Abstract
The multifunctional cytokine transforming growth factor (TGF) β1 is secreted in a latent complex with its processed propeptide (latency-associated peptide [LAP]). TGFβ1 must be functionally released from this complex before it can engage TGFβ receptors. One mechanism of latent TGFβ1 activation involves interaction of the integrins αvβ6 and αvβ8 with an RGD sequence in LAP; other putative latent TGFβ1 activators include thrombospondin-1, oxidants, and various proteases. To assess the contribution of RGD-binding integrins to TGFβ1 activation in vivo, we created a mutation in Tgfb1 encoding a nonfunctional variant of the RGD sequence (RGE). Mice with this mutation (Tgfb1RGE/RGE) display the major features of Tgfb1−/− mice (vasculogenesis defects, multiorgan inflammation, and lack of Langerhans cells) despite production of normal levels of latent TGFβ1. These findings indicate that RGD-binding integrins are requisite latent TGFβ1 activators during development and in the immune system.
Introduction
Signaling by the multifunctional cytokine TGFβ1 is regulated by other secreted proteins that influence TGFβ1's biological activity and localization (Annes et al., 2003). Central to these interactions is the fact that TGFβ1 is secreted in a latent form. TGFβ1 latency arises from a noncovalent interaction between TGFβ1 and its propeptide, latency-associated peptide (LAP). In addition to blocking access of TGFβ1 to TGFβ receptors, LAP interacts via disulfide bonds with proteins of the latent TGFβ-binding protein (LTBP) family (Fig. 1). The trimolecular complex of TGFβ1, LAP, and LTBP is referred to as the large latent complex and is thought to be the major secreted form of latent TGFβ1. LTBPs bind to matrix molecules, thereby anchoring latent TGFβ1 in the extracellular space, and influence the release of TGFβ1 from LAP, a process called latent TGFβ1 activation (Annes et al., 2004). Two other isoforms of TGFβ (TGFβ2 and -3) are secreted in similar latent forms.
Figure 1.
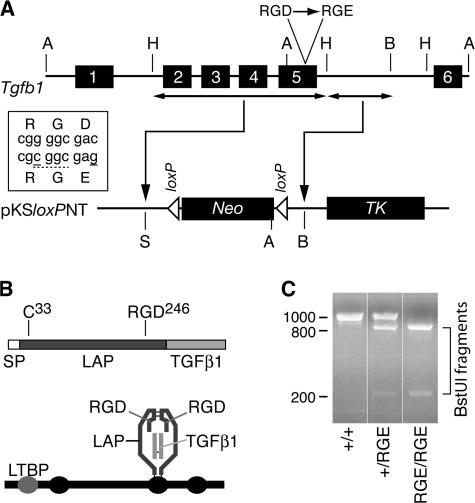
Generation of mice with a targeted mutation of Tgfb1. (A) Schematic of Tgfb1 (top) and fragments thereof used for insertion in targeting vector (bottom). Box shows mutations (solid underlines) introduced in exon 5 to encode a BstUI restriction site (dashed underline) and the D-to-E mutation. Restriction sites: A, ApaI; B, BamHI; H, HindIII; S, SalI. Neo, neomycin resistance cassette; TK, thymidine kinase cassette. (B) The TGFβ1 mRNA encodes a protein sequence consisting of a signal peptide (SP), propeptide (LAP), and the TGFβ1 cytokine (top). The integrin-binding motif RGD is near the C terminus of LAP. The processed latent factor consists of noncovalently associated disulfide-linked homodimers of the LAP and TGFβ1 monomers (bottom). LAP can be disulfide linked via cys-33 to one of the cysteine-repeat domains (ovals) of LTBP-1, -3, or -4. (C) PCR genotyping results from Tgfb1+/+, Tgfb1+/RGE, and Tgfb1RGE/RGE mice.
Proposed TGFβ1 activators include thrombospondin-1 (TSP1), proteases such as plasmin and matrix metalloproteinases (MMPs), the integrins αvβ6 and αvβ8 (which recognize an RGD sequence in LAP), and reactive oxygen species (Sato et al., 1990; Barcellos-Hoff and Dix, 1996; Crawford et al., 1998; Munger et al., 1999; Ribeiro et al., 1999; Yu and Stamenkovic, 2000; Mu et al., 2002). By degrading LAP or changing its conformation, these activators permit TGFβ1 to engage TGFβ receptors. Because TGFβ1 regulates numerous processes (immune function, cell proliferation, apoptosis, extracellular matrix formation, and vascular development, among others), one can speculate that multiple TGFβ1 activation mechanisms are used to activate TGFβ1 in diverse contexts. However, we lack a comprehensive picture that connects specific biological effects of TGFβ1 with specific TGFβ1 activators.
In some cases, deletion of the gene encoding a putative TGFβ1 activator results in a phenotype consistent with a TGFβ1 deficit. For example, mice lacking TSP1 or the integrin β6 subunit (Tsp1−/− and Itgb6−/− mice) develop inflammation, although it is not as severe as that in Tgfb1−/− mice, which develop marked infiltrates of activated T cells in multiple organs and die soon after weaning (Shull et al., 1992; Huang et al., 1996; Crawford et al., 1998). In addition, Itgb6−/− mice are protected from TGFβ-dependent fibrotic tissue reactions (Munger et al., 1999). Some embryos lacking αvβ8 (Itgb8−/−) have defective vasculogenesis that appears identical to defects observed in a subset of Tgfb1−/− embryos (Dickson et al., 1995; Zhu et al., 2002). However, knockouts of genes encoding other putative TGFβ1 activators, such as plasminogen and various MMPs, do not result in phenotypes clearly related to TGFβ1 deficits. Although Itgb6−/−;Tsp1−/− mice have a more severe inflammatory phenotype than either single-null animal (Ludlow et al., 2005), these mice do not fully phenocopy Tgfb1−/− mice.
To delineate the role of RGD-binding integrins in generation of TGFβ1 activity, we created mice with a selective loss of integrin-mediated TGFβ1 activation. To do this, we made a TGFβ1 gene mutation that encodes an inactive version of LAP's integrin-binding site (RGE instead of RGD) and used embryonic stem (ES) cells containing the mutant gene to generate mice. These mice produce latent TGFβ1 that cannot interact with RGD-binding integrins.
Results and discussion
A targeting vector was made by inserting two contiguous fragments of Tgfb1 DNA, with appropriate mutations in the fragment containing exon 5, into the cloning sites of the pKSloxPNT vector (Fig. 1 A). One mutation creates a conservative single amino acid change (D246E) within the RGD motif located near the C terminus of LAP (Fig. 1 B), and the other creates a new restriction site that can be used for identification of the mutant allele. The vector was used to transfect ES cells, and cells with correct targeting of Tgfb1 were injected into blastocysts to generate mice carrying the Tgfb1 mutation (Fig. 1 C). These mice were crossed with Cre-deleter mice to remove the loxP-flanked Neo cassette from the targeted gene.
Tgfb1−/− mice display several abnormalities (Shull et al., 1992), the most prominent of which is T cell–mediated multiorgan inflammation that leads to death soon after weaning. In addition, a strain-dependent fraction of Tgfb1−/− embryos dies around embryonic day (E) 10 because of failure of yolk sac vasculogenesis and/or hematopoiesis (Dickson et al., 1995). Also, Tgfb1−/− mice lack Langerhans cells (LCs), which are dendritic cells residing in the epidermis (Borkowski et al., 1997). We predicted that Tgfb1RGE/RGE mice would have an incomplete version of the Tgfb1−/− phenotype, as occurs in Tsp1−/−, Itgb6−/−, and Itgb8−/− mice (see Table I for a comparison of knockout phenotypes). However, Tgfb1RGE/RGE mice display the cardinal features of Tgfb1−/− mice.
Table I.
Phenotypes of mice with mutations in genes encoding integrin subunits, TSP1, TGFβ1, or TGFβ3
| Phenotype | Gene
|
|||||
|---|---|---|---|---|---|---|
| Itgav−/− | Itgb6−/− | Itgb8−/− | Tsp1−/− | Tgfb1−/−, Tgfb1RGE/RGE | Tgfb3−/− | |
| Autoimmune syndrome | NT | Mild inflammation (lung and skin) | NT | Inflammation (lung and pancreas) | Lethal autoimmune syndrome | NT |
| Abnormal vasculogenesis (∼E10) | 80% of embryos | 0% | 60% of embryos | 0% | ∼50% of embryos | 0% |
| LC deficit | NT | Reduced numbers | NT | NA | Absent | NT |
| Abnormal central nervous system vascular development | 100% at birth | 0% | 100% at birth | 0% | 0% | 0% |
| Cleft palate | 100% | 0% | 10% at birth | 0% | 0% | 100% |
NT, not tested because of early lethality; NA, data not available.
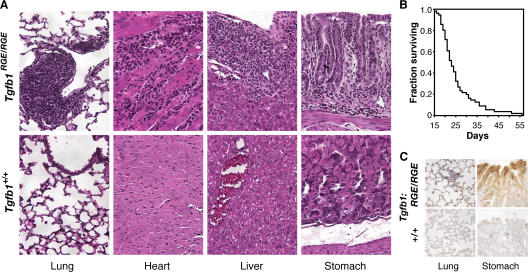
Tgfb1RGE/RGE mice appear normal at birth but are smaller than littermates by 14 d and have markedly reduced survival (Fig. 2 B). Histologic examination of Tgfb1RGE/RGE mice between the ages of 18 and 28 d revealed marked mononuclear cell infiltration of multiple tissues, in particular, the heart, lung, liver, stomach, and pancreas (which were abnormal in >85% of mice examined), and occasionally in the CNS (Fig. 2 A and not depicted). In the lung, inflammatory lesions are usually localized around bronchi and larger vessels and sometimes diffusely within alveolar walls; in the liver, lesions are localized within portal canals. We did not note inflammation in the skin or kidney. These findings are similar to those reported for Tgfb1−/− mice. A side-by-side comparison of the inflammatory lesions in Tgfb1RGE/RGE and Tgfb1−/− mice reveals them to be indistinguishable (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200611044/DC1). Because the β6-integrin subunit, which is expressed predominantly in epithelial cells, is up-regulated by injury and inflammation, we assessed β6 expression in 3-wk-old Tgfb1RGE/RGE mice. β6 protein is markedly increased in lung and gastric epithelium (Fig. 2 C) and is occasionally increased in biliary epithelium (not depicted). We did not detect β6 protein expression in the heart.
Figure 2.
Tgfb1RGE/RGE mice develop fatal multiorgan inflammation. (A) Histology of inflammatory lesions in lung, heart, liver, and stomach of Tgfb1RGE/RGE mice (hematoxylin and eosin staining). (B) Kaplan-Meier survival curve for Tgfb1RGE/RGE mice (n = 54). (C) Increased expression of β6 protein in stomach and lung epithelium of Tgfb1RGE/RGE mice.
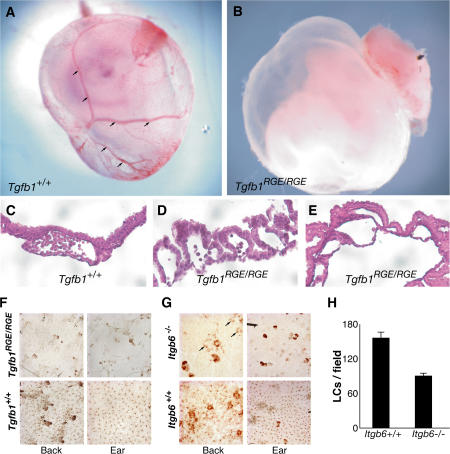
We also compared the developmental consequences of the RGE mutation with the developmental abnormalities observed in Tgfb1−/− mice. We determined the genotype of mice born to parents heterozygous for the mutant Tgfb1 allele. Of 378 births, 36.4% were Tgfb1−/−, 50.2% were Tgfb1+/RGE, and 13.4% were Tgfb1RGE/RGE. Thus, there is a substantial difference between the Mendelian ratios 1:2:1 (+/+:+ /RGE:RGE/RGE) and the observed ratios of ∼1:1.4:0.4, indicating an embryonic lethality of ∼50% associated with the Tgfb1RGE/RGE genotype and ∼25% for the Tgfb1+/RGE genotype. To determine when lethality occurs, we collected embryos for genotyping and histologic analysis. Of 146 E10.5 embryos, 24.3% were Tgfb1−/−, 50% were Tgfb1+/RGE, and 25.7% were Tgfb1RGE/RGE. Therefore, embryonic death occurs at or after E10.5. Approximately half of the E10.5 Tgfb1RGE/RGE yolk sacs had grossly evident anemia and/or absence of normal vasculature (Fig. 3, A and B). Histological abnormalities consisted variably of a paucity of vessels, absence of hematopoietic cells, and extensive buckling between the mesodermal and endodermal layers (Fig. 3, C–E). Similar findings have been reported for Tgfb1−/− mice and mice expressing dominant-negative TGFβ receptors (Dickson et al., 1995; Oshima et al., 1996).
Figure 3.
Defects in vascular development and LCs in Tgfb1RGE/RGE mice. (A and B) Vasculature is present in wild-type yolk sac (arrows) but is not identifiable in an E12.5 Tgfb1RGE/RGE embryo. (C–E) Histologic appearance of control and Tgfb1RGE/RGE E12.5 yolk sacs. In the mutant yolk sacs, there is poor contact between endothelial and mesothelial layers and, in E, absence of blood cells. (F) LCs are absent in epidermis from back and ear of Tgfb1RGE/RGE mice. (G) LCs are less abundant in back epidermis (arrows) and absent in ear epidermis of Itgb6−/− mice. (H) Compared with control mice, Itgb6−/− mice have fewer LCs in back epidermis. P = 0.001. Error bars indicate means ± SEM.
We also compared the LC status of Tgfb1RGE/RGE mice with that of Tgfb1−/− mice. Normally, peripheral blood monocytes enter the epidermis and differentiate into LCs, but Tgfb1−/− mice completely lack LCs (Borkowski et al., 1997). The target cell for TGFβ signaling appears to be the LC or a precursor, not epithelial cells, and TGFβ1 production by nonmarrow-derived cells is sufficient for LC production. We compared the presence of LCs in Tgfb1RGE/RGE mice and littermate normal controls by staining for LCs in epidermal sheets obtained from the back or ear. LCs are absent from both sites (Fig. 3 F), aside from very rare small clusters of LCs in skin from ears, typically near the ear edge (not depicted).
Because αvβ6 is functional in the epidermis, in that Itgb6−/− mice develop skin inflammation (Huang et al., 1996), we suspected that this integrin might be the major or sole activator of TGFβ1 involved in LC generation. Therefore, we examined epidermal sheets from Itgb6−/− and Itgb6−/− mice (C57BL/6 strain) for the presence of LCs (Fig. 3 G). LCs are almost completely absent from Itgb6−/− ear epidermis. In contrast, epidermal sheets from the backs of Itgb6−/− mice have about half as many LCs as equivalent samples from Itgb6−/− mice (Fig. 3, G and H). Although some Itgb6−/− mice develop inflammatory skin lesions at sites of tissue trauma, characterized by hair loss and macrophage infiltration, these changes were not present in skin of our Itgb6−/− mice, and the lack of LCs in Tgfb1−/− mice is independent of the inflammatory phenotype (Borkowski et al., 1997). We conclude that the TGFβ1 required for LC generation is activated by RGD-binding integrins, but the integrins involved vary by region. In ear epidermis, the αvβ6 integrin is required for LC generation, whereas in back epidermis, αvβ6 appears to contribute but is not absolutely necessary.
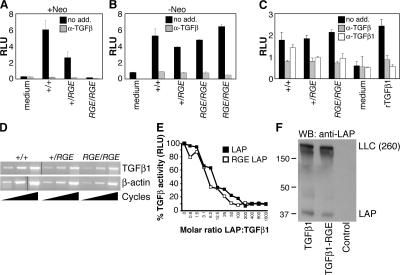
The phenotypes of Tgfb1−/− and Tgfb1RGE/RGE mice are similar to that of Foxp3−/− mice, which lack CD4+CD25+ regulatory T cells (Tregs). However, we detected no difference in the abundance of Tregs among CD4+ cells isolated from spleens of 8–10-d-old Tgfb1−/− and Tgfb1RGE/RGE mice, assessed as the percentage of either CD25+ or Foxp3+ cells (unpublished data). Because the phenotype of Tgfb1RGE/RGE mice is highly similar to that of Tgfb1−/− mice in the processes we examined, we assessed in vivo transcription and translation of the mutated Tgfb1 gene to confirm that the observed phenotype is not due to impaired gene function. The targeting vector introduced a Neo expression sequence into the intron between exons 5 and 6 of Tgfb1, and such sequences often interfere with expression of the targeted gene. Serum levels of TGFβ1 reflect both tissue production of TGFβ1 and release of TGFβ1 by platelets during clotting and can be measured with a luciferase-based bioassay after heating the serum to activate latent TGFβ1 (see Materials and methods). The assay is performed with and without anti-TGFβ antibody to confirm specificity. Serum levels of TGFβ1 were reduced in Tgfb1+/RGE mice and undetectable in Tgfb1RGE/RGE mice when the Neo sequence was present (Fig. 4 A). However, after removal of the Neo sequence, serum levels of latent TGFβ1 in mice heterozygous or homozygous for the RGE mutation were equivalent to those in Tgfb1−/− mice (Fig. 4 B). Levels of latent TGFβ1 in serum-free medium conditioned by lung fibroblasts derived from Tgfb1−/−, Tgfb1+/RGE, or Tgfb1RGE/RGE mice were also equivalent (Fig. 4 C). To confirm that the genetic manipulations had not altered mRNA sequence, we used RNA from Tgfb1RGE/RGE lung fibroblasts to amplify the coding sequence by RT-PCR and found no changes other than the expected mutations introduced in exon 5. Tgfb1 gene expression, measured by semiquantitative RT-PCR using RNA isolated from lung, liver, and heart, was not measurably affected by the Tgfb1 RGE mutation (Fig. 4 D and not depicted).
Figure 4.
The RGE mutation does not affect Tgfb1 gene expression, LAP inhibitory function, or latent TGFβ1 processing and secretion. Serum TGFβ1 levels in Tgfb1−/−, Tgfb1+/RGE, and Tgfb1RGE/RGE mice before (A) and after (B) removal of a Neo sequence within the mutated Tgfb1 allele. (C) TGFβ levels in medium conditioned by lung fibroblasts derived from Tgfb1−/−, Tgfb1+/RGE, and Tgfb1RGE/RGE mice. Effects of an anti-TGFβ antibody active against all three TGFβ isoforms (α-TGFβ) and of an antibody that inhibits only TGFβ1 (α-TGFβ1) are shown. Error bars indicate means ± SEM. (D) Tgfb1 gene expression was assessed by semiquantitative RT-PCR using RNA isolated from Tgfb1−/−, Tgfb1+/RGE, and Tgfb1RGE/RGE lung fibroblasts. Reverse-transcribed β-actin mRNA was amplified as a control. (E) Recombinant wild-type and RGE mutant LAP inhibit equally the activity of recombinant TGFβ1, measured with a cell-based luciferase assay. Luciferase activity, expressed as relative light units (RLU), is proportional to TGFβ activity. (F) Wild-type and RGE mutant TGFβ1 cDNAs were expressed in LTBP1S-expressing CHO cells. Secreted proteins were separated by SDS-PAGE in nonreducing conditions and probed with an anti-LAP antibody. Large latent complex (LLC) indicates migration of the LAP–LTBP1 complex.
The RGD-to-RGE mutation eliminates integrin-mediated TGFβ1 activation in cell culture experiments but does not appear to affect other LAP functions, such as TGFβ inhibition or interaction with LTBP. The D-to-E mutation is conservative, and the location of the RGD sequence is remote from the TSP1-binding site and an MT1-MMP cleavage site (Ribeiro et al., 1999; Mu et al., 2002). Purified recombinant LAP with the RGD-to-RGE mutation does not support adhesion by cells expressing αvβ6 or αvβ8 (Munger et al., 1999; Mu et al., 2002), indicating that these integrins cannot effectively engage the mutated binding site. In contrast, LAP's interaction with TGFβ1 appears unaffected by the RGE mutation. Recombinant LAP binds and inhibits the bioactivity of recombinant TGFβ1, and the dose–response curve for this inhibition is not affected by the RGE mutation (Fig. 4 E); also, cells transfected with TGFβ1-RGE cDNA secrete normal amounts of latent TGFβ1 that is bioactive after activation by heating (not depicted).
We also tested the ability of the RGE mutant form of LAP to form disulfide linkages with LTBP1 and to be incorporated into ECM, because these functions are critical for activation. CHO cells expressing LTBP1 (Annes et al., 2004) were transfected with cDNAs encoding wild-type or RGE TGFβ1, and conditioned media were immunoblotted with anti-LAP antibody. Equal amounts of high molecular weight bands consistent with LTBP1–LAP complexes (Annes et al., 2004) are seen, along with small amounts of monomeric LAP (∼37 kD). We previously showed that cells expressing αvβ6 do not activate soluble latent TGFβ1-RGE (Annes et al., 2002). In cell culture conditions similar to those in previous reports (Annes et al., 2002, 2004), the RGD and RGE forms of latent TGFβ1 are equivalently incorporated into cell-derived matrix, but only the RGD form of matrix-bound latent TGFβ1 is activated by αvβ6- or αvβ8-expressing cells (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200611044/DC1).
In summary, loss of latent TGFβ1 activation by RGD-binding integrins in vivo recapitulates the major features of Tgfb1−/− mice. We conclude that in these biological contexts, essentially all active TGFβ1 is generated in conjunction with integrin–LAP interactions. Issues requiring clarification include the identity of the RGD-binding integrins involved, the role of RGD-binding integrins in activation of TGFβ3, and the role of nonintegrin mechanisms of TGFβ1 activation.
Integrins are heterodimers of α and β subunits. Of 24 mammalian integrins, 8 (αvβ1, αvβ3, αvβ5, αvβ6, αvβ8, αIIbβ3, α5β1, and α8β1) bind RGD sequences in their respective ligands. Of these, 2 (αvβ6 and αvβ8) clearly activate TGFβ1 in vitro and in vivo (Munger et al., 1999; Mu et al., 2002). αvβ5 binds LAP and, when expressed by scleroderma fibroblasts, activates latent TGFβ; however, activation has not been noted in other αvβ5-expressing cells (Asano et al., 2005; Araya et al., 2006). Three other RGD-binding integrins (αvβ1, αvβ3, and α8β1) bind LAP but have not been shown to activate latent TGFβ1 when expressed in cultured mammalian cells (Munger et al., 1998; Lu et al., 2002; Ludbrook et al., 2003). The phenotypes of αv-, β6-, and β8-null mice (Huang et al., 1996; Bader et al., 1998; Zhu et al., 2002) are somewhat similar to those of Tgfb1−/− mice (Table I), but the phenotypes of other integrin-null mice are not (α5- and β1-null mice die early in embryogenesis, precluding comparison). Nevertheless, it is plausible that these LAP-binding but non–TGFβ-activating integrins promote TGFβ1 activation in vivo, perhaps by concentrating latent TGFβ1 at cell surfaces where it might be activated by another process.
TGFβ3-LAP contains an RGD sequence, and latent TGFβ3 is activated by αvβ6 and αvβ8 expressed in cultured cells (Annes et al., 2002; Araya et al., 2006). The phenotypes of Itgb6−/− and Tgfb3−/− mice do not overlap, but 10% of Itgb8−/− mice have cleft palate (Zhu et al., 2002), which occurs in all Tgfb3−/− mice (Kaartinen et al., 1995; Proetzel et al., 1995; Table I). Therefore, αvβ8 may be partially responsible for TGFβ3 activation during palate formation.
There are numerous reports of physiologic TGFβ1 activation by nonintegrin mechanisms, such as TSP1, proteases, and oxidants. Although our results suggest that RGD-binding integrins play a dominant role in TGFβ1 activation, several caveats exist. First, our results are limited to three phenotypes that are evident within the first few weeks of life, and it is possible that other TGFβ1-mediated processes occur independently of integrin-mediated activation. Second, integrins likely act in parallel or in series with nonintegrin mechanisms to generate TGFβ1 signaling in vivo. For example, αvβ8 and MT1-MMP cooperatively activate TGFβ1 at cell surfaces (Mu et al., 2002), and proteolytic activity releases latent TGFβ1 from the ECM (Annes et al., 2003), thereby potentially enhancing access of latent TGFβ1 to integrins for activation under some circumstances. The phenotype of TSP1-null mice partially overlaps that of Tgfb1−/− mice (Table I), suggesting that TSP1 may cooperate with RGD-binding integrins in TGFβ1 activation.
Our findings reveal a critical role for RGD-binding integrins in the generation of TGFβ1 signaling activity in three disparate processes (control of inflammation, vasculogenesis, and LC genesis) and illustrate the utility of a subtle genetic mutation approach to complement gene knockout studies. Further work is needed to establish which RGD-binding integrins are involved in specific TGFβ1 effects and how integrins interact with other molecules involved in TGFβ1 activation.
Materials and methods
Creation of targeting vector containing a Tgfb1 exon 5 mutation
A 1.4-kb mouse TGFb1 cDNA was used to screen a 129/SvEv mouse Lambda FIX II Library. A 13-kb clone containing Tgfb1 exons 1–6 was restriction mapped. HindIII digestion generated a 6-kb fragment containing exons 2–5 and a contiguous 5-kb fragment containing most of the intron between exons 5 and 6. These were subcloned into pBluescriptSK. Exon 5 encodes the RGD246 in TGFβ1-LAP. We used the QuikChange site-directed mutagenesis kit (Stratagene) to mutate the codon encoding D246 to a codon encoding E and to create an adjacent silent mutation creating a BstUI site (cg/cg). The mutagenesis primers were ggatcagccccaaacgtcgcggcgagctgggcaccatccatgac and gtcatggatggtgcccagctcgccgcgacgtttggggctgatcc.
A targeting vector was made using the pKSloxPNT plasmid (a gift from A. Joyner, New York University School of Medicine, New York, NY). The plasmid containing the 5-kb HindIII fragment was digested with BamHI to generate a 3.3-kb BamHI fragment (one BamHI site is derived from the pBluescript cloning site), which was inserted at the BamHI cloning site. The 6-kb HindIII fragment containing the RGD-to-RGE mutation was excised with SalI and XhoI and ligated into the targeting vector at the SalI site. A clone with correct insert orientation (SalI site closest to the Neo cassette destroyed) was identified. The vector was extensively sequenced to confirm that genomic sequences and loxP sites were intact and correctly oriented.
Transfection of ES cells and identification of targeted clones
The targeting vector was linearized by SalI digestion. 50 μg of DNA was used to transfect 5 × 106 W4 ES cells by electroporation. ES cells were selected with 200 μg/ml G418 and 2 μM gancyclovir. Correctly targeted clones were identified by Southern blot using external probes (exons 1 and 6). With ApaI-digested genomic DNA, an exon 6 probe revealed an 8-kb fragment of the wild-type Tgfb1 gene and a 7.4-kb band in the targeted locus (RGE). With EagI-digested genomic DNA, an exon 1 probe revealed a 20-kb fragment of the wild-type Tgfb1 gene and an 11-kb band in the targeted locus. Three ES clones with a homologous recombination event were identified, two of which included the RGE mutation.
Generation of homozygous mutant mice
An ES clone was used for injecting blastocysts of C57BL/6J mice. The chimeric mice were bred with C57BL/6J mice to achieve germline transmission. Tgfb1+/RGE mice were bred to generate homozygous mice. TGFβ1 protein levels were undetectable in these mice because of the presence of Neo. Cre-deleter mice (a gift of A. Joyner) were crossed with Tgfb1+/RGE mice. Two Cre lines were used, one of Swiss-Webster background and the other C57BL/6J. Removal of Neo was confirmed by PCR showing undetectable product using primers for Neo (gaacaagatggattgcacgc and gaagaactcgtcaagaagggc) and an appropriate-sized fragment using primers flanking the Neo insertion site (gaggagacaagatctctcaga and caatggccctacacacacacagag).
Mice were bred on the two mixed backgrounds determined by the background of the Cre mouse: C57BL/6 + 129SvEvTac and C57BL/6 + 129SvEvTac + Swiss-Webster. Phenotypes on the two backgrounds were indistinguishable. Results reported here are from the C57BL/6 + 129SvEvTac background.
Primary fibroblasts
Lungs from newborn mice were minced and placed in tissue culture dishes with high-glucose DME containing 10% FCS, glutamine, penicillin, and streptomycin to allow lung fibroblasts to proliferate. Within the first three passages, cells were transfected with an expression plasmid containing cDNA for the SV40 large T antigen.
TGFβ bioassay
Sera from wild-type and mutant mice were heated to 80°C for 10 min to activate latent TGFβ and then diluted 1:4 in DME. Bioactive TGFβ was measured by adding samples to mink lung epithelial cells stably transfected with a TGFβ-responsive luciferase reporter construct and then assaying cell lysates for luciferase activity (Abe et al., 1994). Conditioned media were obtained by incubating equal numbers of cells in DME containing 0.1% BSA overnight, heat activated as described, diluted with an equal volume of DME, and assayed as described. Samples were tested in the presence and absence of a TGFβ1-specific inhibitory antibody (R&D Systems) or an inhibitory antibody against all three TGFβ isoforms (1D11; R&D Systems). Results are the means of triplicates ± SEM.
TGFβ inhibitory activity of LAP
Recombinant simian wild-type and RGE LAP were produced as described previously (Munger et al., 1999). Active recombinant TGFβ1 was added to DME supplemented with 0.1% BSA. Aliquots of this stock were combined with varying concentrations of recombinant LAP and then added to mink lung epithelial reporter cells and assayed as described.
TGFβ1 transfections and LAP immunoblotting
CHO-K7 cells stably expressing LTBP1 (a gift from J. Annes and D. Rifkin, New York University) were transfected with expression vectors containing cDNA encoding different forms of TGFβ using Lipofectamine Plus (Invitrogen) or with empty plasmid as described previously (Annes et al., 2002, 2004). The next day, cells were used to condition serum-free medium (DME supplemented with 0.1% BSA and glutamine) for 24 h. Immunoblotting with anti-LAP mAb VB3A9 was done as described previously (Annes et al., 2004).
TSP1 activation of TGFβ1
The three types of serum-free conditioned media obtained as described above were incubated with 11 nM of purified, stripped TSP1 (a gift from J. Murphy-Ullrich, University of Alabama, Birmingham, AL; Ribeiro et al., 1999) for 1 h at 37°C and then assayed with mink lung epithelial reporter cells as described, with or without addition of a TGFβ-inhibitory antibody (1D11).
RT-PCR
Primary cultures of lung fibroblasts (see Primary fibroblasts) were used as the source of RNA. RNA was extracted and reverse transcribed by standard techniques. PCR was performed using primers based on the 5′ and 3′ ends of the TGFβ1 coding sequence (5′ primer, tactgccgcttctgctcccac; and 3′ primer, caggagcgcacaatcatgttg). The resulting PCR product was the expected size and was completely sequenced. Semiquantitative RT-PCR was done with RNA isolated from lungs, liver, and heart of Tgfb1−/− and Tgfb1RGE/RGE mice (forward primer, exon 4: cggaatacagggctttcgatt; and reverse primer, exon 6: cttgctgtactgtgtgtccaggc). The PCR program was 94°C for 4 min; 94°C for 45 s, 60°C for 45 s, and 72°C for 90 s for 20, 25, or 29 cycles; and 72°C for 10 min. Amplification of β-actin cDNA (primers used: atctggcaccacaccttctacaatgagctgcg and cgtcatactcctgcttgctgatccacatctgc) was done as a control.
Immunohistochemistry
5-μm sections of formalin-fixed tissue were used for detection of the β6-integrin subunit. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide in methanol for 15 min. Antigen retrieval was with Digest-All 3 Pepsin (Zymed Laboratories) for 5–7 min. Blocking was with avidin/biotin block solution (Vector Laboratories) followed by 0.5% casein solution for 15 min. Anti-β6 monoclonal antibody ch2A1 (a gift from S. Violette, Biogen Idec, Cambridge, MA), a humanized version of a mouse anti-β6 IgG mAb, was used at 0.5 μg/ml in 0.1% BSA for 1 h at room temperature. A Vectastain ABC kit (Vector Laboratories) with anti-human secondary antibody was used according to directions, with 3,3-diaminobenzidine/hydrogen peroxide as chromogen (Sigma Fast tablets; Sigma-Aldrich) and hematoxylin counterstaining. Specificity was confirmed by absent staining on lung sections from Itgb6−/− mice.
LC immunostaining
Ia+ LCs in epidermal sheets were detected as described previously (Thomas et al., 2001). For counts, epidermal sheets from four Itgb6−/− and three wild-type mice were stained (6–12 sheets per mouse; mean 8.6). A digital image (400×) of each stained sheet was captured, and all LCs were counted. The mean cell count for each mouse was the average count of all sheets for that mouse. Statistical significance of the difference between means for wild-type and Itgb6−/− mice was assessed by two-tailed t-test.
Flow cytometry analysis of CD4+CD25+ and Foxp3+ splenocytes
CD4+ splenocytes were isolated from total splenocytes stained by negative selection with immunomagnetic beads (CD4+ T Cell isolation kit; Miltenyi Biotec). Flow cytometry was performed after labeling with phycoerythrin-conjugated anti-CD4 and FITC-conjugated anti-CD25 mAbs (BD Biosciences) or isotype controls to determine the fraction of CD4+ cells that were CD25+. To determine the fraction of CD4+ cells that were Foxp3+, cells were labeled with FITC-conjugated anti-CD4 antibody, and intracellular staining with a phycoerythrin-conjugated anti-Foxp3 antibody was performed (eBioscience).
Protocol for genotyping RGE mice
Primers are CGGAATACAGGGCTTTCGATT and GGTACGGGCATTCTGGATAC. PCR program is 94°C for 4 h and then 30 cycles of 94°C for 30 min, 60°C for 30 min, and 72°C for 50 min, followed by 72°C for 10 h. PCR products are digested with BstUI and electrophoresed on agarose gels.
Image acquisition and manipulation
A light microscope (DM LB; Leica) captured images with a 10, 20, or 40× objective lens at room temperature. Permount imaging medium was used. The camera used was a Spot Insight Color (model 3.2.0), and the acquisition software was the Spot program version 4.0.9, both by Diagnostic Instruments.
Online supplemental material
Fig. S1 shows the similarity of inflammatory lesions in Tgfb1RGE/RGE and Tgfb1−/− mice. Fig. S2 shows data from transfection experiments demonstrating that the normal and RGE forms of latent TGFβ1 can be incorporated into ECM and that cells expressing αvβ6 or αvβ8 can activate the normal form but not the RGE form of ECM-bound latent TGFβ1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200611044/DC1.
Acknowledgments
The authors thank Alexandra Joyner and Sema Sgaier for advice on ES cell targeting; Shelia Violette and colleagues at Biogen Idec for anti-β6 mAbs; Joanne Murphy-Ullrich for purified TSP1; Dean Sheppard for Itgb−/− mice; Polo Black-Golde and Samantha Streicher for additional experiments; and Dan Rifkin and Justin Annes for helpful discussions.
The work was funded by National Institutes of Health grant RO1 HL063786 (J.S. Munger).
Abbreviations used in this paper: E, embryonic day; ES, embryonic stem; LAP, latency-associated peptide; LC, Langerhans cell; LTBP, latent TGFβ-binding protein; MMP, matrix metalloproteinase; TSP1, thrombospondin-1.
References
- Abe, M., J.G. Harpel, C.N. Metz, I. Nunes, D.J. Loskutoff, and D.B. Rifkin. 1994. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 216:276–284. [DOI] [PubMed] [Google Scholar]
- Annes, J.P., D.B. Rifkin, and J.S. Munger. 2002. The integrin αvβ6 binds and activates latent TGFβ3. FEBS Lett. 511:65–68. [DOI] [PubMed] [Google Scholar]
- Annes, J.P., J.S. Munger, and D.B. Rifkin. 2003. Making sense of latent TGFβ activation. J. Cell Sci. 116:217–224. [DOI] [PubMed] [Google Scholar]
- Annes, J.P., Y. Chen, J.S. Munger, and D.B. Rifkin. 2004. Integrin αvβ6-mediated activation of latent TGF-β requires the latent TGF-β binding protein-1. J. Cell Biol. 165:723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya, J., S. Cambier, A. Morris, W. Finkbeiner, and S.L. Nishimura. 2006. Integrin-mediated transforming growth factor-β activation regulates homeostasis of the pulmonary epithelial-mesenchymal trophic unit. Am. J. Pathol. 169:405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, Y., H. Ihn, K. Yamane, M. Jinnin, Y. Mimura, and K. Tamaki. 2005. Involvement of αvβ5 integrin-mediated activation of latent transforming growth factor-β1 in autocrine transforming growth factor-β signaling in systemic sclerosis fibroblasts. Arthritis Rheum. 52:2897–2905. [DOI] [PubMed] [Google Scholar]
- Bader, B.L., H. Rayburn, D. Crowley, and R.O. Hynes. 1998. Extensive vasculogenesis, angiogenesis, and organogenesis precede lethality in mice lacking all αv integrins. Cell. 95:507–519. [DOI] [PubMed] [Google Scholar]
- Barcellos-Hoff, M.H., and T.A. Dix. 1996. Redox-mediated activation of latent transforming growth factor-β1. Mol. Endocrinol. 10:1077–1083. [DOI] [PubMed] [Google Scholar]
- Borkowski, T.A., J.J. Letterio, C.L. Mackall, A. Saitoh, X.J. Wang, D.R. Roop, R.E. Gress, and M.C. Udey. 1997. A role for TGFβ1 in Langerhans cell biology. Further characterization of the epidermal Langerhans cell defect in TGFβ1 null mice. J. Clin. Invest. 100:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, S.E., V. Stellmach, J.E. Murphy-Ullrich, S.M. Ribeiro, J. Lawler, R.O. Hynes, G.P. Boivin, and N. Bouck. 1998. Thrombospondin-1 is a major activator of TGF-β1 in vivo. Cell. 93:1159–1170. [DOI] [PubMed] [Google Scholar]
- Dickson, M.C., J.S. Martin, F.M. Cousins, A.B. Kulkarni, S. Karlsson, and R.J. Akhurst. 1995. Defective haematopoiesis and vasculogenesis in transforming growth factor-β1 knock out mice. Development. 121:1845–1854. [DOI] [PubMed] [Google Scholar]
- Huang, X.Z., J.F. Wu, D. Cass, D.J. Erle, D. Corry, S.G. Young, R.V. Farese, and D. Sheppard. 1996. Inactivation of the integrin β6 subunit gene reveals a role of epithelial integrins in regulating inflammation in the lung and skin. J. Cell Biol. 133:921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen, V., J.W. Voncken, C. Shuler, D. Warburton, D. Bu, N. Heisterkamp, and J. Groffen. 1995. Abnormal lung development and cleft palate in mice lacking TGF-β3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 11:415–421. [DOI] [PubMed] [Google Scholar]
- Lu, M., J.S. Munger, M. Steadele, C. Busald, M. Tellier, and L.M. Schnapp. 2002. Integrin α8β1 mediates adhesion to LAP-TGFβ1. J. Cell Sci. 115:4641–4648. [DOI] [PubMed] [Google Scholar]
- Ludbrook, S.B., S.T. Barry, C.J. Delves, and C.M. Horgan. 2003. The integrin αvβ3 is a receptor for the latency-associated peptides of transforming growth factors β1 and β3. Biochem. J. 369:311–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow, A., K.O. Yee, R. Lipman, R. Bronson, P. Weinreb, X. Huang, D. Sheppard, and J. Lawler. 2005. Characterization of integrin β6 and thrombospondin-1 double-null mice. J. Cell. Mol. Med. 9:421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, D., S. Cambier, L. Fjellbirkeland, J.L. Baron, J.S. Munger, H. Kawakatsu, D. Sheppard, V.C. Broaddus, and S.L. Nishimura. 2002. The integrin αvβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-β1. J. Cell Biol. 157:493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger, J.S., J.G. Harpel, F.G. Giancotti, and D.B. Rifkin. 1998. Interactions between growth factors and integrins: latent forms of transforming growth factor-β are ligands for the integrin αvβ1. Mol. Biol. Cell. 9:2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger, J.S., X. Huang, H. Kawakatsu, M.J. Griffiths, S.L. Dalton, J. Wu, J.F. Pittet, N. Kaminski, C. Garat, M.A. Matthay, et al. 1999. The integrin αvβ6 binds and activates latent TGFβ1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 96:319–328. [DOI] [PubMed] [Google Scholar]
- Oshima, M., H. Oshima, and M.M. Taketo. 1996. TGF-β receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev. Biol. 179:297–302. [DOI] [PubMed] [Google Scholar]
- Proetzel, G., S.A. Pawlowski, M.V. Wiles, M. Yin, G.P. Boivin, P.N. Howles, J. Ding, M.W. Ferguson, and T. Doetschman. 1995. Transforming growth factor-β3 is required for secondary palate fusion. Nat. Genet. 11:409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro, S.M., M. Poczatek, S. Schultz-Cherry, M. Villain, and J.E. Murphy-Ullrich. 1999. The activation sequence of thrombospondin-1 interacts with the latency-associated peptide to regulate activation of latent transforming growth factor-β. J. Biol. Chem. 274:13586–13593. [DOI] [PubMed] [Google Scholar]
- Sato, Y., R. Tsuboi, R. Lyons, H. Moses, and D.B. Rifkin. 1990. Characterization of the activation of latent TGF-β by co-cultures of endothelial cells and pericytes or smooth muscle cells: a self- regulating system. J. Cell Biol. 111:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull, M.M., I. Ormsby, A.B. Kier, S. Pawlowski, R.J. Diebold, M. Yin, R. Allen, C. Sidman, G. Proetzel, D. Calvin, et al. 1992. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. 359:693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, R.M., D.V. Belsito, C. Huang, L.-z. Chen, I. Ormsby, W.J. Simmons, P. Cowin, J. Shaw, T. Doetschman, and G.J. Thorbecke. 2001. Appearance of Langerhans cells in the epidermis of Tgfb1 −/− SCID mice: paracrine and autocrine effects of transforming growth factor-β1 and -β2. J. Invest. Dermatol. 117:1574–1580. [DOI] [PubMed] [Google Scholar]
- Yu, Q., and I. Stamenkovic. 2000. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev. 14:163–176. [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., K. Motejlek, D. Wang, K. Zang, A. Schmidt, and L.F. Reichardt. 2002. β8 integrins are required for vascular morphogenesis in mouse embryos. Development. 129:2891–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]