. Soderholm, for helpful discussions and S. Zhou for mass spectrometry analysis.
This work was supported by National Institutes of Health (NIH) grants to R. Heald (GM057839 and DP1OD00081) and R.M. Harland (GM66684). M.D. Blower is supported by the Damon Runyon Cancer Research Foundation.
K.S. Brown, M.D. Blower, and T.J. Maresca contributed equally to this paper.
M.D. Blower's present address is Dept. of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114.
Abbreviations used in this paper: CSF, cytostatic factor; HCG, human chori-onic gonadotropin; NuMA, nuclear mitotic apparatus protein; Xnf7, Xenopus nuclear factor 7.
References
- Birsoy, B., L. Berg, P.H. Williams, J.C. Smith, C.C. Wylie, J.L. Christian, and J. Heasman. 2005. XPACE4 is a localized pro-protein convertase required for mesoderm induction and the cleavage of specific TGFbeta proteins in Xenopus development. Development. 132:591–602. [DOI] [PubMed] [Google Scholar]
- Blower, M.D., M. Nachury, R. Heald, and K. Weis. 2005. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 121:223–234. [DOI] [PubMed] [Google Scholar]
- Brown, D.D. 2004. A tribute to the Xenopus laevis oocyte and egg. J. Biol. Chem. 279:45291–45299. [DOI] [PubMed] [Google Scholar]
- Chang, P., M.K. Jacobson, and T.J. Mitchison. 2004. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 432:645–649. [DOI] [PubMed] [Google Scholar]
- Chretien, D., B. Buendia, S.D. Fuller, and E. Karsenti. 1997. Reconstruction of the centrosome cycle from cryoelectron micrographs. J. Struct. Biol. 120:117–133. [DOI] [PubMed] [Google Scholar]
- Dasso, M., and J.W. Newport. 1990. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. Cell. 61:811–823. [DOI] [PubMed] [Google Scholar]
- Desai, A., P.S. Maddox, T.J. Mitchison, and E.D. Salmon. 1998. Anaphase A chromosome movement and poleward spindle microtubule flux occur at similar rates in Xenopus extract spindles. J. Cell Biol. 141:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., A. Murray, T.J. Mitchison, and C.E. Walczak. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., and A.W. Murray. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424. [DOI] [PubMed] [Google Scholar]
- Goshima, G., R. Wollman, N. Stuurman, J.M. Scholey, and R.D. Vale. 2005. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988. [DOI] [PubMed] [Google Scholar]
- Hannak, E., and R. Heald. 2006. Xorbit/CLASP links dynamic microtubules to chromosomes in the Xenopus meiotic spindle. J. Cell Biol. 172:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, N., L.B. Zimmerman, and R.M. Grainger. 2002. Xenopus, the next generation: X. tropicalis genetics and genomics. Dev. Dyn. 225:422–433. [DOI] [PubMed] [Google Scholar]
- Kapoor, T.M., and T.J. Mitchison. 2001. Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 154:1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and T. Kapoor. 2005. Microtubule flux: what is it good for? Curr. Biol. 15:R966–R968. [DOI] [PubMed] [Google Scholar]
- Klein, S.L., R.L. Strausberg, L. Wagner, J. Pontius, S.W. Clifton, and P. Richardson. 2002. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev. Dyn. 225:384–391. [DOI] [PubMed] [Google Scholar]
- Maresca, T.J., B.S. Freedman, and R. Heald. 2005. a. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J. Cell Biol. 169:859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca, T.J., H. Niederstrasser, K. Weis, and R. Heald. 2005. b. Xnf7 contributes to spindle integrity through its microtubule-bundling activity. Curr. Biol. 15:1755–1761. [DOI] [PubMed] [Google Scholar]
- Merdes, A., K. Ramyar, J.D. Vechio, and D.W. Cleveland. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458. [DOI] [PubMed] [Google Scholar]
- Murray, A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605. [PubMed] [Google Scholar]
- Nicklas, R.B., and G.W. Gordon. 1985. The total length of spindle microtubules depends on the number of chromosomes present. J. Cell Biol. 100:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani, H., H. Kobayashi, M. Ohtsubo, and T. Nishimoto. 1990. Cloning of Xenopus RCC1 cDNA, a homolog of the human RCC1 gene: complementation of tsBN2 mutation and identification of the product. J. Biochem. (Tokyo). 107:228–235. [DOI] [PubMed] [Google Scholar]
- Srayko, M., E.T. O'Toole, A.A. Hyman, and T. Muller-Reichert. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. [DOI] [PubMed] [Google Scholar]
- Verde, F., M. Dogterom, E. Stelzer, E. Karsenti, and S. Leibler. 1992. Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 118:1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]