Abstract
Centromeres direct chromosomal inheritance by nucleating assembly of the kinetochore, a large multiprotein complex required for microtubule attachment during mitosis. Centromere identity in humans is epigenetically determined, with no DNA sequence either necessary or sufficient. A prime candidate for the epigenetic mark is assembly into centromeric chromatin of centromere protein A (CENP-A), a histone H3 variant found only at functional centromeres. A new covalent fluorescent pulse-chase labeling approach using SNAP tagging has now been developed and is used to demonstrate that CENP-A bound to a mature centromere is quantitatively and equally partitioned to sister centromeres generated during S phase, thereby remaining stably associated through multiple cell divisions. Loading of nascent CENP-A on the megabase domains of replicated centromere DNA is shown to require passage through mitosis but not microtubule attachment. Very surprisingly, assembly and stabilization of new CENP-A–containing nucleosomes is restricted exclusively to the subsequent G1 phase, demonstrating direct coupling between progression through mitosis and assembly/maturation of the next generation of centromeres.
Introduction
Kinetochores mediate the interaction between chromosomes and spindle microtubules, thereby enabling mitotic chromosome movement, and produce a mitotic checkpoint signal that ensures bipolar attachment of all chromosomes before anaphase onset (Cleveland et al., 2003; Chan et al., 2005). Assembly of the kinetochore during mitosis takes place at the centromere, a megabase-sized specialized chromatin region typically formed on arrays of α satellite DNA (Cleveland et al., 2003; Amor et al., 2004b; Carroll and Straight, 2006).
Despite the prevalence of centromeres at adenine-thymine–rich repetitive α satellite DNA, the DNA sequences themselves appear to play a nonessential role in centromere specification. This is most clearly exemplified by the characterization of human neocentromeres. In these rare but naturally occurring patient cases, a specific centromere has relocated to another site on the chromosome without any apparent DNA rearrangements, concomitant with vacating the original α satellite–containing locus (Amor and Choo, 2002; Amor et al., 2004a; Ventura et al., 2004). This shows that DNA sequences normally associated with centromeres are neither necessary nor sufficient to promote centromere propagation and that maintenance of centromeres is determined predominantly in an epigenetic manner.
Centromere protein A (CENP-A) is a conserved histone H3 variant that replaces canonical H3 specifically at centromeres in all known eukaryotes (Palmer et al., 1987; Meluh et al., 1998; Henikoff et al., 2000; Oegema et al., 2001) and has been shown to be required for the localization of nearly all other centromere and kinetochore components (Howman et al., 2000; Oegema et al., 2001; Goshima et al., 2003; Amor et al., 2004a; Regnier et al., 2005; Foltz et al., 2006; Liu et al., 2006). We have recently shown that the loop1 and α2 helix of the CENP-A histone fold domain is responsible for forming a rigid/inaccessible interface with histone H4 and that this region, when transplanted into canonical histone H3, confers centromere targeting (Black et al., 2004, 2007a) and provides an essential function of CENP-A (Black et al., 2007b). CENP-A chromatin directly recruits a six-component CENP-A nucleosome-associated complex (CENP-ANAC) that forms the foundation for the assembly of other centromere components and the kinetochore during mitosis (Foltz et al., 2006). The existence of a CENP-A–directed centromere-specific chromatin structure makes CENP-A a prime candidate for the epigenetic propagation of centromere identity. This directly implies that CENP-A propagation at the centromere is a partially or completely self-directed process. It is, however, unknown how CENP-A is discriminated from canonical histone H3 and how its specific incorporation at centromeric nucleosomes is achieved.
Earlier models have suggested that differences in timing of replication of centromeric DNA versus the genome overall may provide a temporal window permissive for CENP-A loading (O'Keefe et al., 1992; Csink and Henikoff, 1998). However, this appears not to be the case, as replication of centromeric DNA is not restricted to a specific time during S phase (Shelby et al., 2000; Sullivan and Karpen, 2001). Alternatively, CENP-A loading could be separate from assembly of canonical histones altogether by allowing CENP-A loading outside S phase. Indeed, DNA replication is not required for CENP-A assembly and CENP-A mRNA, and protein levels peak only after S phase during late G2 phase, consistent with a disconnect between the timing of CENP-A and H3 assembly (Shelby et al., 1997, 2000). Whether propagation of centromeric chromatin and general chromatin is indeed temporally distinct and how and when CENP-A nucleosomes turn overis not known. This we now test by developing and exploiting a novel, covalent fluorescent pulse-labeling strategy with SNAP tagging.
Results
Timing of assembly and turnover of CENP-A at centromeres using the SNAP tag
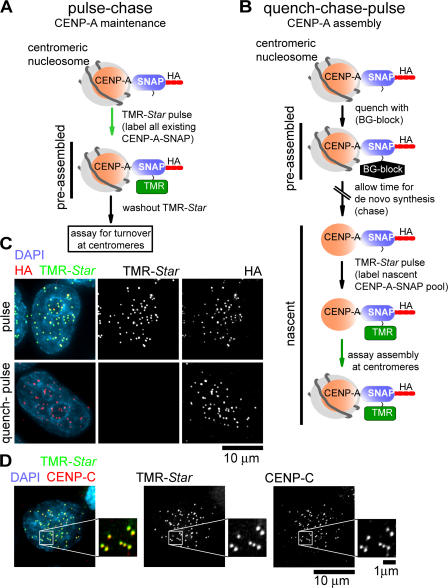
The SNAP tag, a modified variant of the suicide enzyme O6-alkylguanine-DNA alkyltransferase, whose normal function is in DNA repair, has been extensively engineered to covalently and irreversibly modify (and inactivate) itself through acceptance of the cell-permeable guanine derivative O6-benzylguanine (BG; or fluorescent derivatives thereof). In effect, this allows labeling of SNAP fusion proteins at will in vivo (Keppler et al., 2003, 2004, 2006). We applied pulse labeling with this methodology to determining CENP-A turnover specifically at centromeres (Fig. 1 A) as well as quench-chase-pulse labeling to follow the fate of newly synthesized CENP-A (Fig. 1 B). We established cell lines stably expressing centromere-localized CENP-A–SNAP at near endogenous levels in HeLa cells (Fig. 1 D and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200701066/DC1).
Figure 1.
Principle of SNAP tag–based pulse labeling. (A and B) Schematic of labeling strategies for pulse-chase labeling (A) or quench-chase-pulse labeling (B) of CENP-A–SNAP fusion protein with BG (BG-block; quench) or TMR-Star (pulse). (C) The SNAP tag can be efficiently labeled in vivo using fluorescent TMR-Star (top) and quenched using nonfluorescent BG (bottom). CENP-A–SNAP cells were labeled with TMR-Star for 15 min or were treated with BG-block for 30 min before TMR-Star labeling and processing for immunofluorescence with anti-HA. (D) TMR-Star–labeled CENP-A–SNAP is centromere localized. Cells were TMR-Star labeled for 15 min and processed for immunofluorescence with anti–CENP-C.
Multiple lines of evidence indicated that CENP-A with the SNAP tag substituted functionally for CENP-A in centromere maintenance. We have previously reported that transgene-encoded CENP-A expression leads to reduction of the endogenous CENP-A pool through competition at the protein level (Foltz et al., 2006). Here, a similar reduction in endogenous CENP-A in response to CENP-A–SNAP expression resulted in an unchanged overall CENP-A pool, the majority of which was SNAP tagged (Fig. S1 A; line 23 is used for all pulse-labeling experiments). Because chronic reduction of CENP-A to <50% is a cell-autonomous lethal event (Black et al., 2007b), the SNAP-tagged CENP-A pool in the stable CENP-A–SNAP cell lines not only competed for assembly at centromeres with authentic CENP-A but also provided essential aspects of CENP-A function in centromere maintenance. (Retention of substantial CENP-A function by CENP-A–SNAP [219-aa tag] is in agreement with what has been shown for N-terminally YFP-tagged [240 aa] or C-terminally tandem affinity–tagged [TAP; 172 aa] CENP-A, which, respectively, rescue CENP-A lethality and incorporate into bona fide centromeric nucleosomes that are associated with a six-member complex of centromere components [CENP-A–TAP; Foltz et al., 2006; Black et al., 2007b].) 15-min pulse labeling with the tetramethylrhodamine (TMR)-conjugated SNAP substrate, TMR-Star, specifically identified CENP-A–SNAP already assembled into centromeric chromatin (Fig. 1 C, top). Preincubation of CENP-A–SNAP–expressing cells with the nonfluorescent SNAP substrate (BG-block) led to complete quenching of SNAP and rendered CENP-A undetectable with TMR-Star (Fig. 1 C, bottom).
CENP-A is stably associated with centromeres across the cell cycle
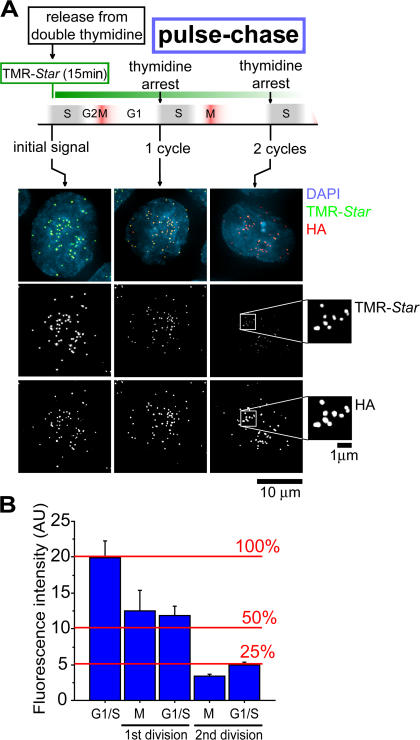
To determine turnover of CENP-A at centromeres, cells were synchronized at the G1–S boundary by tandem treatments with thymidine. CENP-A bound to unreplicated centromeres was pulse labeled with TMR-Star and chased for up to two cell cycles (Fig. 1 A and Fig. 2 A). Consistent with earlier reports on total CENP-A levels that had indicated slow protein turnover (Shelby et al., 2000; Regnier et al., 2005), centromere duplication in the initial round of DNA synthesis produced a 60 ± 14% reduction in intensity of TMR-Star–labeled CENP-A–SNAP at individual centromeres by the first mitosis and through the subsequent G1 (Fig. 2). After a second cycle of DNA replication, the previously labeled, centromere-bound CENP-A–SNAP was diminished to 25 ± 5% of its initial level, whereas the total number of fluorescent centromeres positive per cell remained unchanged throughout the experiment (Fig. 2). Thus, despite continued synthesis of both SNAP-tagged and endogenous CENP-A, CENP-A already loaded into centromeric chromatin by late G1 is redistributed to, and retained by, daughter centromeres.
Figure 2.
CENP-A turnover at centromeres. (A) Outline of cell synchronization and labeling regimen for CENP-A turnover. Cells were synchronized and labeled as depicted followed by fixation and immunofluorescence with anti-HA. Representative images for each time point are shown. After one and two cell cycles, pulse chase–labeled CENP-A–SNAP remaining was detected (insets after two cell cycles [50 h] are magnified an additional 3× and intensity scaled to visualize remaining centromere fluorescence). (B) Quantification of mean TMR-Star intensity at indicated time points. Reduction of signal at each division became apparent in mitosis, during which sister centromeres split and can be resolved individually. A minimum of 500 centromeres in 10 different cells were quantified for each measurement. Error bars represent SEM of centromere intensity. AU, arbitrary units.
Loading of newly synthesized CENP-A initiates in telophase
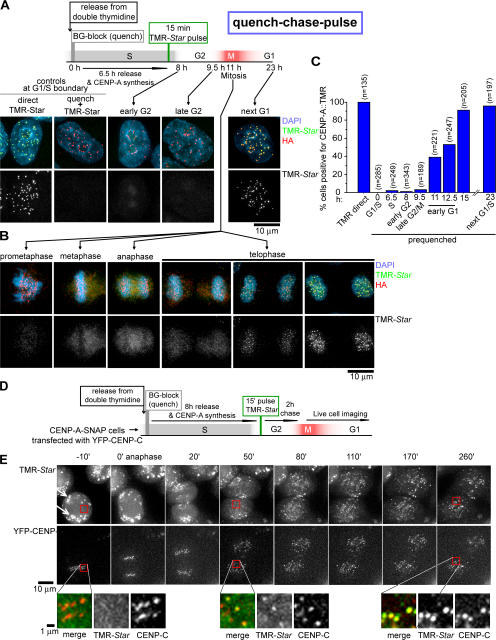
CENP-A must be replenished at centromeres after DNA replication to complete duplication of new centromeres. To determine the timing of CENP-A incorporation into chromatin of newly replicated centromeres, cells stably expressing CENP-A–SNAP were synchronized at the G1–S boundary by double thymidine block, and both centromere-associated and any free pool of CENP-A–SNAP were quenched with nonfluorescent BG (Fig. 1 B and Fig. 3 A). The cells were then released into S phase for 6.5 h, nascent CENP-A–SNAP was pulse labeled for 15 min by reaction with TMR-Star, and incorporation of the fluorescent CENP-A was examined in late S, G2, M, and the subsequent G1. CENP-A synthesized during S phase was diffusely localized in the nucleus (Fig. 3 A and Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200701066/DC1) but did not appear at centromeres during any phase of G2 or M. Rather, only after passage through mitosis and entry into the G1 phase of the next cell cycle did CENP-A assemble at centromeres (Fig. 3 A). Quantification of the number of cells positive for CENP-A loading confirmed that the initially synchronized cell population did not load substantial levels of CENP-A before ∼11 h after release from thymidine and concomitant with entry into G1 (Fig. 3 C).
Figure 3.
CENP-A loading initiates in telophase/early G1. (A) Schematic of cell synchronization and labeling protocol. Cell cycle stages are estimates based on time elapsed after release from double thymidine–induced arrest at G1–S. Representative images for each time point are shown. (B) Different stages of mitosis are shown at 11 h after thymidine release. (C) Percentages of cells positive for TMR-Star fluorescence at indicated time points (with estimated cell cycle stage below) after release from thymidine. Numbers in parentheses represent the number of cells counted for each time point. (D and E) Live-cell time lapse of CENP-A–SNAP assembly at centromeres in early G1. (D) Schematic of cell synchronization and labeling protocol for live-cell time-lapse imaging of CENP-A–SNAP assembly at centromeres in early G1. CENP-A–SNAP cells were transiently transfected with YFP–CENP-C–expressing construct 48 h before SNAP-labeling regimen. (E) Representative stills of a metaphase cell exiting mitosis and assembling CENP-A–SNAP across an ∼4-h time course (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200701066/DC1). A portion of TMR-Star dye is nonspecifically retained near the cell periphery (presumably in internal membranes; white arrows), but no TMR-Star signal is detected at centromeres at this time (colocalizing with YFP–CENP-C [red boxes]). Time points are with respect to anaphase onset. Boxed regions highlight the initial absence of CENP-A–SNAP at centromeres and its earliest detection by 50 min after anaphase onset and continued assembly out to 260 min. All images are equally depth scaled across time.
Close examination of cells just before and after mitotic entry revealed that the earliest time CENP-A loading could be detected at centromeres was concomitant with nuclear envelope reformation and completion of furrow ingression as indicated by midbody formation in late telophase/early G1 (Fig. 3 B and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200701066/DC1). No newly made CENP-A was observed at centromeres at any time during mitosis before telophase. The absence of CENP-A at centromeres at these earlier time points cannot be attributed to a pool of newly synthesized CENP-A too small to be detected because the prelabeled pool size is the same for all time points. The pattern of loading restricted to late mitosis/early G1 was not a result of thymidine treatment per se because randomly cycling cells were also dependent on mitotic progression to permit CENP-A loading (Fig. S1).
To time the arrival of CENP-A–SNAP at the centromere more accurately, we followed live cells containing a pool of TMR-Star–labeled but nonassembled CENP-A–SNAP from metaphase through early G1 (Fig. 3 D). TMR-Star labeling of live cells resulted in the nonspecific retention near the cell periphery (presumably in internal membranes) of a proportion of the dye, a proportion that is removed during normal fixation and washing conditions. Nevertheless, no TMR-Star signal could be detected specifically at centromeres in metaphase (Fig. 3 E). However, assembly of nascent CENP-A–SNAP could be detected as early as ∼50 min after anaphase onset and continued for several hours in early G1 (Fig. 3 E and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200701066/DC1).
CENP-A assembly occurs exclusively during G1
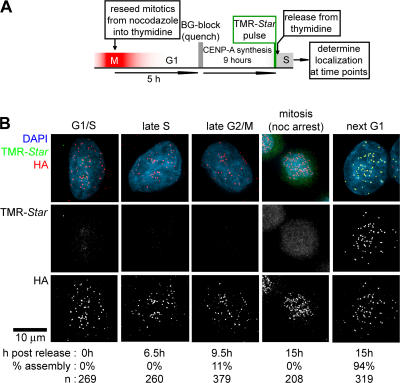
Next, we determined whether loading of CENP-A is unique to the early hours of G1 or whether loading is also permissive at any other point in the cell cycle, including the possibility of a secondary CENP-A loading stage, as has been suggested in fission yeast (Takahashi et al., 2005). CENP-A–SNAP in mitotic cells arrested with nocodazole treatment was initially quenched with nonfluorescent BG, and a G1 phase cell population was generated by release from nocodazole arrest. The CENP-A–SNAP pool produced during mid-to-late G1 was labeled and monitored for timing of centromeric deposition (Fig. 4). DNA content was assayed by FACS to verify cell cycle position (unpublished data). No assembly of CENP-A–SNAP–TMR-Star was detectable at centromeres during the subsequent S, G2, or M phases (Fig. 4 B). However, fluorescent CENP-A from the prior G1 assembled into the new daughter centromeres after exit from this subsequent mitosis (Fig. 4 B). Thus, despite the presence of a stable noncentromeric CENP-A pool, no loading occurred at any stage of the cell cycle before the following G1 phase (10–18 h after CENP-A synthesis and labeling).
Figure 4.
Nascent CENP-A loads exclusively during G1. (A) Schematic of cell synchronization and late G1 labeling protocol. (B) Representative images at indicated time points. Percentages of cells that have loaded CENP-A are indicated below. Note that the 11% of cells that have loaded CENP-A by 9.5 h represent cells that have already entered the next G1 phase, as evident from the absence of CENP-A loading in cells blocked in mitosis by nocodazole.
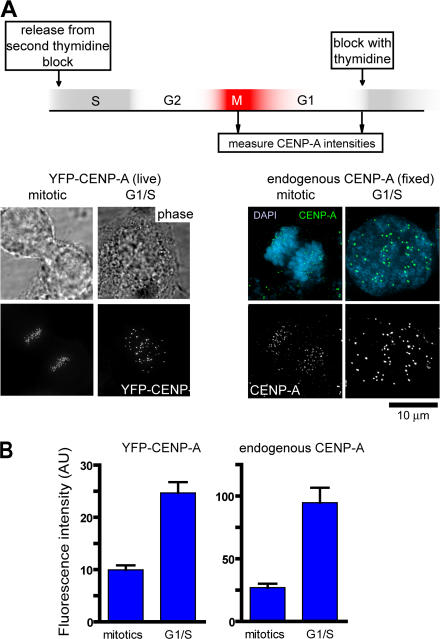
Although SNAP-tagged CENP-A faithfully tracks to centromeres and provides an essential function of CENP-A in centromere maintenance, there remained the possibility that the SNAP tag or the cell synchronization methods we have used would interfere with the timing of CENP-A loading. If CENP-A loading is normally restricted to early G1, levels of CENP-A (tagged or endogenous) on individual centromeres should double from early mitosis to when loading is completed, in late G1. On the other hand, if CENP-A loading occurs before mitosis, as previously proposed (Shelby et al., 2000), CENP-A levels in mitosis and G1 would be similar. Examination using indirect immunofluorescence to track endogenous CENP-A or direct fluorescence measurement of a cell line stably expressing YFP–CENP-A revealed that in both cases CENP-A levels increased from M to late G1 (∼3.4- and ∼2.5-fold, respectively; Fig. 5), findings only consistent with CENP-A loading in G1 rather than before mitosis.
Figure 5.
Centromeric levels of endogenous CENP-A increases during G1 phase. (A) HeLa cells stably expressing YFP–CENP-A (Kops et al., 2004) or parental HeLa cells were synchronized by tandem treatments with thymidine and released, and cells in mitosis or at the G1–S boundary were either imaged directly (YFP fluorescence) in live cells or processed for indirect immunofluorescence with anti–CENP-A (cell cycle times not drawn to scale). (B) Quantification of mean CENP-A intensity. For YFP–CENP-A, >600 centromeres from >10 cells, and for endogenous CENP-A, >200 centromeres from five different cells were quantified for each measurement. Error bars represents SEM of centromere intensity. AU, arbitrary units.
Passage trough mitosis is critical for CENP-A assembly in early G1
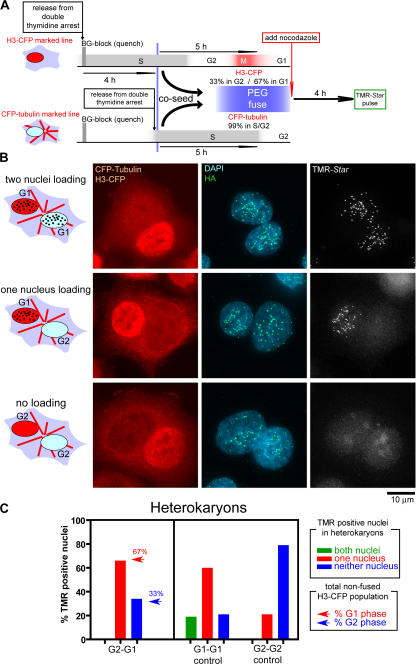
The discrete, abrupt onset of CENP-A assembly as cells exit from mitosis suggested that passage through mitosis is a prerequisite for CENP-A assembly. Alternatively, entering the G1 cell cycle state may be triggering CENP-A assembly without any mechanistic involvement of mitosis per se. To distinguish these possibilities, the G1 cell cycle phase was disconnected from mitotic passage by combining the SNAP-based CENP-A assembly assay with a classic cell–cell fusion approach (Rao and Johnson, 1970). Heterophasic heterokaryons were generated by fusing G1 cells with G2 cells, each expressing CENP-A–SNAP and each uniquely marked by stable expression of CFP-tagged histone H3 or tubulin, respectively, to mark nuclei or microtubules (Fig. 6 A). The two differentially marked CENP-A–SNAP cell populations were synchronized by double thymidine treatment. Previously deposited CENP-A–SNAP was quenched, and each population was released for differing lengths of time so as to produce two synchronized populations, one of which was at mitosis/early G1 (H3-CFP cells) and the other at late S/early G2 phase (CFP-tubulin cells). The two populations were mixed, and cell fusion was induced with polyethylene glycol (PEG). Nocodazole was added to prevent any further passage through mitosis. 4 h after fusion, TMR-Star labeling was used to assay in both nuclei of the heterokaryons for assembly at centromeres of the unloaded, newly synthesized CENP-A–SNAP pool that was present in all nuclei (Fig. 6 A).
Figure 6.
Passage through mitosis is critical for CENP-A loading in early G1. (A) Schematic of cell synchronization, labeling, and PEG-mediated cell–cell fusion protocol. CENP-A–SNAP cells marked with H3-CFP or CFP-tubulin were sequentially released from a double thymidine block, whereas prior assembled CENP-A–SNAP was quenched. At the time of PEG-mediated fusion H3-CFP and CFP-tubulin cells were in G1 or G2 at the indicated frequencies based on the fraction of cells that had loaded CENP-A–SNAP. After PEG fusion, nocodazole was added to prevent any additional passage through mitosis, and CENP-A–SNAP loading in binucleate heterokaryons was determined after 4 additional hours by TMR-Star labeling. (B) Representative images of binucleate heterokaryons double labeled with H3-CFP and CFP-tubulin, in which both, one, or none of the nuclei has assembled CENP-A–SNAP at centromeres. (C) Frequency of binucleate heterokaryons in which both, one, or none of the nuclei have loaded CENP-A–SNAP at centromeres (TMR-positive nuclei) in fusions of the indicated populations. Arrows in bar graph for G2–G1 cell fusion experiment represent the fraction of H3-CFP cells that were in G1 phase (red) or G2 phase (blue) at the time of fusion. At least 30 binucleate heterokaryons were analyzed in each experiment.
After cell–cell fusions, nuclei originating from G2 and G1 cells share the same cytoplasm and, in principle, the same cell cycle state. Control fusions revealed that, as expected, loading of CENP-A in both nuclei occurred exclusively in G1 cell to G1 cell fusions. In contrast, no binucleate heterokaryons derived from fusion of two G2 populations could be found in which both nuclei loaded CENP-A (Fig. 6, B and C), and the vast majority (86%) loaded it in neither nucleus.
It should be noted that because of the short time cells spend in mitosis (∼1 h) and the inherent spread in synchrony as cells transverse across the cell cycle, an early G1 phase cell population will invariably contain a fraction of cells that are in G2. (In this case, 33% of the H3-CFP G1 population had in fact not yet reached G1 by the time cells were fused.) Therefore, in all fusions, a spread of heterokaryons loading CENP-A–SNAP at centromeres at one, both, or neither of the nuclei is expected. Nevertheless, despite this inherently imperfect synchrony, a striking finding was that in binucleate heterokaryons derived from fusion between cell populations enriched in G1 and G2, most (66%) G1 cell–derived nuclei (H3-CFP marked) recruited CENP-A–SNAP to centromeres to levels indistinguishable from surrounding nonfused G1 cells. In contrast, no heterokaryons were found that had assembled CENP-A in both nuclei, indicating that G2-derived nuclei, although sharing the same cytoplasm with a CENP-A–assembling G1-derived nucleus, did not assemble CENP-A (Fig. 6, B and C), despite the presence of fluorescently labeled CENP-A–SNAP. The frequency of heterokaryons loading CENP-A–SNAP in one or neither nucleus corresponded to the frequency of H3-CFP G1 and G2 cells at the time of fusion (Fig. 6 C, arrows), indicating that in heterokaryons the G1- and G2-derived nuclei are neither inducing nor inhibiting CENP-A assembly in the other nucleus. Therefore, the early G1 cell cycle state that is permissive for CENP-A assembly does not directly dictate the ability to load CENP-A. Rather, passage through mitosis is crucial to allow CENP-A assembly as cells enter G1.
Microtubule attachment is not required for CENP-A assembly in G1
Our experiments suggest that mitosis is a key cell cycle determinant in initiating CENP-A loading. To exclude the possibility that proficiency for CENP-A loading is determined by a “timing” mechanism rather than actual mitotic passage and G1 entry, cells were arrested using nocodazole to produce a nascent unloaded pool of CENP-A–SNAP in mitosis. Nocodazole-treated cells never assembled CENP-A–SNAP, even by the time 94% of control cells had reentered G1 and loaded CENP-A–SNAP (Fig. 4 B), reaffirming the notion that exit from mitosis is required for CENP-A loading.
Multiple processes occur during mitosis that might act to trigger new CENP-A nucleosome recruitment. These include chromatin stretching, which occurs during metaphase and has been proposed as a mechanism for the exchange of histone H3 for CENP-A–containing nucleosomes (Ahmad and Henikoff, 2002; Mellone and Allshire, 2003; Carroll and Straight, 2006). Although the concept of functional reinforcement of centromere location that is part of this model is appealing, no experimental evidence has been generated in support for such a mechanism. Alternatively, DNA decondensation or the presence of other mitotic kinetochore components may be integral to triggering the process of centromere assembly.
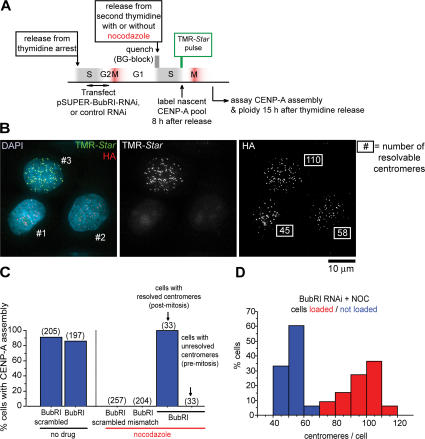
To test the tension-dependent CENP-A loading model, cells were produced that completed mitosis in the absence of microtubule attachment (and therefore microtubule-mediated chromatin stretching). To do this, cells were depleted of BubR1 with transcription-mediated short hairpin RNA and treated with nocodazole to block microtubule assembly, and CENP-A loading was assessed (Fig. 7 A). Under these conditions, cells enter mitosis without spindle assembly or kinetochore attachment, but quickly exit without the BubR1-dependent mitotic checkpoint (Kops et al., 2004).
Figure 7.
Microtubule attachment in mitosis is not required for CENP-A assembly at centromeres. (A) Schematic of cell synchronization, transfection, and labeling protocol. (B) Representative image of cells transiently transfected with a transcription-mediated BubR1 RNAi gene during late S/G2 phase, after which cells were arrested again with thymidine. After quenching CENP-A–SNAP with BG, thymidine inhibition was released, followed by addition of nocodazole to block spindle microtubule assembly. Newly made CENP-A–SNAP was pulse labeled with TMR-Star in the subsequent S/G2 (8 h after release from thymidine). 15 h after thymidine release, two of three cells (1 and 2) were premitotic (based on the centromere number consistent with unresolved sister centromere pairs) and had not loaded CENP-A–SNAP, whereas the third cell had exited mitosis without chromosome segregation and cytokinesis (centromere number consistent with an 8N DNA content in which sister centromeres are resolved) and had loaded CENP-A–SNAP. Centromere number per cell is indicated in the HA image. (C) Percentages of cells that had loaded CENP-A after the manipulations in A with the siRNAs as indicated. Bracketed numbers represent number of cells counted for each condition. (D) Frequency distribution of centromere numbers for cells with (TMR positive; red) or without (TMR negative; blue) CENP-A loading.
Depletion of BubRI alone did not affect the ability of cells to load CENP-A, whereas nocodazole treatment of cells with normal BubR1 levels prevented mitotic exit and any loading (Fig. 7 C). Nocodazole treatment of cells depleted of BubR1 (Fig. 7, B and C) or another mitotic checkpoint component Mad2 (not depicted) produced CENP-A loading to levels comparable to that seen in untreated cells, along with normal interphase nuclei with twice the number of resolved centromeres. This was indicative of a successful mitotic exit where sister chromatids had disjoined but failed to segregate and complete cytokinesis because of the absence of microtubules. Conversely, cells that did not load CENP-A were either arrested in mitosis (i.e., not depleted in BubRI) or had not yet entered mitosis (Fig. 7 D; as indicated by a centromere number consistent with unresolved sister centromeres, as is the case in G2 phase). Thus, passage through mitosis is critical for CENP-A loading, but microtubule attachment or microtubule-generated tension across centromeric chromatin is not.
Discussion
The SNAP tag, a self-labeling enzyme enabling in vivo fluorescent pulse-chase imaging
Our effort validates the SNAP tag (Keppler et al., 2003, 2004, 2006), coupled with indirect immunofluorescence or live-cell imaging, as an approach capable of visualizing and tracking intracellular dynamics of protein pools synthesized at different times. SNAP technology also stands out from other cell biological tools to determine protein dynamics, such as FRAP experiments, in that it allows the determination of protein turnover on a much longer time scale and is therefore well suited for proteins with long half-lives. To earlier efforts that had shown CENP-A to be long lived (Shelby et al., 2000; Regnier et al., 2005), with the SNAP tag approach we have demonstrated that, once assembled at centromeres, CENP-A does not turn over measurably within the ∼50-h time frame of our experiments. (An added benefit of this outcome is demonstration that the covalent SNAP-BG binding is indeed irreversible.) Moreover, the ability to differentially label SNAP protein pools synthesized at different times allows direct assessment of the fate of nascent proteins, including the turnover rates of proteins at the same cellular location, but assembled at different times.
Little CENP-A turnover across the cell cycle, but assembly in G1 phase
Using the SNAP tag approach, nearly all centromeric CENP-A is shown to remain centromere associated even during centromeric DNA replication, consistent with a role for CENP-A as an epigenetic marker maintaining centromere identity though cell division (Vafa and Sullivan, 1997; Warburton et al., 1997; Black et al., 2004, 2007a). More surprisingly, loading of newly synthesized CENP-A occurs in a discrete cell cycle window in early G1. A mitosis intervening between centromere DNA replication and new CENP-A loading is a prerequisite for CENP-A assembly. Although earlier work suggested that CENP-A may load in G2 phase based on an increase in overall CENP-A protein levels at this time (Shelby et al., 2000), our direct visualization with the SNAP tag has demonstrated that, despite its continued expression throughout the cell cycle, newly made CENP-A is accumulated in a nuclear, but noncentromeric, form before mitosis. The abrupt onset of CENP-A assembly at centromeres initiating at the end of mitosis firmly supports a model in which loading of CENP-A requires one or more key events during mitosis that may include nuclear envelope breakdown or chromatin decondensation, thereby allowing potential CENP-A assembly factors access to centromeric chromatin. Alternatively, assembly may be dependent on mitotic modification of CENP-A itself, which creates an environment that is permissive for subsequent CENP-A loading.
Although passage through mitosis itself is a strict requirement for CENP-A loading, microtubule attachment at kinetochores has no apparent role in CENP-A assembly, in contrast to previous proposals (Ahmad and Henikoff, 2002; Mellone and Allshire, 2003; Carroll and Straight, 2006). It remains possible, however, that components of the greater centromere/kinetochore affect CENP-A loading or stabilization after loading. Defects in structural centromere proteins have been shown to affect CENP-A levels (Kline et al., 2006; Okada et al., 2006). It is therefore possible that components of the centromere (which themselves are dependent on CENP-A for their localization) recruit new CENP-A or parts of the loading machinery. This would serve a direct epigenetic feedback between active centromeres and the propagation of new centromeric chromatin.
Finally, propagation of CENP-A chromatin may await the availability of an active loading factor or an adaptor molecule at centromeres. The recently identified hMis18α, hMis18β, and M18BP1/hsKNL2 proteins, of which the M18BP1/hsKNL2 Myb domain–containing protein is an evolutionary conserved component, have been proposed to be required for CENP-A loading (Fujita et al., 2007; see Maddox et al. on p. 757 of this issue). Strikingly, all these proteins display a pattern of centromere localization coincident with CENP-A assembly (from anaphase through early G1). Thus, recruitment of hMis18α, hMis18β, and M18BP1/hsKNL2 at the centromere during late anaphase could be dictated by a modification of centromeric chromatin coincident with mitotic exit or may represent a component of the CENP-A loading machinery that is itself activated during mitotic exit.
Implications for CENP-A assembly and centromeric chromatin structure
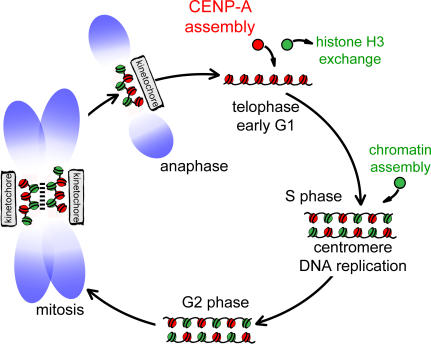
The sudden onset of CENP-A assembly exclusively after reentry into G1, but not in mitosis, carries with it two important implications for epigenetic centromere inheritance. First, a requirement for a subsequent mitosis as a prerequisite for loading of CENP-A onto previously replicated centromeric DNA intrinsically couples centromere replication and maturation to cell cycle progression. Second, loading of new CENP-A after mitosis dictates that centromeres and the kinetochores assembled on them proceed through mitosis with only half the complement of CENP-A. During S phase, CENP-A protein is redistributed among sister centromeres, leaving vacant DNA sequences that are not replenished by CENP-A but are most likely occupied by typical histone H3.1–containing nucleosomes, which are available in excess during DNA replication. Indeed, histone H3–containing nucleosomes have been detected on mitotic centromeres interspersed with CENP-A–containing nucleosomes and have been shown to occupy centromeric chromatin when CENP-A levels are depleted (Blower et al., 2002; Sullivan and Karpen, 2004). Our work now indicates that the mixed chromatin state generated in S phase does not represent an intermediate state of centromeric chromatin where canonical nucleosomes serve as transient placeholders but that in fact this centromeric chromatin composition is what promotes kinetochore formation during mitosis (Fig. 8).
Figure 8.
Schematic depicting centromeric chromatin composition in relation to the cell cycle. CENP-A–containing nucleosomes (red) are interspersed with canonical H3-containing nucleosomes (green) after replication in S phase, and this mixed set of nucleosomes is the substrate for nucleating kinetochore assembly in mitosis and is maintained as cells exit in anaphase. CENP-A assembly initiates in telophase and proceeds through early G1 (presumably concurrent with removal of H3 nucleosomes). CENP-A– and H3-containing nucleosomes are stylized as single nucleosomes but may represent continuous alternating arrays of one or the other type. In mitosis, CENP-A nucleosomes may coalesce to form a rigid interface for kinetochore formation as proposed previously (Zinkowski et al., 1991; Blower et al., 2002; Black et al., 2004, 2007a,b).
Materials and methods
Constructs and cell lines
CENP-A–SNAP–3XHA was constructed by inserting a PCR-generated fragment carrying the human CENP-A open reading frame flanked by KpnI and XhoI sites into the corresponding sites of pSS26m (Covalys) in frame with SNAP26m. A triple HA tag was introduced in frame at the SNAP26m C terminus, resulting in an 371-amino-acid open reading frame producing a 41-kD fusion protein (referred to as CENP-A–SNAP throughout this paper). HeLa cells and their derivatives were cultured in DME supplemented with 10% newborn calf serum (from here onward referred to as complete medium). HeLa monoclonal cell lines expressing CENP-A–SNAP, H3-CFP, or CFP–α-tubulin were generated by stable integration via Moloney murine leukemia retroviral delivery essentially as described previously (Shah et al., 2004; CFP–α-tubulin retroviral construct was provided by J. Shah, Harvard Medical School, Boston, MA). Cells stably expressing the CENP-A–SNAP fusion protein were selected by blasticidin S (5 μg/ml; Calbiochem) and were isolated and individually sorted by flow cytometry. The resulting monoclonal lines were expanded and examined by fluorescence microscopy after TMR-Star labeling and by Western blot to identify lines expressing proper levels of the CENP-A–SNAP fusion protein. Clone 23 (Fig. S1) was used for all experiments in this paper unless stated otherwise. Ratio of CENP-A–SNAP levels to endogenous CENP-A in parental HeLa cells is ∼0.7:1. H3-CFP or CFP–α-tubulin cell lines were isolated by puromycin selection (1 μg/ml; Calbiochem). BubRI or control short hairpin RNA producing pSUPER plasmids and transfection procedures were as described previously (Kops et al., 2004).
SNAP quench and pulse labeling
SNAP tag activity in cells was quenched by addition of 20 μM O6-BG (BG-block; Covalys) in complete growth medium for 30 min at 37°C or pulse labeled with 2 μM TMR-Star (Covalys) in complete growth medium for 15 min at 37°C. After quenching or pulse labeling, cells were washed twice with prewarmed PBS, after which cells were reincubated in complete medium to allow excess compound to diffuse from cells. After 30 min, cells were washed again twice in PBS followed by reincubation in complete medium.
Cell synchronization
Unless stated otherwise in figures or legends, HeLa cells were treated with 2 mM thymidine in complete medium for 17 h, washed twice in PBS, and released in complete medium containing 24 μM deoxycytidine for 9 h followed by addition of thymidine to a final concentration of 2 mM for 17 h, after which cells were released again into complete medium containing 24 μM deoxycytidine and assayed. Nocodazole was used at 100 ng/ml.
Immunofluorescence
Cells were grown and SNAP assayed on glass coverslips followed by fixation and processed for immunofluorescence using standard procedures. Cells were not preextracted before fixation. Anti–CENP-A (a gift from K. Yoda, Nagoya University, Nagoya, Japan) was used at a dilution of 1:100, anti–CENP-C (a gift from W. Earnshaw, University of Edinburgh, Edinburgh, UK) sera was used at a dilution of 1:1,000, and anti-Mad1 (a gift from A. Musacchio, European Institute of Oncology, Milan, Italy) tissue culture supernatant was used at a dilution of 1:20. YL1/2 α-tubulin (Serotec) was used at a dilution of 1:2,500. Anti-HA11 (Covance Research Products, Inc.) was used at a dilution of 1:1,000. Donkey secondary antibodies (anti-mouse Cy5- or FITC-conjugated and anti-rabbit FITC-conjugated) were obtained from Jackson Immunoresearch Laboratories. Samples were stained with DAPI before mounting in ProLong (Invitrogen).
Cell–cell fusions
Double thymidine–arrested H3-CFP and CFP-tubulin–expressing CENP-A–SNAP cells were SNAP-quenched with BG-block followed by release either directly or after 4 h to generate out-of-phase populations. After release of the trailing population, cells were coseeded on 18 × 18 mm uncoated coverslips. 5 h after seeding, coverslips were washed once in prewarmed PBS and incubated cell-side down in a 100 μl PEG-1500 (Roche) for 30 s followed by addition of 500 μl PBS and three subsequent washes in PBS. Coverslips were returned to complete medium containing 100 ng/ml nocodazole to prevent G2 cells from entering G1 and were TMR-Star labeled and fixed 4 h after PEG fusion.
Microscopy
Digital images were captured using a DeltaVision RT system (Applied Precision) controlling an interline charge-coupled device camera (Coolsnap; Roper) mounted on an inverted microscope (IX-70; Olympus). For each sample, images were collected at 1× binning using a 100× oil objective at 0.2 μm z sections spanning the entire nucleus and were subsequently deconvolved, and maximum signals were projected as 2D images using softWoRx (Applied Precision; all images are deconvolved except those shown in Fig. S2). For quantification, images were converted to unscaled TIFF images. Centromere signal intensity was determined using MetaMorph (Molecular Devices) by measuring integrated fluorescence intensity within an 8 × 8 pixel square. Background signal was subtracted from an area within the nucleus not containing centromeres. For live-cell imaging, cells were grown on 22 × 22 mm glass coverslips transfected with YFP–CENP-C (Shah et al., 2004) using Effectene (QIAGEN) 48 h before SNAP labeling, after which coverslips were mounted on a slide separated in a double-stick tape chamber in phenol red–free CO2 independent DME (Invitrogen) containing 0.5 U/ml of the oxygen-scavenging enzyme, Oxyrase (Oxyrase, Inc.), and sealed with a 1:1:1 mixture of vasalin, lanolin, and paraffin.
Images were acquired at 2× binning using a 60× oil objective for TMR-Star and YFP, as well as differential interference contrast at 10-min intervals. For each time point, 5 × 1 μm z sections were acquired for fluorescence images, and a single differential interference contrast image was acquired at the middle z position. Stacks were deconvolved, and maximum intensity was projected using softWoRx and assembled into a paneled video using MetaMorph.
Immunoblots
Whole cell extracts equivalent to 50,000 cells were separated by SDS-PAGE and transferred to nitrocellulose. Blots were probed by human anti-centromere serum (Antibodies, Inc.) at a dilution of 1:300.
Online supplemental material
Fig. S1 shows the cell cycle–dependent CENP-A assembly of independently established cell lines expressing different levels of CENP-A–SNAP. Fig. S2 shows the diffuse nuclear localization of noncentromere-loaded CENP-A–SNAP in G2 cells. Fig. S3 shows evidence for CENP-A–SNAP loading coincident with cytokinesis and nuclear envelope reformation. Video 1 shows a time lapse of early G1 CENP-A–SNAP assembly at centromeres corresponding to stills shown in Fig. 3 E. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200701066/DC1.
Acknowledgments
The authors thank Kinya Yoda, Bill Earnshaw, Jagesh Shah, and Andrea Musacchio for generously providing reagents. We thank Paul Maddox and Arshad Desai for communicating their results before publication. We specifically acknowledge Kevin Sullivan for support to L.E.T. Jansen during the initial stages of this project.
This work has been supported by grants GM29513 and GM74150 from the National Institutes of Health (NIH) to D.W. Cleveland. The Neuroscience Microscopy Shared Facility is supported by grant NS047101 from the National Institute of Neurological Disorders and Stroke. L.E.T. Jansen has been supported by a postdoctoral fellowship from Philip Morris USA, Inc. B.E. Black has been supported by a postdoctoral fellowship from the American Cancer Society and by a Career Award in the Biomedical Sciences from the Burroughs Welcome Fund. D.R. Foltz has been supported by a postdoctoral fellowship from NIH and The Leukemia & Lymphoma Society. D.W. Cleveland receives salary support from the Ludwig Institute for Cancer Research.
Abbreviations used in this paper: BG, benzylguanine; CENP-A, centromere protein A; PEG, polyethylene glycol; TMR, tetramethylrhodamine.
References
- Ahmad, K., and S. Henikoff. 2002. Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA. 99(Suppl. 4):16477–16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., and K.H. Choo. 2002. Neocentromeres: role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 71:695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., K. Bentley, J. Ryan, J. Perry, L. Wong, H. Slater, and K.H. Choo. 2004. a. Human centromere repositioning “in progress.” Proc. Natl. Acad. Sci. USA. 101:6542–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., P. Kalitsis, H. Sumer, and K.H. Choo. 2004. b. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 14:359–368. [DOI] [PubMed] [Google Scholar]
- Black, B.E., D.R. Foltz, S. Chakravarthy, K. Luger, V.L. Woods Jr., and D.W. Cleveland. 2004. Structural determinants for generating centromeric chromatin. Nature. 430:578–582. [DOI] [PubMed] [Google Scholar]
- Black, B.E., M.A. Brock, S. Bédard, V.L. Woods, and D.W. Cleveland. 2007. a. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. In press. [DOI] [PMC free article] [PubMed]
- Black, B.E., L.E. Jansen, P.S. Maddox, D.R. Foltz, A.B. Desai, J.V. Shah, and D.W. Cleveland. 2007. b. Centromere identity maintained by nucleosomes assembled with Histone H3 containing the CENP-A targeting domain. Mol. Cell. 25:309–322. [DOI] [PubMed] [Google Scholar]
- Blower, M.D., B.A. Sullivan, and G.H. Karpen. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, C.W., and A.F. Straight. 2006. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 16:70–78. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., S.T. Liu, and T.J. Yen. 2005. Kinetochore structure and function. Trends Cell Biol. 15:589–598. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Y. Mao, and K.F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. [DOI] [PubMed] [Google Scholar]
- Csink, A.K., and S. Henikoff. 1998. Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 14:200–204. [DOI] [PubMed] [Google Scholar]
- Foltz, D.R., L.E. Jansen, B.E. Black, A.O. Bailey, J.R. Yates III, and D.W. Cleveland. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. [DOI] [PubMed] [Google Scholar]
- Fujita, Y., T. Hayashi, T. Kiyomitsu, Y. Toyoda, A. Kokubu, C. Obuse, and M. Yanagida. 2007. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 12:17–30. [DOI] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad, J.S. Platero, and B. van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA. 97:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman, E.V., K.J. Fowler, A.J. Newson, S. Redward, A.C. MacDonald, P. Kalitsis, and K.H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA. 97:1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler, A., S. Gendreizig, T. Gronemeyer, H. Pick, H. Vogel, and K. Johnsson. 2003. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 21:86–89. [DOI] [PubMed] [Google Scholar]
- Keppler, A., H. Pick, C. Arrivoli, H. Vogel, and K. Johnsson. 2004. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. USA. 101:9955–9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler, A., C. Arrivoli, L. Sironi, and J. Ellenberg. 2006. Fluorophores for live cell imaging of AGT fusion proteins across the visible spectrum. Biotechniques. 41:167–170, 172, 174–175. [DOI] [PubMed] [Google Scholar]
- Kline, S.L., I.M. Cheeseman, T. Hori, T. Fukagawa, and A. Desai. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops, G.J., D.R. Foltz, and D.W. Cleveland. 2004. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA. 101:8699–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.T., J.B. Rattner, S.A. Jablonski, and T.J. Yen. 2006. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 175:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P.S., F. Hyndman, J. Monen, K. Oegema, and A. Desai. Functional genomics identifies a Myb domain–containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 176:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone, B.G., and R.C. Allshire. 2003. Stretching it: putting the CEN(P-A) in centromere. Curr. Opin. Genet. Dev. 13:191–198. [DOI] [PubMed] [Google Scholar]
- Meluh, P.B., P. Yang, L. Glowczewski, D. Koshland, and M.M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 94:607–613. [DOI] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe, R.T., S.C. Henderson, and D.L. Spector. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 116:1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, M., I.M. Cheeseman, T. Hori, K. Okawa, I.X. McLeod, J.R. Yates III, A. Desai, and T. Fukagawa. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. [DOI] [PubMed] [Google Scholar]
- Palmer, D.K., K. O'Day, M.H. Wener, B.S. Andrews, and R.L. Margolis. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, P.N., and R.T. Johnson. 1970. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 225:159–164. [DOI] [PubMed] [Google Scholar]
- Regnier, V., P. Vagnarelli, T. Fukagawa, T. Zerjal, E. Burns, D. Trouche, W. Earnshaw, and W. Brown. 2005. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell. Biol. 25:3967–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J.V., E. Botvinick, Z. Bonday, F. Furnari, M. Berns, and D.W. Cleveland. 2004. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr. Biol. 14:942–952. [DOI] [PubMed] [Google Scholar]
- Shelby, R.D., O. Vafa, and K.F. Sullivan. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby, R.D., K. Monier, and K.F. Sullivan. 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B., and G. Karpen. 2001. Centromere identity in Drosophila is not determined in vivo by replication timing. J. Cell Biol. 154:683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.A., and G.H. Karpen. 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K., Y. Takayama, F. Masuda, Y. Kobayashi, and S. Saitoh. 2005. Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:595–606. (discussion 606–607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa, O., and K.F. Sullivan. 1997. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7:897–900. [DOI] [PubMed] [Google Scholar]
- Ventura, M., S. Weigl, L. Carbone, M.F. Cardone, D. Misceo, M. Teti, P. D'Addabbo, A. Wandall, E. Bjorck, P.J. de Jong, et al. 2004. Recurrent sites for new centromere seeding. Genome Res. 14:1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton, P.E., C.A. Cooke, S. Bourassa, O. Vafa, B.A. Sullivan, G. Stetten, G. Gimelli, D. Warburton, C. Tyler-Smith, K.F. Sullivan, et al. 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7:901–904. [DOI] [PubMed] [Google Scholar]
- Zinkowski, R.P., J. Meyne, and B.R. Brinkley. 1991. The centromere-kinetochore complex: a repeat subunit model. J. Cell Biol. 113:1091–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]