Abstract
Cellular pathways that contribute to adult human mammary gland architecture and lineages have not been previously described. In this study, we identify a candidate stem cell niche in ducts and zones containing progenitor cells in lobules. Putative stem cells residing in ducts were essentially quiescent, whereas the progenitor cells in the lobules were more likely to be actively dividing. Cells from ducts and lobules collected under the microscope were functionally characterized by colony formation on tissue culture plastic, mammosphere formation in suspension culture, and morphogenesis in laminin-rich extracellular matrix gels. Staining for the lineage markers keratins K14 and K19 further revealed multipotent cells in the stem cell zone and three lineage-restricted cell types outside this zone. Multiparameter cell sorting and functional characterization with reference to anatomical sites in situ confirmed this pattern. The proposal that the four cell types are indeed constituents of an as of yet undescribed stem cell hierarchy was assessed in long-term cultures in which senescence was bypassed. These findings identify an adult human breast ductal stem cell activity and its earliest descendants.
Introduction
The mammary gland is a highly dynamic organ that undergoes a series of changes from intrauterine life to senescence. In humans, growth in adulthood commences at puberty where the parenchymal cells branch from a few blunt ending primary and secondary ducts into an elaborate tree with multiple terminal ducts and lobules. With each menstrual cycle, breast proliferation fluctuates, and, in the luteal phase, the growth fraction can become as high as 35% (Potten et al., 1988; Shetty et al., 2005). Accordingly, during pregnancy, there is both a 10-fold increase in the number of alveoli per lobule as well as de novo formation of lobules by lateral budding from existing terminal ductules, leaving behind a small amount of connective tissue space (for review see Russo and Russo, 2004). These cellular dynamics led Taylor-Papadimitriou et al. (1983) to postulate the existence of a population of precursor cells in the adult human breast that are capable of giving rise to new lobules.
From studies mainly in other species, it is known that adult stem cells are generally focal in distribution and not necessarily colocalized with the bulk of transiently amplifying cells (for review see Fuchs et al., 2004). In mice, the location of immature mammary gland stem cells was narrowed down to the peripheral cap cells of the terminal end buds (for review see Woodward et al., 2005). In humans, however, in which end buds are not prominent structures (Howard and Gusterson, 2000), the identification of a candidate stem cell zone has had to rely on a detailed and microscopically directed sampling of well-defined segments of the organ followed by functional assays. By combining microdissection with colony-forming ability, candidate stem cells in the human hair follicle were prospectively identified in the bulge region more than a decade before the bulge was unequivocally proven to be the epithelial stem cell niche in the skin (Yang et al., 1993; Moll, 1995; Tumbar et al., 2004). The proximal ducts of the prostate were also shown to harbor stem cells by this method (Tsujimura et al., 2002). Recently, a side population exhibiting Hoechst dye efflux properties was isolated from the human mammary gland (for review see Smalley and Clarke, 2005), and self-renewing cells were enriched for by the use of nonadherent mammosphere cultures (Dontu et al., 2003). Whereas in mice, the ultimate evidence for the existence of mammary stem cells is the clonal repopulating ability and greater morphogenic capacity within the cleared fat pad (Stingl et al., 2006a; Shackleton et al., 2006), such experiments cannot be performed in humans. Fortunately, surrogate assays conducted with mice and human cells demonstrate that putative mammary stem cells in 3D laminin-rich ECM (lrECM) gels function as they do in vivo in terms of several morphogenetic criteria across the species (Gudjonsson et al., 2002b; Dontu et al., 2003; Clarke et al., 2005; Stingl et al., 2006a). However, the existence of a hierarchy as well as a correspondence between stem cell markers and activity with specific regions of the gland has not been described.
In this study, we examine in situ whether the resting human mammary gland exhibits stem cell markers, which could identify a stem cell zone, as has been done for other tissues (Blanpain et al., 2004; Trosko et al., 2004; Burger et al., 2005; Kim et al., 2005; Moore and Lemischka, 2006). We took advantage of Ki-67 and laminin-2/4 as biomarkers for cellular turnover and differentiation (Simon-Assmann et al., 1994; Belair et al., 1997; Feuerhake et al., 2000; Fleischmajer et al., 2000; Maslov et al., 2004; Kingsbury et al., 2005; Shetty et al., 2005). Further dissection of regions of interest and microcollection allowed us to interrogate functional stem cell properties in culture. These included high clonal proliferative capacity on tissue culture plastic, self-renewal in suspension, and mammary morphogenesis in 3D lrECM (Petersen et al., 1992; Hudson et al., 2000; Gudjonsson et al., 2002b; Tsujimura et al., 2002; Dontu et al., 2003; Benitah et al., 2005; Kim et al., 2005; Ohyama et al., 2006; Stingl et al., 2006a).
Multipotency was determined based on fluorescence imaging in situ and in clonal primary cultures using two lineage markers, keratins K14 and K19, in which K14 marks the myoepithelium and K19 is hypothesized to be a neutral switch keratin that permits the changeover of one type of cytoskeleton to the other (Stasiak et al., 1989). Importantly, double-positive K14 and K19 transitional cells are known also to codistribute with the stem cell zone in the prostate (Hudson et al., 2001).
To compare mouse and human markers, we isolated mammary stem cell activity on the basis of surface markers CD49f and EpCAM (Stingl et al., 2006b). The cells thus isolated were further characterized using additional putative stem and progenitor markers. Finally, we took advantage of human papilloma virus (HPV) 16 E6/E7 to bypass cellular senescence (Band et al., 1990; Gudjonsson et al., 2002b) and to interrupt the normal differentiation of stem and progenitor cells to generate cell lines that maintain their phenotypes in culture. The absence of senescence allowed us to follow the developmental hierarchy of the progenitor cells in the breast and made it possible to isolate stable cell lines representing the different stem and progenitor stages, as done previously for other cell types (Reznikoff et al., 1994; Roecklein and Torok-Storb, 1995; Okamoto et al., 2002; Osyczka et al., 2002; Akimov et al., 2005; Mori et al., 2005). Our results have defined a human breast epithelial stem cell zone in vivo and a progenitor hierarchy both inside and outside this zone. Additionally, we have developed several stem and progenitor cell lines that will aid our understanding of the possible role of these cells in breast cancer.
Results
A candidate stem cell zone resides in ducts
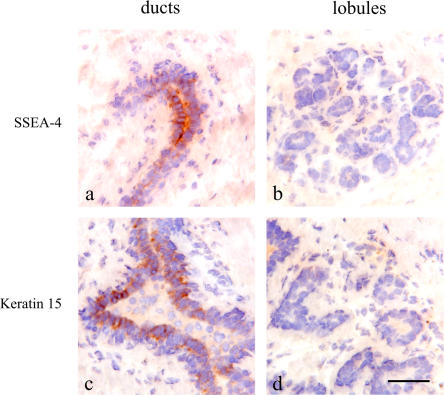
To test whether a stem cell zone could be defined in the human breast, we stained for several postulated surrogate stem cell markers, including stage-specific embryonal antigen-4 (SSEA-4), keratin K6a, keratin K15, keratin K5, Bcl-2, and chondroitin sulfate (Bocker et al., 2002; Legg et al., 2003; Luna-More et al., 2004; Dravida et al., 2005; Schmelz et al., 2005; Ohyama et al., 2006). Three of the markers examined, SSEA-4 (Fig. 1), K15 (Fig. 1) and K6a (see Fig. 5 a), localized focally to discrete clusters of cells in ducts, including terminal ducts, and were essentially absent from lobules. Specifically, although 73–78% of ducts stained with K6a, SSEA-4, and K15, the frequency of lobules with stained cells ranged from 6 to 13% (n = 19). Keratin K5, Bcl-2, and chondroitin sulfate were more widely distributed but still were more prominent in ducts than lobules (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200611114/DC1). The frequency of SSEA-4–positive cells was further determined by FACS analysis of trypsinized uncultured organoids. Single cells identified as epithelial and myoepithelial by dual staining with epithelial antigen EpCAM and β4-integrin in the SSEA-4hi gate comprised 0.5–0.7% of the total population, which also included the stromal cells (Fig. S1 B). Importantly, SSEA-4hi cells were highly enriched for keratin K6a- and K15-positive cells (Fig. S1 B).
Figure 1.
A candidate stem cell zone resides in ducts. Immunoperoxidase staining of ducts (left) and lobules (right) in cryostat sections of the human breast with SSEA-4 (a and b) and keratin K15 (c and d). Staining is strongest in clusters of cells in ducts (brown). Nuclei are counterstained with hematoxylin (blue). Bar, 50 μm.
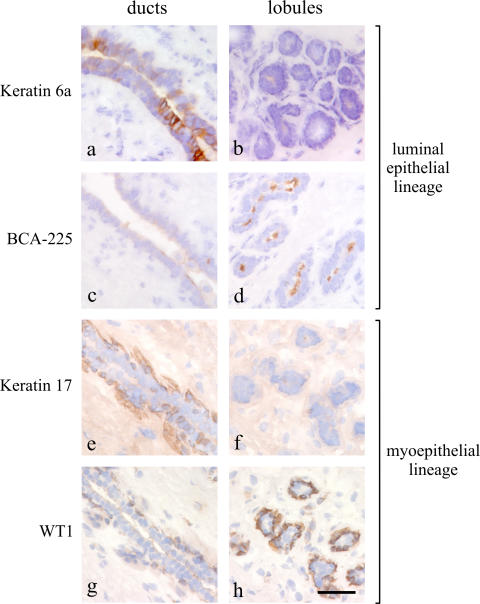
Figure 5.
Markers of distinct anatomical locations within the human breast in situ. Immunoperoxidase staining of ducts (left) and lobules (right) in cryostat sections of the human breast with keratin 6a (a and b), BCA-225 (c and d), keratin K17 (e and f), and WT1 (g and h). Each marker exhibits a restricted, albeit for BCA-225 and WT1 somewhat biopsy-dependent, location (brown) in either the luminal or myoepithelial cells of the ducts or lobules, respectively. Nuclei are counterstained with hematoxylin (blue). Bar, 50 μm.
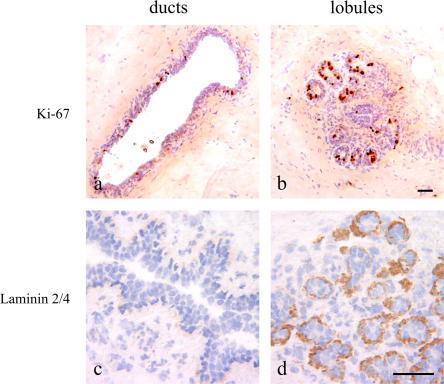
Because quiescence is a general property of stem cells in their niche, we stained ducts and lobules with Ki-67. The level of staining in lobules varied markedly between biopsies, as 6/12 had moderate to strong staining (up to 50% of cells per lobule), and the other six were negative. However, ducts showed a low but constant level of staining: a mean of 2.8% (range of 1.3–4.7%) of cells stained in 10 biopsies (Fig. 2, a and b). The Ki-67 staining pattern was confirmed by staining for minichromosome maintenance protein 2 (Gonzalez et al., 2003; unpublished data). Furthermore, lobules rather than ducts stained specifically for the α-2 chain of laminin-2/4, a basement membrane component surrounding proliferating epithelial cells (Fig. 2, c and d; Simon-Assmann et al., 1994; Chenard et al., 2000; Fleischmajer et al., 2000; Laine et al., 2004). RT-PCR for the different chains of laminin using purified populations of myoepithelial cells and fibroblasts revealed that whereas myoepithelial cells contributed α-1, α-3, and α-5 chains of laminin in addition to α-2 chains of laminin-2/4, fibroblasts expressed α-2 chains of laminin-2/4 only (Fig. S1 C). These data show that a candidate stem cell zone resides in ducts that are enriched in cells identified as being SSEA-4hi/K5+/ K6a+/K15+/Bcl-2+ cells, which are generally quiescent and are surrounded by chondroitin sulfate. The more frequently proliferating progenitors are found outside this region and are often surrounded by laminin-2/4.
Figure 2.
A latent or actively dividing proliferative compartment is delineated by laminin-2/4 in lobules. Immunoperoxidase staining of ducts (left) and lobules (right) in cryostat sections of the human breast with Ki-67 (a and b) and laminin-2/4 (c and d). Staining with Ki-67 can approach 50% of the nuclei in lobules as opposed to a mean of 2.8% in ducts. Staining with laminin-2/4 is confined to lobules (brown). Nuclei are counterstained with hematoxylin (blue). Bars, 50 μm.
Cells with a capacity for clonal growth, self-renewal, and bipotency are derived only from ducts
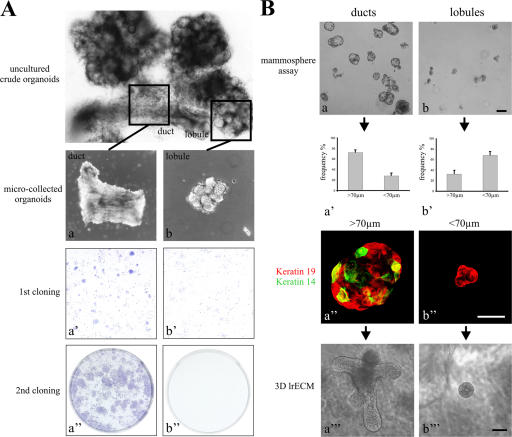
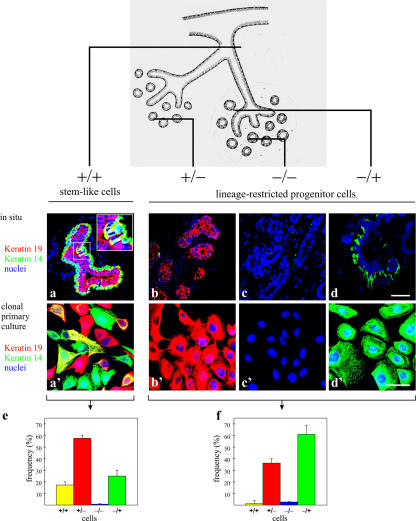
We assessed the proliferative and morphogenic capacity of primary cells on tissue culture plastic, in mammosphere suspension cultures, and in a 3D lrECM assay. The growth of the cells derived from ducts or lobules were compared on tissue culture plastic. Only ductal-derived cells formed colonies that were considerably larger than 100 cells/colony (60 ± 2% of colonies had >100 cells; n = 3 × 50 colonies) compared with lobular-derived cells (0% of colonies had >100 cells), indicating that as expected for stem cell activity in culture, the duct-derived cells have a higher proliferative potential than those derived from lobules (Fig. 3 A, a′–b′′). Similar patterns were obtained from two additional biopsies when placed in culture. To further test for self-renewal, ducts were compared with lobules using the mammosphere assay (Fig. 3 B). Whereas ducts gave rise to relatively large mammospheres (>70 μm), those derived from lobules were small and irregular (<70 μm; Fig. 3 B, a–b′). Primary mammospheres were then trypsinized and replated to derive secondary mammospheres. In our hands, the frequency of primary and secondary mammosphere formation was ∼3/1,000 cells, a figure comparable with that described originally by Dontu et al. (2003). We assessed the presence of prospective multipotent cells in mammospheres by multicolor imaging based on the combined staining for myoepithelial keratin K14 and the luminal or switch keratin K19 (Stasiak et al., 1989). The K14+/19+ phenotype was most often observed in duct-derived mammospheres (26/48 colonies; 54%) as opposed to lineage-restricted lobule-derived mammospheres (5/48 colonies; 10%; Fig. 3 B, a′′ and b′′). Similarly, mammospheres trypsinized and plated at clonal density from ducts and lobules gave rise to K19/K14 double-stained cells in 56% of those derived from ducts (n = 92 colonies) and only 6% of those derived from lobules (n = 92).
Figure 3.
Cells with a capacity for clonal growth, self-renewal, and differentiation are derived only from ducts. (A) Duct-derived clonal growth on tissue culture plastic. Phase-contrast micrograph of an uncultured breast organoid released after collagenase digestion. The anatomical composition into ducts and lobules is readily identifiable. (top) Low magnification phase-contrast micrographs of organoids dissected and microcollected into ducts (a) and lobules (b) and plated in separate culture flasks. Structure and morphology are still preserved. (a′ and b′) Low magnification photos of cells in primary culture plated at clonal density and stained with hematoxylin. (a′′ and b′′) Low magnification photos of cells in secondary culture plated at clonal density and stained with hematoxylin. Extended proliferative capacity is seen in cells from ducts. (B) Duct-derived clonal growth and morphogenesis in mammosphere culture and 3D lrECM gels. Phase-contrast micrographs of mammosphere cultures derived from ducts (a) and lobules (b). (a′ and b′) Two morphologies were recorded in each outgrowth from ducts and lobules: large spheres (>70 μm) and small irregular cell clusters (<70 μm). Large spheres are more frequent in duct-derived cultures. (a′′ and b′′) Multicolor imaging of smeared duct- and lobule-derived spheres stained with keratins K19 (red) and K14 (green). Yellow indicates double-stained cells. (a′′′ and b′′′) Morphogenic potential of duct- and lobule-derived spheres upon culture in 3D lrECM. Whereas large, solid spheres had the capacity to develop into budding (TDLU-like) structures, small cell clusters essentially maintained their morphology. Error bars represent SD. Bars: (a, b, a′′′, and b′′′) 100 μm; (a′′ and b′′) 50 μm.
The morphogenic potential of mammospheres was assessed in a 3D lrECM assay in which 2/49 duct-derived second passage mammospheres developed into terminal duct lobular unit (TDLU)–like structures. The formation of TDLUs was not observed from lobule-derived mammospheres (0/45); these only gave rise to spherical structures (Fig. 3 B, a′′′ and b′′′). A similar pattern was observed with or without a feeder layer of primary epithelial cells. Essentially, this behavior was recorded in mammospheres derived from three randomly collected biopsies from reduction mammaplasty. Finally, immunostaining of plated mammospheres for SSEA-4 from a representative biopsy further revealed that 24% of duct-derived colonies contained positive cells compared with 10% of those derived from lobules. Thus, ducts are enriched for a subpopulation of epithelial cells that, in culture, exhibit a highly proliferative, self-renewing, and morphogenic capacity indicative of stem cell activity.
Keratin profiles reveal that K19+/K14+ cells reside in ducts
When tissue sections were double stained for keratins K19 and K14, four different populations could be distinguished: K19+/K14+ (+/+), K19+/K14− (+/−), K19−/K14− (−/−), and K19−/K14+ (−/+). The +/+ cells were found as scattered single cells or small groups of cells in ducts (Fig. 4, a and inset). The +/− cells were present both in ducts (not depicted) and in lobules (Fig. 4 b). The −/− cells were rare, but, when present, they appeared generally throughout an entire TDLU (Fig. 4 c), indicating their clonal origin. The −/+ cells were observed in the myoepithelial cell layer as expected (Fig. 4 d). These profiles reflect the unique subsets of breast epithelial cells that reside in the mammary gland, as was subsequently revealed in cloned primary cultures from crude collagenase digests (Fig. 4, a′–d′). Furthermore, this pattern was confirmed in microcollected ducts and lobules. Thus, whereas lobules gave rise almost exclusively to colonies of +/−, −/+, and a few −/− cells, only ducts also gave rise to colonies of +/+ cells (Fig. 4, e and f). The predominance of +/+ cells in cultures of ducts was observed in three experiments from two independent biopsies. Similar data were obtained after recloning. Thus, whereas cells from lobules either did not clone out or formed small abortive +/− clones, the ducts formed large +/+ clones (9/10 clones in an experiment from one biopsy; Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200611114/DC1). We conclude that ducts house a subpopulation of epithelial cells that exhibit four important attributes of epithelial stem cells: they express stem cell markers in situ; they are slow cycling in vivo; they exhibit a high proliferative, self-renewal, and morphogenic capacity in culture; and they are bipotent.
Figure 4.
Keratin profiles reveal that K19+/K14+ cells reside in ducts. (top) Drawing illustrating a segment of the breast, including the branching ductal-lobular system, to indicate the approximate location of the cryostat sections. (a–d) Multicolor confocal imaging of cryostat sections of the stained human breast. The sections represent different parts of the ductal-alveolar system as indicated in the drawing. (a′–d′) Cloned primary breast epithelial cells stained for keratin K19 (red), keratin K14 (green), and nuclei (blue). Four epithelial cell types could be identified in the human breast based exclusively on the keratin staining pattern: a, +/+; b, +/−; c, −/−; and d, −/+. Cells double stained for keratins K19 and K14 (+/+) appeared as scattered yellow single cells or small groups of cells in ducts (high magnification inset in a). Yellow pseudocolor represents cells in which the two markers colocalized. These were essentially restricted to duct-derived cultures. Four types of epithelial clones with phenotypes similar to those defined in situ were recorded in cloned primary culture. (e and f) This was substantiated further by quantification of +/+, +/−, and −/+ cells in cultures from microcollected ducts (e) and microcollected lobules (f). Considerable numbers of yellow cells are present in cultures from ducts. Error bars represent SD. Bars, 50 μm.
Enrichment of K19+/K14+ cells in a Lin−CD49f+EpCAMhi population of ductal origin
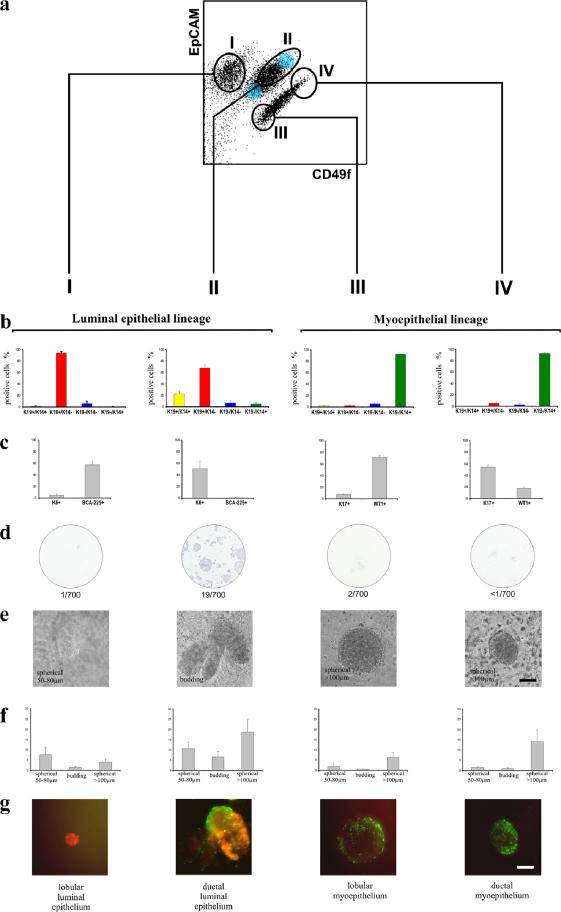
To find the in situ equivalent of ductal/lobular cells isolated by FACS analysis, we defined a set of anatomical markers. Specifically, within the luminal lineage, we used keratin K6a for ductal cells and BCA-225 for lobular cells; within the myoepithelial lineage, we used keratin K17 for ductal cells and WT1 against lobular cells (Fig. 5). Before FACS analysis, we first removed the substantial stromal component of the human breast by depleting endothelial, fibroblastic, lymphocytic, and monocytic lineages using a CD31/1B10/CD34/CD45 immunomagnetic column, allowing a flow through of lineage-negative (Lin−) epithelial cells (Shackleton et al., 2006). Primary cells were then sorted based on their expression of CD49f (α6-integrin) and EpCAM (Stingl et al., 2006b). Four subpopulations, which were identified by gates I–IV, were sorted and then analyzed by immunofluorescence for the expression of anatomical selective markers (Fig. 6 and Fig. S3 C; see also Materials and methods). Similar FACS profiles were revealed in cells from six independent biopsies. Subpopulations isolated in gates I and II represented the luminal epithelial lineage, and gates III and IV contained the myoepithelial lineage (Fig. 6, a and b). A further subdivision into lobular cells (gates I and III) and ductal cells (gates II and IV) could be established by the anatomical markers (Fig. 6, b and c). As expected, the K19+/K14+ cells cosorted with ductal Lin−CD49f+EpCAMhi cells within the luminal epithelial lineage (gate II; Fig. 6, a and b). In addition, triple color staining showed that SSEA-4hi expression was observed in the +/+, Lin−CD49f+EpCAMhi population (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200611114/DC1).
Figure 6.
Enrichment of K19+/K14+ cells in a Lin−CD49f+EpCAMhi population of ductal origin. Molecular and functional characterization of subsets of human breast cells. (a) FACS dot plot showing the distribution of Lin− epithelial cells according to their expression of CD49f and EpCAM and the gating strategy used to select four subpopulations (designated I–IV); the Lin−CD49f+EpCAMhi stem cells reside in population II. The blue shading indicates two other gates in which the keratin staining profiles were nearly identical and were, therefore, combined into the larger gate (shown) to maximize cell yield. (b and c) Each population was analyzed in parallel using cell type–specific protein expression analysis. Sorted uncultured cells were allowed to attach directly to glass slides or to a plate in culture to recover from trypsination (for BCA-225) and were then fixed and stained by immunofluorescence (left) to determine the incidence of K14 and K19 expression in the same cell or by immunocytochemistry (right) to determine the proportion of cells within each sorted population that express K6, BCA-225, K17, and WT1. The colors of the bars used in the left bar graph match the pseudocolors assigned to K19 (red) and K14 (green) used throughout this study. Therefore, K19+/K14+ cells are denoted by yellow bars, and −/− cells are denoted by blue bars. Cells were characterized functionally by colony-forming ability on 2D collagen-coated culture dishes (d), clonal density morphogenesis inside 3D lrECM, as expressed in absolute numbers per 10,000 cells seeded (e and f), and bipotency as revealed by staining for K19/K14 (g). This experimental method was repeated on three individually collected reduction mammoplasty specimens. Only cells from population II demonstrated a high incidence of K19+/K14+ (20%) cells. Importantly, this population exhibited substantial colony-forming ability (19/700) and the ability to generate TDLU-like budding structures in 3D lrECM. Error bars represent SD. Bars, 100 μm.
Functional assays for stem cell activity demonstrated that colony-forming ability was significantly restricted to the Lin−CD49f+EpCAMhi population (gate II; Fig. 6 d), also suggesting the isolation of multipotent progenitor activity. Furthermore, single-cell sorting into 96-well dishes based on SSEA-4 staining within a CD49f/EpCAM context revealed that the SSEA-4hi subpopulation had a cloning efficiency of three colonies per 96-well dish as opposed to zero from the SSEA-4neg gate, which suggests that the SSEA-4hi subpopulation represents the majority of colony-forming activity. However, our experimental design so far does not allow us to account for the considerable variation in cloning efficiency between donor samples, so we cannot conclude definitively that the SSEA-4hi subpopulation is significantly different from the total Lin−CD49f+EpCAMhi population. Nevertheless, multicolor imaging revealed that all three clones were +/+ (Fig. S3). Most importantly, when cells from gates I–IV were cultured in 3D lrECM at clonal density, only the Lin−CD49f+EpCAMhi population (gate II) formed budding (TDLU-like) structures (gate II; Fig. 6, e and f). Thus, whereas the cells isolated from other gates morphologically consisted mainly of small spheres (acinus-like) or large, solid spheres as previously described (Fig. 6, e and f; Petersen et al., 1992) and were lineage restricted, those isolated from gate II were TDLU-like budding structures and formed +/+, +/−, and −/+ as revealed by staining the whole mounts of gels (Fig. 6 g). Collectively, analysis of primary breast tissue reveals that all detectable stem cell–like or stemlike activities are restricted to duct-derived cells.
Immortalization stabilizes the differentiation repertoire in accordance with a breast stem cell hierarchy
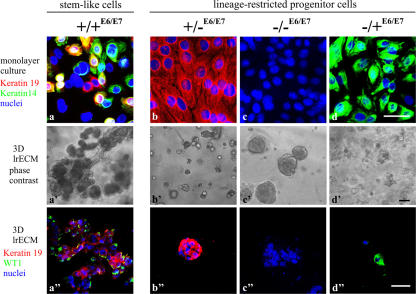
We reasoned that if the mammary epithelial cell types identified above by K14 and K19 expression were connected in a hierarchy, only one cell type should give rise to the other lineage-restricted progenitors. To examine this we used HPV16 E6/E7 (Band et al., 1990) to bypass senescence so cells could be followed for long periods of time. After transducing cells only once with retroviruses harboring E6/E7, we successfully recovered several sublines and clones of the four cell types that could be distinguished by K19/K14 expression as described in Fig. 4. We designated these cell lines +/+E6/E7, +/−E6/E7, −/−E6/E7, and −/+E6/E7 (Fig. 7). HPV16 E6/E7 conferred extended life span or immortality to all four subtypes, although the −/+ myoepithelial cells senesced after passage 30. +/+E6/E7 cells' unique ability to give rise to the other three subtypes (Fig. 7 a) and their ability to form TDLU-like structures in 3D lrECM cultures (Fig. 7 a′) confirmed that +/+E6/E7 cells exhibited stemlike activity. Quantification of morphogenesis using the 3D lrECM assay revealed 54 ± 2.5% (n = 3) of TDLU-like structures in cultures of +/+E6/E7 cells, whereas +/−E6/E7, −/−E6/E7, and −/+E6/E7 gave rise to cells of their own subtypes only (Fig. 7, b–d) and did not form TDLU in lrECM (Fig. 7, b′–d′). Relative to the other E6/E7 cell types, the multipotent +/+E6/E7 cells were unique in their combined expression of SSEA-4, keratin K15, Oct-4 (Tai et al., 2005), and Musashi-1 (Clarke et al., 2005) as well as by their capacity to differentiate into more restricted luminal-like and myoepithelial-like cells in the presence of serum. The latter were characterized by a comprehensive panel of lineage markers (Fig. S4, A–C; and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200611114/DC1). These data provide evidence that a stem cell zone in ducts marked by K19+/K14+ can give rise to K19+/K14−, K19−/K14−, and K19−/K14+ lineage-restricted progenitors.
Figure 7.
Immortalization stabilizes the differentiation repertoire in accordance with a breast stem cell hierarchy. (a–d) HPV16 E6/E7 transduced cell lines on tissue culture plastic were stained for keratin K19 (red), keratin K14 (green), and nuclei (blue). Whereas the +/−E6/E7, −/−E6/E7, and −/+E6/E7 cell lines are homogenous with respect to keratin staining, the +/+E6/E7 cell line contains cells representative of each of the other cell lines as well as doubly stained cells (green, blue, red, and yellow). (a′–d′) +/+E6/E7, +/−E6/E7, −/−E6/E7, and −/+E6/E7 cells embedded within 3D lrECM. Whereas TDLU-like structures in cultures from +/+E6/E7 cells were characterized by organoid-like branching morphogenesis, cultures from +/−E6/E7, −/−E6/E7, and −/+E6/E7 cells exclusively formed rounded or irregular cellular clusters. (a′′–d′′) K19 (red) and WT1 (green) expression in structures in 3D lrECM. Bars: (a–d and a′′–d′′) 50 μm; (a′–d′) 100 μm.
HPV16 E6/E7 transduction reveals that lineage-restricted cells are progenitors
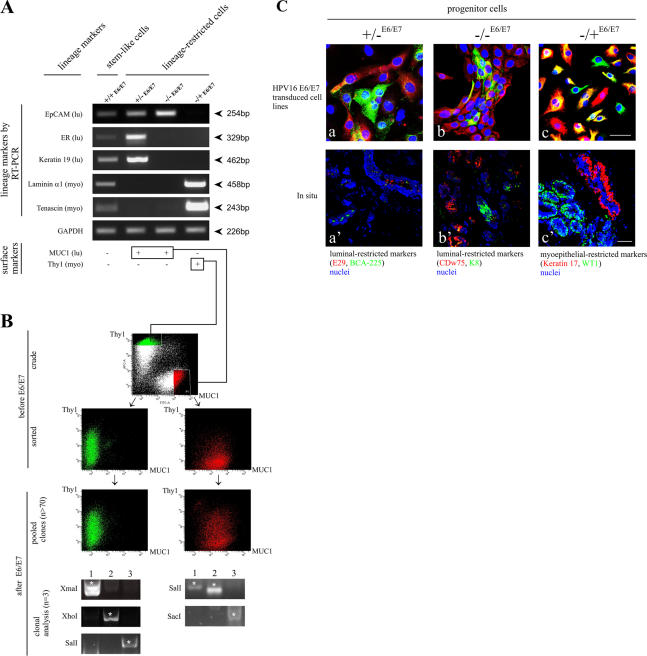
We asked two questions: whether the integration site of the viral genes affected the stability or the expression of the cells and whether lineage-restricted cells represented the end of their differentiation repertoire or whether they could specialize further. We performed a mass transduction of lineage-restricted flow-sorted mammary epithelial cells based on their expression of surface markers. As in other hierarchical tissues, specialization of the lineages in the human breast is defined by the acquisition of certain differentiation markers that are characteristic of luminal epithelial and myoepithelial cells. RT-PCR of five lineage-specific markers revealed that this general pattern carried through to all of the isolated cell lines (Fig. 8 A). Thus, whereas the stemlike cells (+/+E6/E7) had a low expression of three luminal and two myoepithelial markers, lineage-restricted progenitors (+/−E6/E7, −/−E6/E7, and −/+E6/E7) displayed a strong expression of these markers in a mutually exclusive manner (Fig. 8 A). Furthermore, staining of the lineage-restricted progenitors for surface markers revealed that they stained with either the luminal marker (MUC1) or the myoepithelial marker (Thy1). As such, we could use these markers in a large-scale cell sorting to examine the robustness of lineage maintenance after E6/E7 immortalization (acquired self-renewal). Primary cultures were sorted into luminal epithelial (+/− and −/−) and myoepithelial (−/+) lineages based on staining with MUC1 and Thy1 (Fig. 8 B; before E6/E7, crude). The profile of primary cultures was identical to that of uncultured cells (unpublished data). Further plating and cultivation in a second passage of sorted cells did not shift this phenotype appreciably (Fig. 8 B; before E6/E7 transduction, sorted). Sorted cells were transduced with HPV16 E6/E7 and selected in the presence of G418. More than 70 different clones emerged during selection, and these were pooled and reanalyzed by the same criteria. The general patterns of MUC1+/Thy1− and MUC1−/Thy1+ were sustained, albeit with a slight drifting in the population as a whole in spite of the different proviral insertion sites and the large number of pooled clones (Fig. 8 B; after E6/E7, pooled clones). This was confirmed directly by restriction site (RS) PCR and sequence analysis of chromosomal DNA in the unpooled +/−E6/E7 (MUC1+/Thy1−) and −/+E6/E7 (MUC1−/Thy1+) clones to identify individual integrated proviruses (Fig. 8 B, clonal analysis). These data demonstrate that lineage phenotype is essentially sustained irrespective of the retroviral integration site in these lineage-restricted pooled clones.
Figure 8.
HPV16 E6/E7 transduction reveals that lineage-restricted cells are progenitors. (A) Lineage-restricted cells exhibit a distinct subset of the markers in +/+ cells. RT-PCR of three luminal epithelial-specific markers (EpCAM, estrogen receptor [ER], and keratin K19) and two myoepithelial-specific markers (laminin-α1 and tenascin). All cell lines were further stained for MUC1 and Thy1. Whereas stemlike cells (+/+E6/E7) express low levels of all markers recorded by RT-PCR and no staining for MUC1 and Thy1, lineage-restricted cells exhibit strong expression of the markers for their respective lineages: +/−E6/E7 and −/−E6/E7 are positive for MUC1, and −/+E6/E7cells are positive for Thy1. (B) Robustness of lineages irrespective of retroviral integration site. (before E6/E7, crude) Primary human breast epithelial cells sorted into luminal epithelial MUC1+/Thy1− (red) and myoepithelial MUC1−/Thy1+ (green) fractions and plated in second-passage cultures. (before E6/E7, sorted) Cells from second-passage cultures grown for 14 d and reanalyzed by the same MUC1/Thy1 staining protocol as in the crude panel. There is little or no drifting in the MUC1/Thy1 profiles. (after E6/E7, pooled clones) Cells from flow-sorted second-passage culture reanalyzed by the same MUC1/Thy1 staining protocol as in the before E6/E7 panel but after transduction with HPV16 E6/E7 and selection in the presence of G418 to generate >70 transduced clones per culture. Phenotypic drifting across the lineages was still very limited. (after E6/E7, clonal analysis) RS-PCR of three clones (1–3) of MUC1−/Thy1+ or MUC1+/Thy1− lineage-restricted cells. Different sets of primers were designed based on known genomic RSs (e.g., XmaI). Note the unique RS-PCR profile for each clone (asterisks). (C) Lineage-restricted cells are progenitor cells. Multicolor imaging of cell lines (a–c) and cryostat sections (a′–c′) stained with two luminal epithelial markers of +/−E6/E7 cells, E29 (red) and BCA-225 (green; a and a′); two luminal epithelial markers of −/−E6/E7 cells, CDw75 (red) and keratin K8 (green; b and b′); and two myoepithelial markers of −/+E6/E7 cells, keratin K17 (red) and WT1 (green; c and c′), as well as nuclei (blue). The differentiation program is clearly bimodal, generating both red and green cells. In situ, this bimodality translates into the differentiation of distinct cell types within the luminal epithelial and myoepithelial compartments. Bars, 50 μm.
The fact that retroviral transduction also immortalizes the lineage-restricted cells suggested the possibility that these cells may be derived from the lineage-restricted progenitor cells. To show that this is the case, we searched for lineage-restricted markers in cells on the ductal-lobular junction. The search yielded new intralineage combinations of markers, demonstrating clearly that the three lineage-restricted cell lines were able to differentiate into additional cellular phenotypes within their respective lineages. Accordingly, +/− luminal progenitors can give rise to all combinations of the descendant cells expressing the luminal markers E29 and BCA-225 (Fig. 8 C, a). Similarly, −/− and −/+ cells give rise to descendants expressing combinations of either luminal markers K8 and CDw75 or myoepithelial markers WT1 and K17, respectively (Fig. 8 C, b and c). In all three cases, the cellular phenotypes were similar to their counterparts in situ (Fig. 8 C, a′–c′). These findings support the hypothesis that the lineage-restricted cells are progenitors rather than the ultimate end of their respective differentiation lineages. The data presented in this study provide the first evidence for the existence of a stem cell zone in mammary ducts marked by K19+/K14+, which can give rise to K19+/K14−, K19−/K14−, and K19−/K14+ lineage-restricted progenitors.
Discussion
The existence of stem and progenitor cells in the human mammary gland has been widely postulated; however, until now, neither the location nor the candidate cellular entities have been definitively identified and functionally characterized. Previously, we had described two cell lines that were derived from the human breast using magnetic sorting and HPV16 E6/E7 immortalization (Gudjonsson et al., 2002b). One of the immortalized clones, MUC1−/ESA+(E6/E7), was able to generate itself as well as luminal epithelial and myoepithelial cells; it further expressed keratin K19 and formed TDLU-like structures inside a 3D lrECM. The other, MUC1+/ESA+(E6/E7), was lineage restricted, keratin K19 negative, and formed acinus-like spheres within the 3D lrECM. Although this was an important advance, several essential questions remained: did these virally transduced cells have identical counterparts in vivo, or were these a result of immortalization? If counterparts existed, where were they located in situ? Were there additional progenitor cells we had not been able to immortalize? Could we develop a procedure whereby one could isolate the untransduced counterparts reproducibly? Were there differences between ducts and lobules, and, intriguingly, was there a hierarchy?
In this study, we have answered all of the aforementioned questions: we have demonstrated the existence of four distinct human mammary epithelial cell types in situ, two of which most certainly are precursors to the two immortalized cell lines we had isolated previously using E6/E7. The cells are distributed in a stem cell zone in ducts and outside this zone in lobules. The size of the stem cell zone varies somewhat with the marker used, the most restricted and presumably most specific being the presence of keratin K15 and SSEA-4. Ducts and lobules were dissected, collected under the microscope, cloned, and characterized. We believe it is critical to meticulously dissect lobules distal to the intralobular terminal duct. Failure to do so could explain previously published data on the reported absence of a difference in growth patterns between ducts and lobules (O'Hare et al., 1991). Permanent cell lines were established, cloned, and characterized further with respect to the profiles defined in situ and in primary cultures. The procedures we have developed, while painstaking and time consuming by necessity because they are for normal human tissues, are nevertheless robust. We show that these four cell types are hierarchically connected such that only one cell type can give rise to all others, which themselves are lineage-restricted progenitors.
An important implication of these findings is that for the first time, a stem cell zone containing one or more mammary stem cells was identified in the human breast. Mouse mammary stem cells have recently been isolated prospectively based on various FACS strategies (Shackleton et al., 2006; Stingl et al., 2006a). However, so far, FACS profiles have not translated into the cell of origin or location in situ. In this study, by the use of anatomical markers, we demonstrate that the previously described gates for bipotent progenitors, colony-forming cells, and mammary repopulating units in mice (Shackleton et al., 2006; Stingl et al., 2006a) enrich specifically for cells of ductal origin in the human breast. Older studies had suggested that the mammary ducts of rodents may harbor such a stem cell niche. For instance, in the mammary gland from virgin mouse, candidate stem cells (stained with antibodies JB6 and JsE3) were found in ducts rather than in alveoli, and it was postulated that these served to regenerate ductal epithelium as well as forming new alveolar buds (Sonnenberg et al., 1986). This postulate was born out recently in an experiment with rudimentary ducts from postgestational mice transplanted to cleared fat pads of TGF-β1 transgenic mice, which retained the capacity to reactivate lobular structures at late pregnancy (Boulanger et al., 2005). Apparently, the mouse mammary gland ductal niche responds specifically to the MMTV–c-myc transgene by amplification of the stem cell compartment (Chepko et al., 2005). This implied strongly that in mice, an entire TDLU at any time represents the progeny from a single early ductal progenitor. Indeed, seminal studies of X chromosome inactivation in the human breast had demonstrated the presence of contiguous patches of normal mammary epithelium suggestive of being derived from single stem cells (Tsai et al., 1996). These patches were explained to be the result of some developmentally important long-term primitive stem cells, which were presumed to be estrogen receptor negative, as opposed to shorter term estrogen receptor–positive stem cells important for adult tissue homeostasis (Clarke, 2005). The hierarchy we describe here is essentially compatible with the aforementioned findings. However, it is important to recognize that the present identification of a stem cell zone in ducts does not necessarily exclude the existence of stem cells at other locations, although despite an extensive search, we have not observed such additional candidates as of yet.
The possibility of one stemlike cell population in an organ, which precedes all of the other stemlike cells during development, appears to be a general phenomenon because such multiple cell-type niches were also previously described for the hair follicle (Blanpain et al., 2004). The division of labor into that of the maintenance of tissue homeostasis and the more elaborate development or regenerative remodelling between stem cell compartments may also be a general phenomenon because in the skin, homeostasis is maintained exclusively by the epidermal proliferative unit, but wound repair depends on bulge cells from hair follicles (Ito et al., 2005). In analogy to hair follicles, we would propose that mammary tissue homeostasis is maintained by proliferative units in the TDLUs, whereas more elaborate structures, including the formation of new TDLUs from ductal alveolar buds, depend on the recruitment of cells from ducts.
Until recently, very few markers were assigned directly to a candidate mammary stem cell pool in humans or, indeed, even in rodents, but scattered single cells within the ducts of adult mice were shown to be positive for keratin K6a (Buono et al., 2006), a marker originally found in putative stem cells in the terminal end bud (Smith et al., 1990). In the present study, we found that the ductal stem cell zone was characterized by the accumulation of K19+/K14+ cells. If these traits were even indirectly related to stem cell activity, we would expect them to fluctuate with the size of the stem cell compartment. One of the most important pathways responsible for mammary stem cell activity is canonical Wnt signaling (Lindvall et al., 2006). Interestingly, Wnt1- or β-catenin–induced mammary hyperplasia and tumorigenesis in mice correlate with the accumulation of K6-positive cells even though K6 in itself does not appear to be essential for mammary gland development, at least in embryonic knockouts (Grimm et al., 2006). It has been shown that caveolin-1 deficiency in mice, which conveys mammary hyperplasia and tumorigenesis along with an increased stem cell activity, is characterized by the accumulation of keratin K6-positive cells (Sotgia et al., 2005). However, the mammary glands of mice that are unable to signal through the Notch pathway most closely mimic the profile of the human mammary stem cell zone (Buono et al., 2006; Wang et al., 2006). Under these conditions, there is a remarkable ductal accumulation of cells expressing luminal keratins coordinately with keratins K14 and K6 (Buono et al., 2006). A similar stem cell–related profile has been recorded in the prostate (Hudson et al., 2001; Wang et al., 2006). The appearance of +/+ cells in the suprabasal position has been interpreted as an accumulation of intermediate immature luminal cells (Buono et al., 2006). Thus, it cannot be excluded that the stem cell zone described here contains bona fide ductal basal cells, which, under the current available culture conditions, are not colony forming.
In mass cultures derived from reduction mammoplasty, we and others had shown previously that a subpopulation of the primary breast luminal epithelial compartment was capable of giving rise to both luminal epithelial and myoepithelial cells (Kao et al., 1995; Kang et al., 1997; Péchoux et al., 1999; Clarke et al., 2005). Furthermore, in culture and occasionally in suprabasal cells in the epithelial layer in situ, single cells could exhibit dual staining for these two cell types (Péchoux et al., 1999). Cells in this position were shown later to be K19 positive and could be immortalized occasionally in mass culture with HPV E6/E7 (Gudjonsson et al., 2002b). The discovery in the present study of a ductal stem cell zone in the human breast opens the possibility of the existence of a hierarchy among differentiated progeny at other locations. In the mammary gland, the ultimate level of differentiation is reached during lactation. Although we have not addressed the consequence of lactation on ductal differentiation, a previous study strongly suggested that ducts are indeed protected from hormone-induced differentiation (Bocker et al., 2002). This is in agreement with the presumed function of candidate stem cells in mature ducts of humans and rodents as a source of lateral branching during the formation of milk-producing lobules (Taylor-Papadimitriou et al., 1983; Cardiff and Wellings, 1999). Functionally, one of the hallmarks of stem cells is the ability to self-renew (for reviews see Watt, 1998; Mackenzie, 2006). A culture assay for self-renewal in human breast epithelial cells was successfully adopted from the field of neurobiology combined with the use of 3D laminin-rich gels (Petersen et al., 1992). Thus, cells prevented from attachment grew in suspension to form the so-called mammospheres (Dontu et al., 2003) reminiscent of the originally described neurospheres. Self-renewing stem cells were characterized by the ability to form new multipotent mammospheres even after passaging (Dontu et al., 2003). The other widely used culture assay of self-renewal in epithelial tissues is growth after plating at clonal density (for reviews see Watt, 1998; Mackenzie, 2006). We used both assays in the present study and showed that both mammospheres and multipotent clones could self-renew, although we do not yet know how the fraction of the self-renewing clones compare between the assays.
Our findings are pertinent to two poorly understood aspects of breast cancer evolution. For example, we know that breast cancers comprise at least two well-defined subtypes with distinct molecular profiles reminiscent of the luminal and basal lineages (Perou et al., 2000). This has led to a resurgence of speculation that breast cancers may arise in the different compartments within a stem cell hierarchy (Taylor-Papadimitriou et al., 1983; Rudland, 1987; Al-Hajj et al., 2003; Behbod and Rosen, 2005; Clarke, 2005). The hierarchy described here may guide the design of experiments to test the possible role of these progenitors in the origin of breast cancer subtypes, a possibility that is under investigation.
Materials and methods
Breast tissue and cell cultures
Normal breast biopsies (n = 54) were obtained from patients undergoing reduction mammoplasty for cosmetic reasons. The use of human material has been reviewed by the Regional Scientific Ethical Committees for Copenhagen and Frederiksberg and approved with reference to (KF) (11) 263995.
Normal breast tissue was prepared as previously described (Rønnov-Jessen and Petersen, 1993). Upon collagenase treatment, epithelial organoids were either cultured as crude preparations or first manually separated under an inverted phase-contrast microscope (TMS-F; Nikon) into ducts and lobules before explantation (microcollected). Microcollected ducts and lobules were plated in collagen-coated (8 μg/cm2; Vitrogen100; Cohesion) T-25 flasks (Nunc) in the presence of chemically defined medium (CDM3; Petersen and van Deurs, 1988) and were allowed to spread for 8 d. Cells were trypsinized and cloned by limited dilution in the presence of serum-supplemented growth medium consisting of Ham's F12 medium (Invitrogen) supplemented with 2 mM glutamine, 50 mg/ml gentamycin (Biological Industries), 5% FCS (PAA Laboratories), 5 μg/ml insulin (Roche), 1 μg/ml hydrocortisone (Sigma-Aldrich), 0.1 μg/ml cholera toxin (Sigma-Aldrich), and 10 ng/ml EGF (PeproTech) at a density of 60 cells/cm2 (referred to below as F12 medium). After 7 d, some of the cultures were fixed in methanol and stained with hematoxylin. For secondary cloning experiments, cells were first plated at a density of 400 cells/cm2 and cultured for 12 d and then were subcloned at a density of 50 cells/cm2. Cultures were fixed and stained after 10 d, and the number of colonies was quantified. K19/K14 profiles were assessed by fluorescence in parallel cultures and in cloned primary cultures seeded at a density of 350 cells/cm2. The K19/K14 profiles of primary cultures derived from microcollected ducts or lobules were assessed at day 7 at densities of 800 cells/cm2 and in cultures of secondary clones at a density of 60 cells/cm2 at day 10.
For the nonadherent mammosphere assay, large ducts, terminal ducts (identified by connecting alveoli), and lobules were isolated and trypsinized for 10–15 min at 37°C on an orbital shaker to obtain a single cell suspension. Nonadherent mammosphere cultures were prepared as previously described (Dontu et al., 2003). In brief, cells were plated at a concentration of 5,000–20,000 cells/ml. The cultures were monitored for up to 12 d for the appearance of mammospheres. After 8 d, cultures were photographed, and structures derived from ducts (large and terminal) and lobules, respectively, were quantified and separated into two categories: >70 μm and <70 μm (n = 3 × 200 structures). For analysis of keratin expression, duct- and lobule-derived mammospheres were either smeared onto a glass slide and stained or trypsinized at day 9, plated at clonal density (200 cells/cm2), and propagated for 5 d in F12 medium before immunocytochemical staining. A total of 92 colonies from each segment were quantified using a fluorescence microscope (Dialux 20; Leitz) equipped with a 10× objective. Mammosphere populations derived from ducts and lobules were assessed for morphogenic potential by inoculation for 3 wk of each population in 300 μl lrECM (Matrigel; Becton Dickinson). Some cultures were conditioned by a feeder layer of primary human breast epithelial cells separated from the top gel by 200 μl of cell-free gel. The number of mammosphere-derived budding structures was assessed by phase-contrast microscopy.
To passage mammosphere cultures, mammospheres were collected, trypsinized for 10 min at 37°C while shaking, and single cells were plated at a concentration of 1,000 cells/ml (Dontu et al., 2003). Secondary mammospheres were sampled and plated in monolayer culture and analyzed for SSEA-4 expression by immunocytochemical staining (see supplemental Materials and methods, available at http://www.jcb.org/cgi/content/full/jcb.200611114/DC1). For RT-PCR analysis of laminins (see supplemental Materials and methods), myoepithelial cells and fibroblasts were purified and cultured as previously described before RNA extraction (Rønnov-Jessen and Petersen, 1993; Gudjonsson et al., 2002a).
Cell lines and clones
Cells representing K19+/K14+, K19+/K14−, K19−/K14−, and K19−/K14+ profiles were recovered in primary culture and were HPV16 E6/E7 transduced using a previously described protocol (Gudjonsson et al., 2002b). Fig. S4 D shows the culture history of each of the representative cell lines. For 3D culture, single cell suspensions of 105 cells were inoculated in lrECM as previously described (Petersen et al., 1992; Gudjonsson et al., 2002b) and were observed for a period of 12 d.
FACS analysis and cloning
For analysis of the robustness of transduced lineages, uncultured epithelial organoids or normal breast epithelial cells grown in primary monolayer cultures in chemically defined medium (CDM3; Petersen et al., 1990) were trypsinized and filtered through a 20-μm cell strainer (Miltenyi Biotec) and resuspended in Hepes buffer supplemented with 0.5% BSA (bovine fraction V; Sigma-Aldrich) and 2 mM EDTA (Merck), pH 7.5. The suspended cells were incubated for 30 min at 4°C in the presence of monoclonal primary antibodies recognizing MUC1 (CD227; 1:100) and Thy1 (CD90; 1:50). Control samples were incubated without primary antibody. Upon incubation, the cells were washed twice in Hepes/BSA/EDTA buffer and incubated for 15 min at 4°C with secondary isotype-specific fluorescent antibodies, AlexaFluor488 goat anti–mouse IgG2b, and AlexaFluor633 goat anti–mouse IgG1 antibodies (1:500; Invitrogen). After incubation, the cells were washed twice in Hepes/BSA/EDTA buffer and resuspended in a volume of 1 ml of buffer. Propidium iodide (Invitrogen) was added at a concentration of 1 μg/ml, and the cells were analyzed and sorted using a flow cytometer (FACSAria; BD Biosciences). The sorted populations were subsequently plated in collagen-coated T-25 culture flasks in chemically defined media (CDM6; Péchoux et al., 1999) for MUC1+/Thy1− cells and in CDM4 for MUC1−/Thy1+ cells. The sorted cell populations were transduced as previously described (Gudjonsson et al., 2002b). Untransduced sorted cell cultures were run in parallel as controls. Secondary cultures were analyzed for the expression of MUC1 and Thy1 by FACS analysis as described above.
To isolate putative stem cells within the lineage-negative epithelial cell population, breast organoids (n = 3 biopsy samples) were trypsinized for ∼10 min at 37°C under rotation. The solution was robustly agitated a few times during trypsination. Trypsination was stopped with FCS, and the cells were filtered through a 30-μm filter followed by filtration through a 10-μm filter to obtain a single-cell suspension. Cells were incubated with a cocktail of antibodies against stromal cells, which included CD31 (JC70A; 1:50), CD34 (QBEnd/10; 1:50), CD45 (Bra-55; 1:250), and fibroblast surface protein (1B10; 1:50). Cells were incubated at 4°C for 30 min. After incubation, cells were washed twice in Hepes/BSA/EDTA buffer and incubated for 15 min at 4°C with goat anti–mouse IgG and rat anti–mouse IgM microbeads (Miltenyi Biotec). Cells were then washed again twice and further applied to a column for immunomagnetic cell sorting (MACS; Miltenyi Biotec). The flowthrough from this column was collected and incubated with CD49f (GoH3; 1:500) and EpCAM (VU1D9; 1:100). A control solution was also prepared in which the primary antibodies were excluded. The cells were incubated at 4°C for 30 min followed by two washes and were further incubated with the secondary fluorescent antibodies AlexaFluor488 rabbit anti–rat IgG and AlexaFluor633 goat anti–mouse IgG1 (1:500; Invitrogen). Some degree of cross-reaction between CD49f and AlexaFluor633 was observed. For comparison with a directly conjugated EpCAM antibody, please see Fig. S3 C. The four FACS-sorted populations in gates I–IV were analyzed for luminal and myoepithelial markers as described below and were tested for self-renewal by limited dilution cloning in collagen-coated six-well plates (70 cells/cm2) in the presence of F12 medium. Morphogenic potential was analyzed in 24 wells in the presence of CDM3 by inoculation in lrECM at a density of 2 × 104 cells in 300 μl lrECM placed on top on a 200-μl cell-free gel solidified on top of a feeder layer of primary breast epithelial cells. The cultures were observed for 3 wk and assessed in triplicate for morphogenesis by phase-contrast microscopy in 50–80-μm spherical acinus-like structures, budding structures, and spherical colonies (>100 μm).
Immunohistochemistry and cytochemistry
Monolayer cultures and cells derived directly from collagenase digested tissue, cryostat sections of biopsies, mammospheres, or cell lines cultured in 3D lrECM were prepared and stained by immunoperoxidase or immunofluorescence as previously described (Petersen and van Deurs, 1988; Rønnov-Jessen et al., 1992; Gudjonsson et al., 2002b). For antibodies and further details, also see Table I.
Table I.
List of antibodies, suppliers, and dilutions used
| Antigen | Clone | Isotype | Company | Dilution | Fixation |
|---|---|---|---|---|---|
| 1B10 | 1B10 | IgM | Sigma-Aldrich | 1:50 | * |
| BCA-225 | CU-18 | IgG1 | Signet | 1:10 | M |
| Bcl-2 | 124 | IgG1 | DakoCytomation | 1:75 | M |
| Bcl-2 | 8C8 | IgG1 | Neomarkers | 1:50 | M |
| CALLA | 56C6 | IgG1 | Novocastra | 1:50 | M |
| CD31 | JC70A | IgG1 | DakoCytomation | 1:50 | * |
| CD34 | QBEnd/10 | IgG1 | Novocastra | 1:50 | * |
| CD45 | Bra-55 | IgG1 | Oncogene Research Products | 1:250 | * |
| CD49f | GoH3 | IgG2a (rat) | BD Biosciences | 1:500 | ** |
| CDw75 | LN1 | IgM | Neomarkers | 1:30–1:50 | M |
| Chondroitin sulphate | 9.2.27 | IgG2a | BD Biosciences | 1:50 | M/F |
| Collagen IV | 1042 | IgG2b | Monosan | 1:5 | M |
| E-cadherin | HECD-1 | IgG1 | Zymed Laboratories | 1:25 | M |
| EMA | E29 | IgG2a | DakoCytomation | 1:10 | M |
| EpCAM | Ber-EP4 | IgG1 | DakoCytomation | 1:100 | ** |
| EpCAM | VU1D9 | IgG1 | Novocastra | 1:25–1:100 | M** |
| EpCAM-FITC | VU1D9 | IgG1 | Abcam | 1:100 | ** |
| Integrin-β4 | 3E1 | IgG1 | Chemicon | 1:500–1:1,000 | M** |
| Keratin 14 | LL002 | IgG3 | Novocastra | 1:25–1:100 | M |
| Keratin 15 | LHK15 | IgG2a | Neomarkers | 1:10–1:100 | M |
| Keratin 17 | E3 | IgG2b | DakoCytomation | 1:10–1:25 | M/F |
| Keratin 19 | A53-B/A2 | IgG2a | Abcam | 1:50–1:100 | M |
| Keratin 5 | XM26 | IgG1 | Novocastra | 1:250–1:500 | M/F |
| Keratin 5 | Rabbit | Covance | 1:1,000 | M | |
| Keratin 5/6 | D5/16B4 | IgG1 | Boehringer | 1:250 | M |
| Keratin 6a | Ks6.KA12 | IgG1 | Monosan | 1:10 | M/F |
| Keratin 7 | RCK105 | IgG1 | Abcam | 1:100 | M |
| Keratin 8 | LE41 | IgG1 | Lane, 1982 | 1:10 | M |
| Keratin 8 | 35βH11 | IgM | DakoCytomation | 1:25–1:100 | M |
| Ki-67 | MIB1 | IgG1 | DakoCytomation | 1:100 | M |
| Laminin-α1 | 161 EB7 | IgG | Maatta et al., 2001 | 1:5 | M |
| Laminin-α2 | 5H2 | IgG1 | Engvall et al., 1990 | 1:1 | M/F |
| MCM2 | CRCT2.1 | IgG1 | Abcam | 1:25–1:100 | F |
| MCSP | LHM2 | IgG1 | Abcam | 1:50 | M/F |
| MUC1 | 115D8 | IgG2b | Biogenesis | 1:10–1:50 | M** |
| Occludin | OC-3F10 | IgG1 | Zymed Laboratories | 1:50–1:100 | M |
| p63 | 7JUL | IgG1 | Novocastra | 1:10–1:25 | M |
| Smooth muscle actin | HHF35 | IgG1 | Enzo Diagnostics | 1:25 | M |
| SSEA-4 | MC-813-70 | IgG3 | Chemicon | 1:25–1:100 | M/F** |
| SSEA-4 | 5C1 | IgG3 (rat) | US Biological | 1:25–1:100 | M |
| Thy1 | AS02 | IgG1 | Dianova | 1:25–1:100 | M** |
| Vimentin | Vim3B4 | IgG2a | DakoCytomation | 1:25–1:100 | M |
| WT1 | 6F-H2 | IgG1 | DakoCytomation | 1:10–1:50 | M/F |
Staining was performed after fixation with methanol (M) and/or formaldehyde (F). Single asterisks indicate use in immunomagnetic cell sorting (MACS). Double asterisks indicate use in FACS analysis.
The four CD49f/EpCAM cell populations were smeared onto glass slides and stained for BCA-255, K6a, K17, and WT1 by immunoperoxidase and for K19/K14 by fluorescence staining with isotype-specific AlexaFlour488 goat anti–mouse IgG3 and AlexaFlour568 goat anti–mouse IgG2a and were quantified (n = 3 × 100 cells; in triplicate from a representative biopsy). As controls, sorted cells were smeared without further staining to check for residual background fluorescence, or cells were stained after having switched the secondary antibodies to use different colors than were used previously. For staining of whole mounts of 3D cultures of cloned FACS-sorted cells, gels were fixed in methanol/acetone (1:1) for 30 min at −20°C followed by a 2-h incubation with blocking buffer and were incubated with 400 μl of antibody solution (K14 [1:25] and K19 [1:50]) overnight at 4°C, washed for 3–4 h, and incubated with secondary antibodies at 4°C overnight. Immunofluorescence stainings were evaluated using a laser-scanning microscope (LSM 510; Carl Zeiss MicroImaging, Inc.).
RNA isolation and RT-PCR
RNA isolation and PCR reactions were performed as previously described (Gudjonsson et al., 2002a,b). Specifically, primers included EpCAM forward (AGTGTACTTCAGTTGGTGCACAAA) and EpCAM reverse (AGTGTCCTTGTCTGTTCTTCTGAC; TA 56°C for 29 cycles); estrogen receptor forward (CCCTACTGCATCAGATCCAAGG) and estrogen receptor reverse (CTGCAGGAAAGGCGACAGC; TA 60°C for 40 cycles); and tenascin forward (TCCTGCTGACTGTCACAATC) and tenascin reverse (TGCTCACATACACATTTGCC; TA 60°C for 30 cycles).
RS-PCR
To determine the integration sites of the HPV16 E6/E7–expressing vector, an RS-PCR assay was used (Sarkar et al., 1993; Ragin et al., 2004) The HPV-specific primers were modified to locate to the E6/E7 ORF, and additional RS oligonucleotides were added to the assay. In brief, genomic DNA was extracted from each cell line using the DNeasy tissue kit (QIAGEN). Previously used HPV-specific primers (Gudjonsson et al., 2002b) were supplemented with the HPV765-24D/HPV790-25D primer sets (Thorland et al., 2000). The RS oligonucleotides contain a T7 phage promoter, 10 random nucleotides, and a specific RS recognition sequence.
Amplification of vector-genome hybrid sequence was used by a seminested PCR reaction. The PCR reactions were performed using the Expand High Fidelity PCR System (Roche) in a 20-μl volume using 1 U polymerase enzyme, 1× PCR buffer, 200 μM deoxynucleotide triphosphate, 2 pmol HPV primer, and 20 pmol RS oligonucleotide primer. 1 μl from the first round of PCR was used as a template for the second round of amplification. Cycling conditions for the first round of PCR were 94°C for 2 min followed by 30 cycles of 94°C for 30 s, annealing for 30 s at 65°C descending 0.5°C each cycle, and extension at 72°C for 2 min. This was followed by 15 more cycles at a fixed TA of 55°C. The conditions for the second round of PCR were similar, and products were run on agarose gels, bands of interest were cut out, and specific products were extracted using a QIAquick Gel Extraction kit (QIAGEN). The presence of an E6- or E7-specific sequence in the selected products was verified by PCR reamplification using internal E6/E7 primers. Sequencing using a genetic analyzer (ABI PRISM 310; Applied Biosystems) was performed as previously described (Rønnov-Jessen et al., 2002).
Online supplemental material
Fig. S1 shows a more detailed characterization of the ductal stem cell zone. Fig. S2 shows that clonal colonies from ducts, in contrast to lobule-derived colonies, are multipotent. Fig. S3 shows that SSEA-4hi cells cosort with the Lin−CD49f+EpCAMhi population and that these cells give rise to multipotent clonal colonies. Fig. S4 shows that the stemlike cells (+/+E6/E7) express surrogate stem cell markers and are bipotent. A flow chart describing the isolation of one stemlike cell and three lineage-restricted progenitor cells is also shown. Table S1 shows that bypass of senescence maintains the differentiation repertoire of target cells with respect to a panel of markers for differentiated luminal and myoepithelial cells. Supplemental materials and methods provides further details about cell lines and clones, FACS analysis and cloning, immunohistochemistry and cytochemistry, and RNA isolation and RT-PCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200611114/DC1.
Supplementary Material
Acknowledgments
We thank Tove Marianne Lund for expert technical assistance, Naja Slot Bundgaard and Mimi Birkelund for help with immunostainings and RT-PCR, and Keld Ottosen for photographic assistance. We also thank Søllerød Privathospital, the Private Clinic, and Dr. A. Aasted for providing the normal breast biopsy material.
This work was supported by the Danish Cancer Society, Dansk Kræftforskningsfond, Danish Research Council, Fru Astrid Thaysens Legat for Lægevidenskabelig Grundforskning, The Meyer Foundation, The Toyota Foundation, the Novo Nordic Foundation, the European Commission Research Directorates (contract HPRN-CT-2002-00246), Bernhard Rasmussen og hustru Meta Rasmussens fond (to L. Rønnov-Jessen), the US National Cancer Institute (grant CA-64786 to M.J. Bissell and O.W. Petersen), and the Office of Biological and Environmental Research (OBER) of the US Department of Energy (grant to M.J. Bissell). M.J. Bissell is the recipient of an Innovator award from the US Department of Defense and a Distinguished Fellow award from the OBER office of the US Department of Energy. M.A. LaBarge was supported by a postdoctoral fellowship from the American Cancer Society. T. Gudjonsson is supported by the Research Grant of Excellence from the Icelandic Research Council.
R. Villadsen and A.J. Fridriksdottir contributed equally to this paper.
Abbreviations used in this paper: HPV, human papilloma virus; lrECM, laminin-rich ECM; RS, restriction site; SSEA-4, stage-specific embryonal antigen-4; TDLU, terminal duct lobular unit.
References
- Akimov, S.S., A. Ramezani, T.S. Hawley, and R.G. Hawley. 2005. Bypass of senescence, immortalization, and transformation of human hematopoietic progenitor cells. Stem Cells. 23:1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj, M., M.S. Wicha, A. Benito-Hernandez, S.J. Morrison, and M.F. Clarke. 2003. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 100:3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band, V., D. Zajchowski, V. Kulesa, and R. Sager. 1990. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proc. Natl. Acad. Sci. USA. 87:463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbod, F., and J.M. Rosen. 2005. Will cancer stem cells provide new therapeutic targets? Carcinogenesis. 26:703–711. [DOI] [PubMed] [Google Scholar]
- Belair, C.D., T.R. Yeager, P.M. Lopez, and C.A. Reznikoff. 1997. Telomerase activity: a biomarker of cell proliferation, not malignant transformation. Proc. Natl. Acad. Sci. USA. 94:13677–13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitah, S.A., M. Frye, M. Glogauer, and F.M. Watt. 2005. Stem cell depletion through epidermal deletion of Rac1. Science. 309:933–935. [DOI] [PubMed] [Google Scholar]
- Blanpain, C., W.E. Lowry, A. Geoghegan, L. Polak, and E. Fuchs. 2004. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 118:635–648. [DOI] [PubMed] [Google Scholar]
- Bocker, W., R. Moll, C. Poremba, R. Holland, P.J. Van Diest, P. Dervan, H. Burger, D. Wai, R. Ina Diallo, B. Brandt, et al. 2002. Common adult stem cells in the human breast give rise to glandular and myoepithelial cell lineages: a new cell biological concept. Lab. Invest. 82:737–746. [DOI] [PubMed] [Google Scholar]
- Boulanger, C.A., K.U. Wagner, and G.H. Smith. 2005. Parity-induced mouse mammary epithelial cells are pluripotent, self-renewing and sensitive to TGF-beta1 expression. Oncogene. 24:552–560. [DOI] [PubMed] [Google Scholar]
- Buono, K.D., G.W. Robinson, C. Martin, S. Shi, P. Stanley, K. Tanigaki, T. Honjo, and L. Hennighausen. 2006. The canonical Notch/RBP-J signaling pathway controls the balance of cell lineages in mammary epithelium during pregnancy. Dev. Biol. 293:565–580. [DOI] [PubMed] [Google Scholar]
- Burger, P.E., X. Xiong, S. Coetzee, S.N. Salm, D. Moscatelli, K. Goto, and E.L. Wilson. 2005. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proc. Natl. Acad. Sci. USA. 102:7180–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff, R.D., and S.R. Wellings. 1999. The comparative pathology of human and mouse mammary glands. J. Mammary Gland Biol. Neoplasia. 4:105–122. [DOI] [PubMed] [Google Scholar]
- Chenard, M., J.R. Basque, P. Chailler, E. Tremblay, J.F. Beaulieu, and D. Menard. 2000. Expression of integrin subunits correlates with differentiation of epithelial cell lineages in developing human gastric mucosa. Anat. Embryol. (Berl.). 202:223–233. [DOI] [PubMed] [Google Scholar]
- Chepko, G., R. Slack, D. Carbott, S. Khan, L. Steadman, and R.B. Dickson. 2005. Differential alteration of stem and other cell populations in ducts and lobules of TGFalpha and c-Myc transgenic mouse mammary epithelium. Tissue Cell. 37:393–412. [DOI] [PubMed] [Google Scholar]
- Clarke, R.B. 2005. Isolation and characterization of human mammary stem cells. Cell Prolif. 38:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, R.B., K. Spence, E. Anderson, A. Howell, H. Okano, and C.S. Potten. 2005. A putative human breast stem cell population is enriched for steroid receptor-positive cells. Dev. Biol. 277:443–456. [DOI] [PubMed] [Google Scholar]
- Dontu, G., W.M. Abdallah, J.M. Foley, K.W. Jackson, M.F. Clarke, M.J. Kawamura, and M.S. Wicha. 2003. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 17:1253–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravida, S., R. Pal, A. Khanna, S.P. Tipnis, G. Ravindran, and F. Khan. 2005. The transdifferentiation potential of limbal fibroblast-like cells. Brain Res. Dev. Brain Res. 160:239–251. [DOI] [PubMed] [Google Scholar]
- Engvall, E., D. Earwicker, T. Haaparanta, E. Rouslahti, and J.R. Sanes. 1990. Distribution and isolation of four laminin variants: tissue restricted distribution of heterotrimers assembled from five different subunits. Cell Regul. 1:731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrieres, G., M. Cuny, J. Simony-Lafontaine, J. Jacquemier, C. Rouleau, F. Guilleux, J. Grenier, P. Rouanet, H. Pujol, P. Jeanteur, and C. Escot. 1997. Variation of bcl-2 expression in breast ducts and lobules in relation to plasma progesterone levels: overexpression and absence of variation in fibroadenomas. J. Pathol. 183:204–211. [DOI] [PubMed] [Google Scholar]
- Feuerhake, F., W. Sigg, E.A. Hofter, T. Dimpfl, and U. Welsch. 2000. Immunohistochemical analysis of Bcl-2 and Bax expression in relation to cell turnover and epithelial differentiation markers in the non-lactating human mammary gland epithelium. Cell Tissue Res. 299:47–58. [DOI] [PubMed] [Google Scholar]
- Fleischmajer, R., K. Kuroda, A. Utani, E. Douglas MacDonald, J.S. Perlish, E. Arikawa-Hirasawa, K. Sekiguchi, N. Sanzen, R. Timpl, and Y. Yamada. 2000. Differential expression of laminin alpha chains during proliferative and differentiation stages in a model for skin morphogenesis. Matrix Biol. 19:637–647. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., T. Tumbar, and G. Guasch. 2004. Socializing with the neighbors: stem cells and their niche. Cell. 116:769–778. [DOI] [PubMed] [Google Scholar]
- Gonzalez, M.A., S.E. Pinder, G. Callagy, S.L. Vowler, L.S. Morris, K. Bird, J.A. Bell, R.A. Laskey, and N. Coleman. 2003. Minichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancer. J. Clin. Oncol. 21:4306–4313. [DOI] [PubMed] [Google Scholar]
- Grimm, S.L., W. Bu, M.A. Longley, D.R. Roop, Y. Li, and J.M. Rosen. 2006. Keratin 6 is not essential for mammary gland development. Breast Cancer Res. 8:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson, T., L. Rønnov-Jessen, R. Villadsen, F. Rank, M.J. Bissell, and O.W. Petersen. 2002. a. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J. Cell Sci. 115:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson, T., R. Villadsen, H.L. Nielsen, L. Rønnov-Jessen, M.J. Bissell, and O.W. Petersen. 2002. b. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 16:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, B.A., and B.A. Gusterson. 2000. Human breast development. J. Mammary Gland Biol. Neoplasia. 5:119–137. [DOI] [PubMed] [Google Scholar]
- Hudson, D.L., M. O'Hare, F.M. Watt, and J.R. Masters. 2000. Proliferative heterogeneity in the human prostate: evidence for epithelial stem cells. Lab. Invest. 80:1243–1250. [DOI] [PubMed] [Google Scholar]
- Hudson, D.L., A.T. Guy, P. Fry, M.J. O'Hare, F.M. Watt, and J.R. Masters. 2001. Epithelial cell differentiation pathways in the human prostate: identification of intermediate phenotypes by keratin expression. J. Histochem. Cytochem. 49:271–278. [DOI] [PubMed] [Google Scholar]
- Ito, M., Y. Liu, Z. Yang, J. Nguyen, F. Liang, R.J. Morris, and G. Cotsarelis. 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11:1351–1354. [DOI] [PubMed] [Google Scholar]
- Kang, K.-S., I. Morita, A. Cruz, Y.J. Jeon, J.E. Trosko, and C.-C. Chang. 1997. Expression of estrogen receptors in a normal human breast epithelial cell type with luminal and stem cell characteristics and its neoplastically transformed cell lines. Carcinogenesis. 18:251–257. [DOI] [PubMed] [Google Scholar]
- Kao, C.-Y., K. Nomata, C.S. Oakley, C.W. Welsch, and C.-C. Chang. 1995. Two types of normal human breast epithelial cells derived from reduction mammoplasty: Phenotypic characterization and response to SV40 transfection. Carcinogenesis. 16:531–538. [DOI] [PubMed] [Google Scholar]
- Kim, C.F., E.L. Jackson, A.E. Woolfenden, S. Lawrence, I. Babar, S. Vogel, D. Crowley, R.T. Bronson, and T. Jacks. 2005. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 121:823–835. [DOI] [PubMed] [Google Scholar]
- Kingsbury, S.R., M. Loddo, T. Fanshawe, E.C. Obermann, A.T. Prevost, K. Stoeber, and G.H. Williams. 2005. Repression of DNA replication licensing in quiescence is independent of geminin and may define the cell cycle state of progenitor cells. Exp. Cell Res. 309:56–67. [DOI] [PubMed] [Google Scholar]
- Klimanskaya, I., Y. Chung, L. Meisner, J. Johnson, M.D. West, and R. Lanza. 2005. Human embryonic stem cells derived without feeder cells. Lancet. 365:1636–1641. [DOI] [PubMed] [Google Scholar]
- Laine, M., I. Virtanen, T. Salo, and Y.T. Konttinen. 2004. Segment-specific but pathologic laminin isoform profiles in human labial salivary glands of patients with Sjogren's syndrome. Arthritis Rheum. 50:3968–3973. [DOI] [PubMed] [Google Scholar]
- Lane, E.B. 1982. Monoclonal antibodies provide specific intramolecular markers for the study of epithelial tonofilament organization. J. Cell Biol. 92:665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg, J., U.B. Jensen, S. Broad, I. Leigh, and F.M. Watt. 2003. Role of melanoma chondroitin sulphate proteoglycan in patterning stem cells in human interfollicular epidermis. Development. 130:6049–6063. [DOI] [PubMed] [Google Scholar]
- Lindvall, C., N.C. Evans, C.R. Zylstra, Y. Li, C.M. Alexander, and B.O. Williams. 2006. The Wnt signaling receptor Lrp5 is required for mammary ductal stem cell activity and Wnt1-induced tumorigenesis. J. Biol. Chem. 281:35081–35087. [DOI] [PubMed] [Google Scholar]
- Locke, M., M. Heywood, S. Fawell, and I.C. Mackenzie. 2005. Retention of intrinsic stem cell hierarchies in carcinoma-derived cell lines. Cancer Res. 65:8944–8950. [DOI] [PubMed] [Google Scholar]
- Luna-More, S., B. Weil, D. Bautista, E. Garrido, P. Florez, and C. Martinez. 2004. Bcl-2 protein in normal, hyperplastic and neoplastic breast tissues. A metabolite of the putative stem-cell subpopulation of the mammary gland. Histol. Histopathol. 19:457–463. [DOI] [PubMed] [Google Scholar]
- Maatta, M., I. Virtanen, R. Burgeson, and H. Autio-Harmainen. 2001. Comparative analysis of the distribution of laminin chains in the basement membranes in some malignant epithelial tumors: the alpha1 chain of laminin shows a selected expression pattern in human carcinomas. J. Histochem. Cytochem. 49:711–726. [DOI] [PubMed] [Google Scholar]
- Mackenzie, I.C. 2006. Stem cell properties and epithelial malignancies. Eur. J. Cancer. 42:1204–1212. [DOI] [PubMed] [Google Scholar]
- Maslov, A.Y., T.A. Barone, R.J. Plunkett, and S.C. Pruitt. 2004. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. J. Neurosci. 24:1726–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, I. 1995. Proliferative potential of different keratinocytes of plucked human hair follicles. J. Invest. Dermatol. 105:14–21. [DOI] [PubMed] [Google Scholar]
- Moore, K.A., and I.R. Lemischka. 2006. Stem cells and their niches. Science. 311:1880–1885. [DOI] [PubMed] [Google Scholar]
- Mori, T., T. Kiyono, H. Imabayashi, Y. Takeda, K. Tsuchiya, S. Miyoshi, H. Makino, K. Matsumoto, H. Saito, S. Ogawa, et al. 2005. Combination of hTERT and bmi-1, E6, or E7 induces prolongation of the life span of bone marrow stromal cells from an elderly donor without affecting their neurogenic potential. Mol. Cell. Biol. 25:5183–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare, M.J., M.G. Ormerod, P. Monaghan, B.E. Lane, and B.A. Gusterson. 1991. Characterization in vitro of luminal and myoepithelial cells isolated from the human mammary gland by cell sorting. Differentiation. 46:209–221. [DOI] [PubMed] [Google Scholar]
- Ohyama, M., A. Terunuma, C.L. Tock, M.F. Radonovich, C.A. Pise-Masison, S.B. Hopping, J.N. Brady, M.C. Udey, and J.C. Vogel. 2006. Characterization and isolation of stem cell-enriched human hair follicle bulge cells. J. Clin. Invest. 116:249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, T., T. Aoyama, T. Nakayama, T. Nakamata, T. Hosaka, K. Nishijo, T. Nakamura, T. Kiyono, and J. Toguchida. 2002. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 295:354–361. [DOI] [PubMed] [Google Scholar]
- Osyczka, A.M., U. Noth, J. O'Connor, E.J. Caterson, K. Yoon, K.G. Danielson, and R.S. Tuan. 2002. Multilineage differentiation of adult human bone marrow progenitor cells transduced with human papilloma virus type 16 E6/E7 genes. Calcif. Tissue Int. 71:447–458. [DOI] [PubMed] [Google Scholar]
- Péchoux, C., T. Gudjonsson, L. Rønnov-Jessen, M.J. Bissell, and O.W. Petersen. 1999. Human mammary luminal epithelial cells contain progenitors to myoepithelial cells. Dev. Biol. 206:88–99. [DOI] [PubMed] [Google Scholar]
- Perou, C.M., T. Sorlie, M.B. Eisen, M. van de Rijn, S.S. Jeffrey, C.A. Rees, J.R. Pollack, D.T. Ross, H. Johnsen, L.A. Akslen, et al. 2000. Molecular portraits of human breast tumours. Nature. 406:747–752. [DOI] [PubMed] [Google Scholar]
- Petersen, O.W., and B. van Deurs. 1988. Growth factor control of myoepithelial-cell differentiation in cultures of human mammary gland. Differentiation. 39:197–215. [DOI] [PubMed] [Google Scholar]
- Petersen, O.W., B. van Deurs, K.V. Nielsen, M.W. Madsen, I. Laursen, I. Balslev, and P. Briand. 1990. Differential tumorigenicity of two autologous human breast carcinoma cell lines, HMT-3909S1 and HMT-3909S8, established in serum-free medium. Cancer Res. 50:1257–1270. [PubMed] [Google Scholar]
- Petersen, O.W., L. Rønnov-Jessen, A.R. Howlett, and M.J. Bissell. 1992. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc. Natl. Acad. Sci. USA. 89:9064–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten, C.S., R.J. Watson, G.T. Williams, S. Tickle, S.A. Roberts, M. Harris, and A. Howell. 1988. The effect of age and menstrual cycle upon proliferative activity of the normal human breast. Br. J. Cancer. 58:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragin, C.C., S.C. Reshmi, and S.M. Gollin. 2004. Mapping and analysis of HPV16 integration sites in a head and neck cancer cell line. Int. J. Cancer. 110:701–709. [DOI] [PubMed] [Google Scholar]
- Reznikoff, C.A., C. Belair, E. Savelieva, Y. Zhai, K. Pfeifer, T. Yeager, K.J. Thompson, S. DeVries, C. Bindley, M.A. Newton, et al. 1994. Long-term genome stability and minimal genotypic and phenotypic alterations in HPV16 E7-, but not E6-, immortalized human uroepithelial cells. Genes Dev. 8:2227–2240. [DOI] [PubMed] [Google Scholar]
- Roecklein, B.A., and B. Torok-Storb. 1995. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 85:997–1005. [PubMed] [Google Scholar]
- Rønnov-Jessen, L., and O.W. Petersen. 1993. Induction of α-smooth muscle actin by transforming growth factor-β1 in quiescent human breast gland fibroblasts. Implications for myofibroblast generation in breast neoplasia. Lab. Invest. 68:696–707. [PubMed] [Google Scholar]
- Rønnov-Jessen, L., J.E. Celis, B. van Deurs, and O.W. Petersen. 1992. A fibroblast-associated antigen: characterization in fibroblasts and immunoreactivity in smooth muscle differentiated stromal cells. J. Histochem. Cytochem. 40:475–486. [DOI] [PubMed] [Google Scholar]
- Rønnov-Jessen, L., R. Villadsen, J.C. Edwards, and O.W. Petersen. 2002. Differential expression of a chloride intracellular channel gene, CLIC4, in transforming growth factor-beta1-mediated conversion of fibroblasts to myofibroblasts. Am. J. Pathol. 161:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudland, P.S. 1987. Stem cells and the development of mammary cancers in experimental rats and humans. Cancer Metastasis Rev. 6:55–83. [DOI] [PubMed] [Google Scholar]
- Russo, J., and I.H. Russo. 2004. Development of the human breast. Maturitas. 49:2–15. [DOI] [PubMed] [Google Scholar]
- Sarkar, G., R.T. Turner, and M.E. Bolander. 1993. Restriction-site PCR: a direct method of unknown sequence retrieval adjacent to a known locus by using universal primers. PCR Methods Appl. 2:318–322. [DOI] [PubMed] [Google Scholar]
- Schmelz, M., R. Moll, U. Hesse, A.R. Prasad, J.A. Gandolfi, S.R. Hasan, M. Bartholdi, and A.E. Cress. 2005. Identification of a stem cell candidate in the normal human prostate gland. Eur. J. Cell Biol. 84:341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton, M., F. Vaillant, K.J. Simpson, J. Stingl, G.K. Smyth, M.L. Asselin-Labat, L. Wu, G.J. Lindeman, and J.E. Visvader. 2006. Generation of a functional mammary gland from a single stem cell. Nature. 439:84–88. [DOI] [PubMed] [Google Scholar]
- Shetty, A., M. Loddo, T. Fanshawe, A.T. Prevost, R. Sainsbury, G.H. Williams, and K. Stoeber. 2005. DNA replication licensing and cell cycle kinetics of normal and neoplastic breast. Br. J. Cancer. 93:1295–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Assmann, P., B. Duclos, V. Orian-Rousseau, C. Arnold, C. Mathelin, E. Engvall, and M. Kedinger. 1994. Differential expression of laminin isoforms and alpha 6-beta 4 integrin subunits in the developing human and mouse intestine. Dev. Dyn. 201:71–85. [DOI] [PubMed] [Google Scholar]
- Smalley, M.J., and R.B. Clarke. 2005. The mammary gland “side population”: a putative stem/progenitor cell marker? J. Mammary Gland Biol. Neoplasia. 10:37–47. [DOI] [PubMed] [Google Scholar]
- Smith, G.H., T. Mehrel, and D.R. Roop. 1990. Differential keratin gene expression in developing, differentiating, preneoplastic, and neoplastic mouse mammary epithelium. Cell Growth Differ. 1:161–170. [PubMed] [Google Scholar]
- Sonnenberg, A., H. Daams, M.A. van der Valk, J. Hilkens, and J. Hilgers. 1986. Development of mouse mammary gland: identification of stages in differentiation of luminal and myoepithelial cells using monoclonal antibodies and polyvalent antiserum against keratin. J. Histochem. Cytochem. 34:1037–1046. [DOI] [PubMed] [Google Scholar]
- Sotgia, F., T.M. Williams, A.W. Cohen, C. Minetti, R.G. Pestell, and M.P. Lisanti. 2005. Caveolin-1-deficient mice have an increased mammary stem cell population with upregulation of Wnt/beta-catenin signaling. Cell Cycle. 4:1808–1816. [DOI] [PubMed] [Google Scholar]
- Stasiak, P.C., P.E. Purkis, I.M. Leigh, and E.B. Lane. 1989. Keratin 19: predicted amino acid sequence and broad tissue distribution suggest it evolved from keratinocyte keratins. J. Invest. Dermatol. 92:707–716. [DOI] [PubMed] [Google Scholar]
- Stingl, J., P. Eirew, I. Ricketson, M. Shackleton, F. Vaillant, D. Choi, H.I. Li, and C.J. Eaves. 2006. a. Purification and unique properties of mammary epithelial stem cells. Nature. 439:993–997. [DOI] [PubMed] [Google Scholar]
- Stingl, J., A. Raouf, P. Eirew, and C.J. Eaves. 2006. b. Deciphering the mammary epithelial cell hierarchy. Cell Cycle. 5:1519–1522. [DOI] [PubMed] [Google Scholar]
- Tai, M.H., C.C. Chang, L.K. Olson, and J.E. Trosko. 2005. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 26:495–502. [DOI] [PubMed] [Google Scholar]
- Taylor-Papadimitriou, J., E.B. Lane, and S.E. Chang. 1983. Cell lineages and interactions in neoplastic expression in the human breast. In Understanding Breast Cancer: Clinical and Laboratory Concepts. M.A. Rich, J.C. Hager, and P. Furmanski, editors. Dekker, New York. 215–246.
- Thorland, E.C., S.L. Myers, D.H. Persing, G. Sarkar, R.M. McGovern, B.S. Gostout, and D.I. Smith. 2000. Human papillomavirus type 16 integrations in cervical tumors frequently occur in common fragile sites. Cancer Res. 60:5916–5921. [PubMed] [Google Scholar]
- Trosko, J.E., C.C. Chang, B.L. Upham, and M.H. Tai. 2004. Ignored hallmarks of carcinogenesis: stem cells and cell-cell communication. Ann. NY Acad. Sci. 1028:192–201. [DOI] [PubMed] [Google Scholar]
- Tsai, Y.C., Y. Lu, P.W. Nichols, G. Zlotnikov, P.A. Jones, and H.S. Smith. 1996. Contiguous patches of normal human mammary epithelium derived from a single stem cell: implications for breast carcinogenesis. Cancer Res. 56:402–404. [PubMed] [Google Scholar]
- Tsujimura, A., Y. Koikawa, S. Salm, T. Takao, S. Coetzee, D. Moscatelli, E. Shapiro, H. Lepor, T.T. Sun, and E.L. Wilson. 2002. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. J. Cell Biol. 157:1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar, T., G. Guasch, V. Greco, C. Blanpain, W.E. Lowry, M. Rendl, and E. Fuchs. 2004. Defining the epithelial stem cell niche in skin. Science. 303:359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.D., C.C. Leow, J. Zha, Z. Tang, Z. Modrusan, F. Radtke, M. Aguet, F.J. de Sauvage, and W.Q. Gao. 2006. Notch signaling is required for normal prostatic epithelial cell proliferation and differentiation. Dev. Biol. 290:66–80. [DOI] [PubMed] [Google Scholar]
- Watt, F.M. 1998. Epidermal stem cells: markers, patterning and control of stem cell fate. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353:831–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, W.A., M.S. Chen, F. Behbod, and J.M. Rosen. 2005. On mammary stem cells. J. Cell Sci. 118:3585–3594. [DOI] [PubMed] [Google Scholar]
- Yang, J.S., R.M. Lavker, and T.T. Sun. 1993. Upper human hair follicle contains a subpopulation of keratinocytes with superior in vitro proliferative potential. J. Invest. Dermatol. 101:652–659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.