Abstract
An assay based on the consumption of nitrilotriacetate (NTA) was developed to measure the activity of NTA monooxygenase (NTA-Mo) in cell extracts of "Chelatobacter" strain ATCC 29600 and to purify a functional, NTA-hydroxylating enzyme complex. The complex consisted of two components that easily dissociated during purification and upon dilution. Both components were purified to more than 95% homogeneity, and it was possible to reconstitute the functional, NTA-hydroxylating enzyme complex from pure component A (cA) and component B (cB). cB exhibited NTA-stimulated NADH oxidation but was unable to hydroxylate NTA. It had a native molecular mass of 88 kDa and contained flavin mononucleotide (FMN). cA had a native molecular mass of 99 kDa. No catalytic activity has yet been shown for cA alone. Under unfavorable conditions, NADH oxidation was partly or completely uncoupled from hydroxylation, resulting in the formation of H2O2. Optimum hydroxylating activity was found to be dependent on the molar ratio of the two components, the absolute concentration of the enzyme complex, and the presence of FMN. Uncoupling of the reaction was favored in the presence of high salt concentrations and in the presence of flavin adenine dinucleotide. The NTA-Mo complex was sensitive to sulfhydryl reagents, but inhibition was reversible by addition of excess dithiothreitol. The Km values for Mg(2+)-NTA, FMN, and NADH were determined as 0.5 mM, 1.3 microM, and 0.35 mM, respectively. Of 26 tested compounds, NTA was the only substrate for NTA-Mo.
Full text
PDF


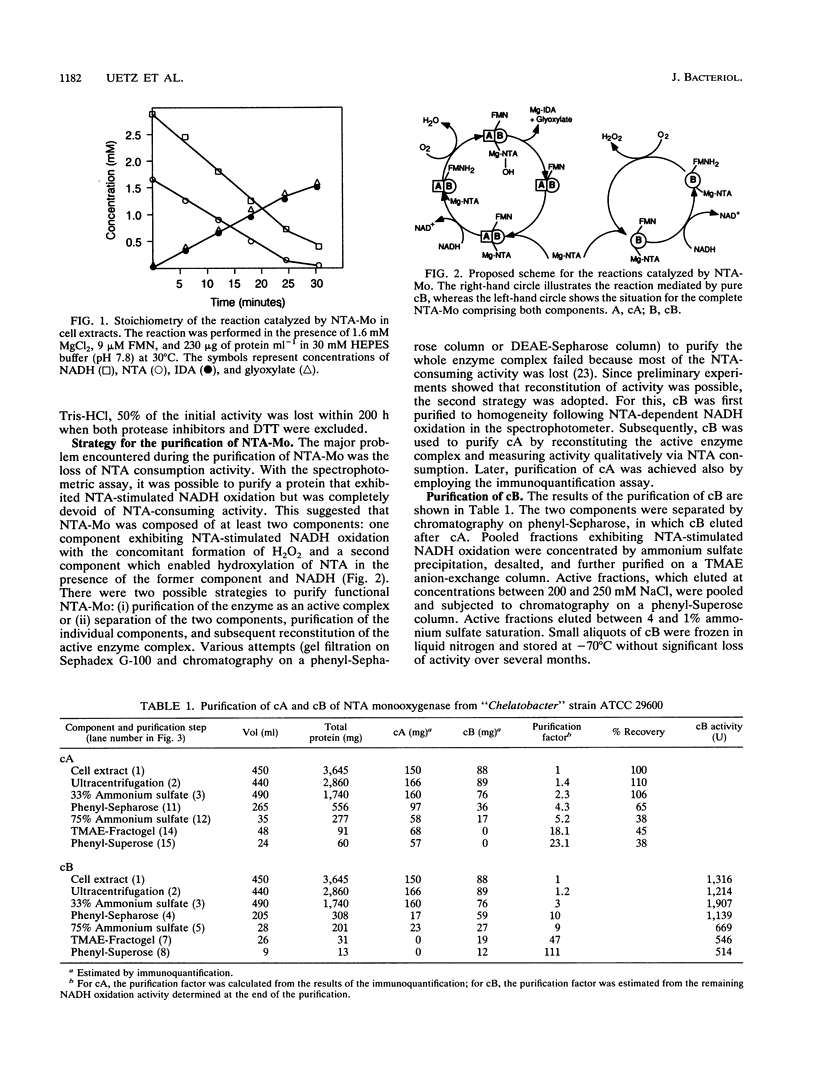
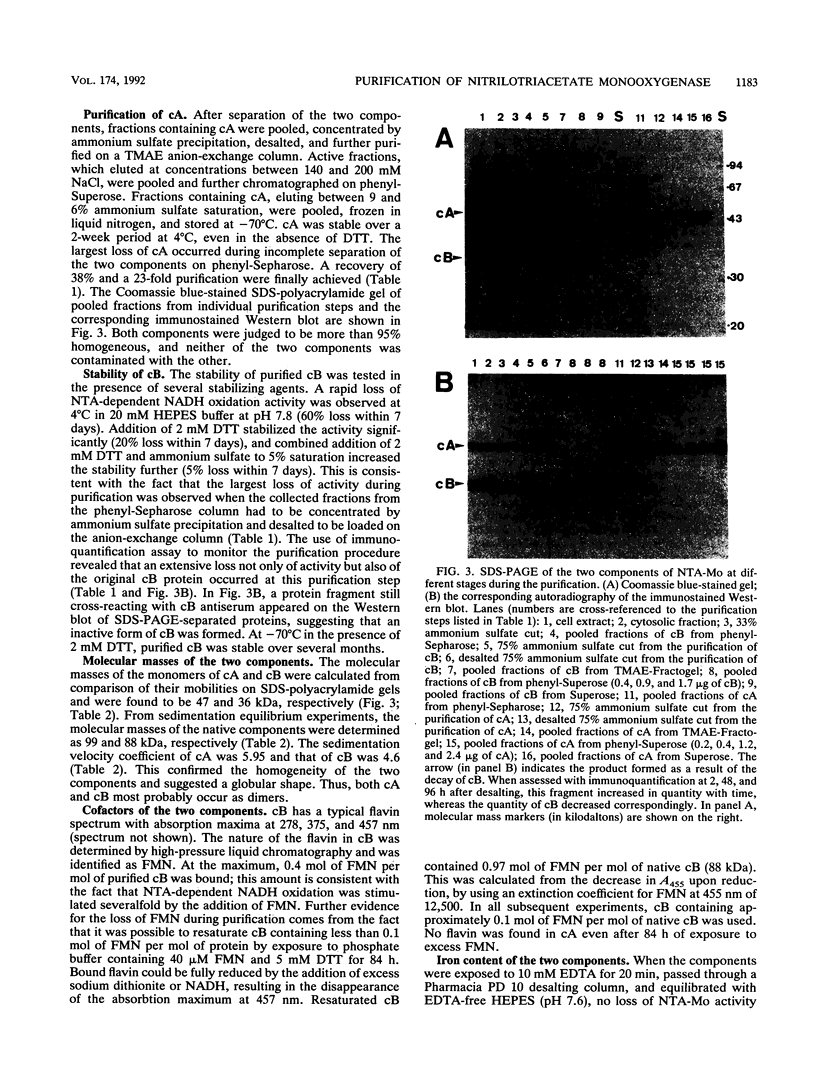
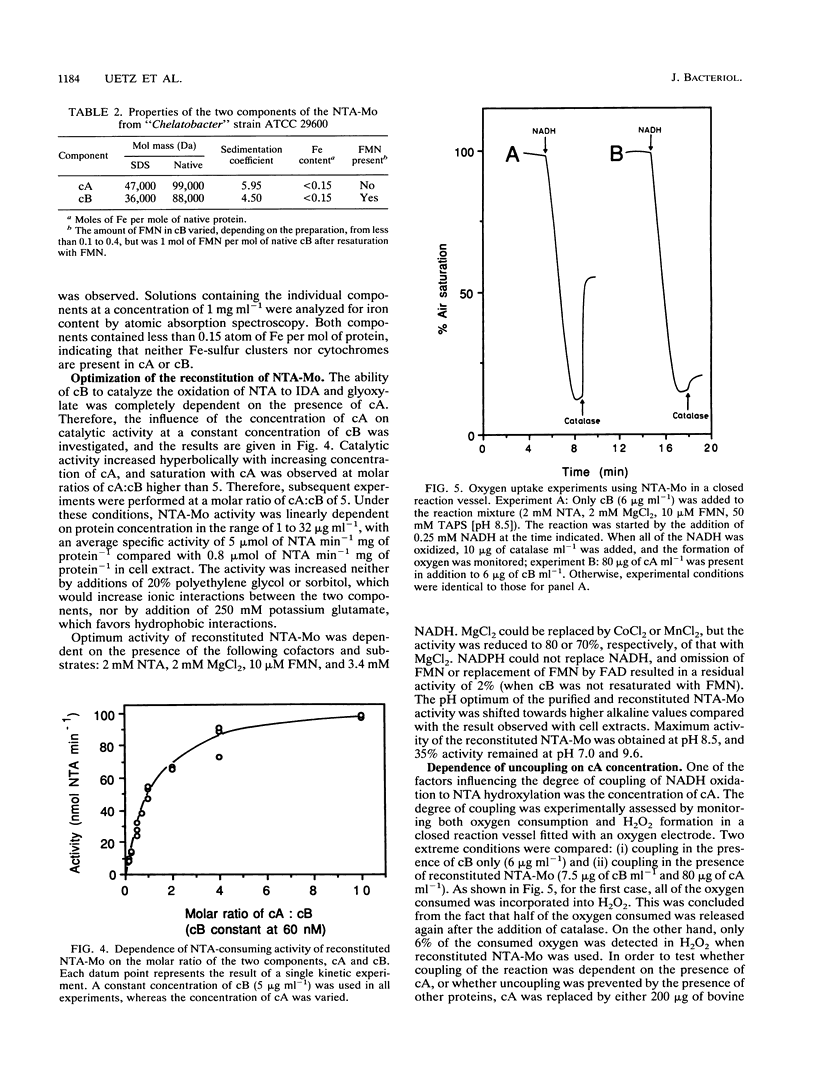



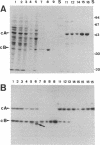
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cripps R. E., Noble A. S. The metabolism of nitrilotriacetate by a pseudomonad. Biochem J. 1973 Dec;136(4):1059–1068. doi: 10.1042/bj1361059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka L. N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989 Mar;53(1):121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli T., Bally M., Uetz T. Microbial degradation of chelating agents used in detergents with special reference to nitrilotriacetic acid (NTA). Biodegradation. 1990;1(2-3):121–132. doi: 10.1007/BF00058831. [DOI] [PubMed] [Google Scholar]
- Entsch B., Husain M., Ballou D. P., Massey V., Walsh C. Oxygen reactivity of p-hydroxybenzoate hydroxylase containing 1-deaza-FAD. J Biol Chem. 1980 Feb 25;255(4):1420–1429. [PubMed] [Google Scholar]
- Firestone M. K., Aust S. D., Tiedje J. M. A nitrilotriacetic acid monooxygenase with conditional NADH-oxidase activity. Arch Biochem Biophys. 1978 Oct;190(2):617–623. doi: 10.1016/0003-9861(78)90318-1. [DOI] [PubMed] [Google Scholar]
- Firestone M. K., Tiedje J. M. Pathway of degradation of nitrilotriacetate by a Pseudomonas species. Appl Environ Microbiol. 1978 May;35(5):955–961. doi: 10.1128/aem.35.5.955-961.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focht D. D., Joseph H. A. Bacterial degradation of nitrilotriacetic acid (NTA). Can J Microbiol. 1971 Dec;17(12):1553–1556. doi: 10.1139/m71-247. [DOI] [PubMed] [Google Scholar]
- Fox B. G., Froland W. A., Dege J. E., Lipscomb J. D. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J Biol Chem. 1989 Jun 15;264(17):10023–10033. [PubMed] [Google Scholar]
- Green J., Dalton H. Protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath). A novel regulatory protein of enzyme activity. J Biol Chem. 1985 Dec 15;260(29):15795–15801. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McFeters G. A., Egli T., Wilberg E., Alder A., Schneider R., Suozzi M., Giger W. Activity and adaptation of nitrilotriacetate (NTA)-degrading bacteria: field and laboratory studies. Water Res. 1990 Jul;24(7):875–881. doi: 10.1016/0043-1354(90)90137-u. [DOI] [PubMed] [Google Scholar]
- Neujahr H. Y. Effect of anions, chaotropes, and phenol on the attachment of flavin adenine dinucleotide to phenol hydroxylase. Biochemistry. 1983 Feb 1;22(3):580–584. doi: 10.1021/bi00272a009. [DOI] [PubMed] [Google Scholar]
- Neujahr H. Y., Gaal A. Phenol hydroxylase from yeast. Sulfhydryl groups in phenol hydroxylase from Trichosporon cutaneum. Eur J Biochem. 1975 Oct 15;58(2):351–357. doi: 10.1111/j.1432-1033.1975.tb02381.x. [DOI] [PubMed] [Google Scholar]
- Ribbons D. W., Ohta Y. Uncoupling of electron transport from oxygenation in the mono-oxygenase, orcinol hydroxylase. FEBS Lett. 1970 Dec 28;12(2):105–108. doi: 10.1016/0014-5793(70)80574-9. [DOI] [PubMed] [Google Scholar]
- Ryerson C. C., Ballou D. P., Walsh C. Mechanistic studies on cyclohexanone oxygenase. Biochemistry. 1982 May 25;21(11):2644–2655. doi: 10.1021/bi00540a011. [DOI] [PubMed] [Google Scholar]
- Schneider R. P., Zürcher F., Egli T., Hamer G. Determination of nitrilotriacetate in biological matrices using ion exclusion chromatography. Anal Biochem. 1988 Sep;173(2):278–284. doi: 10.1016/0003-2697(88)90190-x. [DOI] [PubMed] [Google Scholar]
- Taylor D. G., Trudgill P. W. Camphor revisited: studies of 2,5-diketocamphane 1,2-monooxygenase from Pseudomonas putida ATCC 17453. J Bacteriol. 1986 Feb;165(2):489–497. doi: 10.1128/jb.165.2.489-497.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje J. M., Mason B. B., Warren C. B., Malec E. J. Metabolism of nitrilotriacetate by cells of Pseudomonas species. Appl Microbiol. 1973 May;25(5):811–818. doi: 10.1128/am.25.5.811-818.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trijbels F., Vogels G. D. Degradation of allantoin by Pseudomonas acidovorans. Biochim Biophys Acta. 1966 Feb 14;113(2):292–301. doi: 10.1016/s0926-6593(66)80068-1. [DOI] [PubMed] [Google Scholar]
- Tsuji H., Ogawa T., Bando N., Sasaoka K. Purification and properties of 4-aminobenzoate hydroxylase, a new monooxygenase from Agaricus bisporus. J Biol Chem. 1986 Oct 5;261(28):13203–13209. [PubMed] [Google Scholar]
- Wende P., Bernhardt F. H., Pfleger K. Substrate-modulated reactions of putidamonooxin. The nature of the active oxygen species formed and its reaction mechanism. Eur J Biochem. 1989 Apr 15;181(1):189–197. doi: 10.1111/j.1432-1033.1989.tb14710.x. [DOI] [PubMed] [Google Scholar]
- White-Stevens R. H., Kamin H. Studies of a flavoprotein, salicylate hydroxylase. I. Preparation, properties, and the uncoupling of oxygen reduction from hydroxylation. J Biol Chem. 1972 Apr 25;247(8):2358–2370. [PubMed] [Google Scholar]
- Ziegler D. M. Flavin-containing monooxygenases: enzymes adapted for multisubstrate specificity. Trends Pharmacol Sci. 1990 Aug;11(8):321–324. doi: 10.1016/0165-6147(90)90235-z. [DOI] [PubMed] [Google Scholar]