Abstract
The mRNA encoding the Drosophila Zn-finger transcription factor Nerfin-1, required for CNS axon pathfinding events, is subject to post-transcriptional silencing. Although nerfin-1 mRNA is expressed in many neural precursor cells including all early delaminating CNS neuroblasts, the encoded Nerfin-1 protein is detected only in the nuclei of neural precursors that divide just once to generate neurons and then only transiently in nascent neurons. Using a nerfin-1 promoter controlled reporter transgene, replacement of the nerfin-1 3’ UTR with the viral SV-40 3’ UTR releases the neuroblast translational block and prolongs reporter protein expression in neurons. Comparative genomics analysis reveals that the nerfin-1 mRNA 3’ UTR contains multiple highly conserved sequence blocks that either harbor and/or overlap 21 predicted binding sites for 18 different microRNAs. To determine the functional significance of these microRNA-binding sites and less conserved microRNA target sites, we have studied their ability to block or limit the expression of reporter protein in nerfin-1 expressing cells during embryonic development. Our results indicate that no single microRNA is sufficient to fully inhibit protein expression but rather multiple microRNAs that target different binding sites are required to block ectopic protein expression in neural precursor cells and temporally restrict expression in neurons. Taken together, these results suggest that multiple microRNAs play a cooperative role in the post-transcriptional regulation of nerfin-1 mRNA, and the high degree of microRNA-binding site evolutionary conservation indicates that all members of the Drosophila genus employ a similar strategy to regulate the onset and extinction dynamics of Nerfin-1 expression.
Keywords: microRNAs, Post-transcriptional regulation, Nervous system development
Introduction
The Drosophila nerfin-1 gene encodes a transcription factor that is essential for the wild-type expression pattern of a subset of axon guidance genes in nascent neurons (Kuzin et al., 2005). The Nerfin-1 protein belongs to a highly conserved subfamily of transcriptional regulators that are identified by their unique set of tandem Zn-fingers called the EIN-domain (Stivers et al., 2000). EIN-domain Zn-finger genes are present in all metazoans, including nematodes (Desai and Horvitz, 1989) and man (Goto et al., 1992), and expression studies reveal that, as in Drosophila, they are expressed in the developing nervous system (Wu et al., 2001; Breslin et al., 2003). Although nerfin-1 mRNA expression is detected in most, if not all, CNS neuroblasts (NBs), ganglion mother cells (GMCs) and nascent neurons, the nuclear located Nerfin-1 protein is restricted to those neural precursor cells that divide only once to yield neurons and then found just transiently in the new born neurons (Kuzin et al., 2005). For example, during early CNS lineage development nerfin-1 mRNA expression is detected in many neural precursor cells including most NBs and GMCs, however, significant levels of Nerfin-1 protein are detected only in MP NBs and in GMCs that undergo a single cell division to generate neurons (Figure 1; also see Kuzin et al., 2005). The observation that prolonged ectopic expression of Nerfin-1 protein in neurons is lethal also suggests that temporal regulation of Nerfin-1 expression is critical (Kuzin et al., 2005).
Fig. 1.
nerfin-1 mRNA and protein expression during embryonic stages 10, 13 and 14. Whole-mount in situ hybridizations and immunostains reveal nerfin-1 mRNA (panels A, C & E) and Nerfin-1 protein (panels B, D & F) distributions in stage 10 (panels A & B) and stage 13 (panels C & D) embryos. A & B) During embryonic stage 10, most if not all ventral cord neuroblasts express nerfin-1 mRNA albeit at varying levels. However, during this developmental period only a single neuroblast per hemisegment, the MP2, expresses detectable levels of Nerfin-1 protein. Shown are ventral views of the 4th thoracic and 1-3 abdominal segments (arrowheads indicate the ventral midline, anterior is up). Panel A inset) Higher magnification reveals that the nerfin-1 mRNA is not evenly distributed in neuroblasts. The MP2 neuroblasts (asterisk) have higher in situ hybridization signals while flanking neuroblasts contain less in situ staining that is asymmetrically located in a patchy subcellular distribution. C & D) During stage 13, most if not all nerfin-1 mRNA expressing secondary PNS precursor cells and nascent neurons are also immunopositive for Nerfin-1 protein. Shown are flattened left side embryo fillets that reveal nerfin-1 mRNA (C) and protein (D) distribution within the PNS and the left half of the ventral cord (on the right, anterior up). E & F) By late stage 14 there is a significant reduction of both the mRNA and protein steady state levels of nerfin-1 in the developing nervous system.
Recent in vitro studies in cultured Drosophila S2 cells indicate that the block or inhibition of nerfin-1 mRNA translation in the developing nervous system may be the result of regulation by microRNAs (miRNAs) (Stark et al., 2005; Rehwinkel et al., 2006). nerfin-1 is targeted by miR-9b, but not miR-9a in cultured cells, and repression was relieved in Drosha- and AGO1-depleted cells, indicating that nerfin-1 is a genuine target of the miRNA pathway (Rehwinkel et al., 2006). miRNA genes encode small regulatory RNAs (21–22nt) that are incorporated into silencing complexes that cleave and/or inhibit the translation of specific target mRNAs. Nucleotide base-pairing between miRNAs and target sites found in the 3’ UTR of mRNA triggers the translational inhibition and/or cleavage of the mRNA (reviewed by Lai et al., 2003; Bartel et al., 2004; Filipowicz, 2005; Carthew, 2006). mRNA-miRNA base-pairing to as few as eight bases is sufficient for binding and silencing (Brennecke, et al., 2005; Ma, et al., 2005). In Drosophila there are ~100 miRNA genes, and bioinformatics screens have identified numerous potential mRNA targets for miRNAs (Enright, et al., 2003). Previous studies have demonstrated that Bearded gene family members (Lai et al., 2005), the Delta gene (Kwon et al., 2005), and apoptosis pathway genes (Leaman et al., 2005) all undergo miRNA-mediated post-transcriptional regulation. miRNAs have also been shown to regulate neural development in C. elegans (Johnston et al., 2005), in Drosophila (Li et al., 2006) and in mammals (Conaco et al., 2006).
Different miRNA target prediction methods, that utilize overlapping sets of binding site selection criteria, have all identified multiple miRNA binding sites within the nerfin-1 1.6 kb 3’ UTR, many of which are conserved in multiple Drosophila species (see below; discussed in Stark et al., 2005 supplemental data). To determine the in vivo significance of the predicted miRNA target sites within the highly conserved 3’ UTR sequences, we employed nerfin-1 promoter regulated transgenes to test their ability to inhibit reporter protein expression in neural precursor cells and nascent neurons. Here, we show that multiple 3’ UTR binding sites for different miRNAs, but not single miRNA binding sites, are required for nerfin-1 post-transcriptional regulation in both the developing CNS and PNS. These studies reveal that in the developing CNS miRNA mediated post-transcriptional regulation primarily limits the spatial distribution of Nerfin-1 while in the developing PNS it plays an important role in restricting its temporal expression in nascent neurons.
Materials and methods
Generation of nerfin-1 miRNA-binding site reporter constructs
To test if the different conserved nerfin-1 miRNA-binding sites play a role in in vivo post-transcriptional regulation, a nerfin-1 reporter transgene was prepared by modifying a nerfin-1 rescue construct (Kuzin et al., 2005) that contained both upstream and downstream transcriptional cis-regulatory regions. Assembled using standard molecular biology methods, the initial pCaSpeR-3 based transgene, P[nerfin-1.GFP-NLS.SV-40], contains the following three DNA fragments in 5’ to 3’ order: 1) 5,947 bp of nerfin-1 genomic DNA that includes 5,762 bp of upstream genomic transcriptional regulatory sequences, the nerfin-1 transcription start site and its 5’ UTR; 2) 1,104 bp DNA fragment from the pH-Stinger plasmid (Barolo et al., 2004) that contains an ORF encoding a nuclear targeted green fluorescent protein linked to the SV-40 3’ UTR; and 3) 2,124 bp of 3’ flanking nerfin-1 genomic DNA that contains additional transcriptional regulatory sequences (details of the cloning steps are available upon request). To generate the iA (1668 bp), iB (354 bp), iF (514 bp), iH (852 bp), iJ (138 bp) and iL (266 bp) nerfin-1 3’ UTR reporter constructs (see Figure 2B), PCR amplified fragments were inserted into a unique Not-I site within the P[nerfin-1.GFP-NLS.SV-40] vector located in the SV-40 3’ UTR, 14 bp 3’ of the GFP ORF translation stop codon. The following PCR primers were used to amplify the different 3’ UTR regions:
Fig. 2.

The nerfin-1 3’ UTR contains multiple highly conserved miRNA-binding sites. A) Shown is an EvoPrint of the 1.6 kb nerfin-1 3’ UTR. DNA sequences from the following Drosophila species were used to generate the EvoPrint: D. melanogaster (reference sequence), D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. willistoni, D. mojavensis and D. grimshawi. Conserved sequence blocks, present in all, or in all but one of the species, are indicated by uppercase black letters and non- or less-conserved sequences are indicated as lowercase gray letters. Predicted RNA-binding sites for 18 different miRNAs, as identified previously by Stark et al. (2005) and Enright et al. (2003), are underlined. Note, D. simulans and D. virilis were not used in the analysis due to sequencing gaps in their DNAs. B) A linear map of the nerfin-1 3’UTR miRNA-binding sites aligned with the different DNA fragments (iA through iL) that were tested for miRNA responsive activity within the P[nerfin-1.GFP-NLS.sv-40] transgenic reporter (see Materials and Methods for reporter construct details).
iA: 5’-CCATGGCCCACTGAAATCGAGTGAG-3’ & 5’-CCCTGACAACCCAAAGAGAACCCAACAAG-3’
iB: 5’-CCATGGCCCACTGAAATCGAGTGAG-3’ & 5’-GCCTTCGTGGTACACAAGAGACACTC-3’
iF: 5’-GAGTGTCTCTTGTGTACCACGAAGGC-3’ & 5’-CCTGAATATGACTAAAGCTGTATCCG-3’
iH: 5’-CGGATACAGCTTTAGTCATATTCAGG-3’ & 5’-CCCTGACAACCCAAAGAGAACCCAACAAG-3’
iJ: 5’-CTCAGTTTAGTTTAGTTAGTT-3’ & 5’-GCTTTAAGTACACAACCGCC-3’
iL: 5’-GTGTAAATTGGTTGTAACCGC-3’ & 5’-GGTCTTCAAGAGTTTGTTTTTG-3’.
For the shorter inserts iC (35bp), iD (28bp), iE (33bp), iG (30 bp), iI (32bp) and iK (29 bp) and the following doubled-stranded synthetic oligos were cloned into the Not-I site (upper strand shown):
iC: 5’-CGATGCAGCTGAAACAGACCAAAGAATTAGTTATA-3’
iD: 5’-CGCAAAATGAGTTCAATTCTAGTCAGAC-3’
iE: 5’-TTGGCACTAGTCAGCTTCAATACGATCTCGAAA-3’
iG: 5’-AAAATTCAGGCAAAATTGTGCAGTAATAGT-3’
iI: 5’-AACCAAAGTCGAGTGTGAGCTCTAGTCATTTT-3’
iK: 5’-ACAATATCACAGCGCTATTGTTCCTTAGC-3’.
Drosophila P-element transformants and stocks
Germ-line transformants were generated using standard techniques based on the methodology described in Rubin and Spradling (1982). Constructs were injected into Df(1)w67c,y, y w, or w118 strains using delta 2-3 helper DNA (Rubin and Spradling, 1982). Standard animal husbandry procedures were used in the care and handling of Drosophila stocks (Ashbruner 1989).
Immunohistochemistry and mRNA localization
Embryo fixation and whole-mount immunostaining of 8 and 16 hr embryo collections were carried out according to the procedures described in Patel (1994). Rabbit anti-GFP was obtained from Invitrogen and used at a 1 to 2,000 dilution. Nerfin-1 immunostains were performed according to the procedure described in Kuzin et. al. (2005). Vectastain ABC second antibody avidin/biotin HRP visualization reagents were used according to the manufacturer’s protocol (Vector Labs). For in situ hybridization, a nerfin-1 single-strand riboprobe was prepared from a full-length cDNA (Stivers et al., 2000) as previously described (Kopczynski et al., 1998), with the exception that the riboprobe was prepared from PCR amplified cDNA with a labeling mix containing Fluorescein-12-UTP (Roche) and visualized using anti-FITC Fab fragments coupled to alkaline phosphatase. To detect P[nerfin-1.GFP-NLS.SV-40] transgene mRNA expression, a GFP single-strand riboprobe was prepared from the PCR amplified GFP-NLS ORF of pH-Stinger vector (Barolo, 2004). Detailed protocols for the immunostains and in situ hybridization are available upon request. Embryos and dissected fillets were viewed in 70% glycerol with 30% phosphate-buffered saline (PBS) and photographed using a Nikon Optiphot microscope equipped with DIC/Nomarski optics. Embryo developmental staging was determined by morphological criteria (Campos-Ortega and Hartenstein 1985).
Results and Discussion
Nerfin-1 protein expression is blocked in most CNS neuroblasts
During embryonic stage 10, nerfin-1 mRNA is detected in all early delaminating CNS NBs albeit at differing levels; and within many of the NBs the message appears to be asymmetrically distributed (Figure 1A and Stivers et al., 2000). Although nerfin-1 mRNA expression is pan-neural during this early stage in CNS development, immunostains using different Nerfin-1 specific polyclonal antibodies detect significant levels of Nerfin-1 protein only in the ventral cord MP2 NBs (Figure 1B and Kuzin et al., 2005). The punctate/irregular distribution of the nerfin-1 mRNA in NBs lacking detectable levels of Nerfin-1 protein (Figure 1A insert) is reminiscent of that observed for mRNAs targeted for miRNA mediated cleavage in mammals (for example see Liu, 2005 and references therein), suggesting that nerfin-1 message in many of the NBs may likewise be targeted for degradation.
Although the dynamics of nerfin-1 mRNA and protein expression differ considerably during the early stages of nervous system development, by stage 13 the pattern of Nerfin-1 expression closely matches that of its mRNA and close inspection of nerfin-1 message distribution in the Nerfin-1 protein expressing cells revealed an even cytoplasmic distribution (Figures 1C and 1D and data not shown). Expression of both the message and protein in the new born CNS and PNS neurons is short lived; levels of both rapidly decline such that by late stage 14 both message and protein levels are significantly lower throughout the nervous system (Figures 1E and 1F; also see Stivers et al., 2000; Kuzin et al., 2005).
Predicted miRNA-binding sites overlap highly conserved nerfin-1 3’ UTR sequences
miRNA target prediction programs have identified multiple putative miRNA binding sites within the nerfin-1 1,622 bp 3’ UTR and many of these sites are conserved in other nerfin-1 Drosophila orthologues (Stark et al., 2003; Grun et al., 2005; reviewed in Stark et al., 2005). For example, a Drosophila EvoPrint (Odenwald et al., 2005) of the nerfin-1 locus (using D. melanogaster as the reference sequence and D. sechellia, D. yakuba, D. erecta, D. ananassae, D. persimilis, D. pseudoobscura, D. willistoni, D. mojavensis and D. grimshawi as test sequences) revealed conserved sequence blocks within the 3’ UTR that contain or overlap 21 predicted miRNA binding sites for 18 different miRNAs (Figure 2A). The conserved sequences (shown in upper case black) are present in all, or all but one, species used in the analysis and represent over 100 million years of collective evolutionary divergence. The partial and/or interrupted conservation within the predicted miRNA binding sites may reflect the fact that initial base-pairing of an miRNA and its mRNA target sequence requires only eight bases to initiate translational regulation (Lewis et al., 2003 and 2005; Lai, et al., 2005). EvoPrint analysis of the nerfin-1 3’ UTR also identified additional conserved sequence blocks that do not contain or overlap predicted miRNA binding sites and their role(s) in gene function are currently unknown (Figure 2A). In vivo cis-regulatory analysis of the nerfin-1 3’ UTR failed to detect any transcriptional enhancer activity (unpublished results; Kuzin et al.). In addition to the conserved miRNA target sites, less conserved predicted binding sites have been identified within the 3’ UTR (Stark et al., 2003; Grun et al., 2005; reviewed in Stark et al., 2005). For example, the central miR-279/miR-286 and miR-279 target sites are present in the species that are evolutionarily close to D. melanogaster but not conserved in the more distant D. persimilis, D. pseudoobscura, D. willistoni, D. mojavensis and D. grimshawi species (Figure 2A).
The conservation of nerfin-1 miRNA sites suggests that miRNA mediated post-transcriptional regulation of nerfin-1 occurs in all members of this genus. MicroRNAs most likely regulate other EIN-domain containing zinc finger genes. For example, multiple miRNA binding sites have also been detected in the vertebrate IA-1 3’ UTR (Griffiths-Jones et al., 2006), and EvoPrint analysis reveals that one of sites within the human IA-1 gene is highly conserved (data not shown).
Multiple miRNA binding sites are required to block Nerfin-1 expression in CNS neuroblasts
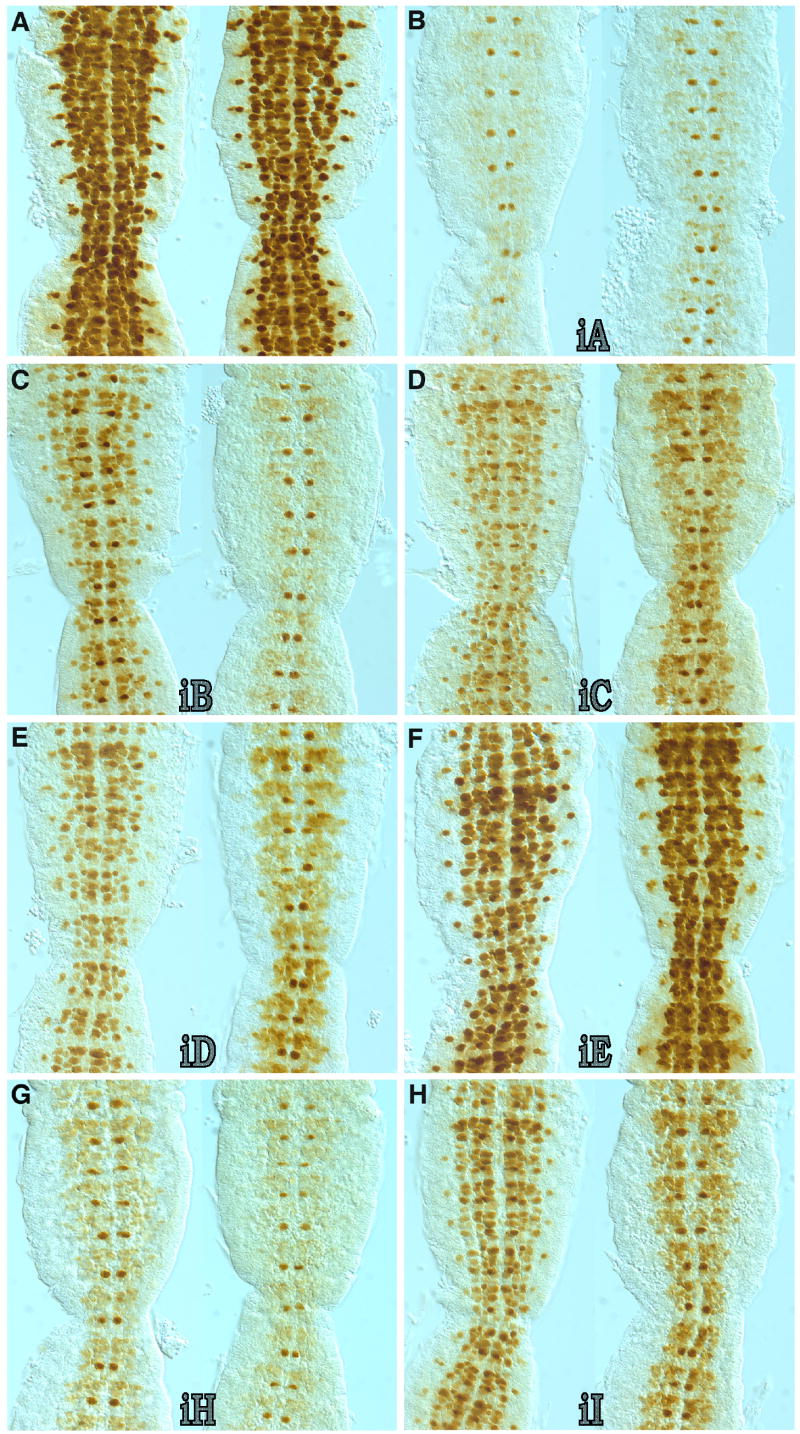
To determine if the conserved nerfin-1 3’ UTR sequences are required for the embryonic NB post-transcriptional regulation, a series of reporter transgene constructs were generated that tested the silencing activity of different regions of its 3’ UTR. The starting construct was prepared by replacing the nerfin-1 ORF and 3’ UTR in an 11 kb nerfin-1 genomic rescue construct (Kuzin et al., 2005) with a sequence that contains the ORF for a nuclear targeted Green Fluorescent Protein (GFP-NLS) linked to the viral SV-40 3’ trailer that lacks any predicted miRNA binding sites. As expected, transformants that contain the P[nerfin-1.GFP-NLS.SV-40] construct expressed GFP in all early delaminating CNS NBs and no translational block of GFP expression was detected when compared to nerfin-1 mRNA expression (Figure 3A and data not shown). The full-length or different sub-regions of the nerfin-1 3’ UTR containing the conserved sequence blocks (Figure 2B) were then inserted into a unique restriction site within the vector’s SV-40 3’ UTR. Embryo GFP-immunostains were performed on multiple independent transformant lines for each construct (results summarized in Table 1). As controls, multiple independent transformant lines that contain the nerfin-1 3’ UTR sequences in the opposite orientation were also generated for each construct and embryo GFP-immunostains revealed that in all cases the translational block in GFP expression was orientation dependent (data not shown).
Fig. 3.
The nerfin-1 3’ UTR contains multiple sequences that block protein expression in CNS neuroblasts. Shown are embryonic stages 10 (left side of each panel) and 11 (right side of each panel) embryos that were immunostained in whole-mount for GFP expression and then filleted to reveal most of the ventral cord (anterior is up). Embryos were collected from transformant lines that harbor different nerfin-1 miRNA-binding site reporter transgenes (see figure 2 for the locations of the different nerfin-1 3’ UTR inserts). A) Vector alone (no nerfin-1 3’ UTR sequences); No block in GFP expression was observed. GFP expression matches endogenous nerfin-1 mRNA expression. B) Vector with the iA fragment (complete nerfin-1 3’ UTR); GFP expression mirrors that of the endogenous Nerfin-1 protein. The only neuroblast to express significant levels of GFP during this phase of development is the MP2 neuroblast. C) Vector plus iB (contains the miR-9A, miR-9B, miR-9C, miR-279/miR-286 and bantam miRNA-binding sites); GFP expression is partially blocked at stage 10, however, by stage 11 GFP expression pattern is close to that observed in transformants containing the full-length nerfin-1 3’ UTR iA construct. D) Vector plus iC (contains the overlapping miR-9A, miR-9B and miR-9C binding sites); GFP expression is only partially blocked during stages 10 and 11. E) Vector plus iD (contains just the miR-279/mir-286 binding sites); like iC, GFP expression is only partially blocked during stages 10 and 11. F) Vector plus iE (contains the bantam binding site); no block in GFP expression was detected at any time during embryonic development. GFP immunostaining was indistinguishable to that observed in embryos with the vector alone. G) Vector plus iH (contains miR-92A, miR-92B, miR-310-313, miR-279/286, miR-34, miR-315, miR-305, miR-307, miR-5 and miR-13B sites); GFP expression in neuroblasts other than the MP2 is blocked but not as complete as with the full-length 3’ UTR iA construct. H) Vector plus iI (contains the overlapping miR-279 and miR-286 binding sites); only a slight reduction in GFP expression is detected during both developmental stages.
Table 1.
Results summary for the different nerfin-1 miRNA-binding site reporter transgenes
| CNS
|
PNS (Stage 15)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Constructs | Transformant lines(a) | Stage 10 | Stage 11 | Dorsal | Lateral | Ventral’ | Ventral | Figure(s) |
| Vector | 5 | − (b) | − | − | − | − | − | 3A, 4A |
| + iA | 5 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | 3B, 4B |
| + iB | 5 | ++ | +++ | ++ | ++ | ++ | ++ | 3C, 4C |
| + iC | 5 | ++ | ++ | − | − | − | − | 3D |
| + iD | 6 | ++ | ++ | +++ | ++ | ++ | ++ | 3E, 4D |
| + iE | 7 | − | − | − | − | − | − | 3F |
| + iF | 6 | − | − | − | − | − | − | not shown |
| + iG | 3 | − | − | − | − | − | − | not shown |
| + iH | 4 | +++ | +++ | ++++ | ++++ | ++++ | +++ | 3G, 4E |
| + iI | 7 | + | ++ | + | + | ++ | ++ | 3H, 4F |
| + iJ | 6 | − | − | ++ | + | + + | + + | 4G |
| + iK | 3 | − | − | − | − | − | − | not shown |
| + iL | 3 | − | − | ++ | + | ++ | ++ | 4H |
Number of independent transformant lines tested.
Ranking of reduction in GFP expression (-, no effect; ++++, full block similar to Nerfin-1 protein expression pattern)
Insertion of the full-length 3’ UTR into the P[nerfin-1.GFP-NLS.SV-40] reporter (construct iA) recapitulated the silencing of nerfin-1 mRNA translation (Figure 3B). Similar to the endogenous Nerfin-1 protein expression during embryonic stages 10 and 11, significant levels of GFP expression in the ventral cord were observed only in the MP2 NBs. However, reporter transgenes that contained sub-regions of the 3’ UTR gave only partial or no block in NB GFP expression (Figure 3 and summarized in Table 1). For example, although the iB and iH constructs, consisting respectively of the conserved 5’ and 3’ multiple miRNA binding site sub-regions, significantly reduced GFP expression in stage 11 NBs, both of these sub-regions only partially blocked expression during stage 10 (Figure 3C and 3G). Further sub-division of the 5’ conserved miRNA binding site cluster (constructs iC, iD and iE) revealed that the overlapping miR-9A, miR-9B and miR-9C binding sites and the miR-279/mir-286 both contributed to the partial inhibition observed with the iB construct, but the conserved Bantam miRNA-binding site did not (Figure 3D – 3F). It is worth noting that the 5’ predicted miR-279/mir-286 target site within our nerfin-1 rescue construct (Kuzin et al., 2005) contains the sequence TCTAGTCA that agrees with the predicted miR-279/mir-286 binding site. This sequence differs in the second to last base from that of the D. melanogaster genomic sequence (FlyBase BLAST), in which there is a T in place of C. cDNA sequence analysis of all ESTs in the database reveals a C instead of a T at this position.
Given that only the full-length insert recapitulates the silencing of endogenous expression in the CNS, it is concluded the miRNAs act in a cooperative fashion to regulate the onset of Nerfin-1 protein expression. The block in translation by sub-regions of the 3’ UTR was more effective at stage 11 than at stage 10; this could reflect time of onset of miRNA expression or the possibility that the level of mRNA expression is too high for a complete block at the earlier stage. Previous studies have shown that ectopic expression of nerfin-1 outside the wild-type temporal/spatial boundaries during CNS development results in axon guidance defects (Kuzin et al., 2005). The requirement for multiple miRNA binding sites may reflect the need for tight spatial control of Nerfin-1 expression.
Dissection of the sub-regions reveals that the miR-9A, miR-9B and miR-9C combined site, as well as the miR-279/mir-286 site, contribute to silencing, but the Bantam site did not show an effect. The conservation of the Bantam site suggests that it is functionally important, but no effect on embryonic CNS expression of nerfin-1 was observed. Consistent with this, no effect on Nerfin-1 protein expression was detected in bantam minus embryos (data not shown). Bantam has been shown to have developmental roles in post-embryonic development (Brennecke et al., 2003). Analysis of the 3’ sub-region sites (constructs iG, iI, iJ, iK and iL) indicates that in the CNS, the combined miR-279/miR-286 site (construct iI) exhibits partial silencing, with no effect observed for the other miRNA binding sites. Interestingly, the less conserved centrally located miR-279/miR-286 and miR-279 sites, contained in construct iF, did not promote silencing (data not shown). These two sites share less homology to the miR-279 and miR-286 binding sites than the other conserved miR-279/mir-286 target sites (http://www.microrna.org/drosophila/targetsv2.html). Construct iG, which contains the overlapping miR-92A, miR-92B, and miR-310-313 sites revealed no detectable miRNA silencing. In addition, construct iK that contains a predicted miR-5 binding site did not affect the reporter mRNA translation in the embryonic CNS and PNS (data not shown). The other sites in the 3’ sub-region exhibited an effect in PNS silencing (see below), suggesting spatial specificity for microRNA effects on nerfin-1 expression.
miRNA mediated post-transcriptional regulation limits the temporal expression of Nerfin-1 in PNS neurons
During embryonic PNS development, nerfin-1 mRNA and protein are transiently expressed in secondary precursor cells that divide once to generate neurons, and then both its transcript and encoded protein are only transiently detected in nascent neurons (Figure 1C and 1D; Stivers et al., 2000; Kuzin et al., 2005). Unlike the post-transcriptional regulation observed in the developing CNS, when the full-length nerfin-1 3’ UTR was included in the reporter transgene the onset of GFP expression was not blocked in precursor cells but the duration of GFP expression in the nascent neuron was significantly reduced (Figure 4B and data not shown). The rapid extinction of detectable GFP expression mirrored that of the endogenous Nerfin-1 transient expression; the short-lived expression was observed throughout the PNS in the ventral, lateral and dorsal neurons such that by stage 15 little or no GFP immunostaining was detected. Similar to the reporter results obtained in the CNS for the different 3’ UTR sub-regions, no one sub-region or single miRNA binding site was able to fully limit GFP expression in older stage 14 and 15 neurons (Figure 4C – 4H). However, except for the predicted Bantam miRNA-binding site that showed no detectable effect on silencing GFP expression, all of the other 3’ UTR sub-regions exhibited different degrees of silencing. Each of the constructs had differential effects on reporter expression in different cells of the PNS (Figure 4), suggesting an involvement of miRNAs in cell-type regulation of Nerfin-1 expression. For example, construct iB, containing the 5’ end of the 3’ UTR, exhibited a higher levels of silencing in individual cells of the dorsal (d) and lateral (l) clusters (Figure 4C); construct iH, containing the 3’ end of the 3’ UTR, exhibited a higher level of silencing in the chordotonal neurons in the lateral cluster than in other cells of the lateral and ventral clusters; construct iJ, containing a subset of sites in the 3’ UTR, exhibited a higher level of silencing in a subset of cells in the dorsal and lateral clusters than in other cells of the same clusters (Figure 4G).
Fig. 4.
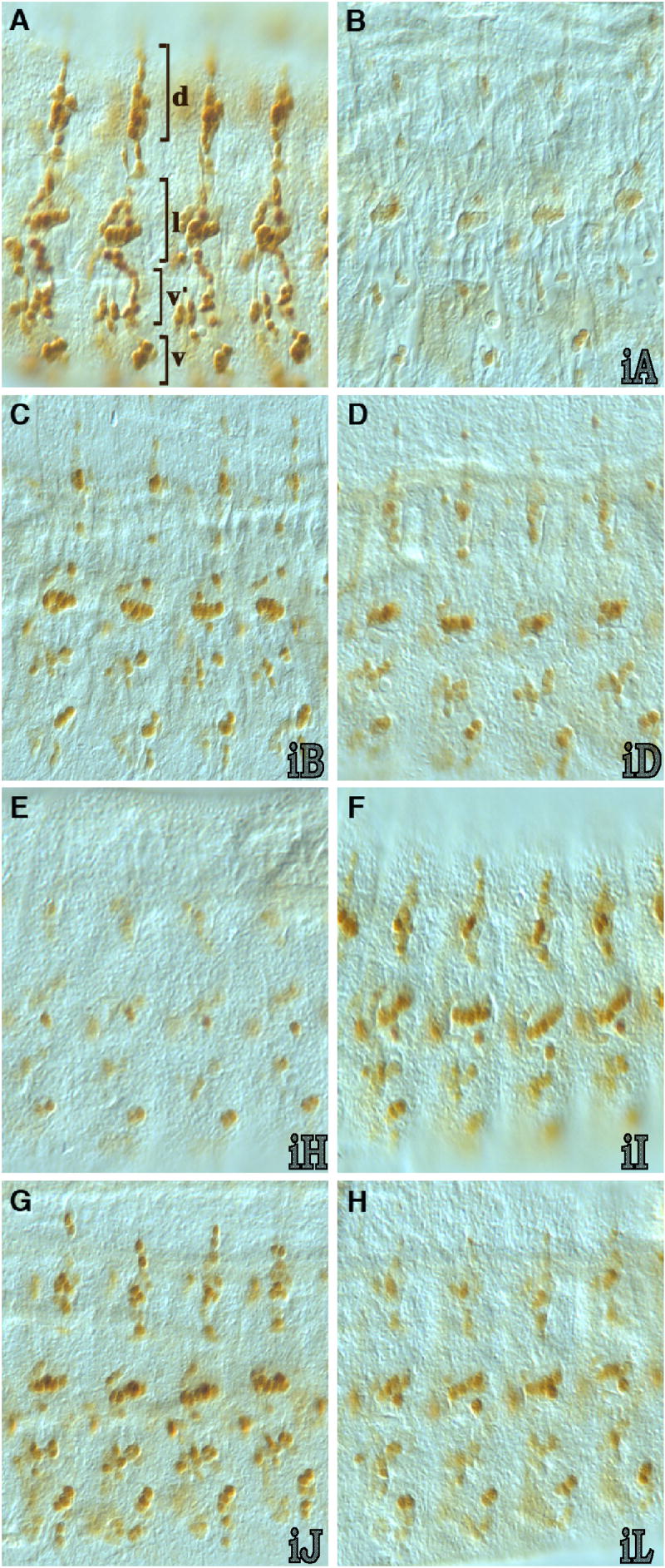
nerfin-1 3’ UTR miRNA-binding sites reduce reporter protein expression in nascent PNS neurons. Shown are lateral, flattened views of abdominal 1-4 segments (dorsal up) from late stage 14/early stage 15 whole-mount GFP immunostained embryos from nerfin-1 miRNA-binding site reporter transformant lines (see figure 2 for a linear map of the tested sequences). A) Vector alone; most, if not all, PNS neurons express GFP (brackets v, v’, l and d indicate the two ventral, lateral and dorsal neuronal cell clusters as described by Breuster and Bodmer, 1995). B) Vector plus iA (complete nerfin-1 3’ UTR); low level of GFP immunostaining through out the PNS matches that of Nerfin-1 immunostains at this stage. C) Vector with iB (contains the miR-9A, miR-9B, miR-9C, miR-279/286 and bantam miRNA-binding sites); only partial reduction of GFP expression in a subset of neurons in all clusters with the most significant reduction observed in the dorsal cluster. D) Vector plus iD; similar to iB partial reduction in GFP expression was observed when just the miR-279/286 binding sites was included in the reporter vector. E) Vector plus iH (3’ half of the nerfin-1 3’ UTR containing the miR-92A, miR-92B, miR-310-313, miR-279/286, miR-34, miR-315, miR-305, miR-307, miR-5 and miR-13B sites); reduction in GFP expression in the d and v’ clusters is similar to that observed with the full-length iA construct but the reduction in immunostaining was less than that observed with iA in the v and l PNS cell clusters. F) Vector with iI (contains the overlapping miR-279 and miR-286 binding sites); a partial reduction in GFP expression was observed in all clusters. G) Vector plus iJ (contains overlapping miR-34/miR-315/miR-305 binding sites and the miR-307 binding site); partial reduction in GFP expression was detected in subsets of d and v’ cluster neurons. H) Vector plus iL (contains the miR-305/miR-13B miRNA-binding site); partial reduction in GFP expression was observed in all PNS clusters.
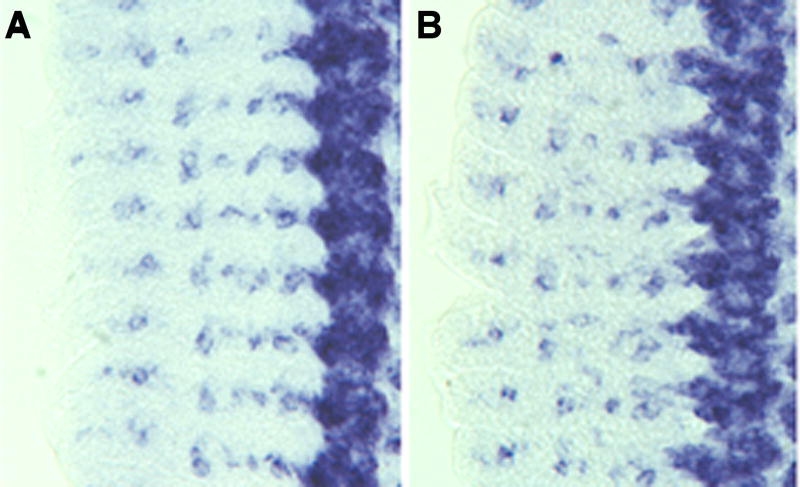
Taken together, the data suggest that the miRNA binding sites in the 3’ UTR are required to restrict the onset (CNS) and extinction (PNS) dynamics of Nerfin-1 protein expression. The limited expression of Nerfin-1 protein may be the result of translational inhibition and/or enhanced miRNA mediated degradation of the nerfin-1 mRNA. To determine whether mRNA expression dynamics were different for different constructs and thus were affected by the presence of different combinations of nerfin-1 miRNA binding sites, we compared the mRNA expression dynamics of the nerfin-1.GFP-NLS.SV-40 transgene to mRNA expression dynamics of this transgene containing the various nerfin-1 3’UTR fragments. Our in situ hybridization mRNA study of embryos containing these different nerfin-1 3’ UTR transgene constructs revealed that none of the nerfin-1 miRNA binding site constructs exhibited a marked alteration of the PNS or CNS expression dynamics of the reporter transgene during embryonic development (Figure 5 and data not shown). However, because our in situ hybridizations only reveal relative steady state mRNA levels, we cannot definitively rule out the possibility that the miRNAs may be promoting nerfin-1 mRNA degradation.
Fig. 5.
nerfin-1 3’ UTR miRNA-binding sites do not significantly alter the steady state levels of the reporter transgene mRNA. Transgene mRNA expression was detected using a GFP-specific riboprobe. Shown are late stage 13 embryo fillets (left side with anterior up). A) Vector alone. B) Vector with the iA fragment (the complete nerfin-1 3’ UTR).
Whereas the overlapping miR-9A, miR-9B, and miR-9C target sites showed partial silencing of nerfin-1 expression in the CNS, no effect was observed in the PNS. Interestingly, mutational analysis of a miR-9a mutant reveals that it is required for embryonic PNS development, and it has been shown to silence expression of senseless mRNA (Li et al. 2006). However, our studies show that miR-9A is unlikely to be a dominant regulator of embryonic Nerfin-1 protein expression; analysis with a number of cell fate markers reveal that nerfin-1 mutation is not likely to effect embryonic PNS cell fate (Kuzin et al. 2005) and staining miR-9a mutants (Li et al. 2006) with antibody to Nerfin-1 reveals no alteration in the number or positions of Nerfin-1 positive cells (data not show). In contrast, the miR-305 and miR-13B sites within construct iL partially reduced reporter expression in the PNS but not in the CNS, and the combined miR-34/315/305, miR-307 sites also exhibited partial silencing in the PNS but not in the CNS. This observation suggests that part of the reason for the complexity of miRNA binding sites in the nerfin-1 3’UTR could be due to tissue specificity of miRNA expression.
Summary
We have examined the ability of the predicted miRNA binding sites within the Drosophila nerfin-1 3’ UTR to silence mRNA translation in vivo. The principle finding of this study is that multiple miRNAs act cooperatively to regulate the spatial and temporal expression of Nerfin-1 in the developing embryonic nervous system. Indeed, no single miRNA-binding site is sufficient to recapitulate the endogenous post-transcriptional regulation in either the embryonic CNS or PNS. In the CNS, mRNA binding sites for multiple miRNAs are required to regulate the spatial expression of Nerfin-1 by silencing expression in all but the MP NBs. In the developing PNS, these studies indicate that miRNA mediated regulation does not restrict the onset of Nerfin-1 expression but rather it helps accelerate the rate of disappearance of Nerfin-1 in nascent neurons.
Whereas the whole 3’UTR was required for wild-type expression of nerfin-1, three individual sites had a partial effect of silencing in the CNS and four individual sites had only a partial effect in silencing in the PNS. The incomplete silencing in the CNS was stronger at a later stage of development than at an earlier stage, pointing to temporal effects of individual miRNAs. In two instances, partial silencing of nerfin-1 expression is accomplished by different sites in the CNS and PNS pointing to a potential tissue specificity of miRNA effects. miR-9A, miR-9B and miR-9C showed an effect in the CNS but not in the PNS, and, in contrast, the combined miR-34/315/305, miR-307 sites (construct iJ) exhibited partial silencing in the PNS but not in the CNS. The same differential effect was observed for combined miR-305 and miR-13B binding sites (construct iL). In addition, in the PNS, partial effects exhibited a degree of cell type specificity, suggesting that individual miRNAs exhibit cellular specificity even within a single tissue. Our results suggest that the high number of conserved miRNA binding sites in the nerfin-1 3’ RNA are likely to reflect differential temporal and spatial specificity of miRNA function. Further confirmation of this awaits in depth studies of the tissue specificity of miRNA expression.
Acknowledgments
We thank Jermaine Ross and Anthony Ekatomatis for their technical support, members of the NINDS central sequencing facility and Vladislav Panin for critiquing the manuscript. We also acknowledge the editorial expertise of Judith Brody. This research was supported by the Intramural Research Program of the NIH, NINDS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Barolo S, Castro B, Posakony JW. New Drosophila transgenic reporters: insulated P-element vectors expressing fast-maturing RFP. Biotechniques. 2004;36:436–442. doi: 10.2144/04363ST03. [DOI] [PubMed] [Google Scholar]
- Bartel F, Harris LC, Wurl P, Taubert H. MDM2 and its splice variant messenger RNAs: expression in tumors and down-regulation using antisense oligonucleotides. Mol Cancer Res. 2004;2:29–3. [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin MB, Zhu M, Lan MS. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem. 2003;278:38991–7. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Springer; Berlin, Heidelberg: 1985. [Google Scholar]
- Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C, Garriga G, McIntire SL, Horvitz HR. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature. 1988;336:638–46. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Goto Y, De Silva MG, Toscani A, Prabhakar BS, Notkins AL, Lan MS. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with “zinc-finger” DNA-binding motifs. J Biol Chem. 1992;267:15252–7. [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski CC, Noordermeer JN, Serano TL, Chen WY, Pendleton JD, Lewis S, Goodman CS, Rubin GM. A high throughput screen to identify secreted and transmembrane proteins involved in Drosophila embryogenesis. Proc Natl Acad Sci. 1998;95:9973–8. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin A, Brody T, Moore AW, Odenwald WF. Nerfin-1 is required for early axon guidance decisions in the developing Drosophila CNS. Dev Biol. 2005;277:347–65. doi: 10.1016/j.ydbio.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci. 2005;102:18986–91. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–80. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih I, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5’-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–70. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci. 2005;102:14700–5. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Natalin P, Stark A, Brennecke J, Cohen SM, Izaurralde E. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol Cell Biol. 2006;26:2965–75. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–52. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 2005;123:1133–46. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stivers C, Brody T, Kuzin A, Odenwald WF. Nerfin-1 and -2, novel Drosophila Zn-finger transcription factor genes expressed in the developing nervous system. Mech Dev. 2000;97:205–10. doi: 10.1016/s0925-4773(00)00409-3. [DOI] [PubMed] [Google Scholar]
- Wu J, Duggan A, Chalfie M. Inhibition of touch cell fate by egl-44 and egl-46 in C. elegans. Genes Dev. 2001;15:789–802. doi: 10.1101/gad.857401. [DOI] [PMC free article] [PubMed] [Google Scholar]