Abstract
TrmD and Trm5 are respectively the bacterial and eukarya/archaea methyl transferases that catalyze transfer of the methyl group from S-adenosyl methionine (AdoMet) to the N1 position of G37 in tRNA to synthesize m1G37-tRNA. The m1G37 modification prevents tRNA frameshifts on the ribosome by assuring correct codon-anticodon pairings, and thus is essential for the fidelity of protein synthesis. Although TrmD and Trm5 are derived from unrelated AdoMet families and recognize the cofactor using distinct motifs, the question of whether they select G37 on tRNA by the same, or different, mechanism has not been answered. Here we address this question by kinetic analysis of tRNA truncation mutants that lack domains typically present in the canonical L shaped structure, and by evaluation of the site of modification on tRNA variants with an expanded or contracted anticodon loop. With both experimental approaches, we show that TrmD and Trm5 exhibit separate and distinct mode of tRNA recognition, suggesting that they evolved by independent and non-overlapping pathways from their unrelated AdoMet families. Our results also shed new light onto the significance of the m1G37 modification in the controversial quadruplet-pairing model of tRNA frameshift suppressors.
Keywords: tRNA(m1G37) methyl transferase, anticodon stem-loop, frameshift suppressor tRNA, m1G37
tRNAs in all organisms contain extensive site-specific base and backbone modifications that are important for their overall structural stability and functions 1; 2; 3. Many of these modifications are in the anticodon region to improve tRNA-ribosome interactions and enhance translational fidelity. A notable example is the m1G37 modification that is created by introducing a methyl group to the N1 position of the G37 base 3′ to the anticodon. This modification occurs almost universally in tRNA species that contain G37 and is found in all three domains of life. It is also present in organelles (mitochondria and chloroplasts) and in the bacteria Mycoplasma spp, which have the smallest genomes known to date 4. The presence of the m1G37 modification has been reported by previous genetic studies to increase ribosomal selectivity of tRNAs at the A site 5 and to reduce frameshift errors during translation 6; 7. A structural analysis suggests that the m1G37 modification imposes constraints on the anticodon loop dynamics 8, providing a rationale for the ability of the modified base to enforce codon-anticodon pairings. The importance of the m1G37 modification in the overall quality of protein synthesis emphasizes its conservation throughout evolution.
The m1G37 modification is synthesized post-transcriptionally by the enzyme tRNA(m1G37) methyl transferase. The bacterial enzyme is encoded by the trmD gene 9, which is essential for growth in E. coli10, Salmonella typhimurium6, and Streptococcus pneumonia11. The enzyme in the eukaryotic and archaeal domains is encoded by the trm5 gene, which is required for growth in Saccharomyces cerevisiae4. Although present in different domains of life, TrmD and Trm5 catalyze the same biochemical reaction, both using AdoMet as the methyl donor and recognizing G37 in tRNA to synthesize the modified m1G37-tRNA and to release AdoHcy (S-adenosyl homocysteine) as a product. In steady-state kinetics, the two enzymes exhibit similar parameters with respect to the AdoMet and tRNA substrates 12; 13. Unexpectedly, TrmD and Trm5 possess distinct structural motifs for recognition of AdoMet and thus are considered unrelated in their origins 12; 14; 15.
A combination of extensive bioinformatics, biochemical, and structural studies has identified five classes of AdoMet-binding fold 14; 15; 16; 17. Crystal structures of TrmD reveal a deep trefoil knot for binding AdoMet at the interface of the dimeric enzyme 18; 19; 20. This trefoil knotted structure is the signature of the class IV fold 16, and is formed by folding back the C-terminus of the AdoMet-binding domain into the catalytic cleft. The class IV fold is present only in the SPOUT family of methyl transferases 14, which consists of the SpoU (RNA ribose 2′-O-methyl transferases) and TrmD enzymes. In contrast, although no crystal structures are available yet, Trm5 exhibits sequence homology to the larger family of the class I fold 12; 21, which utilizes the ancient Rossmann-dinucleotice fold to bind AdoMet. Mutations in the predicted class I fold of Trm5 severely reduce cofactor binding and enzyme activity 12; 21. In the class I family, Trm5 is most closely related to the RsmC and ErmC’ methyl transferases, which introduce the m2G1207 and m6A2058 modifications to the 16S and 23S rRNAs of the E. coli ribosome, respectively 22; 23. Of interest is the ErmC’ methyl transferase, which confers the bacterial resistance to erythromycin and functions as a monomeric enzyme 23, consistent with the biochemical finding that Trm5 is also a monomer 12; 13.
Both TrmD and Trm5 are modular enzymes that contain, besides the AdoMet domain, a domain for recognition of G37 in tRNA for methyl transfer. An important question is how these two enzymes approach tRNA in search of G37. This question has two major ramifications. First, it addresses the evolutionary development of two unrelated enzymes that recognize the same position in tRNA and catalyze the same reaction. In principle, while TrmD and Trm5 use distinct AdoMet folds, this does not preclude them from approaching tRNA in the same way. Alternatively, the two enzymes could have evolved separately from their respective structural origins to develop different strategies of tRNA recognition. In the latter case, the lack of commonality in tRNA recognition, together with their distinct AdoMet folds, would make TrmD-Trm5 an attractive pair to explore the potential for selective inhibition of TrmD for medical purposes. Second, because G37 is embedded within tRNA sequences that contain many other G residues, the precise identification of G37 by these enzymes is intriguing. For example, although the m1G modification is also found at position 9 in eukaryotic tRNAs, the synthesis of m1G9 is specifically catalyzed by the enzyme Trm10p in S. cerevisiae24, suggesting that TrmD or Trm5 does not recognize G9. Also, although Trm5 shares sequence homology to the enzyme Trm1 25, it does not overlap with Trm1 to synthesize (m2)2G26-tRNA. Clearly, a challenge for both TrmD and Trm5 is how to localize the anticodon loop from the large and folded tRNA structure, and once in the anticodon loop, how to search for G37. Insights into this recognition and search mechanism are necessary to resolve questions concerning the function of tRNA frameshift suppressors, which have been isolated from both bacteria and yeast in response to +1 frameshift mutations 26; 27; 28; 29. Although these frameshift suppressors contain one extra nucleotide in the anticodon loop, their ability to decode four-base codons has been controversial 30.
The uniform folding of tRNA from a cloverleaf secondary structure to the L shaped tertiary structure provides a molecular framework to study tRNA recognition by TrmD and Trm5. In the L structure, the acceptor stem domain and TΨC (T) stem-loop domain coaxially stack to form the top helix, while the dihydrouridine (D) stem-loop domain and anticodon stem-loop domain coaxially stack to form the bottom helix 31; 32. The two helices join in a tertiary core made up of long-range interactions between the D, T, and V (variable) loops. Because each domain in tRNA is well defined by crystal structures 31; 32, this provides the basis to probe the TrmD and Trm5 recognition mechanism by determining the domains that are required for the methyl transfer reaction. Here we use the E. coli TrmD and the archaeal Methanocaldococcus jannaschii Trm5 as an exemplary pair to address their similarities and differences in tRNA recognition. E. coli TrmD has a well-defined high-resolution crystal structure that has been tested by mutational analysis 19. M. jannaschii Trm5 has been subjected to extensive sequence alignment, structural modeling, and mutational analysis 12; 21. Our data show that TrmD and Trm5 use distinct modes of tRNA recognition to search for G37, which sheds new light onto their distinct origins and developments. These results also provide the basis to exploit the differences between these two enzymes to improve the utility of frameshift suppressors in protein engineering.
Results
Recognition of tRNA truncation mutants
We created tRNA truncation mutants based on the transcript of M. jannaschii tRNAPro (Fig.1). This tRNA transcript, while lacking all natural modifications, is correctly and efficiently methylated by E. coli TrmD and M. jannaschii Trm5 12. It contains G36-G37, which are known determinants for E. coli and other bacterial TrmD enzymes 33; 34. Truncation mutants of the tRNA were synthesized by in vitro transcription of DNA templates that were made by primer extension of overlapping oligonucleotides 35. One truncation mutant lacked the single-stranded CCA sequence at the 3′ end (denoted as ΔCCA) but maintained all the short- and long-range H-bonding and stacking interactions of a normal tRNA, such that it should have the capacity to fold into the canonical L shape. Four other truncation mutants were created, which lacked specific domains but retained the anticodon stem-loop domain (Fig.1). For example, the Acc-ASL-T mutant retained the acceptor stem (Acc), the anticodon stem-loop (ASL), and the T stem-loop, the Acc-D-ASL mutant retained the acceptor stem, the D stem-loop, and the anticodon stem-loop, the D-ASL mutant retained the D and anticodon stem-loops, and the Acc-ASL mutant retained the acceptor stem and the anticodon stem-loop. All of these truncation mutants were gel purified, heat denatured and reannealed in the presence of 10 mM MgCl2 to assume their most stable structures before they were tested as substrates for the m1G37 methyl transfer reaction.
Fig.1.
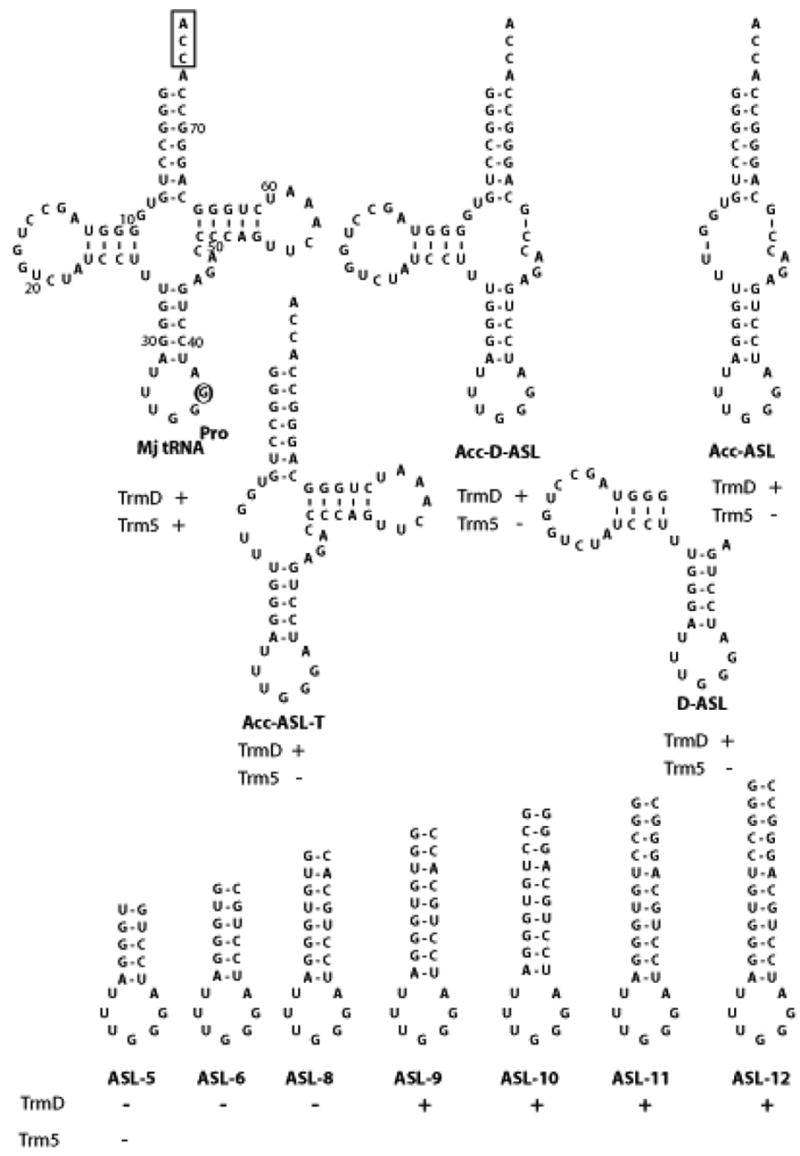
(Top) Sequences and predicted secondary structures of the wild-type and truncation mutants of M. jannaschii tRNAPro transcript. The position of G37 in the wild-type tRNA is circled and the conserved CCA end is boxed. Numbering of the wilt-type tRNA is according to that established for yeast tRNAPhe. (Bottom) Sequences and predicted secondary structures of the wild-type and extension variants of the ASL of M. jannaschii tRNAPro. The activity of each variant is marked “+” when the kcat/Km is at least 10% of the value obtained for the full-length tRNA.
Activities of the tRNA truncation mutants were determined by steady-state kinetic analysis at 37 °C for E. coli TrmD and at 52 °C for M. jannaschii Trm5 (Table 1). The assay for methyl transfer used 3H-labeled AdoMet as the substrate and monitored incorporation of the labeled methyl group into tRNA as acid-precipitable counts on filter pads 12. The steady-state parameter Km was determined with respect to tRNA under saturating AdoMet conditions (50 μM, legend to Table 1), while the parameter kcat was determined from Vmax/[E] assuming that the enzyme was fully active. Because of the uncertainty of the enzyme active fraction, the truncation mutants were evaluated by their kcat/Km parameter relative to that of the full-length tRNA transcript. With the full-length M. jannaschii tRNAPro transcript, E. coli TrmD exhibited Km of 7.3 μM and kcat of 0.6 min−1, while M. jannaschii Trm5 exhibited Km of 1.2 μM and kcat of 0.5 min−1 (Table 1), which are similar to values published previously 12; 21. In addition, kinetics of E. coli TrmD with its homologous E. coli tRNAPro was also determined, which revealed Km of 2.8 μM and kcat of 1.0 min−1 (Table 1). It appears that the Km values for tRNA of E. coli TrmD are generally higher than that of M. jannaschii Trm5. If Km values are an indication of enzyme affinity to tRNA, this result is consistent with the finding that E. coli TrmD binds tRNA with a rather poor affinity 36.
Table 1.
Kinetic analysis of tRNA truncation mutants1.
| E. coli TrmD | M. jannaschii Trm5 | |||||||
|---|---|---|---|---|---|---|---|---|
| tRNAPro | kcat (min−1) | Km (μM) | kcat/Km (min−1μM−1) | Rel | kcat (min−1) | Km (μM) | kcat/Km (min−1μM−1) | Rel |
| Ec tRNA | 1.0 ± 0.4 | 2.8 ± 0.7 | 0.3 ± 0.2 | 3.7 | ||||
| Mj tRNA | 0.6 ± 0.2 | 7.3 ± 0.6 | 0.08 ± 0.07 | 1.0 | 0.52 ± 0.01 | 1.2 ± 0.1 | 0.4 ± 0.1 | 1.0 |
| ΔCCA | 0.5 ± 0.03 | 11 ± 2 | 0.05 ± 0.01 | 0.63 | 0.23 ± 0.03 | 1.2 ± 0.1 | 0.19 ± 0.02 | 0.48 |
| Acc-ASL-T2 | 0.17 ± 0.07 | 11 ± 3 | 0.02 ± 0.01 | 0.25 | 0.01 ± 0.00 | 13 ± 2 | 0.001 ± 0.001 | 0.003 |
| Acc-D-ASL | 0.13 ± 0.06 | 7.5 ± 1.1 | 0.02 ± 0.01 | 0.25 | 0.02 ± 0.00 | 12 ± 1 | 0.001 ± 0 | 0.005 |
| Acc-ASL | 0.27 ± 0.1 | 6.6 ± 4.9 | 0.04 ± 0.07 | 0.5 | 0.01 ± 0.00 | 13 ± 3 | 0.001 ± 0.001 | 0.003 |
| D-ASL | 0.31 ± 0.1 | 14 ± 1 | 0.02 ± 0.01 | 0.25 | 0.03 ± 0.02 | 16 ± 1 | 0.002 ± 0.001 | 0.005 |
| ASL-5bp | 0.03 ± 0 | 20 ± 4 | 0.002 ± 0.001 | 0.01 | 0.02 ± 0.01 | 25 ± 2 | 0.001 ± 0.001 | 0.003 |
| ASL-6bp | 0.01 ± 0 | 8.1 ± 2 | 0.001 ± 0.001 | 0.03 | ||||
| ASL-8bp | 0.01 ± 0 | 9.1 ± 4 | 0.001 ± 0.001 | 0.01 | ||||
| ASL-9bp | 0.44 ± 0.04 | 13 ± 1 | 0.034 ± 0.001 | 0.43 | ||||
| ASL-10bp | 0.68 ± 0.2 | 24 ± 6 | 0.03 ± 0.01 | 0.38 | ||||
| ASL-11bp | 0.9 ± 0.2 | 28 ± 1 | 0.032 ± 0.006 | 0.4 | ||||
| ASL-12bp | 0.8 ± 0.1 | 21 ± 4 | 0.04 ± 0.008 | 0.5 | ||||
All tRNA truncation mutants were constructed based on the sequence of M. jannaschii tRNAPro (Fig.1) and their kinetics were compared relative to that of the full-length M. jannaschii tRNAPro. Full-length and truncated tRNA transcripts were made by run-off transcription of synthetic DNA templates by T7 RNA polymerase, which was purified from an over-expression clone provided by Dr. William McAllister 55; 56. The synthesized T7 transcripts were purified from a denaturing 12 PAGE/7M urea gel, heat denatured and reannealed before kinetic analysis. E. coli TrmD (with an N-terminal His-tag) and M. jannaschii Trm5 (with a C-terminal His-tag) were purified as described 12; 33. The assay for E. coli TrmD was performed at 37°C in 0.1 M Tris-HCl (pH 8.0), 24 mM NH4Cl, 4 mM DTT, 0.1 mM EDTA, 6 mM MgCl2, and 0.024 mg/mL BSA, while that for M. jannaschii Trm5 was performed at 52 °C in the same buffer but with100 mM KCl instead of 24 mM NH4Cl. AdoMet was included in these assays at 50 μM (Perkin Elmer, specific activity of 867 dpm/pmole in assay reactions), which is greater than the Km of the cofactor for E. coli TrmD (3.8 μM) and for M. jannaschii Trm5 (8.0 μM) by 13-fold and 6-fold, respectively. This concentration was saturating because further increases of the cofactor concentration showed little improvement in rate. For kinetic analysis, tRNA concentrations ranged from 0.5 to 150 μM, while enzymes were maintained at least 10-fold lower than the lowest concentration of the tRNA substrate and ranged from 50 to 500 nM. The background was subtracted from a control sample without tRNA. Kinetic data were fit to the Michaelis-Menten equation to derive Km and Vmax (= kcat × [E0]) for each tRNA. Values are reported with standard deviations by taking the average of at least three independent measurements.
An M-fold analysis of the Acc-ASL-T mutant predicted a Tm of 72 °C. A direct measurement of Tm was performed by placing a dry pellet of the tRNA (30 μg in 10 mM Tris-HCl, pH 8.0) onto the tip of a capillary tube in an electro-thermal apparatus (MEL-TEMP, model 1201D, Barnstead International, Dubuque, Iowa) equipped with a heating unit. The temperature at which melting of the tRNA was visibly observed in the capillary tube was defined as the Tm, which was determined to be 75 °C.
With respect to the truncation mutants of M. jannaschii tRNAPro, both E. coli TrmD and M. jannaschii Trm5 retained considerable activity with the ΔCCA mutant, suggesting that the conserved single-stranded CCA sequence is not required for these enzymes. In contrast, both enzymes exhibited reduced activity with the other truncation mutants, although the reduction in activity was generally minor (only 2–4 fold in the relative kcat/Km values) for TrmD, but much more significant (250–500-fold) for Trm5 (Table 1). One concern with the much-reduced activity of Trm5 with respect to the truncation mutants could be due to the instability of these mutants at the assay temperature of 52 °C. However, an M-fold analysis 37 of these truncation mutants revealed significantly high Tm values, ranging from the lowest 70 °C (for the ASL-D mutant) to the highest 85 °C (for the Acc-ASL mutant). These Tm values were 20 °C higher than the assay temperature of 52 °C, suggesting that these mutants were likely to maintain the predicted secondary structures at the assay temperature. Indeed, a direct measurement of Tm of the Acc-ASL-T mutant by an electrothermal apparatus revealed a Tm of 75 °C, which is in good agreement with the M-fold predicted Tm of 72 °C for this mutant. Using the Acc-ASL-T mutant as an example, the activity of Trm5 was measured at 37 °C and the ability of the enzyme to discriminate against this mutant was compared to that at 52 °C. However, the Trm5 activity with the Acc-ASL-T mutant was significantly reduced at 37 °C, particularly at lower mutant concentrations, which prevented accurate determination of individual kcat and Km. The mutant was thus evaluated at saturating concentrations to obtain the parameter kcat only. At substrate concentrations (100 and 10 μM, respectively) nearly 10-fold higher than the respective Km of the mutant and full-length tRNAs (13.1 and 1.17 μM, respectively, Table 1), analysis of initial rates yielded a kcat of 0.01 min−1 for the mutant and a kcat of 0.35 min−1 for the full-length tRNA at 37 °C. Thus, Trm5 discriminated against the truncation mutant by 35-fold at 37 °C, which is comparable to the discrimination of 50-fold in kcat by Trm5 at 52 °C, demonstrating that Trm5 is sensitive to tRNA truncations at both temperatures.
The sensitivity of Trm5 to tRNA truncations, and the lack of sensitivity of TrmD to truncations indicates a clear difference in tRNA recognition: while TrmD can accommodate substantial deletions in tRNA, even up to the loss of the entire top half, Trm5 is sensitive to deletion and must require the integrity of the tRNA L shape for activity. For Trm5, elimination of any integral part the L shape results in elevation of the Km value for tRNA by 10–20-fold at 52 °C, and decrease of the kcat value by 25–50-fold. The stronger effect on kcat by deletions suggests that even domains that are distal to the anticodon end, such as those in the acceptor stem and T stem-loop, are important for the proper positioning G37 in the Trm5 active site.
The ability of TrmD to accommodate substantial truncations raised the possibility that the ASL domain might be sufficient for methyl transfer. However, the ASL domain alone, with 5 base pairs in the stem (denoted as ASL-5bp), was a poor substrate for TrmD. It manifested a loss of ~100-fold in kcat/Km due to an increase of ~3-fold in Km and a decrease of 20-fold in kcat (Fig.1, Table 1). As expected, this ASL-5bp domain was also a poor substrate for Trm5. A comparison of substrates that were active for TrmD, however, suggested that the poor activity with the ASL-5bp domain might be improved by lengthening the anticodon stem. For example, the D-ASL and Acc-ASL truncation mutants, which were much more efficient than ASL as substrates for TrmD, can be viewed as extended ASLs in which the D- and acceptor stems are stacked on top of the anticodon stem. To test this possibility, a series of ASL derivatives with increasing lengths of the anticodon stem were constructed. The sequence of the added base pairs was based on that of the acceptor stem (Fig.1), so as to avoid reconstitution of the natural D-anticodon helix in tRNAPro. In this design, the hybrid sequences would shed light on whether it was the sequence or length that was the determinant for TrmD activity. Interestingly, while ASL-6bp and ASL-8bp remained poorly recognized by TrmD, ASL-9bp to ASL-12bp were efficient substrates (Table 1). The improvement in activity from 8 to 9 base pairs in the stem was largely driven by an improved kcat, raising the overall kcat/Km to within ~2-fold of that for the full-length tRNA, and emphasizing the importance of lengths, but not sequences, in the stem region of the ASL domain for recognition by TrmD. The effect of length on kcat suggests that the length requirement is to mediate the proper positioning of G37 in the TrmD active site.
Alteration of the anticodon loop size
Once TrmD and Trm5 localize the ASL domain, the next question is to search for G37. Because these enzymes identify G37 independent of sequence context or flexibility of the ASL domain, it is likely that their search determinant is the positioning of the target base in the anticodon loop. To test this possibility, the size of the anticodon loop of M. jannaschii tRNAPro was altered. Additionally, because previous genetic studies had isolated suppressor mutants of tRNAPro that contained an extra nucleotide in the anticodon loop in response to +1 frameshift mutations 26; 27, it was of interest to determine how TrmD and Trm5 recognize such suppressor tRNAs.
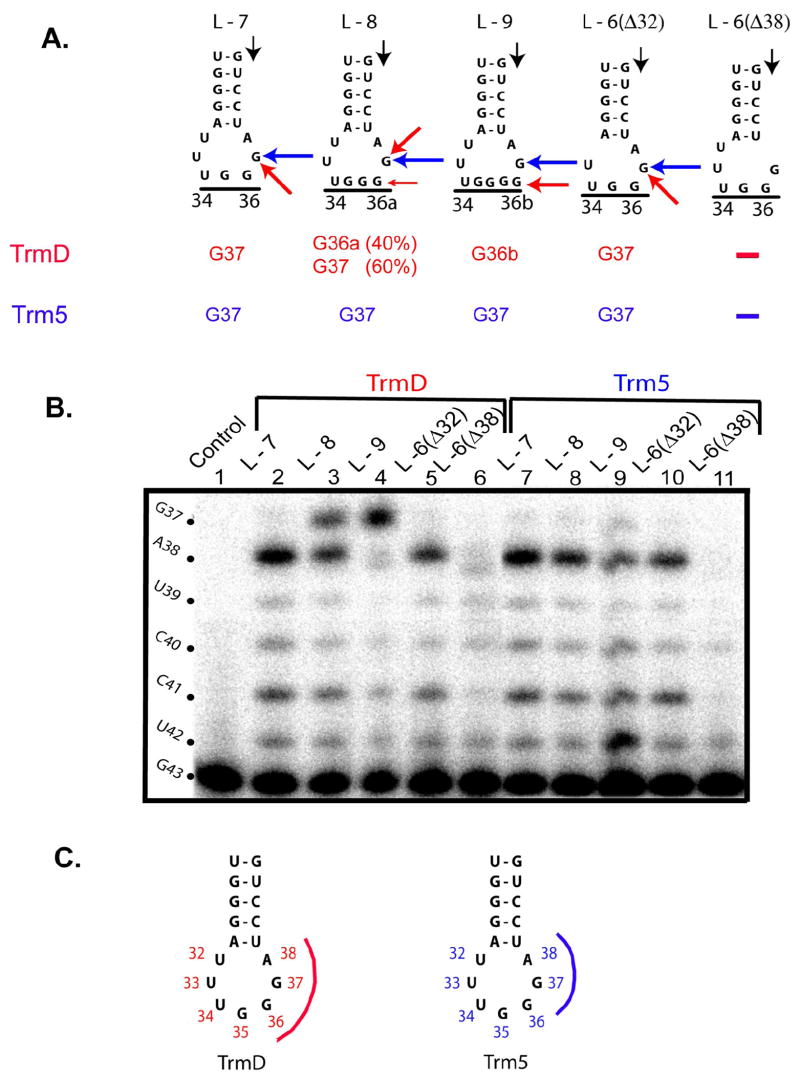
The transcript of M. jannaschii tRNAPro contains the anticodon loop 5′-UUUGGGA-3′, where the underlined nucleotides indicate anticodon positions 34–36. The wild-type sequence was denoted as the L-7 construct (Fig.2A). Two variants with an expanded anticodon loop were created, one containing an insertion of the G36a nucleotide and the other containing two additional Gs (G36a-G36b). They were denoted respectively as the L-8 and L-9 variants. Two variants with a contracted anticodon loop were also generated, with deletion of either U32 or A38 from the normal sequence. They were denoted respectively as the L-6 (Δ32) and L-6 (Δ38) variants. Kinetic analysis showed that, only the L-6 (Δ38) variant showed significantly reduced activity as compared to the wild-type tRNA, exhibiting relative kcat/Km values of 0.03 for TrmD and 0.01 for Trm5 (Table 2).
Fig.2.
(A) Sequences of wild-type and ASL variants of M. jannaschii tRNAPro. Positions 34-36 that encode the anticodon triplet are underlined, where expansion of the anticodon to positions 36a and 36b is indicated. The black arrow indicates the terminus of the primer used for extension analysis, while the sites of m1G modification introduced by E. coli TrmD and M. jannaschii Trm5 are indicated by the red and blue arrows, respectively, and shown at the bottom. The lack of detectable modifications is shown by the symbol “−“. (B) A representative gel analysis of primer extension products. The DNA primer, complementary to A58 to G43 in M. jannaschii tRNAPro, was labeled with 32P at the 5′ end, annealed to the tRNA substrates, and extended by AMV reverse transcriptase (Roche) in 50 mM Tris-HCl (pH 8.5), 8 mM MgCl2, 30 mM KCl, 1 mM DTT, 40 mM potassium borate for 30 min at 42 °C. After termination of the reaction with 7 M urea, extension products were resolved by 12% PAGE/7 M urea and analyzed by a PhosphorImager. Positions for primer extension on tRNAPro are indicated on the left (black). Lanes 1 provides a control, where primer extension was performed with unmodified tRNA transcript. Some non-specific primer extension stops are notable in variant tRNA samples but not in the normal tRNA substrate. These non-specific primer extension stops are likely premature termination stops due to the presence of heterogeneous folding products as a result of alteration of the anticodon loop size. (C) A model showing the approach of TrmD (pink) and Trm5 (blue) to G37 in ASL.
Table 2.
Kinetic analysis of tRNA mutants with alterations to the anticodon loop.
| E. coli TrmD | M. jannaschii Trm5 | |||||||
|---|---|---|---|---|---|---|---|---|
| tRNAPro | kcat (min−1) | Km (μM) | kcat/Km (min−1μM−1) | Rel | kcat (min−1) | Km (μM) | kcat/Km (min−1μM−1) | Rel |
| L-7 (wt) | 0.6 ± 0.2 | 7.3 ± 0.6 | 0.08 ± 0.07 | 1.0 | 0.52 ± 0.01 | 1.2 ± 0.1 | 0.4 ± 0.1 | 1.0 |
| L-8 | 0.13 ± 0.01 | 4.4 ± 1.1 | 0.03 ± 0.01 | 0.38 | 0.5 ± 0.02 | 2 ± 0.3 | 0.25 ± 0.01 | 0.63 |
| L-9 | 0.26 ± 0.06 | 15.5 ± 3.0 | 0.02 ± 0.004 | 0.25 | 0.37 ± 0.04 | 2.8 ± 0.1 | 0.13 ± 0.04 | 0.33 |
| L-6 (Δ32) | 0.18 ± 0.03 | 7.1 ± 1.5 | 0.025 ± | 0.31 | 1.0 ± 0.1 | 2.5 ± 0.1 | 0.4 ± 0.02 | 1.0 |
| L-6 (Δ38) | 0.06 ± 0.02 | 24.3 ± 2.5 | 0.0025 ± 0.0005 | 0.03 | 0.08 ± 0.01 | 15.2 ± 2.4 | 0.005 ± 0.0007 | 0.01 |
The site of methylation in each of these variants was determined by primer extension. To ensure homogeneity of the tRNA substrate for primer extension, the methylated fraction after the TrmD or Trm5 reaction was purified away from the unmodified fraction by our recently developed RNaseH procedure 38. In this procedure, the tRNA product after methylation was subjected to hybridization with a DNA oligonucleotide complementary to the site of modification. The unmodified fraction formed a stable hybrid and was cleaved by RNaseH, whereas the modified fraction did not hybridize and was RNaseH-resistant. The intact methylated tRNA was then purified by PAGE and used as the substrate for primer extension. Two primers were used, one complementary to nucleotides A57 to G43 of M. jannaschii tRNAPro and the other complementary to nucleotides C56 to U42. Results of primer extension were the same with these two primers and are shown only for the A57-G43 primer. Extension of the A57-G43 primer began with G43 (indicated by the short black arrow in Fig.2A) and proceeded to the base 3′ to the methylated base, where the methylation blocked primer extension. Primer extension analysis (Fig.2B) showed that both TrmD and Trm5 correctly methylated G37 in the wild-type tRNA, because the extension stopped at position 38 immediately 3′ to G37 (lanes 2,7). However, TrmD and Trm5 are guided by different rules to select the site of methylation in tRNA variants. For example, with the L-8 variant that contained the extra G36a, TrmD methylated either G36a (~40%) or G37 (~60%) (lane 3), whereas Trm5 only methylated G37 (lane 8). With the L-9 substrate that contained G36a and G36b, TrmD methylated G36b (lane 4) while Trm5 methylated G37 (lane 9). Thus, expansion of the anticodon altered the site of methylation for TrmD, but it did not affect the selectivity of G37 for Trm5.
Deletion of nucleotides on either side of the anticodon had the same effect on TrmD and Trm5. With the L-6 (Δ32) variant that lacked U32, primer extension analysis showed that both TrmD and Trm5 methylated G37 (lanes 5, 10), suggesting that deletion of U32 does not affect tRNA recognition by these enzymes. With the L-6 (Δ38) variant that lacked A38, however, no primer extension products were observed on the tRNA substrate modified by TrmD or Trm5 (lanes 6, 11), suggesting that A38 was an important determinant for G37 recognition by both enzymes. The lack of modification by TrmD and Trm5 upon removal of A38, as observed by the primer extension analysis, is consistent with the significant loss of activity for both enzymes in the methylation assay (Table 2).
Discussion
Previous studies of bioinformatics and structural modeling have suggested that TrmD and Trm5 are a pair of “non-orthologous” enzymes that descended from unrelated protein families and that were developed in different domains of life to catalyze the same biological reaction 12; 14; 15. This concept was proposed based on the notion that these two proteins use unrelated motifs to bind AdoMet for the methyl transfer reaction. While the AdoMet-binding motifs justifiably define the origins of these enzymes, due to the broad and diverse reactions in which this cofactor is involved 16; 17, the question of how these enzymes developed from their respective AdoMet-binding motifs, and whether or not they acquired similar or dissimilar mechanisms for tRNA recognition was not addressed. Here we employed two different methods to examine and compare how E. coli TrmD and M. jannaschii Trm5 recognize tRNA. These methods probe the activity of tRNA truncation mutants, and the selectivity of the target nucleotide for methylation in expanded or contracted anticodon loops. In both cases, the two enzymes recognize different features of tRNA and follow different guides to search for G37. Thus, while TrmD and Trm5 originated from unrelated motifs for binding AdoMet, they also have developed different strategies and mechanisms for tRNA recognition, suggesting that they evolved independently and in non-overlapping paths.
Studies of tRNA truncation mutants show that the activity of Trm5 is sensitive to truncations of the tRNA main body at 52 °C. This sensitivity has also been demonstrated for the Acc-ASL-T mutant at 37 °C. Although Trm5 exhibits stronger discrimination against the Acc-ASL-T mutant at 52 °C (50-fold in kcat) than at 37 °C (35-fold in kcat), which might reflect the instability of the truncated tRNA at the higher temperature to some degree, the general trend of discrimination is preserved at both temperatures. This suggests that the integrity of the L shape is the primary determinant for the Trm5 activity. In contrast, TrmD requires just the anticodon loop that is capped with a stem of minimally 9 base pairs. The defined length requirement suggests that TrmD must bind at least 9 base pairs in the stem for sufficient stability. Interestingly, the length requirement of 9 base pairs is approximately the sum of the coaxially stacked D stem (3–4 base pairs) and anticodon stem (5 base pairs) in tRNA, suggesting that TrmD may use the tRNA tertiary core as a point of contact for binding stability and for recognition of G37. It should be pointed out, however, that our result is strictly based on in vitro analysis and cannot be directly compared with the result of an earlier in vivo study that suggested that certain substitutions in the tRNA acceptor stem, D stem, and anticodon stem can reduce TrmD activity 39. Although the use of in vivo genetic studies represents a powerful approach toward appreciating the physiological significance of particular enzyme-tRNA interactions, it is inherently limited because a substantially impaired enzyme activity due to a tRNA mutation may still sustain cellular activity. Second, tRNA maturation in vivo may proceed in a temporal pathway, in which certain modifications occur prior to others. Although the precise sequence of this temporal pathway is unknown at present, it is possible that the isolated mutations in the previous genetic study may actually affect earlier steps of modifications that precede TrmD. Third, the cellular readout of in vivo studies does not usually report on the specific interaction of interest, but rather on a downstream effect, to which the specific interaction may only be part of the contributing factor. For example, the readout of the previous in vivo study of TrmD is the suppression of a frameshift mutation that arose due to the lack of the m1G37 modification 39. In this case, the frequency of frameshift suppression is ultimately determined by the ability of the ribosome to communicate the anticodon-codon interaction in the 30S to the GTPase center in the 50S for interactions with regions of tRNAs distal from the anticodon end. These interactions are necessary to promote accommodation of aminoacyl-tRNA to the A site, and translocation of peptidyl-tRNA from the A to P to E sites on the ribosome. While m1G37 is clearly a determinant for the anticodon-codon interaction, recent studies have shown that regions of the D stem and tertiary core are important for ribosome activities 40; 41. Thus, it is conceivable that mutations that disrupt the tRNA acceptor stem structure may have little effect on TrmD, but have more effect on the communication between the 30S and 50S subunits so as to affect frameshift suppression. This analysis illustrates that the information obtained from in vivo studies is only suggestive of a particular enzyme-tRNA recognition mechanism but it cannot definitively resolve the recognition issue.
TrmD and Trm5 differ in their search for G37 in the anticodon loop. Analysis of variant anticodon loops with altered sizes reveals that TrmD is sensitive to expansion of the anticodon triplet and changes its selectivity of the target base. In the two cases of anticodon expansion (L-8 and L-9), TrmD appears to prefer the 3′ end of the anticodon as the site of modification. However, this selection is dependent on the presence of position 38, because elimination of position 38 prevents tRNA recognition by TrmD. One possibility is that TrmD anchors on positions 38 and approaches the site of methylation by “counting” the number of the nucleotides from position 34 of the anticodon sequence. Thus, both sides of position 37 are important for substrate selection by TrmD (Fig.2C). In contrast, the only element that is critical to the selection of G37 by Trm5 is position 38. Trm5 is insensitive to deletion or insertion on the 5′ side of position 37 but is inactivated upon removal of position 38, suggesting that it approaches from the 3′ side of position 37 (Fig.2C).
Alternatively, the difference between TrmD and Trm5 in the search for G37 may be interpreted from a structural point of view. High-resolution crystal structures of several RNA modification enzymes in complex with their RNA substrates have come to light recently. Analysis of these structures has identified two general mechanisms of RNA recognition. In one mechanism, modification enzymes recognize their RNA substrates by rigidly docking onto the pre-formed RNA structures. Enzymes that use this mechanism include TruA (which converts U38, U39, U40 in tRNA to Ψ) 42 and TruB (which converts U55 in tRNA to Ψ) 43; 44. In the other mechanism, modification enzymes recognize their RNA substrates by unfolding the pre-formed RNA structures and re-folding the RNAs to different structures. Enzymes that use this mechanism include RluA (which converts U32 in tRNA to Ψ) 45, RumA (which converts U1939 in E. coli 23S rRNA to m5U) 46, and ArcTGT (which converts G15 in archaeal tRNAs to archaeosine) 47. Between the two mechanisms, the refolding mechanism is more likely than the rigid docking mechanism to accommodate nucleotide insertions or deletions, while maintaining the capacity to position the target base at the active site. Thus, because the selectivity of TrmD changes upon nucleotide insertion, this enzyme appears to use the rigid docking model. In contrast, because Trm5 maintains its selectivity even with nucleotide insertion, it is likely to use the refolding model. Further structural analysis of TrmD and Trm5 in complex with an RNA substrate is necessary to provide insights into these possibilities.
The tRNA construct with one nucleotide insertion to the anticodon triplet (L-8) is of important biological interest. Previous studies isolated mutations from S. typhimurium on the basis of their ability to suppress single base-pair insertion (+1 frameshift) mutations. Many of these suppressors of frameshift (suf) mutations were mapped to tRNA genes containing insertion of one nucleotide of the anticodon 48; 49; 50. For example, the sufA6 and sufB2 mutations 48; 49 were found in a tRNAPro variant carrying the G36a insertion as in the L-8 construct (Fig.2). The isolation of tRNAs with an 8-nt anticodon loop suggested the possibility that the expanded anticodon might be used to read a “quadruplet” codon, thus restoring the reading frame at the next triplet sequence. However, this concept was called into questions, because analysis of the tRNA suppressors isolated from cells in a previous study identified a m1G modification at position 36a, which would block quadruplet pairing 30. An alternative model was thus proposed, suggesting that sufA6 and sufB2 promote frameshifting by an indirect mechanism 30. Our results show that TrmD introduces the m1G modification either to G36a or to G37, raising the possibility that the modification of G37 would allow G36a to participate in quadruplet pairing, whereas the modification of G36a would block quadruplet pairing. The discrepancy between our results and the previous study is most likely due to our use of tRNA transcripts as substrates rather than tRNAs isolated from cells as in the previous study, which are limited in quantities. Although it is possible that patterns of tRNA modification in transcripts do not reflect those in native tRNAs, this is unlikely because of the observed specificity and accuracy of modification on the wild-type transcript by both TrmD and Trm5 (lanes 2,7). Thus, sufA6 and sufB2 can function as frameshift suppressors due to the modification at G37, but their suppression efficiency would be compromised due to partial modification at G36a. This new insight is consistent with the dominant nature of the sufA6 and sufB2 alleles 48, and with other studies that demonstrate the feasibility of quadruplet pairing at both the A and P sites on the ribosome 51.
The results here offer new perspectives for medical and biotechnology applications. Notably, the partition of TrmD into bacteria and Trm5 into eukaryotes based on their AdoMet-folds had suggested the possibility of selective targeting of TrmD in bacteria. However, the distinction by the AdoMet-fold alone is insufficient for drug design because there exist human enzymes that share the same AdoMet-fold with TrmD (such as the TrmH enzyme that catalyzes the 2′-O-methylation of G18 of tRNA 52). The demonstration that TrmD and Trm5 have distinct determinants for tRNA recognition and for search of the G37 nucleotide should provides new impetus to target the bacterial TrmD. In addition, the results should aid in genetic engineering to incorporate unnatural amino acids into specific positions in proteins during ribosomal translation. One prominent approach has been to use tRNAs with an 8-nt anticodon loop to insert unnatural amino acids in response to a complementary quadruplet in the mRNA sequence 53; 54. The approach is targeted to the bacterial protein synthesis machinery because of its capacity to generate significantly larger quantities of protein relative to the eukaryotic machinery. However, the ability of bacterial TrmD to modify G36a will inevitably compromise the efficiency of suppression, and thus the efficiency of incorporation of unnatural amino acids. Because the eukaryotic/archaeal Trm5 does not modify G36a, we suggest that Trm5 should be substituted for TrmD in the bacterial protein synthesis machinery to improve the efficiency of this approach. Because we have shown that the activity of trm5 can support cell growth in the absence of trmD12, the idea can be implemented by introducing a plasmid-borne trm5 into a trmD-knockout strain. Alternatively, in the presence of TrmD, quadruplet anticodon should be designed to prevent methylation at the 36a position.
Acknowledgments
This work was supported by the NIH grant GM66267 to YMH. We thank Dr. Eric Wickstrom for assistance with the electo thermal melting experiment, Dr. Bill McAllister for the T7 RNA polymerase clone, and Dr. Howard Gamper for discussion and critically reading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bjork GR. Genetic dissection of synthesis and function of modified nucleosides in bacterial transfer RNA. Prog Nucleic Acid Res Mol Biol. 1995;50:263–338. doi: 10.1016/s0079-6603(08)60817-x. [DOI] [PubMed] [Google Scholar]
- 2.Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–38. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 4.Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? Embo J. 2001;20:231–9. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Esberg B, Curran JF, Bjork GR. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J Mol Biol. 1997;271:209–21. doi: 10.1006/jmbi.1997.1176. [DOI] [PubMed] [Google Scholar]
- 6.Bjork GR, Wikstrom PM, Bystrom AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–9. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 7.Hagervall TG, Tuohy TM, Atkins JF, Bjork GR. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J Mol Biol. 1993;232:756–65. doi: 10.1006/jmbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- 8.Stuart JW, Koshlap KM, Guenther R, Agris PF. Naturally-occurring modification restricts the anticodon domain conformational space of tRNA(Phe) J Mol Biol. 2003;334:901–18. doi: 10.1016/j.jmb.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 9.Bystrom AS, Bjork GR. Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA (m1G) methyltransferase in Escherichia coli K-12. Mol Gen Genet. 1982;188:440–6. doi: 10.1007/BF00330046. [DOI] [PubMed] [Google Scholar]
- 10.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Dwyer K, Watts JM, Biswas S, Ambrad J, Barber M, Brule H, Petit C, Holmes DJ, Zalacain M, Holmes WM. Characterization of Streptococcus pneumoniae TrmD, a tRNA methyltransferase essential for growth. J Bacteriol. 2004;186:2346–54. doi: 10.1128/JB.186.8.2346-2354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christian T, Evilia C, Williams S, Hou YM. Distinct origins of tRNA(m1G37) methyltransferase. J Mol Biol. 2004;339:707–19. doi: 10.1016/j.jmb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Brule H, Elliott M, Redlak M, Zehner ZE, Holmes WM. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry. 2004;43:9243–55. doi: 10.1021/bi049671q. [DOI] [PubMed] [Google Scholar]
- 14.Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J Mol Microbiol Biotechnol. 2002;4:71–5. [PubMed] [Google Scholar]
- 15.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–64. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–35. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol. 2002;12:783–93. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 18.Ahn HJ, Kim HW, Yoon HJ, Lee BI, Suh SW, Yang JK. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. Embo J. 2003;22:2593–603. doi: 10.1093/emboj/cdg269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkins PA, Watts JM, Zalacain M, van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J Mol Biol. 2003;333:931–49. doi: 10.1016/j.jmb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Wang W, Shin DH, Yokota H, Kim R, Kim SH. Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 A resolution: a novel methyltransferase fold. Proteins. 2003;53:326–8. doi: 10.1002/prot.10479. [DOI] [PubMed] [Google Scholar]
- 21.Christian T, Evilia C, Hou YM. Catalysis by the second class of tRNA(m1G37) methyl transferase requires a conserved proline. Biochemistry. 2006;45:7463–73. doi: 10.1021/bi0602314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tscherne JS, Nurse K, Popienick P, Ofengand J. Purification, cloning, and characterization of the 16 S RNA m2G1207 methyltransferase from Escherichia coli. J Biol Chem. 1999;274:924–9. doi: 10.1074/jbc.274.2.924. [DOI] [PubMed] [Google Scholar]
- 23.Schluckebier G, Zhong P, Stewart KD, Kavanaugh TJ, Abad-Zapatero C. The 2.2 A structure of the rRNA methyltransferase ErmC’ and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J Mol Biol. 1999;289:277–91. doi: 10.1006/jmbi.1999.2788. [DOI] [PubMed] [Google Scholar]
- 24.Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. Rna. 2003;9:574–85. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Constantinesco F, Motorin Y, Grosjean H. Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J Mol Biol. 1999;291:375–92. doi: 10.1006/jmbi.1999.2976. [DOI] [PubMed] [Google Scholar]
- 26.Sroga GE, Nemoto F, Kuchino Y, Bjork GR. Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNA(Pro)2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res. 1992;20:3463–9. doi: 10.1093/nar/20.13.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cummins CM, Donahue TF, Culbertson MR. Nucleotide sequence of the SUF2 frameshift suppressor gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1982;79:3565–9. doi: 10.1073/pnas.79.11.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bossi L, Smith DM. Suppressor sufJ: a novel type of tRNA mutant that induces translational frameshifting. Proc Natl Acad Sci U S A. 1984;81:6105–9. doi: 10.1073/pnas.81.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaber RF, Culbertson MR. The yeast frameshift suppressor gene SUF16-1 encodes an altered glycine tRNA containing the four-base anticodon 3′-CCCG-5′. Gene. 1982;19:163–72. doi: 10.1016/0378-1119(82)90002-6. [DOI] [PubMed] [Google Scholar]
- 30.Qian Q, Li JN, Zhao H, Hagervall TG, Farabaugh PJ, Bjork GR. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol Cell. 1998;1:471–82. doi: 10.1016/s1097-2765(00)80048-9. [DOI] [PubMed] [Google Scholar]
- 31.Kim SH, Quigley GJ, Suddath FL, McPherson A, Sneden D, Kim JJ, Weinzierl J, Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973;179:285–8. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- 32.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974;250:546–51. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 33.Redlak M, Andraos-Selim C, Giege R, Florentz C, Holmes WM. Interaction of tRNA with tRNA (guanosine-1)methyltransferase: binding specificity determinants involve the dinucleotide G36pG37 and tertiary structure. Biochemistry. 1997;36:8699–709. doi: 10.1021/bi9701538. [DOI] [PubMed] [Google Scholar]
- 34.Takeda H, Toyooka T, Ikeuchi Y, Yokobori S, Okadome K, Takano F, Oshima T, Suzuki T, Endo Y, Hori H. The substrate specificity of tRNA (m1G37) methyltransferase (TrmD) from Aquifex aeolicus. Genes Cells. 2006;11:1353–65. doi: 10.1111/j.1365-2443.2006.01022.x. [DOI] [PubMed] [Google Scholar]
- 35.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. Rna. 1999;5:1268–72. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watts JM, Gabruzsk J, Holmes WM. Ligand-mediated anticodon conformational changes occur during tRNA methylation by a TrmD methyltransferase. Biochemistry. 2005;44:6629–39. doi: 10.1021/bi0481038. [DOI] [PubMed] [Google Scholar]
- 37.Zuker M, Mathews D, Turner D. In: Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide in RNA biochemistry and biotechnology NATO ASI series. Barciszewski J, Clark B, editors. Kluwer Academic; 1999. [Google Scholar]
- 38.Hou YM, Li Z, Gamper H. Isolation of a site-specifically modified RNA from an unmodified transcript. Nucleic Acids Res. 2006;34:e21. doi: 10.1093/nar/gnj018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian Q, Bjork GR. Structural requirements for the formation of 1-methylguanosine in vivo in tRNA(Pro)GGG of Salmonella typhimurium. J Mol Biol. 1997;266:283–96. doi: 10.1006/jmbi.1996.0789. [DOI] [PubMed] [Google Scholar]
- 40.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–80. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan D, Kirillov S, Zhang CM, Hou YM, Cooperman BS. Rapid ribosomal translocation depends on the conserved 18–55 base pair in P-site transfer RNA. Nat Struct Mol Biol. 2006;13:354–9. doi: 10.1038/nsmb1074. [DOI] [PubMed] [Google Scholar]
- 42.Hur S, Stroud RM. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol Cell. 2007;26:189–203. doi: 10.1016/j.molcel.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoang C, Ferre-D’Amare AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–39. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 44.Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc Natl Acad Sci U S A. 2003;100:12648–53. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoang C, Chen J, Vizthum CA, Kandel JM, Hamilton CS, Mueller EG, Ferre-D’Amare AR. Crystal structure of pseudouridine synthase RluA: indirect sequence readout through protein-induced RNA structure. Mol Cell. 2006;24:535–45. doi: 10.1016/j.molcel.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 46.Lee TT, Agarwalla S, Stroud RM. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 47.Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–94. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 48.Riddle DL, Roth JR. Frameshift suppressors. 3. Effects of suppressor mutations on transfer RNA. J Mol Biol. 1972;66:495–506. doi: 10.1016/0022-2836(72)90429-9. [DOI] [PubMed] [Google Scholar]
- 49.Riddle DL, Roth JR. Frameshift suppressors. II. Genetic mapping and dominance studies. J Mol Biol. 1972;66:483–93. doi: 10.1016/0022-2836(72)90428-7. [DOI] [PubMed] [Google Scholar]
- 50.Bossi L, Kohno T, Roth JR. Genetic characterization of the sufj frameshift suppressor in Salmonella typhimurium. Genetics. 1983;103:31–42. doi: 10.1093/genetics/103.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker SE, Fredrick K. Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J Mol Biol. 2006;360:599–609. doi: 10.1016/j.jmb.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nureki O, Watanabe K, Fukai S, Ishii R, Endo Y, Hori H, Yokoyama S. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure (Camb) 2004;12:593–602. doi: 10.1016/j.str.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Anderson JC, Magliery TJ, Schultz PG. Exploring the limits of codon and anticodon size. Chem Biol. 2002;9:237–44. doi: 10.1016/s1074-5521(02)00094-7. [DOI] [PubMed] [Google Scholar]
- 54.Magliery TJ, Anderson JC, Schultz PG. Expanding the genetic code: selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli. J Mol Biol. 2001;307:755–69. doi: 10.1006/jmbi.2001.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lyakhov DL, He B, Zhang X, Studier FW, Dunn JJ, McAllister WT. Mutant bacteriophage T7 RNA polymerases with altered termination properties. J Mol Biol. 1997;269:28–40. doi: 10.1006/jmbi.1997.1015. [DOI] [PubMed] [Google Scholar]
- 56.He B, Rong M, Lyakhov D, Gartenstein H, Diaz G, Castagna R, McAllister WT, Durbin RK. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr Purif. 1997;9:142–51. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]