Abstract
In the yeast Saccharomyces cerevisiae, several components of the septin ring are sumoylated during anaphase and then abruptly desumoylated at cytokinesis. We show that septin sumoylation is controlled by the interactions of two enzymes of the sumoylation pathway, Siz1p and Ulp1p, with the nuclear transport machinery. The E3 ligase Siz1p is imported into the nucleus by the karyopherin Kap95p during interphase. In M phase, Siz1p is exported from the nucleus by the karyopherin Kap142p/Msn5p and subsequently targeted to the septin ring, where it participates in septin sumoylation. We also show that the accumulation of sumoylated septins during mitosis is dependent on the interactions of the SUMO isopeptidase Ulp1p with Kap121p and Kap95p–Kap60p and the nuclear pore complex (NPC). In addition to sequestering Ulp1 at the NPC, Kap121p is required for targeting Ulp1p to the septin ring during mitosis. We present a model in which Ulp1p is maintained at the NPC during interphase and transiently interacts with the septin ring during mitosis.
Introduction
The movement of molecules between the cytoplasm and the nucleoplasm occurs through nuclear pore complexes (NPCs), which form channels across an impermeable nuclear envelope (NE) membrane. The NPCs are elaborate, symmetrical structures consisting of repetitive subunits composed of proteins termed nucleoporins, or nups. Passage of most macromolecules through NPCs involves the recognition of a nuclear transport signal by soluble transport receptors called karyopherins, or kaps, which use their affinity for nups to traverse the NPC with their cargos in tow (for reviews see Suntharalingam and Wente, 2003; Lusk et al., 2004).
There are 14 members of the karyopherin family in yeast. Importins move cargos into the nucleus, whereas exportins move them out, and each kap binds a different, but sometimes overlapping, set of cargos. This redundancy may explain why only five kaps are essential for viability in yeast. Three of these are the importins: Kap121p and the heterodimeric complex of Kap95p–Kap60p. The latter complex binds to what is referred to as a classical NLS (cNLS), which has been identified in a large number of cargo molecules. The number of known cargos imported by Kap121p is more limited but of significant interest, as this kap is required for normal progression through mitosis. Moreover, Kap121p-mediated transport is specifically regulated during mitosis (Makhnevych et al., 2003).
In addition to their function in cargo import, both Kap121p and the Kap95p–Kap60p complex control the association of specific proteins with the NPC (Dilworth et al., 2001; Lusk et al., 2002; Panse et al., 2003). One example is the yeast protein Ulp1p. Both Kap121p and the Kap95p–Kap60p complex bind to Ulp1p and concentrate this protein at the NPC (Panse et al., 2003). However, the functional significance of the association of Ulp1p with the NPC remains unclear. Ulp1p is one of two isopeptidases in yeast that specifically target the small ubiquitin-like modifier, SUMO. One of the functions of Ulp1p is to cleave the SUMO precursor (encoded by the SMT3 gene) to its mature form. Processed SUMO can then be covalently linked to its target proteins in a sequential process that involves an E1 activating enzyme, an E2 conjugating enzyme, and one of several E3 ligases (for review see Johnson, 2004). Importantly, sumoylation is a reversible process, as Ulp1p and another isopeptidase, Ulp2p, are capable of desumoylating proteins (Li and Hochstrasser, 1999, 2000). The regulation of the sumoylation state of a protein can profoundly affect its function, often by dictating its localization, interactions, and stability.
The number of proteins identified as being modified by the addition of SUMO is rapidly growing (Panse et al., 2004; Wohlschlegel et al., 2004; Y. Zhao et al., 2004). Many of the targets are nuclear proteins involved in DNA replication and repair, chromatin remodeling, and transcriptional control (Seeler and Dejean, 2003; Johnson, 2004). However, members of a group of proteins that form the septin ring at the bud neck of budding yeast are also major sumoylation targets (Johnson and Blobel, 1999). The septin ring appears before bud formation in G1 phase of the cell cycle. As the bud grows, the ring extends through the bud neck, forming an hourglass-shaped structure that appears as a double ring, which later divides during cytokinesis. The ring persists in the mother and daughter until disassembling during G1 (for reviews see Lew, 2003; Longtine and Bi, 2003). At least three septins, Cdc3p, Cdc11p, and Shs1p, become sumoylated specifically during mitosis; the modification occurs before anaphase and is removed at cytokinesis (Johnson and Blobel, 1999; Takahashi et al., 1999). The function of this modification has not been studied to the point that it is fully understood. One suggestion is that it plays a role in the disassembly of the septin ring (Johnson and Blobel, 1999).
The sumoylation of the septins during M phase requires the E3 ligase Siz1p (Johnson and Gupta, 2001; Takahashi et al., 2001). Siz1p is located in the nucleus during interphase. At a point before anaphase, and coincident with septin sumoylation, Siz1p is phosphorylated. This is accompanied by an egress of Siz1p from the nucleus and its accumulation at the septin ring (Johnson and Gupta, 2001), where a visible accumulation of this protein remains until again concentrating in the nucleus after cytokinesis. These changes in the localization of Siz1p suggest an active role for the nuclear transport machinery in controlling septin sumoylation by dictating the movement of Siz1p.
Septin desumoylation occurs at cytokinesis (Johnson and Blobel, 1999) and requires Ulp1p (Takahashi et al. 2000; see Results); however, how its activity is regulated is less clear. Ulp1p is localized to the nucleoplasmic face of the NPC (X. Zhao et al., 2004) but presumably must gain access to the cytoplasmic septins to promote desumoylation during cytokinesis. Interestingly, the regions of Ulp1p that function as kap binding sites (Panse et al., 2003) are also required for normal sumoylation of numerous targets (Li and Hochstrasser, 2003), suggesting that, like Siz1p, kaps may regulate the activity of Ulp1p through controlling its localization in the cell.
To further understand the role of the nuclear transport machinery in cell cycle progression, we have focused on the role of kap-mediated movement of Siz1p and Ulp1p in controlling the cell cycle oscillation of septin sumoylation. The kaps Kap95p, Kap121p, and Kap142p/Msn5p play specific roles in regulating septin sumoylation. Kap95p imports Siz1p into the nucleus, and Kap142p/Msn5p exports Siz1p to the cytoplasm at M phase. Ulp1p localization and activity are regulated by Kap121p and the Kap95p–Kap60p complex. We propose that Ulp1p is transiently released from the NPC during mitosis, facilitating the desumoylation of the septins during cytokinesis. This event is controlled by the association of Ulp1p with Kap121p and changes in the interactions of this kap with the NPC. Defects in any of these kap interactions lead to abnormalities in the cycle of septin sumoylation and desumoylation.
Results
Sumoylation of the septin ring requires regulated movement of Siz1p in and out of the nucleus by Kap95p and Kap142p/Msn5p
Siz1p is concentrated in the nucleus throughout most of the cell cycle until mitosis, when nuclear amounts decrease and the protein accumulates at the septin ring. Here it resides until reentering the nucleus after mitosis (Johnson and Gupta, 2001; Takahashi et al., 2001; Fig. 1, wild type [WT]). We investigated the nuclear import and export pathways that control Siz1p localization. The nuclear localization of Siz1-GFP, a chimeric protein that functions similar to WT Siz1p (Johnson and Gupta, 2001), was examined in strains containing temperature-sensitive mutations in either of two essential kaps, Kap121p (kap121-34) and Kap95p (kap95-14). In each mutant, transport of cargos recognized by each kap is strongly inhibited at the nonpermissive temperature for growth (37°C; Marelli et al., 2001; Leslie et al., 2002). We observed that the localization pattern of Siz1-GFP was unaltered in kap121-34 cells at both 23 and 37°C (Fig. 1). In contrast, the nuclear concentration of Siz1-GFP was not observed in the kap95-14 mutant at either temperature and was only restored after introducing a plasmid-born WT copy of KAP95. In the kap95-14 mutant, Siz1-GFP was diffusely distributed throughout the cell with some generally small-budded cells exhibiting a nuclear rim signal. The significance of this localization is unclear. The association of Siz1-GFP with the bud neck in large-budded cells, in either asynchronous or nocodazole-arrested cultures, was not affected by the kap95-14 mutation (Fig. 1). We conclude from these results that Kap95p is required for the import of Siz1p into the nucleus but not for its targeting to the septin ring.
Figure 1.
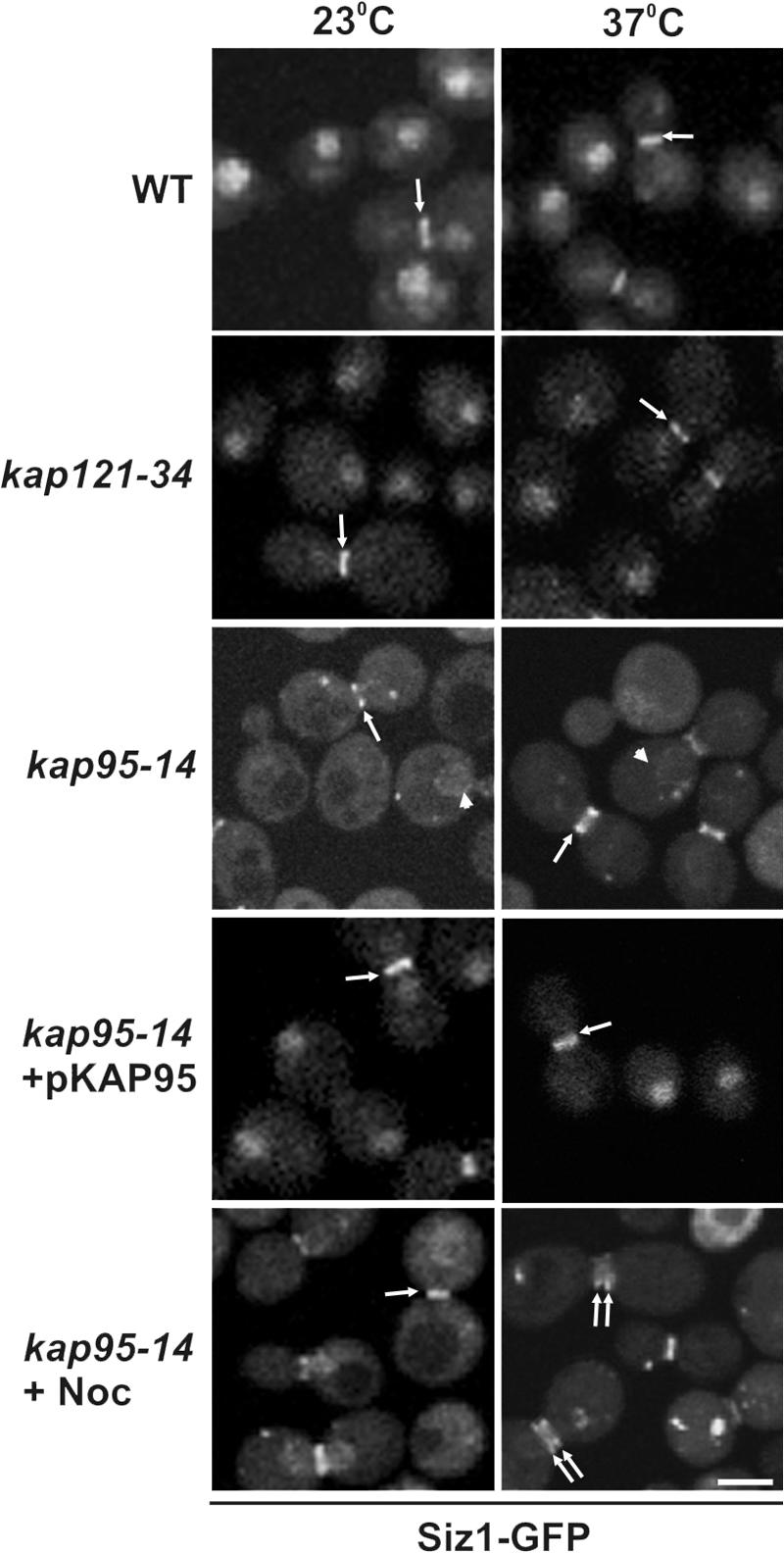
Nuclear import of Siz1-GFP is dependent on Kap95p. The endogenous SIZ1 gene was tagged with GFP in the indicated haploid strains, and the localization of Siz1-GFP was visualized by fluorescence confocal microscopy in cells grown at 23°C or after a 3-h incubation at 37°C. Siz1-GFP localization is also shown in a kap95-14 mutant arrested with nocodazole (Noc) and incubated at the indicated temperatures. Arrows point to the location of the labeled septin rings in representative cells. The arrowheads point to NE-bound Siz1-GFP in the kap95-14 mutant. Bar, 5 μm.
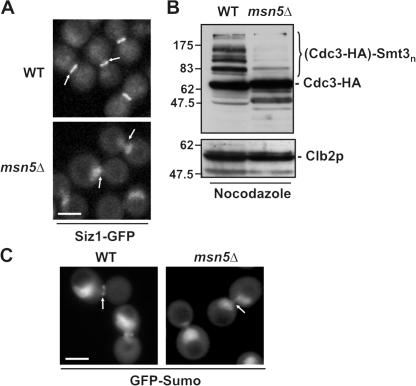
The nuclear pool of Siz1p has been suggested to be the source of Siz1p that is recruited to the septin ring during M phase (Johnson and Gupta, 2001). However, as shown with the kap95-14 mutant, the nuclear localization of Siz1p is not a prerequisite for its septin ring association (Fig. 1). To further define the dynamics of Siz1p recruitment to the septin ring and the role of nuclear export machinery, we examined whether the export kap Kap142p/Msn5p was required for this event. Kap142p/Msn5p was targeted for analysis, as it has been shown to export cargos whose transport is induced by phosphorylation (Kaffman et al., 1998; Boustany and Cyert, 2002; Jaquenoud et al., 2002), and phosphorylation of Siz1p occurs concomitantly with its recruitment to septin rings (Johnson and Gupta, 2001). WT and msn5Δ cells expressing Siz1-GFP were synchronized in M phase, and the subcellular localization of Siz1-GFP was examined by confocal microscopy. As shown in Fig. 2 A, deletion of MSN5 inhibited the export of Siz1-GFP and its concentration at the bud neck.
Figure 2.
Msn5p mediates nuclear export of Siz1-GFP and septin sumoylation. (A) Nuclear export of Siz1p during mitosis requires Msn5p. WT and msn5Δ cells producing Siz1-GFP were arrested in mitosis using nocodazole, and the localization of the GFP fusion was detected by confocal microscopy. Arrows point to the location of the septin ring. (B and C) Septin sumoylation is inhibited in an msn5Δ mutant. (B) The sumoylation state of endogenously tagged Cdc3-HA was evaluated in whole-cell lysates derived from WT and an msn5-null strain (msn5Δ) arrested with nocodazole. Proteins from the indicated strains were analyzed by SDS-PAGE and Western blotting using antibodies directed against HA. The region of the blot containing sumoylated Cdc3-HA is indicated ([Cdc3-HA]-Smt3n). Identical blots were probed with anti-Clb2p antibodies to confirm cell cycle arrest and compare protein levels. The positions of molecular mass markers in this and subsequent figures are show in kilodaltons. (C) The localization pattern of GFP-Sumo was visualized in nocodazole-arrested WT and the msn5-null (msn5Δ) strains using fluorescence confocal microscopy. Bars, 5 μm.
We examined the effect of inhibiting nuclear export of Siz1p on the sumoylation of septin ring components. A GFP-Sumo fusion protein was used to monitor the subcellular localization of SUMO-modified proteins by fluorescence microscopy (Panse et al., 2003; Fig. 2 C). As shown in Fig. 2 C, the GFP-Sumo signal concentrated in the nucleus and a diffuse cytoplasmic signal was also detected. In addition, the septin ring was strongly labeled in large-budded (M phase) WT cells (Fig. 2 C; Panse et al., 2003). Western blotting confirmed that, in addition to the free form, GFP-Sumo was readily incorporated into cellular proteins, including septins (unpublished data). However, no septin signal was detected in the msn5Δ mutant, suggesting that export of Siz1p from the nucleus is required for septin sumoylation. This conclusion was further supported by the results of experiments examining the sumoylation state of a septin, Cdc3-HA, containing a C-terminal HA tag. Consistent with previous reports, various sumoylated species of Cdc3-HA could be detected by Western blotting in cell lysates derived from WT cultures arrested in M phase with nocodazole (Johnson and Blobel, 1999; Fig. 2 B). However, the slower migrating sumoylated Cdc3-HA species were greatly reduced in lysates from msn5Δ cells, further supporting the conclusion that Kap142p/Msn5p-mediated export of Siz1p is required for normal septin sumoylation.
Septin desumoylation by Ulp1p is regulated by Kap121p and Kap95p–Kap60p
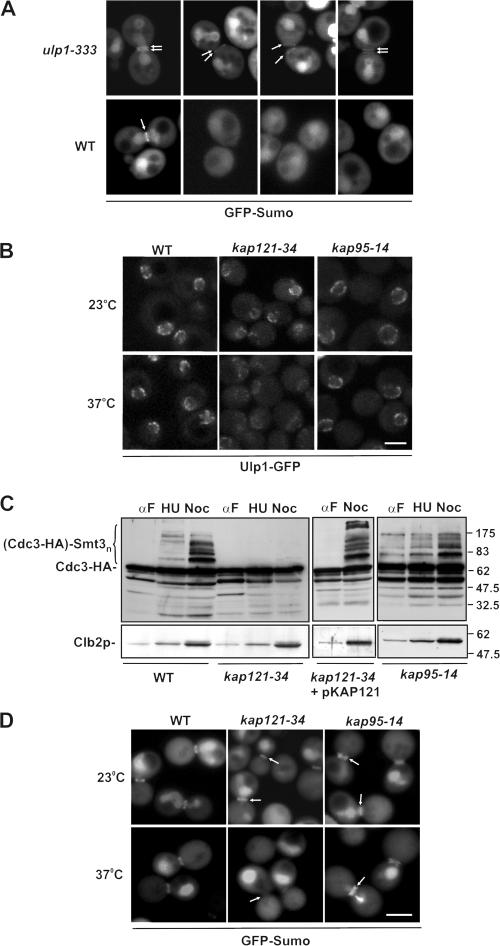
Previously published data suggest that Ulp1p is required for septin desumoylation at cytokinesis (Takahashi et al., 2000). Consistent with this, we observed that a strain (YRW122) containing a mutation (ulp1-333; Li and Hochstrasser, 1999) in the catalytic domain of Ulp1p exhibited abnormal septin sumoylation. Large-budded cells expressing GFP-SMT3 exhibited GFP-Sumo–labeled septins in both the mother and the bud. This was rarely observed in WT cells, where sumoylation was confined to the mother side of the septin ring (Johnson and Blobel, 1999; Fig. 2 A and Fig. 3 A). Moreover, septin sumoylation persisted beyond separation of the ring and cytokinesis in cells expressing the ulp1-333 mutation (Fig. 3 A). This phenotype was not observed in WT cells (Fig. 3 A; Johnson and Blobel, 1999).
Figure 3.
Kap121p is required for septin sumoylation during mitosis. (A) Septin sumoylation is altered in cells expressing the ulp1-333 mutation. Asynchronous cultures of YRW121 (WT) and YRW122 (ulp1-333) cells were grown at 23°C and examined by confocal microscopy. These cells also express GFP-SMT3-ΔC, which encodes mature GFP-Sumo (Wykoff and O'Shea, 2005), bypassing the requirement for Ulp1p-dependent processing. M phase (left) and postcytokinesis mother/daughter pairs (right three columns) are shown. Arrows point to labeled septin rings. (B) Ulp1p localization at the nuclear rim is diminished at the nonpermissive temperature in kap121-34, but not kap95-14, containing mutants. The endogenous ULP1 gene was tagged with GFP in the indicated haploid strains. Strains were grown at 23°C or shifted to 37°C for 3 h, and the localization of Ulp1-GFP was examined by confocal microscopy. (C) Cdc3p sumoylation is inhibited at the nonpermissive temperature in the presence of a kap121-34, but not a kap95-14, mutation. Levels of sumoylated Cdc3-HA in WT, kap121-34, and kap95-14 mutants were detected as in Fig. 2 using anti-HA antibodies. Cells were arrested with α-factor, and G1 phase arrest was confirmed by visual inspection of cell morphology. Cultures were then shifted to 37°C and either maintained in α-factor (αF) or switched to media containing hydroxyurea (HU) or nocodazole (Noc). The region of the blot containing sumoylated Cdc3-HA is indicated ([Cdc3-HA]-Smt3n). Blots were also probed with anti-Clb2p antibodies to confirm cell cycle M phase arrest and compare protein levels. Note that Cdc3-HA sumoylation could be rescued in the kap121-34–containing strain by introducing a plasmid-born copy of KAP121 (pKAP121). (D) GFP-Sumo concentration at the septin ring is inhibited at the nonpermissive temperature in kap121-34, but not kap95-14, mutant cells. WT, kap121-34, and kap95-14 mutants producing GFP-Sumo were arrested in M phase with nocodazole and incubated at 23 and 37°C. The GFP fusion was visualized by confocal microscopy. Arrows point to the positions of septin rings. Bars, 5 μm.
Ulp1p has been localized to the nucleoplasmic side of the NPC (X. Zhao et al., 2004), where it interacts with Kap121p and the Kap95–Kap60p complex (Panse et al., 2003). Because Saccharomyces cerevisiae has a closed mitosis, this localization raised the question as to how Ulp1p could desumoylate the cytoplasmic septins. We hypothesized that the association of Ulp1p with the kaps and the NPC may control its accessibility to the septin ring. We therefore examined the effects of the kap121-34 and kap95-14 alleles on the NPC association of Ulp1p and the sumoylation state of the septins at various points in the cell cycle. As shown in Fig. 3 B, the kap95-14 mutation appeared to have little effect on the NPC association of Ulp1p at either 23 or 37°C. In contrast, shifting the kap121-34–containing strain to the nonpermissive temperature caused a decrease in the level of NPC-associated Ulp1p and a corresponding increase in its cytoplasmic levels. Quantification of these signals revealed a change in the nuclear/cytoplasmic ratio from ∼5 at 23°C to ∼1.5 at 37°C (unpublished data). This observation is consistent with data obtained with other KAP121 mutant alleles (Panse et al., 2003).
Septin sumoylation was also examined in the kap95-14 and kap121-34 strains expressing CDC3-HA or GFP-SMT3. Cells were synchronized in G1 phase at 23°C. The cultures were then split, and cells were placed in fresh medium containing α-factor (to maintain G1 arrest), hydroxyurea (to arrest in S phase), or nocodazole (for M phase arrest). After incubating at 37°C, Cdc3-HA sumoylation was evaluated by Western blot analysis. Consistent with previous results (Johnson and Blobel, 1999), in a WT strain, sumoylated forms of Cdc3-HA were detected predominantly in nocodazole-arrested cells (Fig. 3 C). In contrast, M phase accumulation of sumoylated Cdc3-HA was completely inhibited in a kap121-34 mutant at 37°C, but was normal at 23°C (unpublished data) or upon complementation of the mutant with WT KAP121. Moreover, GFP-Sumo failed to accumulate at septin rings in M phase kap121-34 cells at 37°C, whereas nuclear levels were unaffected (Fig. 3 D). The kap95-14 strain appeared more similar to WT cells both in terms of Cdc3-HA sumoylation and GFP-Sumo labeling of the septins. However, GFP-labeled septin rings were detectable in both mother and daughter cells (Fig. 3 D). kap95-14 cells also exhibited increased amounts of sumoylated Cdc3-HA in α-factor–arrested cultures (Fig. 3 C); however, the relevance of this observation was difficult to assess, as these cells exhibited a slight decrease in the efficiency of arrest (88% of kap95-14 vs. 92% of WT cells).
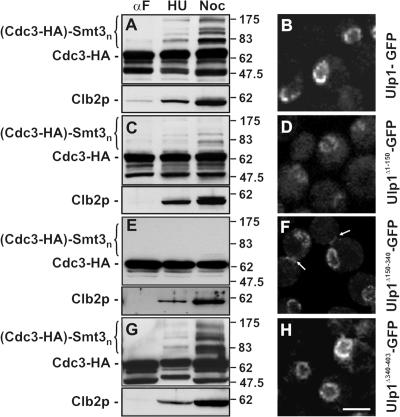
To assess whether changes in septin sumoylation observed in the kap mutants were linked to Ulp1p, we tested the effects of deletion mutations that remove the various kap binding domains of Ulp1p on the sumoylation state of Cdc3-HA and correlated this with their subcellular localization. Previous data from Panse et al. (2003) suggest that the Kap121p and Kap95p–Kap60p binding sites in Ulp1p are positioned within residues 1–150 and 150–340, respectively, and are outside of the region containing the catalytic domain (residues 403–621). In addition, Ulp1p contains a putative nuclear export signal (NES) in a region between residues 340 and 403. Deletion of the Kap121p binding domain of Ulp1p (Ulp1Δ1-150) led to a partial mislocalization of Ulp1p and a localization pattern similar to that observed for Ulp1p in the kap121-34 mutant (Fig. 4 D). Concomitant with the mislocalization of Ulp1p from the NPC was a failure to accumulate sumoylated Cdc3-HA at M phase (induced by nocodazole arrest; Fig. 4 C). These data support the contention that Kap121p-mediated localization of Ulp1p at the NPC is important for efficient septin sumoylation.
Figure 4.
Binding of Ulp1p to kaps is required for septin sumoylation and regulating Ulp1p association with the septin ring. Plasmids containing coding sequences for Ulp1-GFP and various Ulp1p deletion derivatives, including Ulp1Δ1-150-GFP, Ulp1Δ150-340-GFP, and Ulp1Δ340-403-GFP, were introduced into a ulp1Δ deletion mutant expressing CDC3-HA. (A, C, E, and G) Ulp1p deletion mutants lacking kap binding domains alter septin sumoylation. Cells were arrested with α-factor and then either maintained in α-factor (αF) or washed to remove α-factor and incubated in media containing hydroxyurea (HU) or nocodazole (Noc) at 30°C. Levels of Cdc3-HA sumoylation in the indicated strains were detected using anti-HA antibodies as in Fig. 2. The region of the blot containing sumoylated Cdc3-HA is indicated ([Cdc3-HA]-Smt3n). Blots were also probed with anti-Clb2p antibodies to confirm M phase arrest and compare protein levels. (B, D, F, and H) Localization of the various Ulp1-GFP derivatives was visualized in logarithmically growing cells using confocal microscopy. Arrows indicate the localization of Ulp1Δ150-340 to the bud neck. Bar, 5 μm.
Deletion of the NES region (Ulp1Δ340-403) did not affect Cdc3-HA sumoylation or alter Ulp1p association with the nuclear periphery (Fig. 4, G and H). We also examined mutations lacking the Kap95p–Kap60p binding site (Ulp1Δ150-340). Ulp1Δ150-340-GFP exhibited no obvious changes in NE association compared with WT Ulp1p. Surprisingly, however, disruption of the Kap95p–Kap60p binding site led to the accumulation of Ulp1p-GFP at the septin rings in large-budded cells (Fig. 4 F, arrows). Likely as a consequence of this abnormal localization, septin sumoylation was completely inhibited in cells containing Ulp1Δ150−340 (Fig. 4 E). These data suggest that Kap95p–Kap60p negatively regulates Ulp1p binding to the septin ring.
To further understand how Kap121p and Kap95p–Kap60p regulate the localization of Ulp1p, a mutant that lacked both Kap121p and Kap95p–Kap60p binding sites (Ulp1Δ1-340) was examined. This mutant failed to bind to either the septin ring or the NE (Panse et al., 2003; unpublished data). Thus, Kap121p binding to Ulp1p, in addition to its role in targeting Ulp1p to the NPC, appeared to be required for the association of Ulp1Δ150−340 with the septin ring. We therefore directly examined the effect of the kap121-34 mutation on the binding of Ulp1Δ150−340 to the septin ring. In these cells, the Ulp1Δ150−340 mutant failed to accumulate at the NE or septin rings at 37°C and was dispersed throughout the cell (Fig. 5), supporting a role for Kap121p in directing the association of Ulp1p with the septin ring.
Figure 5.
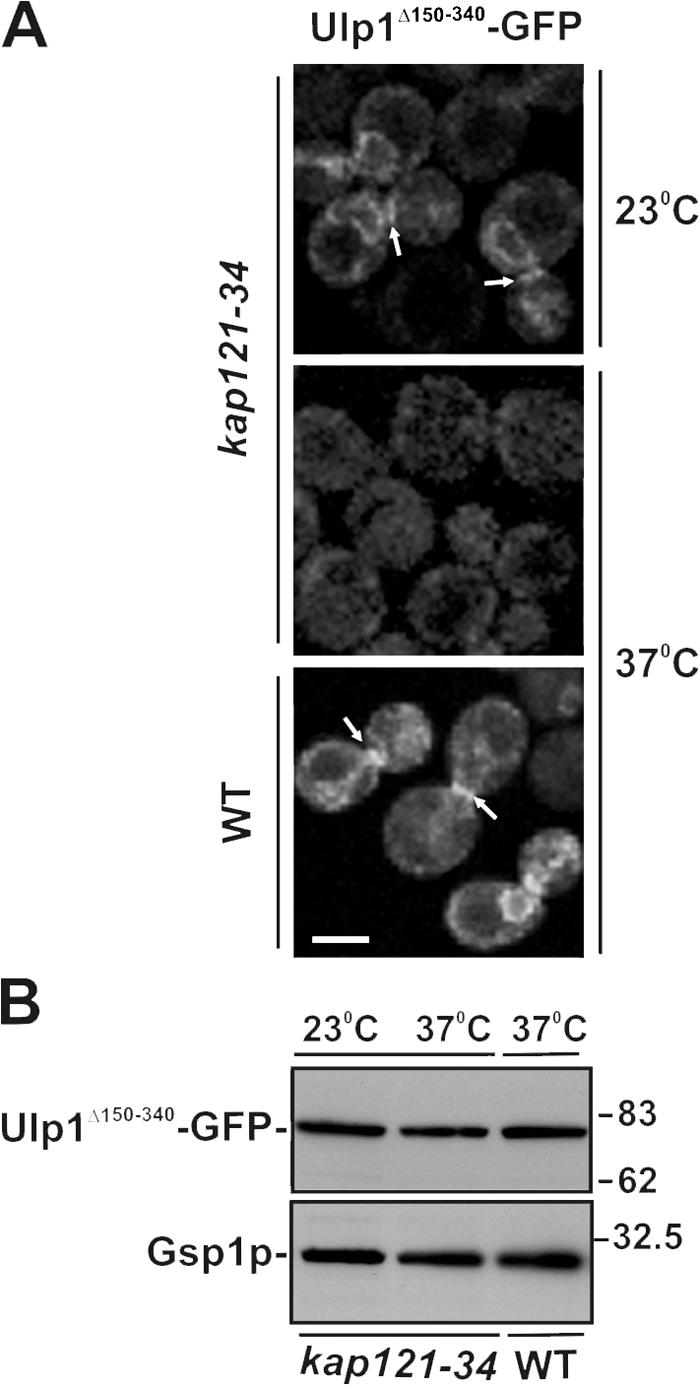
Kap121p is required for the association of Ulp1Δ150-340 with the septin ring. (A) WT and kap121-34 cells were transformed with pULP1Δ150-340-GFP. Logarithmically growing cultures were arrested in M phase with nocodazole. The distribution of Ulp1Δ150-340-GFP was examined by confocal microscopy at 23°C or 2 h after cultures were shifted to 37°C. Arrows indicate the localization of Ulp1Δ150-340 to the bud neck. Bar, 5 μm. (B) Cellular levels of Ulp1Δ150-340-GFP were examined by SDS-PAGE and Western blotting of whole-cell lysates using α-GFP antibodies. Blots were also probed with anti-Gsp1p antibodies to compare protein levels.
Ulp1p is dynamically associated with the NPC
Our data are consistent with a model in which the interaction of the Kap–Ulp1p complexes with the NPC control Ulp1p recruitment from the NPC to the septin rings. To further develop this model, we performed several experiments that focused on the nature of Ulp1p's interactions with the kaps and its relevance to septin sumoylation. For this analysis, plasmid-born gene fusions encoding Ulp11-150-GFP (containing the Kap121p binding domain) or Ulp1150-340-GFP (containing the Kap95p–Kap60p binding domain) were introduced into cells (Fig. 6 A). Variation in plasmid numbers between cells within the culture allowed us to evaluate the localization of the fusion proteins at differing cellular levels. At low levels of expression, both Ulp11-150-GFP and Ulp1150-340-GFP were visible at the NE. These observations support the conclusions of Panse et al. (2003) that both the Kap121p and the Kap95p–Kap60p binding regions of Ulp1p were capable of independently mediating its binding to the NPC, albeit with reduced efficiency relative to the WT protein. However, when levels of Ulp1150-340-GFP were elevated, the chimera began to accumulate in the nucleus, suggesting that the NE binding sites were limited and excess protein was imported into the nucleoplasm. In contrast, Ulp11-150-GFP was generally observed evenly distributed along the NE in all cells. A punctate pattern was only occasionally detected in low-expressing cells (unpublished data). A similar uniform NE distribution was also observed upon overproduction of full-length Ulp1p (X. Zhao et al., 2004). The localization patterns of Ulp11-150-GFP and Ulp1150-340-GFP were dependent on functional Kap121p and Kap95p, respectively (unpublished data). These results suggest that Kap121p and Kap95p mediate the association of Ulp1p with distinct sites at the NPC and perhaps locations on the inner nuclear membrane (INM).
Figure 6.
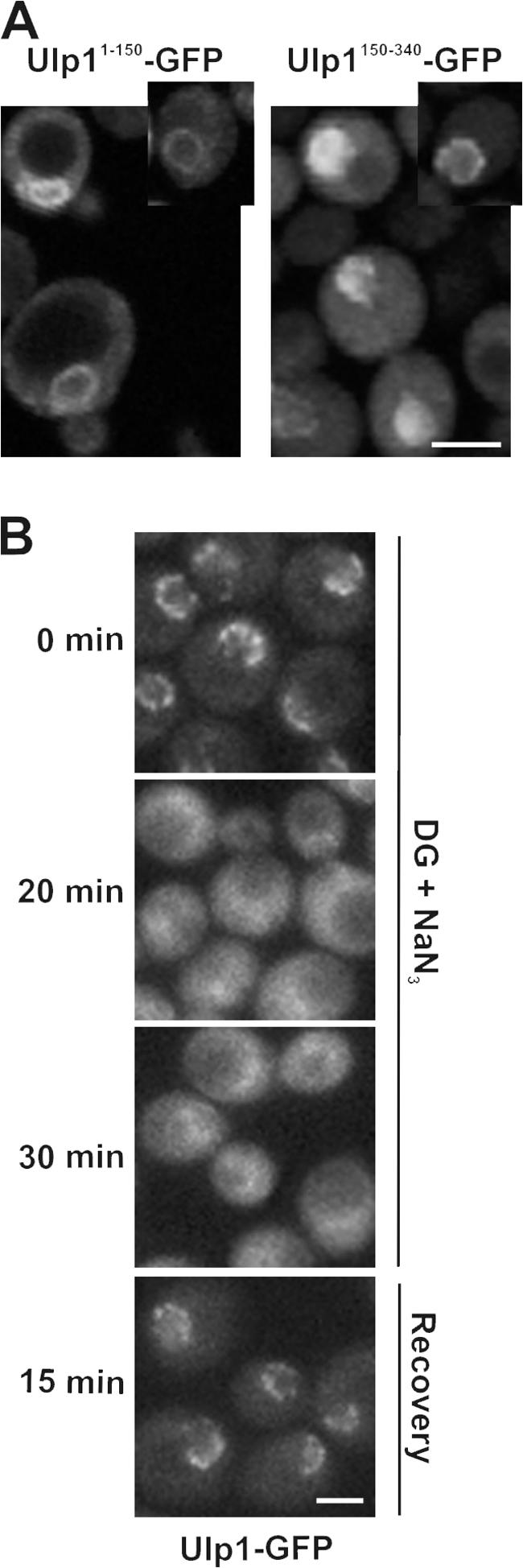
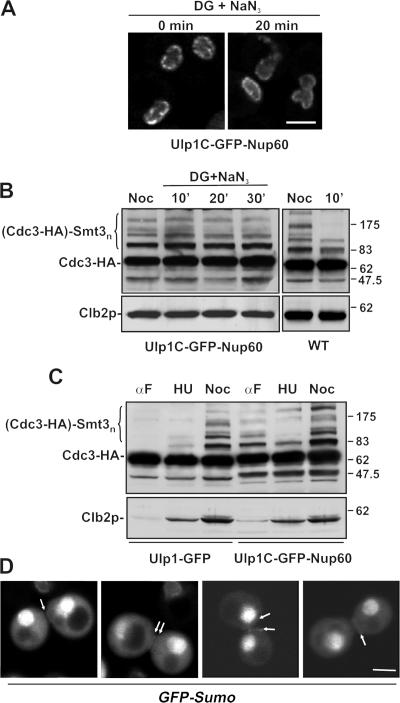
The association of Ulp1p with nuclear pores is energy dependent. (A) The kap binding domains of Ulp1p are targeted to distinct locations. WT cells expressing plasmid-born ULP11-150-GFP or ULP1150-340-GFP were grown to early log phase in selective media, and the distribution of each GFP fusion was examined using confocal microscopy. Individual cells in the culture exhibit different levels of the GFP fusions representative of high and low producers. Insets show cells containing low to moderate levels of the fusion protein. (B) Cells synthesizing Ulp1-GFP (ULP1-GFP integrated at the ULP1 locus) were synchronized in M phase using nocodazole. Cells were washed and placed in media lacking glucose and containing 100 mM 2-deoxyglucose (DG), 10 mM sodium azide (NaN3), and nocodazole. The localization of the GFP fusion was visualized by confocal microscopy after 0, 20, and 30 min of incubation at 30°C. After the 30-min incubation, cells were washed and released into YPD media containing nocodazole (recovery), and the localization of Ulp1-GFP was visualized. Bars, 5 μm.
The basis for the binding of Ulp1p to the NPC was further evaluated by examining the energy dependence of this interaction. Certain NPC-associated proteins, such as Mad1p, Mad2p, the Mlp proteins, and the nups themselves, including those that exhibit mobility, such as Nup2p, remain bound to the NE in the presence of metabolic poisons such as deoxyglucose (Shulga et al., 1996; Dilworth et al., 2001; Shulga and Goldfarb, 2003; unpublished data). Moreover, Kap60p and Kap95p concentrate at the NPCs after treatment of cells with deoxyglucose (Dilworth et al., 2001). In contrast, Kap121p, although still visible in the cytoplasm, shows a marked accumulation in the nucleus under these same conditions (Makhnevych et al., 2003). The localization of Ulp1-GFP, however, was distinct from that observed for its interacting kaps. Within 20 min of treatment of cells with deoxyglucose and sodium azide, the NE concentration of Ulp1-GFP was largely lost and the protein was distributed throughout the cell (Fig. 6 B). These effects were rapidly reversed after removal of the poison.
The apparent mobility of U1p1p led us to conclude that, at some point during mitosis, Ulp1p gains access to the cytoplasm to desumoylate the septins. To further test this idea, experiments were performed to determine whether a ulp1 mutation that alters its mobility affects the sumoylation state of septins. Strains were isolated that express a mutant form of Ulp1p in which the kap binding domains were removed and the catalytic domain was fused to the nucleoporin Nup60p. This fusion protein is predicted to localize to the nucleoplasmic face of the NPC in a region where WT Ulp1p has been reported to reside (X. Zhao et al., 2004). Consistent with this, Ulp1C-GFP-Nup60 localized to the NPC and could rescue the growth of the ulp1 deletion mutant (Fig. 7 A; Panse et al. 2003; unpublished data). However, unlike the WT protein, Ulp1C-GFP-Nup60 does not appear to have access to the cytoplasm. As show in Fig. 7 A, treatment of cells with deoxyglucose and sodium azide did not induce the release of Ulp1C-GFP-Nup60 from the NE; moreover, it did not induce Cdc3-HA desumoylation in M phase–arrested cultures (Fig. 7 B). In contrast, these metabolic poisons induced septin desumoylation in WT cells where Ulp1p can access the cytoplasm. The predicted requirement of Ulp1p release from the NPC for septin desumoylation is also supported by data showing that in Ulp1C-GFP-Nup60–containing cells, septin sumoylation persisted beyond separation of the ring and cytokinesis similar to that observed in the ulp1-333 mutant (Fig. 7 D). This GFP-Sumo ring signal is later lost as cells progress through G1 or in α-factor–arrested cells (unpublished data). Elevated levels of sumoylated Cdc3-HA were, however, detected throughout the cell cycle Ulp1C-GFP-Nup60 cells (Fig. 7 C).
Figure 7.
Anchoring of Ulp1p to the nuclear pores inhibits septin desumoylation. WT Ulp1p was replaced by a plasmid encoding the Ulp1C-GFP-Nup60 fusion protein in a strain expressing CDC3-HA (YRW110). (A) YRW110 cells were arrested in M phase using nocodazole and then treated with 100 mM 2-deoxyglucose (DG) and 10 mM sodium azide (NaN3) for the indicated times, as in Fig. 6 B. Cells were examined by confocal microscopy. (B) In YRW110 and an isogenic WT strain, the sumoylation state of Cdc3-HA was evaluated in nocodazole-arrested cells or after treatment with the metabolic poisons for the indicated times. (C) Cdc3-HA sumoylation in Ulp1C-GFP-Nup60– or Ulp1-GFP–containing cells was compared at various points in the cell cycle. Cells were arrested with α-factor, and G1 phase arrest was confirmed by visual inspection of cell morphology. Cultures were then maintained in α-factor (αF) or switched to media containing hydroxyurea (HU) or nocodazole (Noc). Samples were analyzed by Western blotting of whole-cell lysates using anti-HA antibodies. Anti-Clb2p blots were used to confirm cell cycle arrest and compare protein levels. (D) Ulp1C-GFP-Nup60 is restricted to the NE in YRW110 (A), allowing us to examine GFP-Sumo labeling of septins in this strain background (YMR123). Shown are images of postcytokinesis mother/daughter pairs. Arrows point to the positions of labeled septin rings. Bars, 5 μm.
The NPC regulates the timing of septin desumoylation
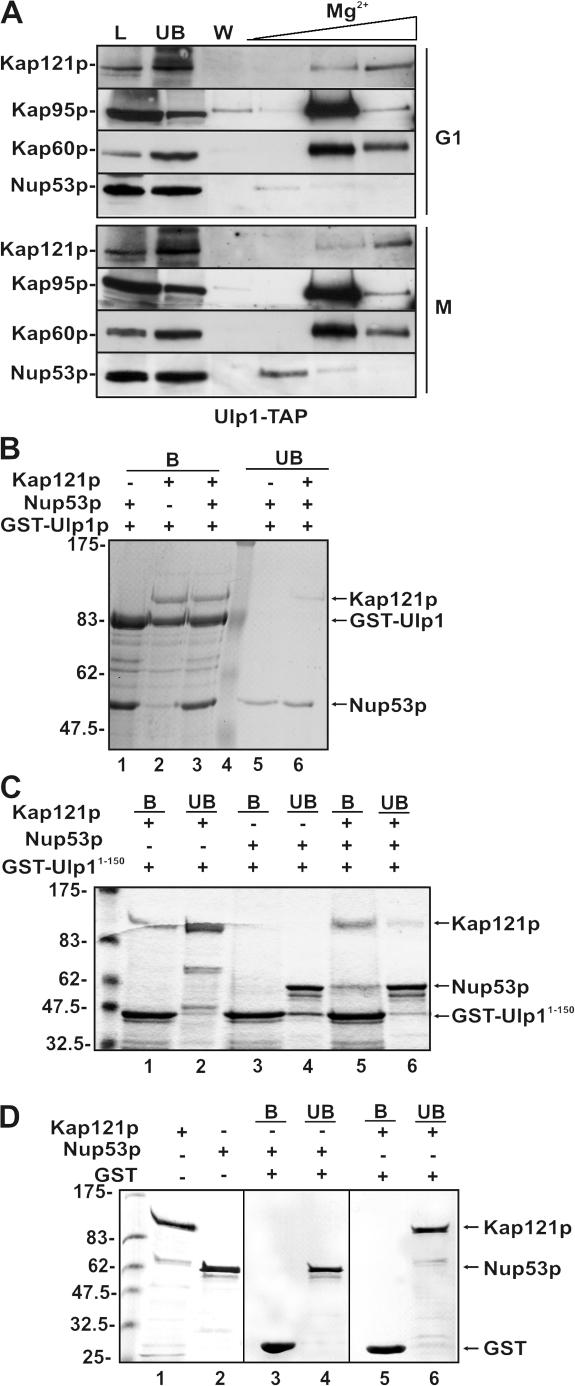
The likelihood that Ulp1p is released from the NPC combined with the observation that recruitment of Ulp1Δ150-340 to the septin ring was dependent on Kap121p linked this kap to the targeting step. This prompted us to examine whether mitosis-specific molecular rearrangements in the NPC that alter the binding of Kap121p to the NPC and inhibit its import pathway (Makhnevych et al., 2003) contribute to releasing Ulp1p into the cytoplasm. In cells arrested in M phase with nocodazole, Kap121p becomes associated with Nup53p (Makhnevych et al., 2003), and we wondered if this change would alter the binding of Ulp1p at the NPC. Using a strain producing tandem affinity purification (TAP)–tagged Ulp1p, we tested whether Ulp1-TAP was recruited with Kap121p to Nup53p. Ulp1-TAP was purified from cells arrested in G1 (with α-factor) or M (with nocodazole) phase. Ulp1-TAP derived from both cultures was associated with Kap121p, Kap95p, and Kap60p. However, an enrichment of Nup53p was visible in the nocodazole-arrested culture, suggesting that both Kap121p and Ulp1p are recruited to Nup53p (Fig. 8 A). Consistent with this observation, we were able to reconstitute interactions between recombinant versions of these proteins (Fig. 8, B and C). Surprisingly, full-length Ulp1p bound Nup53p and Kap121p independently. However, GST-Ulp1-150 bound Kap121p, but not Nup53p. Nonetheless, a preformed complex of GST-Ulp1-150–Kap121p bound to Nup53p, showing that these proteins could form a trimeric complex through Kap121p.
Figure 8.
Ulp1p is associated with Kap121p and Nup53p. (A) Kap121p, Kap95p, Kap60p, and Nup53p copurify with Ulp1p. A yeast strain expressing TAP-tagged ULP1 (ULP1-TAP integrated at the ULP1 locus) and KAP95-GFP (pKAP95-GFP) was synchronized in either G1 (with α-factor) or M phase (with nocodazole). Cell extracts were prepared, and the Ulp1-TAP fusion was affinity purified using IgG-Sepharose. Beads were washed with lysis buffer containing 50 mM MgCl2 (W), and Ulp1-TAP–associated proteins were eluted with a step gradient of 200, 500, and 1,000 mM MgCl2. Proteins were analyzed by Western blotting using antibodies directed against Kap121p, Kap60p, Nup53p, and GFP (to detect Kap95p). Lanes L and UB contain a sample of the fraction loaded on the IgG-Sepharose and the unbound fraction, respectively. (B–D) Kap121p, Ulp1p, and Nup53p form a trimeric complex in vitro. GST-Ulp1 (B), GST-Ulp11-150 (C), or GST (D) was bound to glutathione–Sepharose beads. Bound proteins were then incubated with combinations of purified recombinant Kap121p or Nup53p. These proteins do not bind GST alone (D). In addition, purified recombinant Nup53p was incubated with bead-bound complexes of Kap121p–Ulp1p (B, lane 3) or Kap121p–Ulp11-150 (C, lane 5). In each case, the beads were washed extensively and bound proteins were subsequently eluted from the beads using SDS-PAGE sample buffer. Proteins present in bead-bound (B) and unbound (UB) samples were separated by SDS-PAGE and visualized by Coomassie blue staining.
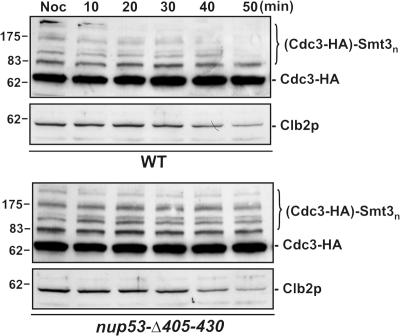
The results described above suggest that the Kap121p–Ulp1p complex is recruited to Nup53p during mitosis. We therefore investigated the relevance of this interaction to septin sumoylation using specific mutations in Nup53p that fail to bind Kap121p (nup53Δ405-430). We speculated that if Nup53p plays a role in regulating the accessibility of Ulp1p to the septins, then the kinetics of septin desumoylation might be altered in this strain. To evaluate this, cells were released from nocodazole arrest and Cdc3-HA sumoylation was monitored over time. As shown in Fig. 9, we reproducibly observed that desumoylation of the Cdc3-HA was delayed in the nup53Δ405-430 strain relative to the WT strain. This effect was not due to a delay in progression out of mitosis, as both strains showed similar kinetics of Clb2p decay (Fig. 9). In addition, Siz1p release from the septin ring occurred at similar time after release from nocodazole arrest in both the WT and the nup53Δ405-430 strain (unpublished data). These results suggest that changes in the NPC association of Kap121p and Ulp1p contribute to timing of septin desumoylation.
Figure 9.
Desumoylation of Cdc3-HA after mitosis is delayed in a nup53 mutant. WT cells or a strain containing the nup53-Δ405-430 mutant and expressing CDC3-HA were arrested in M phase with nocodazole (Noc). Cells were then released from arrest and allowed to exit mitosis. At the indicated times, Cdc3-HA sumoylation and Clb2p degradation were evaluated by Western blotting of whole-cells lysates using anti-HA and anti-Clb2p antibodies.
Discussion
Several components of the septin ring are major cytoplasmic sumoylation targets. Septin sumoylation is cell cycle regulated and reversible, with sumoylated septins accumulating before anaphase and persisting until desumoylation after mitosis (Johnson and Blobel, 1999). Here, we have shown that the nuclear transport machinery regulates septin sumoylation by controlling access of two key SUMO-modifying enzymes, Siz1p and Ulp1p, to the septins. Siz1p is localized to the nucleus during interphase by Kap95p, which would preclude the spurious sumoylation of cytoplasmic targets. Such a role for Kap95p could explain the elevated levels of sumoylated septins seen in the kap95-14 strain (Fig. 3). At the onset of anaphase, Siz1p is phosphorylated (Johnson and Gupta, 2001), and it is exported from the nucleus by Kap142p/Msn5p. Phosphorylation may act as a signal for Siz1p export from the nucleus, as Kap142p/Msn5p has previously been shown to recognize NESs containing phosphorylated amino acid residues (Kaffman et al., 1998; Boustany and Cyert, 2002; Jaquenoud et al., 2002). Kap142p/Msn5p is strictly required for the accumulation of Siz1p at the septin ring, suggesting that export mediated by this kap is required to generate a cytoplasmic pool necessary for the accumulation of Siz1p at the septin ring.
Septin desumoylation is mediated by Ulp1p (Takahashi et al., 2000; Fig. 3 A), whose activity is controlled by kaps. Our data suggest a complex situation in which Kap121p and the Kap95p–Kap60p complex direct Ulp1p to distinct locations, which controls the accessibility of this isopeptidase to septin substrates. Mutations that disrupt the interaction of Ulp1p with Kap121p or the Kap95p–Kap60p complex cause an increase in the cytoplasmic levels of Ulp1p and a loss of septin sumoylation (Figs. 3 and 4). These results are consistent with the hypothesis that these kaps function as negative regulators of Ulp1p activity against cytoplasmic targets. One possibility is that kap binding directly suppresses the isopeptidase activity of Ulp1p. However, this is unlikely, as not all Ulp1p substrates are similarly affected by Ulp1p mutants that inhibit kap binding (Li and Hochstrasser, 2003; see below). Instead, we envisage kaps negatively affecting the cytoplasmic activity of Ulp1p by sequestrating it at the NPC. Once Ulp1p is delivered to the NPC, it associates with the nucleoplasmic face of this structure, where it interacts, directly or indirectly, with the Mlp proteins (X. Zhao et al., 2004). Here, Ulp1p would be sequestered away from cytoplasmic targets. Similarly, the cytoplasmic activity of SENP2, the human counterpart of Ulp1p, is suppressed by interaction with the NPC (Hang and Dasso, 2002). Conversely, the concentration of Ulp1p at the nuclear face of the NPC is likely to stimulate its activity directed against nuclear targets, including those in transit through the NPC. Thus, we predict that mutations in Ulp1p that decrease the efficiency of its targeting to the NPC would lead to higher levels of nuclear protein–SUMO conjugates. Consistent with this idea, Li and Hochstrasser (2003) have shown that deletion mutants of Ulp1p that reduce its NPC association (and binding to the kaps; Panse et al., 2003; Fig. 4) lead to an overall increase in cellular levels of SUMO-modified proteins, the vast majority of which are contained within the nucleus (Johnson, 2004). These observations suggest that kaps control the access of Ulp1p to targets in both the cytoplasm and the nucleus.
Although Ulp1p binds both Kap121p and the Kap95p–Kap60p complex, it remains unclear whether it binds simultaneously to these kaps or forms two complexes. The functional significance of binding multiple kaps is unclear, but it seems unlikely that the kaps play an entirely redundant role. For example, when independently examined, the Kap121p (Ulp11-150) and the Kap95p–Kap60p (Ulp1150-340) binding domains of Ulp1p exhibited distinctly different localization patterns. The Kap121p binding domain is generally visible in a uniform perinuclear pattern, whereas the Kap95p–Kap60p domain displayed a punctuate NE and intranuclear localization pattern. The uniform distribution and the capacity of the NE to accommodate overproduced Ulp11-150 or WT Ulp1p (X. Zhao et al., 2004) is most likely explained by direct binding to the INM. Several observations led us to this conclusion. First, overproduced Ulp11-150 and Ulp1p are restricted to the NE and are not detected in the peripheral ER, as would be expected if they were associated with the outer nuclear membrane. Second, Ulp1p is visible in association with INM-derived intranuclear membranes that are induced by the overproduction of the nucleoporin Nup53p (Marelli et al., 2001; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200608066/DC1). Interestingly, both Nup53p and Ulp1p may follow pathways similar to the INM. In both cases, Kap121p is required for NE accumulation of Ulp11-150 (unpublished data). These observations raise the intriguing possibility that Ulp1p can access the INM through a process that is controlled by Kap121p. This would place Ulp1p in a position to regulate the sumoylation state of INM-associated proteins, including chromatin components.
We have also shown that the steady-state association of Ulp1p with the NE is energy dependent. Energy-dependent association is not a feature of any of the nups or currently known NPC-interacting proteins. Interestingly, the dispersed localization of Ulp1p induced by 2-deoxyglucose/sodium azide is also distinct from that observed for the bulk of Kap121p or Kap95p and Kap60p, which either accumulate in the nucleus or at the NPC, respectively. This suggests that the Ulp1p–kap complexes are specifically dissociated in the absence of energy or that they are functionally distinct from the bulk of the kaps. The former possibility seems remote, as Ulp1–kap complexes can be isolated from cell lysates and reconstituted in vitro in the absence of energy (Panse et al., 2003; Fig. 8). A more likely scenario is that kaps bound to Ulp1p are not part of the cycling pool of kaps involved in the nuclear transport cycle. Several observations support this model. First, the interaction of Ulp1p with Kap121p and the Kap95p–Kap60p complex may prevent these kaps from binding RanGTP (Panse et al., 2003). Panse et al. (2003) have shown that Ulp1p is not dissociated from Kap121p or the Kap60p–Kap95p complex by treatment with RanGTP. Moreover, we have shown that overexpression of Ulp11-150 inhibits Kap121p-mediated transport and induces a nuclear accumulation of Kap121p (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200608066/DC1). Both of these phenotypes can be explained by Ulp11-150 inhibiting the binding of RanGTP to Kap121p, as similar phenotypes are also observed upon depletion of cellular levels of RanGTP (Makhnevych et al., 2003).
On the basis of these observations, including the karyopherin and energy-dependent NPC association of Ulp1p, we suggest that Ulp1p dynamically interacts with the NPC. Although its primary function likely occurs at the NPC, we envisage that Ulp1p can also migrate to other locations in the cell, where it desumoylates specific targets, including those in the INM and the septin rings. In the latter case, this movement appears to be cell cycle regulated, occurring during mitosis. The hypothesis that Ulp1p can leave the NPC can explain previous observations that Ulp1p is required to desumoylate cytoplasmic proteins. For example, in addition to Ulp1p being required for the desumoylation of septins (Takahashi et al., 2000), the Drosophila melanogaster Ulp1 is required to prevent the accumulation of SUMO modifications on the cytoplasmic protein glutamyl-prolyl-tRNA synthetase (Smith et al., 2004). Moreover, human SENP2 has also been shown to shuttle between the nucleus and the cytoplasm (Itahana et al., 2006). Finally, the movement of Ulp1p from the NPC appears to be required for normal septin desumoylation, as this process is inhibited by anchoring of the catalytic domain of Ulp1p on the nucleoplasmic face of the NPC by tethering it to Nup60p (Fig. 7).
Although both Kap121p and the Kap95p–Kap60p complex function in sequestering Ulp1p at the NPC, we also propose that they play a role in regulating the access of Ulp1p to targets in the cytoplasm, specifically, the septins. These events are likely controlled by the interactions of the Ulp1p–kap complexes with the NPC. Importantly, we show that, when unable to bind to the Kap95p–Kap60p complex, Ulp1p (Ulp1Δ150-340) is detected at the septin ring in large-budded cells. This targeting event is dependent on Kap121p, as mutations that abolish Ulp1p–Kap121p binding (Ulp1Δ1-150 [Fig. 4] and Ulp1Δ1-340 [not depicted]) and the kap121-34 mutation (Fig. 5) prevent the association of Ulp1p with the septin ring. Our interpretation is that, although both Kap121p and the Kap95p–Kap60p complex function in concentrating Ulp1p at the NPC during interphase, Kap121p plays a distinct role in targeting Ulp1p to septin rings during mitosis. A key step in this process would likely involve cell cycle–specific changes in the binding of the Ulp1p–kap complex to the NPC that facilitate release and recruitment of Ulp1p to binding sites at the septin ring. Consistent with this idea, the positioning of Ulp1p and Kap121p within the NPC is altered during mitosis, with both being recruited to sites where they interact with Nup53p (Fig. 8). It has previously been shown that the association of Kap121p with Nup53p inhibits its translocation through the NPC and import of its cargos, leading to their accumulation in the cytoplasm (Makhnevych et al., 2003). These, or as-yet-undefined, changes in the NPC could contribute to increased cytoplasmic levels of Ulp1p and facilitate its association with the septin ring.
We speculate that, at points during mitosis, the opposing activities of Kap121p-dependent septin association and Kap95p–Kap60p–mediated NPC targeting allow Ulp1p to cycle between the NPC and the septin ring. The concerted targeting functions of the kaps would presumably tightly regulate the levels of Ulp1p that can associate with septin rings. Thus, binding of Ulp1p to the septin rings is likely to be transient. This, coupled with low cellular levels of Ulp1p, would explain our inability to detect cytoplasmic Ulp1p, except under conditions where its interactions with kaps are altered. Interestingly, targeting of Ulp1p to the septin ring may occur before completion of mitosis, when levels of sumoylated septins drop. We observed that Ulp1Δ150-340 is detected at septin rings in large-budded cells before nuclear division (Fig. 4), coinciding with time points when Siz1p is also associated with the septin ring. Moreover, a catalytic domain mutant of Ulp1p showed a more predominant septin sumoylation pattern during M phase and a delay in septin desumoylation after cytokinesis (Fig. 3 A). This would imply that steady-state levels of sumoylated septins are a product of the relative activities of both Ulp1p and Siz1p, with the balance being shifted to the desumoylated state by the release of Siz1p from the septin ring and its reimport into the nucleus at the end of mitosis.
Materials and methods
Yeast strains and media
Strains used in this study are presented in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200608066/DC1). The gene encoding Cdc3-HA was integrated within the strains indicated in Table S1 using a PCR product derived from a previously described yeast strain (Johnson and Blobel, 1999), provided by E. Johnson (Thomas Jefferson University, Philadelphia, PA). Yeast strains were grown at 30°C unless otherwise indicated in YPD or synthetic media (SM) supplemented with appropriate nutrients and 2% glucose (Sherman et al., 1983). Yeast transformations were performed as described by Delorme (1989).
Plasmids
All of the following plasmids contain inserts synthesized using Expand High Fidelity PCR system (Roche Diagnostics). pRS316 ULP1-GFP, pRS316 ULP1Δ1-150-GFP, pRS316 ULP1Δ150-340-GFP, pRS316 ULP1Δ340-403-GFP, pRS316 ULP11-150-GFP, and pRS316 Ulp1150-340-GFP were constructed as follows. The promoter region of ULP1 (−300 to +3, where +1 corresponds to the A of the start codon) was amplified with Xho1 and EcoR1 linkers and inserted into the corresponding sites of pRS316 (CEN/URA3; Sikorski and Hieter, 1989). Next, the GFP+ ORF (Scholz et al., 2000) was amplified with Sac1 and Not1 linkers and cloned into the corresponding sites of pRS316. The following regions of the ULP1 ORF were then inserted into the resulting plasmid at an EcoR1 site after the ULP1 promoter and in frame with the 5′ end of the GFP ORF: +4 to +1863 (encoding Ulp1p), +4 to +450 (Ulp11-150), +448 to +1020 (Ulp150-340), +451 to +1863 (Ulp1Δ1-150), +4 to +448 linked to +1021 to +1863 (Ulp1Δ150-340), and +4 to +1021 linked to +1210 to +1863 (Ulp1Δ340-403). Note that the latter two inserts contain a BssH II linker between the two ULP1 fragments. pRS316 ULP1-GFP, pRS316 ULP1Δ1-150-GFP, pRS316 ULP1Δ150-340-GFP, and pRS316 ULP1 Δ340-403-GFP were introduced into a ulp1-null mutant by plasmid shuffle. Each plasmid complemented the lethal phenotype of a ulp1-null mutant.
pGULP11-150 and pGULP1 were constructed by cloning nucleotides 1–450 and 1–1863 of the ULP1 ORF plus stop codons into the BamH1 site of pGEX-6P-1 (GE Healthcare). pYULP11-150 was constructed by cloning nucleotides 1–450 of the ULP1 ORF followed by a stop codon into BamH1 and EcoR1 sites of pYEX-BX (CLONTECH Laboratories, Inc.). pcNLS-GFP (Stade et al., 1997), pGNUP53, pGKAP121 (Lusk et al., 2002), pPHO4-NLS-GFP, pKAP121-GFP, and pYNUP53 (Marelli et al., 2001; Lusk et al., 2002) were previously described. pULP1C-GFP-NUP60 and pSMT3-GFP (Panse et al., 2003) were provided by E.C. Hurt (University of Heidelberg, Heidelberg, Germany).
Fluorescence microscopy
Cell pellets used for fluorescence microscopy were suspended in 20 μl of an appropriate SM, and 2 μl of this suspension was spotted onto a slide. Images of GFP fusion proteins were acquired at room temperature as 0.7-μm optical sections using a Plan-Apochromat 63×/1.4 NA oil differential interference contrast objective on an microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) equipped with a confocal scanning system (LSM 510 META; Carl Zeiss MicroImaging, Inc.). All images were acquired using LSM 510 software and viewed using LSM Image Browser. Images were subsequently incorporated into figures using Draw 10 (Corel) and Photoshop (Adobe).
Detection of Cdc3-HA sumoylation
Strains containing the endogenous CDC3 ORF tagged after the last codon with the HA epitope were made as follows. Genomic DNA was isolated from the strain EJY301 containing the CDC3-HA gene (Johnson and Blobel, 1999). The gene was amplified by PCR, and the product was transformed into the indicated strains. WT, kap121-34, kap95-14, and YRW110 cells expressing CDC3-HA were synchronized in G1 phase with 7.5 μg/ml α-factor (Sigma-Aldrich) at 23°C as described previously (Makhnevych et al., 2003). α-Factor was removed by washing cells in YPD. Cells were then resuspended in fresh YPD media containing either 100 mM hydroxyurea (Sigma-Aldrich) or 17 μg/ml nocodazole (Sigma-Aldrich) and shifted to 37°C for 2.5 h. Cell cycle arrest was monitored by examining cell morphology and by Western blotting (see Western blot analysis) to detect levels of the mitotic cyclin Clb2p. Cdc3-HA was detected by Western blotting of whole-cell lysates prepared as described in the following section. Levels of Cdc3-HA sumoylation in strains that were not temperature sensitive were determined as described, except cells were grown at 30°C.
Cdc3-HA desumoylation was also examined after release from nocodazole arrest in DF5 and nup53Δ405-430 cells. Cells were arrested with 17 μg/ml nocodazole for 2.5 h. After this incubation period, cells were washed with YPD to remove nocodazole and resuspended in YPD. At the indicated times, cells were harvested and washed once with water, and whole-cell lysates were prepared for Western blot analysis.
Western blot analysis
Whole-cell lysates were prepared from cell cultures as follows. Cells derived from 2-ml cultures were harvested by centrifugation, washed with water, and sonicated for 20–30 s in 35 μl SDS-PAGE sample buffer. Samples were then incubated at 75°C for 10 min, and proteins were separated by SDS-PAGE. Proteins were then transferred to nitrocellulose membranes, and membranes were blocked with 5% skim milk and 0.1% Tween 20 in PBS. HA moieties were detected using a monoclonal anti-HA antibodies (3F10) conjugated to HRP (Roche Diagnostics) and the ECL system (GE Healthcare). Specific rabbit polyclonal antibodies were used to detect Clb2p (Santa Cruz Biotechnology, Inc.), GFP, Gsp1p (Makhnevych et al., 2003), Nup53p, and Kap121p (Marelli et al., 1998). Binding of primary antibodies was detected using HRP-conjugated donkey anti-rabbit secondary antibodies and ECL.
Purification of TAP-tagged Ulp1p
Cells synthesizing Ulp1-TAP (Ghaemmaghami et al., 2003) were grown in YPD (0.3 OD600/ml) and arrested in G1 or M phase as described previously (Makhnevych et al., 2003). After harvesting, cells were lysed and Ulp1-TAP was isolated using IgG-Sepharose (GE Healthcare) chromatography as previously described (Marelli et al., 1998; Makhnevych et al., 2003). After isolation, bead-bound complexes were washed extensively with lysis buffer containing 25 mM MgCl2 and eluted using a step gradient of MgCl2 (200, 500, and 1,000 mM) followed by a 0.5-M acetic acid, pH 3.4, elution. Proteins in each eluate were TCA precipitated, separated by SDS-PAGE, and transferred to nitrocellulose for Western blotting.
In vitro binding assays
Escherichia coli cells transformed with pGKAP121, pGULP11-150, pGULP1, or pGNUP53 were grown to midlog phase and induced with 1 mM IPTG for 4 h. Cells were lysed, and GST fusions were purified on glutathione–Sepharose beads according to the manufacturer's instructions (GE Healthcare) using 150 mM NaCl, 1 mM MgCl2, 0.1% Tween 20, and 50 mM Tris, pH 7.5, as a lysis and wash buffer. Purified Kap121p and Nup53p were recovered by treatment of the bead-bound fusions with thrombin and Precision Protease (GE Healthcare), respectively. To evaluate binding to Ulp1p, 10 μg Kap121p or Nup53p in 40 μl of lysis buffer were incubated for 45 min at 4°C with 10 μl glutathione–Sepharose beads preloaded with ∼10 μg GST-Ulp1p or ∼5 μg GST-Ulp11-150. Unbound fractions were collected, and beads were eluted with SDS-PAGE sample buffer. Alternatively, the GST-Ulp11-150–Kap121p complex was further incubated with ∼10 μg of recombinant Nup53p for 1 h at 4°C, and bound and unbound fractions were collected. Proteins in the various fractions were separated by SDS-PAGE and detected with Bio-Safe Coomassie (Bio-Rad Laboratories).
Metabolic poisoning assay
Treatment of cell cultures with metabolic poisons was performed essentially as described previously (Shulga et al., 1996). Cell cultures were grown in YPD or SM in the presence or absence of 10 μg/ml nocodazole for 2.5 h. Cultures were then centrifuged, and cell pellets were washed twice with media lacking glucose. Pellets were resuspended at room temperature in medium lacking glucose and supplemented with 100 mM 2-deoxyglucose (Sigma-Aldrich) and 17 mM sodium azide (ICN Biomedicals) and, where indicated, 10 μg/ml nocodazole (Sigma-Aldrich). Localization of GFP-tagged proteins was detected at the indicated time points. For recovery, cells were washed twice with PBS to remove metabolic poisons, resuspended in media containing 2% glucose, and incubated at 30°C for the indicated times.
Induction of Ulp11-150 and Nup53p overexpression
For the induction of CUP1 promoter–controlled expression of ULP11-150 and NUP53, strains harboring the pY ULP11-150 or pY NUP53 plasmids were grown to midlogarithmic phase in SM, and expression was induced for 6 h by the addition of copper sulfate to a final concentration of 0.3 mM.
Online supplemental material
Fig. S1 shows that Ulp1-GFP is associated with the INMs in Nup53p-overproducing cells. Fig. S2 shows that overproduction of the Kap121p binding domain of Ulp1p inhibits the import of Kap121p cargoes and alters Kap121p localization. Table S1 contains a list of the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200608066/DC1.
Supplementary Material
Acknowledgments
We would like to thank Erica Johnson and Ed Hurt for kindly providing us with strains and plasmids.
Funds for this work were provided by the Canadian Institute of Health Research (grant 36519), the Alberta Heritage Foundation for Medical Research, and the National Institutes of Health (grant P50 GM076547).
T. Makhnevych and C. Ptak contributed equally to this paper.
Abbreviations used in this paper: INM, inner nuclear membrane; NE, nuclear envelope; NES, nuclear export signal; NPC, nuclear pore complex; TAP, tandem affinity purification; WT, wild type.
References
- Boustany, L.M., and M.S. Cyert. 2002. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 16:608–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme, E. 1989. Transformation of Saccharomyces cerevisiae by electroporation. Appl. Environ. Microbiol. 55:2242–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth, D.J., A. Suprapto, J.C. Padovan, B.T. Chait, R.W. Wozniak, M.P. Rout, and J.D. Aitchison. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami, S., W.K. Huh, K. Bower, R.W. Howson, A. Belle, N. Dephoure, E.K. O'Shea, and J.S. Weissman. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741. [DOI] [PubMed] [Google Scholar]
- Hang, J., and M. Dasso. 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277:19961–19966. [DOI] [PubMed] [Google Scholar]
- Itahana, Y., E.T. Yeh, and Y. Zhang. 2006. Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol. Cell. Biol. 26:4675–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud, M., F. van Drogen, and M. Peter. 2002. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1). EMBO J. 21:6515–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355–382. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., and G. Blobel. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E.S., and A.A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 106:735–744. [DOI] [PubMed] [Google Scholar]
- Kaffman, A., N.M. Rank, E.M. O'Neill, L.S. Huang, and E.K. O'Shea. 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 396:482–486. [DOI] [PubMed] [Google Scholar]
- Leslie, D.M., B. Grill, M.P. Rout, R.W. Wozniak, and J.D. Aitchison. 2002. Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol. Cell. Biol. 22:2544–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, D.J. 2003. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15:648–653. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature. 398:246–251. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.J., and M. Hochstrasser. 2003. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160:1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., and E. Bi. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13:403–409. [DOI] [PubMed] [Google Scholar]
- Lusk, C.P., T. Makhnevych, M. Marelli, J.D. Aitchison, and R.W. Wozniak. 2002. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk, C.P., T. Makhnevych, and R.W. Wozniak. 2004. New ways to skin a kap: mechanisms for controlling nuclear transport. Biochem. Cell Biol. 82:618–625. [DOI] [PubMed] [Google Scholar]
- Makhnevych, T., C.P. Lusk, A.M. Anderson, J.D. Aitchison, and R.W. Wozniak. 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 115:813–823. [DOI] [PubMed] [Google Scholar]
- Marelli, M., J.D. Aitchison, and R.W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., C.P. Lusk, H. Chan, J.D. Aitchison, and R.W. Wozniak. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panse, V.G., B. Kuster, T. Gerstberger, and E. Hurt. 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5:21–27. [DOI] [PubMed] [Google Scholar]
- Panse, V.G., U. Hardeland, T. Werner, B. Kuster, and E. Hurt. 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279:41346–41351. [DOI] [PubMed] [Google Scholar]
- Scholz, O., A. Thiel, W. Hillen, and M. Niederweis. 2000. Quantitative analysis of gene expression with an improved green fluorescent protein. Eur. J. Biochem. 267:1565–1570. [DOI] [PubMed] [Google Scholar]
- Seeler, J.S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690–699. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1983. Methods In Yeast Genetics: Laboratory Manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY. 120 pp.
- Shulga, N., and D.S. Goldfarb. 2003. Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol. Cell. Biol. 23:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga, N., P. Roberts, Z. Gu, L. Spitz, M.M. Tabb, M. Nomura, and D.S. Goldfarb. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M., V. Bhaskar, J. Fernandez, and A.J. Courey. 2004. Drosophila Ulp1, a nuclear pore-associated SUMO protease, prevents accumulation of cytoplasmic SUMO conjugates. J. Biol. Chem. 279:43805–43814. [DOI] [PubMed] [Google Scholar]
- Stade, K., C.S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 90:1041–1050. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., and S.R. Wente. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 4:775–789. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., M. Iwase, M. Konishi, M. Tanaka, A. Toh-e, and Y. Kikuchi. 1999. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem. Biophys. Res. Commun. 259:582–587. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., J. Mizoi, E. Toh, and Y. Kikuchi. 2000. Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J. Biochem. (Tokyo). 128:723–725. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., T. Kahyo, E. Toh, H. Yasuda, and Y. Kikuchi. 2001. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276:48973–48977. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel, J.A., E.S. Johnson, S.I. Reed, and J.R. Yates III. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279:45662–45668. [DOI] [PubMed] [Google Scholar]
- Wykoff, D.D., and E.K. O'Shea. 2005. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell. Proteomics. 4:73–83. [DOI] [PubMed] [Google Scholar]
- Zhao, X., C.Y. Wu, and G. Blobel. 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167:605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., S.W. Kwon, A. Anselmo, K. Kaur, and M.A. White. 2004. Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J. Biol. Chem. 279:20999–21002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.