Abstract
Homologue segregation during the first meiotic division requires the proper spatial regulation of sister chromatid cohesion and its dissolution along chromosome arms, but its protection at centromeric regions. This protection requires the conserved MEI-S332/Sgo1 proteins that localize to centromeric regions and also recruit the PP2A phosphatase by binding its regulatory subunit, Rts1. Centromeric Rts1/PP2A then locally prevents cohesion dissolution possibly by dephosphorylating the protein complex cohesin. We show that Aurora B kinase in Saccharomyces cerevisiae (Ipl1) is also essential for the protection of meiotic centromeric cohesion. Coupled with a previous study in Drosophila melanogaster, this meiotic function of Aurora B kinase appears to be conserved among eukaryotes. Furthermore, we show that Sgo1 recruits Ipl1 to centromeric regions. In the absence of Ipl1, Rts1 can initially bind to centromeric regions but disappears from these regions after anaphase I onset. We suggest that centromeric Ipl1 ensures the continued centromeric presence of active Rts1/PP2A, which in turn locally protects cohesin and cohesion.
Introduction
During mitosis, a parental cell divides to generate two new cells, each with genome content identical to that of the parental cell. In contrast, during meiosis, a single parental cell generates four gametes, each of which has half the number of chromosomes in the parental cell. The dramatically different pattern of meiotic and mitotic chromosome segregation is achieved in part by the temporal and spatial regulation of sister chromatid cohesion (for review see Petronczki et al., 2003).
Both mitotic and meiotic cells replicate their chromosomes to generate sister chromatids that are held in juxtaposition by cohesion around the centromeres and along the length of the arms. In mitosis, the paired sister chromatids become attached to the mitotic spindle, all cohesion is dissolved, and sister chromatids segregate from each other. In contrast, during meiosis I (MI), homologues become linked as a consequence of reciprocal exchange and the presence of sister chromatid cohesion on the arms. After the linked homologues become attached to the MI spindle, cohesion on the arms is dissolved, allowing homologues to segregate from each other, but sister chromatids remain paired and segregate together to the same pole because cohesion at the centromeres and pericentric regions is protected from dissolution. MI is followed by MII, in which paired sister chromatids attach to the spindle, the remaining centromere and pericentric cohesion is dissolved, and sister chromatids segregate from one another. Therefore, the temporal and spatial regulation of sister chromatid cohesion plays a key role in generating the meiotic pattern of chromosome transmission, distinct from that of mitosis.
Several studies have provided substantial insights into the molecular basis for the meiotic regulation of sister chromatid cohesion (for reviews see Marston and Amon, 2004; Watanabe, 2005). Cohesion is mediated largely by cohesin, a multisubunit protein complex conserved from yeast to human (Guacci et al., 1997; Michaelis et al., 1997). Cohesin is loaded onto the chromosomes at S-phase to generate cohesion between sister chromatids. In order for homologues to dissociate from each other in MI, cohesin is removed from the chromosome arms by two different mechanisms: one involves condensin and Polo kinase and acts before anaphase I (Yu and Koshland, 2005); the other requires separase and Polo kinase and acts at the onset of anaphase I (Buonomo et al., 2000; Kitajima et al., 2003). The separase pathway is able to remove all cohesin and, therefore, to dissolve all sister chromatid cohesion, but the separase pathway and possibly the condensin pathway are blocked from removing cohesin at the centromeric and pericentric regions by a mechanism involving a conserved protein, called MEI-S332/Sgo1 (Kerrebrock et al., 1995; Kitajima et al., 2004).
The mechanism of meiotic cohesion protection by the MEI-S332/Sgo1 pathway is complex. MEI-S332/Sgo1 localizes to the centromeres and pericentric regions (Moore et al., 1998; Katis et al., 2004; Kitajima et al., 2004; Kiburz et al., 2005). The pericentric localization depends on cohesin and Bub1, a component of the spindle assembly checkpoint (Hamant et al., 2005; Kiburz et al., 2005). Subsequently, Sgo1 recruits to the centromere phosphatase PP2A by binding its B regulatory subunit, called Rts1 (Kitajima et al., 2006; Riedel et al., 2006). The centromeric localization of PP2A is thought to enhance the ability to dephosphorylate centromeric cohesin and thus shields it from Polo kinase–dependent removal by the separase. As expected, mutations in Bub1, MEI-S332/Sgo1, and Rts1 cause precocious separation of sister chromatids during MI. These studies show that Bub1, MEI-S332/Sgo1, and PP2A are key components for the retention of centromere and pericentric cohesin.
A recent study of cohesin protection in Drosophila melanogaster has implicated two new players, the Aurora B kinase and its binding partner inner centromere protein (INCENP; Resnick et al., 2006). In vitro MEI-S332 is bound by INCENP and phosphorylated by Aurora B. In mitosis, MEI-S332 localization to the centromeric region is compromised by a mutation in INCENP with reduced activity or by a mutation in MEI-S332 that reduces its Aurora B–dependent phosphorylation as defined by in vitro studies. These observations have led to a model in which Aurora B–INCENP complex protects cohesion by phosphorylating MEI-S332 and thereby increasing MEI-S332 ability to bind to centromeres. The authors suggest that INCENP plays a similar role in meiosis because a partially defective allele of INCENP causes partial precocious separation of sister chromatids in meiosis I and partially compromises MEI-S332 localization. These important studies pose several questions. Is the role of Aurora B in protecting centromere cohesion conserved among eukaryotes? Given the partial defects, how important is Aurora B relative to other components that are known to be essential to protect centromere cohesion? Does Aurora B protect sister chromatid cohesion through cohesin localization? Does Aurora B help recruit MEI-S332 to the centromere in meiosis, as it apparently does in mitosis, or might it have a different or additional function?
Previously, we discovered an essential function of Ipl1, the founding member of Aurora B, in modulating meiotic chromosome transmission in the budding yeast Saccharomyces cerevisiae (Yu and Koshland, 2005). In this work, we show that one function of Ipl1 is to ensure the protection of centromeric cohesin during MI, indicating that this function of Aurora B kinase is conserved between yeast and flies. The role of Ipl1 in protection of meiotic centromere cohesion is as critical as MEI-S332/Sgo1, Bub1, and Rts1. Ipl1 is only marginally required for Sgo1 localization to the centromeres. Rather, Ipl1 is critical to maintaining the PP2A subunit Rts1 at centromeres after but not before the onset of anaphase I. The continued centromeric localization of Rts1/PP2A presumably ensures that centromeric cohesion is protected from separase until MII.
Results
Ipl1 is required for sister chromatid cohesion and homologue disjunction during MI
Previously, we and others have shown that the Aurora B kinase is essential for meiosis in a diversity of organisms (Kaitna et al., 2002; Rogers et al., 2002; Yu and Koshland, 2005). To dissect Aurora B function during meiosis of budding yeast, we generated a meiosis-specific null allele of IPL1 (PCLB2IPL1) by replacing the endogenous promoter with a mitosis-specific promoter from CLB2 (Yu and Koshland, 2005). The expression of IPL1 is preserved in mitosis, and PCLB2IPL1 cells have no detectable mitotic mutant phenotype as judged by cell cycle progression and cell viability (unpublished data), but because no new Ipl1 is made from this promoter in meiosis and preexisting mitotic Ipl1 is degraded at the end of mitosis, cells with this allele have no detectable Ipl1 during meiosis (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200609153/DC1). PCLB2IPL1 cells initiate meiotic nuclear divisions, albeit with a delay (Fig. S1). These cells produce tetrads with unequal nuclei, and <1% spore viability (unpublished data). These observations suggest that Ipl1 has an essential function in meiotic chromosome transmission.
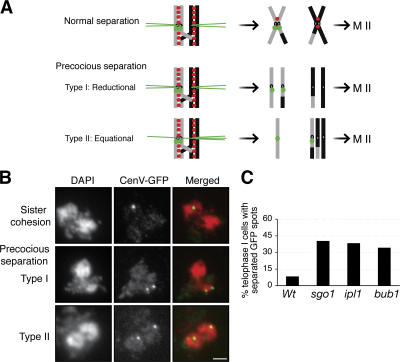
To determine the role of Ipl1 in meiotic chromosome segregation, we examined PCLB2IPL1 cells for changes in several different aspects of chromosome structure. No detectable changes were evident in chromosome compaction or assembly of the synaptonemal complex (unpublished data). To monitor cohesion in a region near the centromere, we used the GFP chromosome-marking system. Tandem arrays of Tet operators were inserted into one homologue of chromosome V at the URA3 locus, which is ∼35 Kb away from the centromere (Michaelis et al., 1997). If cohesion is preserved at the centromeres and in pericentric regions (henceforth referred to collectively as centromeric regions) throughout MI, a single GFP spot should be observed (Fig. 1, A and B, top). Failure to maintain sister chromatid cohesion leads to the appearance of two GFP spots (Fig. 1, A and B, middle and bottom). Before onset of anaphase I, a very small percentage of both wild-type and PCLB2IPL1 cells exhibited two GFP spots, indicating that Ipl1 function is not required for sister chromatid cohesion before anaphase I.
Figure 1.
Ipl1 prevents precocious separation of sister chromatids during meiosis in yeast. Yeast cells were induced for synchronous meiosis and processed for nuclei spread and indirect immunofluorescence. The tetO/tetR-GFP chromosome-marking system was used to assay sister chromatid cohesion at the centromeres. (A) A diagram shows sister chromatid segregation during meiosis I. Chromosomes are shown as gray and black bars, cohesin as red dots, and tetR-GFP as green dots. (B) Representative images show sister chromatid cohesion and precocious separation of sister chromatids during meiosis I. Two categories of precocious sister chromatid separation were observed in PCLB2IPL1 cells: in type I, sisters segregated to the same pole (middle); in type II, sisters segregated to opposite poles (bottom). Bar, 2 μm. (C) Quantitative measures of centromeric cohesion at telophase I in wild type (Wt), ipl1 (PCLB2IPL1), sgo1 (PCLB2SGO1), and bub1 (PCLB2BUB1). Telophase I cells were judged as separated chromosome masses and depolymerized microtubule spindles. Type I and II cells were summarized in C. At least 50 cells were scored for each strain.
We then examined telophase I cells for precocious separation of sister chromatids. Only 8% of wild-type cells exhibited two GFP spots (Fig. 1 C), indicating that the vast majority of sister chromatids maintain cohesion at the centromeres as expected. In contrast, 38% of PCLB2IPL1 cells (27% of type I and 11% of type II; Fig. 1 B) show precocious separation of sister centromeres, about five times as many as in wild-type cells (Fig. 1 C). The severity of precocious chromatid separation in PCLB2IPL1 cells is similar to that in meiotic null alleles of sgo1 (PCLB2SGO1) and bub1 (PCLB2BUB1; Fig. 1 C). These data suggest that Ipl1, like Sgo1 and Bub1, plays a major role in the protection of sister chromatid cohesion during MI of budding yeast.
If the sole function of Ipl1 were to protect centromeric cohesion during MI, then the removal of Ipl1 would lead to segregation of both separated sister chromatids to only one of the two MI products (Fig. 1, A and B, middle), but in PCLB2IPL1, about half of the cells with two GFP spots (the sister chromatids) segregate to opposite poles and thus to different MI products (Fig. 1 B, bottom). Furthermore when we used the same GFP system to mark both homologues of chromosome V, we observed that, in 52% of PCLB2IPL1 cells in MI, both homologues segregated to the same spindle pole, compared with only 3% in wild-type cells (unpublished data). These observations suggest that Ipl1, in addition to protecting centromeric cohesion, is required to ensure the co-oriented attachment of sister kinetochores to the MI spindle (Fig. 1 A, top).
Ipl1 is required for the protection of centromeric cohesin during MI
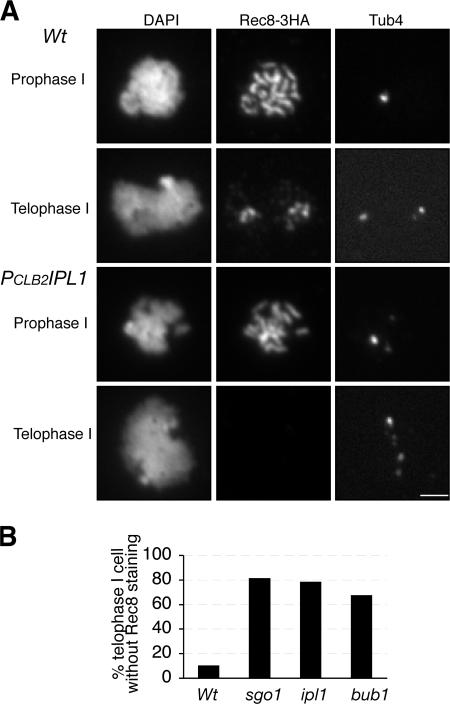
Because sister chromatid cohesion is mediated by cohesin, we addressed whether Ipl1 is required for the maintenance of cohesin in the centromeric region during MI. We followed the localization of different cohesin subunits in wild-type and PCLB2IPL1 cells by indirect immunofluorescence on spread nuclei. Similar results were obtained with cohesin subunits Smc1, Scc3, and Rec8. We show the localization of the representative cohesin subunit Rec8 tagged with a 3xHA epitope (Fig. 2). In prophase I of wild-type and PCLB2IPL1 cells, cohesin is able to associate with the chromosomes along their entire length (Fig. 2 A). By telophase I, 89% of wild-type cells lose cohesin staining on the chromosome arms but retain staining around the spindle poles, where the centromeres are positioned (Fig. 2 A). The remaining 11% show no cohesin staining. In contrast, by telophase I, 78% of PCLB2IPL1 cells lose all cohesin staining, a sevenfold difference (Fig. 2 B). We also examined cohesin localization in cells lacking Ipl1 that were arrested in metaphase I or in telophase I (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200609153/DC1). This analysis confirmed that the absence of Ipl1 eliminates cohesin localization to centromeres in telophase I but not to centromeres or arms in metaphase I. Finally, the severity of cohesin loss in PCLB2IPL1 cells is similar to that of PCLB2SGO1 and PCLB2BUB1 (Fig. 2 B and see the following paragraphs), suggesting that, like Sgo1 and Bub1, Ipl1 plays a major role in the protection of meiotic cohesin. Furthermore, similarity of their mutant phenotypes implies that these proteins act in the same pathway to protect cohesin.
Figure 2.
Ipl1 prevents premature loss of centromeric cohesin during meiosis in yeast. Yeast cells were induced for synchronous meiosis and processed for indirect immunofluorescence as in Fig. 1. The anti-HA antibody (12CA5) was used to localize cohesin subunit Rec8, which was tagged by 3xHA. A polyclonal anti-Tub4 antibody was used to localize the spindle pole bodies. (A) Representative images show meiotic cells at prophase I and telophase I. Note that PCLB2IPL1 mutant cells tend to form multiple spindle pole bodies (bottom right). Bar, 2 μm. (B) Quantitative measures of cohesin retention in wild type (Wt), ipl1 (PCLB2IPL1), sgo1 (PCLB2SGO1), and bub1 (PCLB2BUB1) as in Fig. 1 C. At least 50 telophase I cells were scored for each strain.
Ipl1 is a passenger protein that colocalizes with Sgo1 at the centromeres
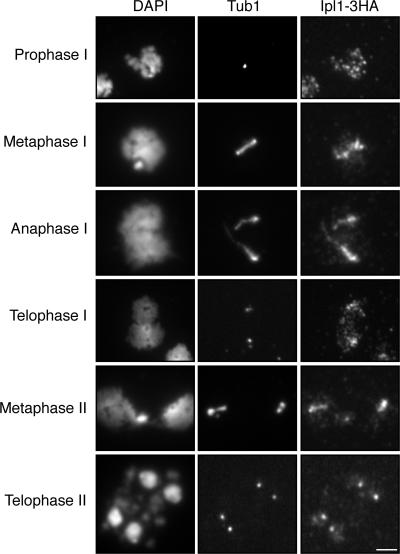
To begin to investigate how Ipl1 protects centromeric cohesin, we examined the localization of Ipl1 through meiosis. We incorporated 3xHA into the C terminus of Ipl1 and visualized the localization of the tagged protein by indirect immunofluorescence in spread nuclei. Ipl1 localizes to prophase chromosomes, as intense dispersed foci superimposed on a diffuse weak general staining (Fig. 3, top). At metaphase and telophase I, Ipl1 is more concentrated around the spindle poles (Fig. 3). This pattern of staining is reminiscent of centromeric proteins (see the following paragraphs). At late anaphase I, a substantial portion of Ipl1 is localized along the microtubule spindle (Fig. 3). The pattern of dynamic localization of Ipl1 during yeast meiosis is very similar to that of Aurora B kinase and INCENP observed in mitosis in many organisms, which led to their designation as passenger proteins (Ducat and Zheng, 2004; Vagnarelli and Earnshaw, 2004).
Figure 3.
Localization of Ipl1 during meiosis. Yeast cells were induced for synchronous meiosis and processed for indirect immunofluorescence. An anti-HA antibody (12CA5) was used to localize Ipl1-3HA; an anti–α-tubulin antibody (YOL134) was used to localize the microtubules. Note that Ipl1 is a passenger protein during meiosis. Bar, 2 μm.
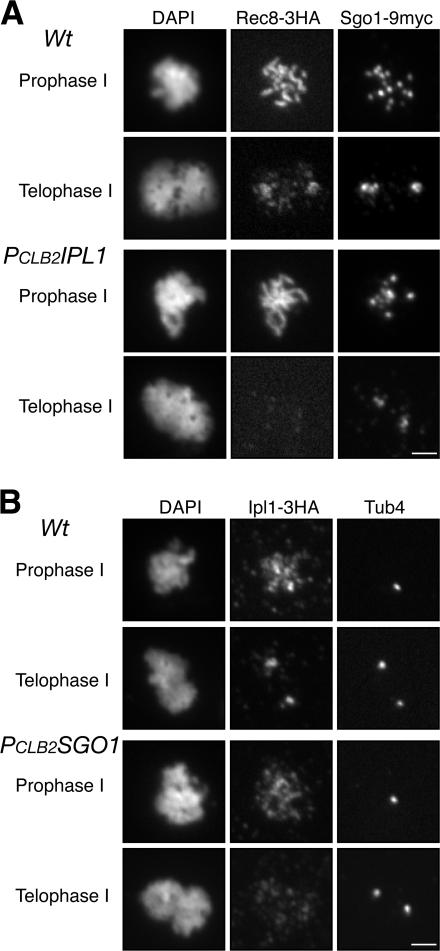
The dispersed foci of Ipl1 in prophase I and spindle pole body–proximal foci in metaphase and telophase I cells are similar to the localization pattern previously reported for Sgo1. Sgo1 immunostaining was subsequently demonstrated to correspond to its binding to centromeric regions by chromatin immunoprecipitation (ChIP; Kiburz et al., 2005), so we tested for colocalization of Ipl1 with the centromeric Sgo1 foci by performing indirect immunofluorescence to localize both Ipl1 and Sgo1 in yeast meiotic cells. At prophase I, when homologues are paired, Sgo1 forms ∼16 foci, corresponding to 16 paired yeast centromeres (Fig. 4 A). The Ipl1 foci colocalize with those of Sgo1 (Fig. 4 A). The colocalization of Ipl1 and Sgo1 at the centromere is also evident during telophase I, when yeast centromeres are clustered around the spindle poles (Fig. 4 A). Collectively, both Ipl1 and Sgo1 are concentrated at the yeast centromeres, supporting the idea that these proteins act together to protect centromeric cohesin and sister chromatid cohesion.
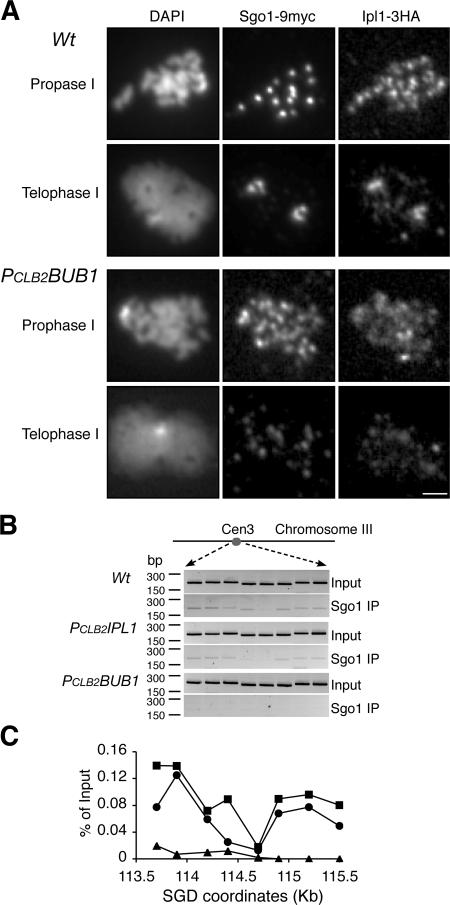
Figure 4.
Bub1 controls the localization of Sgo1 and Ipl1 during meiosis. Yeast cells were induced for synchronous meiosis and processed for indirect immunofluorescence (A) and ChIP (B and C). An anti-MYC antibody (9E10) was used to detect Sgo1-9MYC, and an anti-HA antibody (3F10) was used for Ipl1-3HA. (A) Localization of Ipl1 and Sgo1 in meiotic cells at prophase I and telophase I. Note that both Ipl1 and Sgo1 are concentrated at the poles at telophase I only in wild-type (Wt) cells. Bar, 2 μm. (B) ChIP of Sgo1 in wild type, PCLB2IPL1, and PCLB2BUB1. Representative agarose gels are shown. Total input was diluted at 1:100. (C) Quantitative measurements of Sgo1 binding to yeast centromere III. SGD coordinates at the x axis show chromosomal positions of PCR primers used in the ChIP. All strains harbor the conditional allele of PCLB2CDC20 to arrest the cells at metaphase I. Solid squares indicate wild type, solid circles indicate PCLB2IPL1, and triangles indicate PCLB2BUB1.
The centromeric localization of Ipl1 requires both Bub1 and Sgo1
The centromeric localization of Sgo1 depends on the spindle checkpoint protein Bub1 (Kitajima et al., 2004; Kiburz et al., 2005). We therefore determined whether Bub1 is required for the localization of Ipl1 to yeast meiotic centromeres. We created a PCLB2BUB1 conditional allele that depletes Bub1 protein specifically during meiosis (Kiburz et al., 2005). This mutant bub1 allele fails to protect centromeric cohesin during meiosis I (Fig. 2 B), and as expected, sister chromatids separate precociously (Fig. 1 C). As shown in Fig. 4 A, neither Ipl1 nor Sgo1 is concentrated at the centromeres in PCLB2BUB1 cells. They are dispersed throughout the chromosomes (Fig. 4 A, bottom). We used ChIP to confirm the failure of Sgo1 to associate with centromeric DNA in the absence of Bub1 (Fig. 4 B). Unfortunately, Ipl1 has not been detectable by ChIP even in wild-type cells. Bub1 is therefore required for the proper localization of both Ipl1 and Sgo1 to centromeric regions, lending further support to the idea that Ipl1 and Sgo1 act together in the same pathway to protect centromeric cohesin.
We asked next whether the centromeric localization of Ipl1 depends on Sgo1. We examined Ipl1 localization in cells harboring a conditional sgo1 allele (PCLB2SGO1), which depletes Sgo1 during meiosis (not depicted; a similar SGO1 allele was reported previously by Katis et al. [2004] and Kiburz et al. [2005]). In wild-type cells, Ipl1 forms discrete foci that colocalize in prophase I with centromeric protein Sgo1 (Fig. 4) and in telophase I with Sgo1 and spindle poles (Figs. 3 and 4). In contrast, in >90% of PCLB2SGO1 cells, Ipl1 either fail to form detectable foci (Fig. 5 B) or form a few foci that do not colocalize with Ctf19, a centromere protein (not depicted). The centromeric association of Ipl1 therefore depends on Sgo1, suggesting that Ipl1 acts concomitant with or downstream of Sgo1 in protection of centromeric cohesin.
Figure 5.
Meiotic interaction between Ipl1 and Sgo1. Yeast cells were induced for synchronous meiosis and processed for indirect immunofluorescence with an anti-HA antibody (3F10) and an anti-MYC antibody (9E10). (A) Localization of Sgo1 and Rec8 in wild-type (Wt) and PCLB2IPL1 cells. Centromeres tend to cluster in PCLB2IPL1 cells at prophase I. Note that Sgo1 associates with the centromeres, but Rec8 is lost in PCLB2IPL1 cells at telophase I. (B) Localization of Ipl1 and Tub4 in wild-type and PCLB2SGO1 cells. Note that the Ipl1 signal is concentrated at the poles in wild-type cells but not in PCLB2SGO1 cells. Bars, 2 μm.
Ipl1 is required for centromeric localization of Rts1 in anaphase I
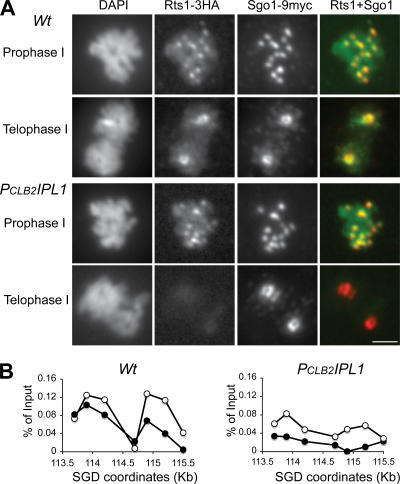
To address whether Ipl1 acts downstream of or concomitant with Sgo1, we asked whether Ipl1 function is needed for the centromeric association of Sgo1 or Rts1. Rts1 is a regulatory subunit of PP2A phosphatase. The binding of Rts1 by Sgo1 recruits PP2A to the centromeric region, where its phosphatase activity is used to protect cohesion (Kitajima et al., 2006; Riedel et al., 2006). In prophase I of wild-type cells, Sgo1 and Rts1 are concentrated at the centromeres (Fig. 4, Fig. 5 A, and Fig. 6). In prophase I of PCLB2IPL1 cells, Sgo1 is able to associate with the centromeric DNA, as shown both by ChIP (Fig. 4 B) and by indirect immunofluorescence of spread nuclei (Fig. 5 A). Similarly, Rts1 is also able to associate with centromeric regions in the Ipl1 mutant (Fig. 6). These results show that Ipl1 function is not necessary to establish localization of Sgo1 and Rts1 to the centromeric regions, but the presence of Sgo1 and Rts1 at centromeric regions in prophase I is clearly not sufficient to protect centromeric cohesin in anaphase I, given the cohesin defect in the ipl1 mutant.
Figure 6.
Localization of Rts1 during meiosis. Yeast cells were induced for synchronous meiosis and processed for indirect immunofluorescence (A) and ChIP (B). The anti-HA antibody (3F10) and anti-MYC antibody (9E10) were used to localize Rts1 and Sgo1 in spread nuclei, respectively. (A) Immunolocalization of Rts1 in meiotic cells. Note that Rts1 signals are missing from the centromeres in PCLB2IPL1 cells at telophase I. Bar, 2 μm. (B) ChIP of Rts1 in meiotic cells. Quantitative measurements of Rts1 binding to yeast centromere III as shown in Fig. 4 C. SGD coordinates at the x axis show chromosomal positions of PCR primers used in the ChIP. All strains harbor the conditional allele of PCLB2CDC20 to arrest the cells at metaphase I. Open circles indicate 3 h of induction of meiosis with the majority of cells at prophase I, and solid circles indicate 8 h of induction of meiosis with the majority of cells arrested at metaphase I by depletion of meiotic Cdc20.
We then examined Sgo1 and Rts1 localization in telophase I cells by indirect immunofluorescence of spread meiotic nuclei. In telophase I of wild type, 94% of cells show robust staining of Sgo1 and Rts1 at centromeric regions. In telophase I of PCLB2IPL1, a similar fraction of cells show Sgo1 staining of centromeric regions, although the intensity is slightly reduced in a subset of cells (Fig. 6 A). Therefore, Ipl1 function appears to play a minor role in maintaining Sgo1 at centromeric regions in telophase I. In contrast, 66% of PCLB2IPL1 cells have no Rts1 staining in centromeric regions in telophase I (Fig. 6 A). In the remaining 34%, the level of Rts1 staining appears reduced relative to wild type. This cytological observation is confirmed by a biochemical analysis of Rts1 association with the centromeric DNA using ChIP (Fig. 6 B). These data, collectively, show that Ipl1 function acts downstream of Sgo1 localization by playing a critical role in the maintenance of Rts1 in centromeric regions in telophase I cells.
Discussion
During MI, sister chromatids segregate together because sister chromatid cohesion is protected at centromeric regions, and sister kinetochores are modulated to co-orient, attaching to microtubules from the same poles. Here, we report that Ipl1, the Aurora B kinase in budding yeast, is required for the protection of centromeric cohesin during meiosis I. In D. melanogaster, mutations in INCENP and potential Aurora B phosphorylation sites of MEI-S332 have also implicated INCENP and Aurora B in the protection of centromeric cohesion in MI (Resnick et al., 2006). Given the evolutionary distance between these organisms, the role of Aurora B in the protection of meiotic cohesin is probably conserved in all eukaryotes. Interestingly, in budding yeast, Ipl1 also plays a critical role for the co-oriented spindle attachment of sister kinetochores (Monje-Casas et al., 2007). Therefore, Aurora B controls both co-orientation of sister kinetochores and protection of centromere cohesion, the two modifications that differentiate chromosome behavior in meiosis I from mitosis. Having these two key features of MI chromosomes under the control of the same kinase may help to ensure they occur in a coordinated fashion.
Our analyses of Ipl1 in budding yeast suggest that it protects centromeric cohesion by controlling the centromeric localization of Rts1, the regulatory subunit of PP2A, and a key component of the MEI-S332/Sgo1 pathway (Kitajima et al., 2006; Riedel et al., 2006). We propose that the spindle-assembly checkpoint protein Bub1 recruits Sgo1 to the centromeres. Sgo1 recruits the Aurora B complex and Rts1/PP2A. Aurora B ensures that Rts1 remains stably bound to centromeric regions in telophase I, thereby ensuring that Rts1/PP2A is properly positioned to protect centromeric cohesion. This model is driven by the following observations. From our phenotypic analyses, Ipl1 is as important as Sgo1, Bub1, and Rts1 in the protection of meiotic cohesion. Ipl1, Bub1, Sgo1, and Rts1 proteins are required to maintain the binding of cohesin to centromeric regions during MI. Sgo1 and Ipl1 are enriched in centromeric regions in a Bub1-dependent manner. Ipl1 depends on Sgo1 to associate with centromeric regions. Finally, we show that Ipl1 is required for the efficient maintenance of Rts1 but not Sgo1 in the centromeric regions. The role of Ipl1 in Rts1 localization is sufficient to explain its role in protecting cohesion, given the essential role of Rts1 in protecting centromeric cohesion (Kitajima et al., 2006; Riedel et al., 2006).
In budding yeast, we observed a minor defect in Sgo1 localization in ipl1 mutant cells, suggesting that Aurora B may also protect centromeric cohesion by facilitating the localization of MEI-S332/Sgo1 to centromeric regions, as has been proposed from a study in D. melanogaster (Resnick et al., 2006). Although this study suggests that phosphorylation of MEI-S332 by Aurora B is important for MEI-S332 centromere localization, our study suggests that Aurora B may also modulate MEI-S332/Sgo1 localization through its effect on Rts1. The dissociation of Rts1 from centromeric regions in Aurora B–defective cells (this study) may partially destabilize Sgo1 localization. In support of this possibility, Rts1 binds directly to Sgo1 family members in several organisms (Kitajima et al., 2006; Riedel et al., 2006; Tang et al., 2006), and in human cells, Sgo1 localization depends on Rts1/PP2A family members (Tang et al., 2006). Importantly, if MEI-S332/Sgo1 recruitment were the only function of Aurora B in the protection of centromeric cohesin/cohesion, then chromosomes in Aurora B mutant cells that manage to recruit MEI-S332/Sgo1 should protect centromeric cohesion, but this is clearly not the case in ipl1 mutants of budding yeast, because chromosomes with Sgo1 (the vast majority) fail to protect cohesin (this study). In this light, it will be interesting to assess whether Aurora B in D. melanogaster also plays a role in Rts1 recruitment as well as stable binding of MEI-S332.
The regulation of Rts1 by Ipl1 provides several important insights into the protection of centromeric cohesion. In recent analyses, mislocalization of Rts1 to different regions of chromosomes gives rise to ectopic cohesion in MI (Kitajima et al., 2006; Riedel et al., 2006). This result suggests that the localization of Rts1 and presumably PP2A to chromosomes is sufficient to protect cohesion, but in this study we showed that, even though Rts1 localization occurs properly in ipl1 cells before anaphase I, failure of cohesion ensues. Therefore, Rts1 localization to centromeric regions before anaphase I is not sufficient to protect cohesion. Either this localized Rts1/PP2A is inactive and must be activated by Ipl1 or Rts1/PP2A is constitutively active but must be maintained at the centromeric region until telophase I and possibly until metaphase II to protect centromeric cohesion. Ipl1 may regulate Rts1 function/localization by phosphorylating Rts1 or Sgo1. In D. melanogaster, MEI-S332 is a target of Aurora B in vitro, and mutations in phosphorylation sites do lead to cohesion defects. In budding yeast, no changes in mobility of either Rts1 or Sgo1 have been detected in the absence of Ipl1, although at least Rts1 is a phosphoprotein (unpublished data). Determining the target of Ipl1 in cohesion protection will therefore require more sensitive studies of Rts1 and Sgo1 or the identification of additional potential targets.
The regulation of cohesion by PP2A is one of the most recently discovered of a long list of processes influenced by this phosphatase. PP2A has been implicated in cell proliferation, gene expression, insulin regulation, and hyperosmotic stress, to name a few. In many cases, the specific function of the PP2A appears to result from association of the core catalytic subunits with a different B regulatory factor, like Rts1, but very few studies have examined the regulation of these regulatory factors. Understanding the regulation of a B subunit (Rts1) by Aurora B kinase (Ipl1) may provide a new paradigm for understanding how PP2A is regulated and how it in turn regulates so many other processes.
Materials and methods
Yeast strains and cultures
Yeast strains used in this study are SK1 derivatives and are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200609153/DC1). The promoter of CLB20 was used to generate meiotic conditional alleles of PCLB2IPL1, PCLB2SGO1, and PCLB2BUB1 as described previously (Yu and Koshland, 2005). The cdc14-1 allele was obtained from A. Amon (Massachusetts Institute of Technology, Cambridge, MA; Marston et al., 2003). A PCR-based method was used to generate gene deletions and C terminus protein tagging (Longtine et al., 1998). Synchronous yeast cultures were induced for meiosis as described previously (Yu and Koshland, 2003). A single colony was inoculated in 5 ml YEPD overnight. This culture was diluted into YEPA medium to reach an OD (λ = 600) of 0.1. When the YEPA culture reached an OD of 1.4, yeast cells were harvested by centrifugation and washed once in prewarmed water. Cells were resuspended in the same volume of 2% KOAC to induce meiosis. All yeast cultures were inoculated at 30°C.
Meiotic nuclei spread and immunofluorescence
Yeast nuclei spread and immunofluorescence were performed as described previously (Yu and Koshland, 2003). Anti-HA antibodies (12CA5 and 3F10; Roche) were used to detect HA epitope–tagged proteins (0.5 μg/ml). An anti-MYC antibody (9E10; Roche) was used to detect MYC epitope–tagged proteins. The α-tubulin antibody (YOL1/34; Serotec) was used to localize the microtubule spindle (1:500 dilution). A polyclonal antibody against yeast γ-tubulin (1:10,000 dilution; a gift from J. Kilmartin, Medical Research Council, Cambridge, UK) was used to localize the spindle pole bodies. A GFP antibody (ab290; 1:5,000; Abcam) was used to detect TetI-GFP in spread nuclei. Secondary antibodies (goat anti-mouse, anti-rat, and anti-rabbit; Jackson ImmunoResearch Laboratories) were used at a dilution of 1:500. Fluorescence images were acquired with a microscope (Axioplan 2 [Carl Zeiss MicroImaging, Inc.]; 100× objective lens [NA = 1.40] or 63× objective lens [NA = 1.40]) equipped with a charge-coupled device camera (Quantix; Photometrics). Displayed images were processed with IP-Lab for contrast adjustment and pseudocoloring.
Immunoblotting and ChIP
Yeast protein extraction and Western blot were performed as previously described (Yu and Koshland, 2003). Yeast synchronous cultures were induced for meiosis, and aliquots were withdrawn at 1-h intervals. Yeast total proteins were separated by SDS-PAGE and followed by immunoblotting with an ECL kit (Pierce Chemical Co.). ChIP was performed as previously described (Laloraya et al., 2000).
Online supplemental material
Table S1 shows yeast strains used. Fig. S1 shows depletion of Ipl1 protein during meiosis. Fig. S2 shows that Ipl1 protects centromeric cohesin at anaphase I. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200609153/DC1.
Acknowledgments
We thank members of the Koshland laboratory for critical reading of the manuscript and for helpful suggestions. We also thank A. Amon for sharing reagents and unpublished data. A. Thistle helped with text editing.
This work was supported by a grant from the Howard Hughes Medical Institute to D. Koshland.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; INCENP, inner centromere protein.
References
- Buonomo, S.B., R.K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann, and K. Nasmyth. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 103:387–398. [DOI] [PubMed] [Google Scholar]
- Ducat, D., and Y. Zheng. 2004. Aurora kinases in spindle assembly and chromosome segregation. Exp. Cell Res. 301:60–67. [DOI] [PubMed] [Google Scholar]
- Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 91:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant, O., I. Golubovskaya, R. Meeley, E. Fiume, L. Timofejeva, A. Schleiffer, K. Nasmyth, and W.Z. Cande. 2005. A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr. Biol. 15:948–954. [DOI] [PubMed] [Google Scholar]
- Kaitna, S., P. Pasierbek, M. Jantsch, J. Loidl, and M. Glotzer. 2002. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12:798–812. [DOI] [PubMed] [Google Scholar]
- Katis, V.L., M. Galova, K.P. Rabitsch, J. Gregan, and K. Nasmyth. 2004. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 14:560–572. [DOI] [PubMed] [Google Scholar]
- Kerrebrock, A.W., D.P. Moore, J.S. Wu, and T.L. Orr-Weaver. 1995. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 83:247–256. [DOI] [PubMed] [Google Scholar]
- Kiburz, B.M., D.B. Reynolds, P.C. Megee, A.L. Marston, B.H. Lee, T.I. Lee, S.S. Levine, R.A. Young, and A. Amon. 2005. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 19:3017–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima, T.S., Y. Miyazaki, M. Yamamoto, and Y. Watanabe. 2003. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 22:5643–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima, T.S., S.A. Kawashima, and Y. Watanabe. 2004. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 427:510–517. [DOI] [PubMed] [Google Scholar]
- Kitajima, T.S., T. Sakuno, K. Ishiguro, S. Iemura, T. Natsume, S.A. Kawashima, and Y. Watanabe. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 441:46–52. [DOI] [PubMed] [Google Scholar]
- Laloraya, S., V. Guacci, and D. Koshland. 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151:1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Marston, A.L., and A. Amon. 2004. Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 5:983–997. [DOI] [PubMed] [Google Scholar]
- Marston, A.L., B.H. Lee, and A. Amon. 2003. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Dev. Cell. 4:711–726. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45. [DOI] [PubMed] [Google Scholar]
- Monje-Casas, F., V.R. Prabhu, B.H. Lee, M. Boselli, and A. Amon. 2007. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 128:477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, D.P., A.W. Page, T.T. Tang, A.W. Kerrebrock, and T.L. Orr-Weaver. 1998. The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J. Cell Biol. 140:1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki, M., M.F. Siomos, and K. Nasmyth. 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 112:423–440. [DOI] [PubMed] [Google Scholar]
- Resnick, T.D., D.L. Satinover, F. MacIsaac, P.T. Stukenberg, W.C. Earnshaw, T.L. Orr-Weaver, and M. Carmena. 2006. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev. Cell. 11:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel, C.G., V.L. Katis, Y. Katou, S. Mori, T. Itoh, W. Helmhart, M. Galova, M. Petronczki, J. Gregan, B. Cetin, et al. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 441:53–61. [DOI] [PubMed] [Google Scholar]
- Rogers, E., J.D. Bishop, J.A. Waddle, J.M. Schumacher, and R. Lin. 2002. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., H. Shu, W. Qi, N.A. Mahmood, M.C. Mumby, and H. Yu. 2006. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev. Cell. 10:575–585. [DOI] [PubMed] [Google Scholar]
- Vagnarelli, P., and W.C. Earnshaw. 2004. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 113:211–222. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y. 2005. Shugoshin: guardian spirit at the centromere. Curr. Opin. Cell Biol. 17:590–595. [DOI] [PubMed] [Google Scholar]
- Yu, H.-G., and D.E. Koshland. 2003. Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J. Cell Biol. 163:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.-G., and D.E. Koshland. 2005. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell. 123:397–407. [DOI] [PubMed] [Google Scholar]