Abstract
Exchanging the glycophosphatidylinositol (GPI) anchor signal sequence of neural cell adhesion molecule (NCAM) for the signal sequence of carcinoembryonic antigen (CEA) generates a mature protein with NCAM external domains but CEA-like tumorigenic activity. We hypothesized that this resulted from the presence of a functional specificity signal within this sequence and generated CEA/NCAM chimeras to identify this signal. Replacing the residues (GLSAG) 6–10 amino acids downstream of the CEA anchor addition site with the corresponding NCAM residues resulted in GPI-anchored proteins lacking the CEA-like biological functions of integrin modulation and differentiation blockage. Transferring this region from CEA into NCAM in conjunction with the upstream proline (PGLSAG) was sufficient to specify the addition of the CEA anchor. Therefore, this study identifies a novel specificity signal consisting of six amino acids located within the GPI anchor attachment signal, which is necessary and sufficient to specify the addition of a particular functional GPI anchor and, thereby, the ultimate function of the mature protein.
Introduction
Glycophosphatidylinositol (GPI) anchorage is a common feature of surface proteins that leads to membrane raft localization. The observation of different functional GPI anchors (Screaton et al., 2000; Nicholson and Stanners, 2006) as well as the fact that different rafts show markedly different lipid and protein profiles (Madore et al., 1999; Wang et al., 2002; Brugger et al., 2004) implies the existence of a heterogeneous set of anchors and matching rafts. Anchor addition is determined by the GPI anchor signal sequence, which consists of a set of small amino acids at the site of anchor addition (the ω site) followed by a hydrophilic spacer and ending in a hydrophobic stretch (Low, 1989). Cleavage of this signal sequence occurs in the ER before the addition of an anchor with conserved central components (Low, 1989) but with variable peripheral moieties (Homans et al., 1988).
Carcinoembryonic antigen (CEA) is a GPI-anchored protein, whereas the closely related CEACAM1 (CEA-related cell adhesion molecule 1 [CC1]) contains a transmembrane (TM) domain. In vitro, both proteins mediate intercellular adhesion (Benchimol et al., 1989; Rojas et al., 1990), but CEA, not CC1, blocks cellular differentiation (Eidelman et al., 1993) and inhibits the apoptotic process of anoikis (Ordonez et al., 2000; Soeth et al., 2001). Exchanging the membrane anchors of these proteins results in a TM version of CEA that does not show CEA-like activity and a GPI-anchored CC1-like protein that now exhibits CEA-like properties, demonstrating the importance of the membrane anchor (Screaton et al., 2000). Replacing the GPI anchor signal sequence of neural cell adhesion molecule (NCAM) for that of CEA results in a functionally CEA-like protein (NCB), demonstrating the existence of functionally specific anchors whose addition is determined by a particular signal sequence (Screaton et al., 2000). The sole role of the external domains is to mediate self-binding and, thereby, concentration-dependent clustering (Taheri et al., 2003; Camacho-Leal et al., 2007), as external domain mutations that disrupt the self-binding of CEA abrogate CEA function, whereas irrelevant self-binding external domains (such as that of NCAM in NCB) suffice for function.
Because the amino acid sequence of various GPI anchor signal sequences radically affects protein function, we hypothesized that a signal existed within the GPI anchor signal sequence specifying the addition of a particular functional GPI anchor. Chimeras were generated by exchanging fragments of the CEA and NCAM GPI anchor signal sequences and were tested for CEA-like biological properties. We identify a specificity signal consisting of five amino acids that is necessary and sufficient in conjunction with an upstream proline for addition of the CEA-specific GPI anchor.
Results and discussion
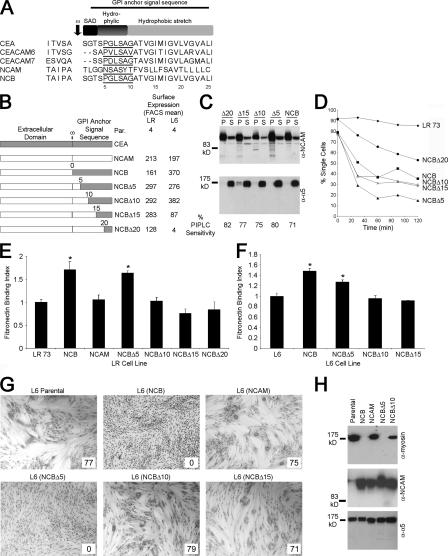
Although the primary sequences of the GPI anchor signal sequences of CEA and CEACAM6 are very similar, mirroring their identical tumorigenic functions, that of NCAM differs greatly (Fig. 1 A). The signal sequence from CEA is capable on its own of specifying the addition of the CEA anchor, so chimeras were generated reducing (in five–amino acid increments) the CEA-derived sequence in NCB to localize the sequence responsible for this effect (Fig. 1 B). These chimeras were tested for biological activity in the CHO-derived LR-73 and the rat myoblast L6 cell lines, although NCBΔ20 was not expressed in L6 transfectants (Fig. 1 B). The sensitivity of these proteins to phosphatidylinositol PLC (PIPLC) and their insolubility in cold Triton X-100, with most of each protein present in the insoluble fraction, confirmed GPI anchorage (Fig. 1 C; Screaton et al., 2000). LR transfectants showed strong intercellular adhesive ability (Fig. 1 D), which is indicative of the retention of the self-binding activity of their external domains (Eidelman et al., 1993; Taheri et al., 2003), although NCBΔ20 adhered somewhat less, likely because of its lower expression level.
Figure 1.
Reducing the CEA-derived sequence in NCB. (A) The amino acid sequences of the GPI anchor signal sequences of CEA, CEACAM6, CEACAM7, NCAM, and the NCAM-CEA chimera NCB. SAD, small amino acid domain; ω, GPI anchor attachment site. The underlined sequences represent the key residues determined in this study. (B) Representation of the CEA-NCAM chimeras, with the GPI anchor attachment site (ω) denoted. FACS means in arbitrary units for cell surface levels of NCAM and chimeric proteins are indicated. (C) The chimeric constructs are GPI anchored on LR transfectants; Western blots of Triton X-100 solubility assays (top) demonstrated that the majority of each chimera localized in the pellet (P) fraction. The integrin α5 was a lysis and gel-loading control. The proteins were also PIPLC sensitive (bottom), and the percent decrease in surface levels are given. (D) LR transfectants showed increased intercellular adhesive ability compared with the parental cell line. (E) Binding of LR parental and transfectant cells to immobilized Fn. (F) Binding of L6 transfectant cells to Fn. (E and F) Values represent means relative to the parental cell ± SEM (error bars) for three independent experiments. *, P < 0.01. (G) Morphological differentiation of L6 parental and transfected myoblasts. Inset values indicate the fusion index expressed as a percentage. (H) Differentiated cultures were lysed, and 5 μg was probed for the biochemical differentiation marker myosin.
CEA and NCB but not NCAM alter the activity of integrin α5β1 (Nicholson and Stanners, 2006; Ordonez et al., 2007) and block differentiation (Eidelman et al., 1993; Screaton et al., 2000), which are characteristics used to determine which chimeras retained the functional activities conferred by the CEA GPI anchor. LR transfectants were tested for binding to the major α5β1 ligand fibronectin (Fn; Fig. 1 E), with NCB expression significantly increasing binding compared with NCAM (P < 0.01). Replacing the first five CEA-derived amino acids of NCB with the equivalent NCAM residues had no effect on CEA-like function (P < 0.005 vs. NCAM), but replacing 10 residues resulted in a protein (NCBΔ10) that no longer altered binding. In L6 myoblasts, NCBΔ5 had a similar effect on binding to Fn compared with NCB (P < 0.005; Fig. 1 F), whereas NCBΔ10 transfectants lost this ability completely. NCB expression completely blocks L6 morphological differentiation, whereas NCAM transfectants differentiate readily to form large multinucleated myotubes (Screaton et al., 2000). NCBΔ5 expression also blocked differentiation, whereas NCBΔ10-expressing cells fused substantially, which is comparable with the parental and NCAM controls (all fused between 75 and 80%; Fig. 1 G). NCAM and NCBΔ10 transfectants up-regulated myosin, a biochemical differentiation marker, but NCB and NCBΔ5 transfectants did not (Fig. 1 H). Thus, addition of the CEA-specific GPI anchor is determined by the residues that differ from NCBΔ5 to NCBΔ10 (GLSAG), as their substitution with the corresponding NCAM amino acids (SASYT) abrogated the CEA-like biological function.
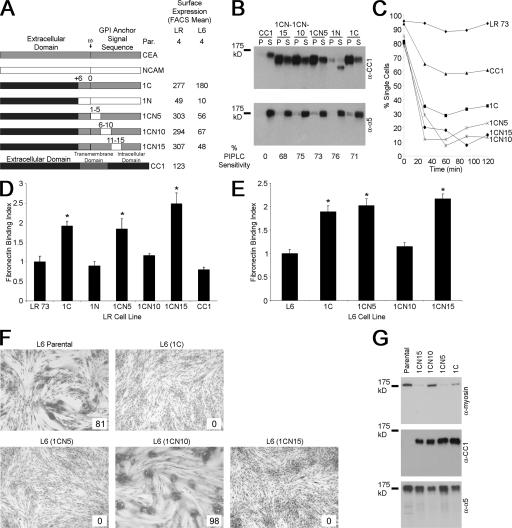
Five amino acid stretches in the CEA GPI anchor signal sequence were next replaced with the corresponding NCAM residues (Fig. 2 A). CC1-CEA (1C) chimeras were used because previous attempts to attach the NCAM anchor to CEA failed (Screaton et al., 2000), whereas the CC1-NCAM chimera (1N) is partially processed (Fig. 2 B), as seen previously in certain CC1 mutants (Naghibalhossaini and Stanners, 2004). These proteins were expressed at high levels with the exception of 1N (Fig. 2 A) and were GPI anchored (Fig. 2 B) and mediated strong intercellular adhesion (Fig. 2 C).
Figure 2.
Replacing five–amino acid stretches in the GPI anchor signal sequence of CEA. (A) Representation of the replacement of CEA amino acids with the corresponding NCAM residues. Expression of the various chimeras is given in terms of FACS mean fluorescence. (B) The CC1 chimeras were GPI anchored, as shown by insolubility in cold Triton X-100 (top, localization to the pellet [P] fraction). Note that the TM CC1 was found entirely in the soluble (S) fraction. This was confirmed by PIPLC treatment, in which surface levels decreased after treatment (bottom), except for CC1. (C) LR transfectants aggregated in suspension, demonstrating that the chimeras mediate intercellular adhesion. (D) Binding of LR cells to Fn. Values represent the mean relative binding compared with parental cells ± SEM (error bars) for four independent experiments (*, P < 0.001). (E) L6 transfectants showed similar effects on Fn binding as in LR. Values represent the mean ± SEM for five independent experiments relative to parental cells (*, P < 0.0001). (F) The effects of these chimeras on morphological differentiation of L6 cells. Percent fusion is given in the insets. (G) Probing 5 μg of differentiated cell lysates by immunoblotting for myosin up-regulation.
In LR cells, the expression of 1C caused a significant increase (P < 0.001) in cellular binding to Fn compared with parental cells (Fig. 2 D). Replacing the first five CEA amino acids (1CN5) or amino acids 11–15 (1CN15) downstream of the ω site resulted in proteins that were still active (P < 0.001); however, replacing amino acids 6–10 (1CN10) completely abrogated the increased binding. In L6 cells, 1C, 1CN5, and 1CN15 expression significantly increased binding to Fn (P < 0.001; Fig. 2 E), whereas 1CN10 transfectants bound the same as parental cells. Although the expression of 1CN10 was lower than 1C in L6 cells, 1CN5 and 1CN15 still showed increased binding despite having expression levels similar to 1CN10 (Fig. 2 A). The expression of 1C, 1CN5, and 1CN15 but not 1CN10 also strongly inhibited L6 morphological (Fig. 2 F) and biochemical (Fig. 2 G) differentiation. Replacing amino acid stretches shorter than five residues in this region had no effect on the binding of LR transfectants to Fn (Fig. S1 D, available at http://www.jcb.org/cgi/content/full/jcb.200701158/DC1). Therefore, these results confirm the importance of the residues GLSAG in determining the addition of the CEA anchor.
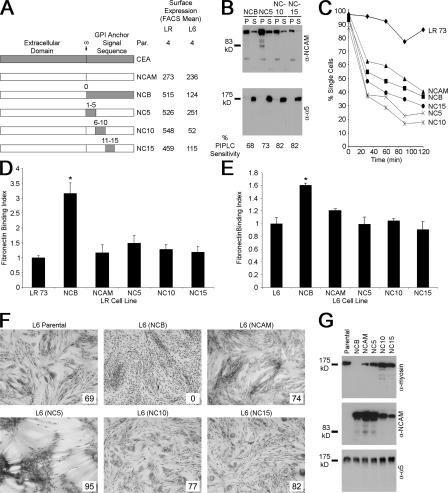
It was next examined whether this sequence was sufficient to confer CEA-like biological properties. CEA amino acids were inserted into NCAM at positions 1–5, 6–10, or 11–15 downstream of the ω site, with NC10 containing the GLSAG sequence (Fig. 3 A), resulting in GPI-anchored proteins (Fig. 3 B). Although the proteins mediated intercellular adhesion (Fig. 3 C), chimera expression did not result in increased Fn binding in either LR or L6 cells (Fig. 3, D and E). Differentiation of L6 cells was not blocked because morphological (Fig. 3 F) and biochemical (Fig. 3 G) differentiation was observed in these transfectants. Thus, inserting the sequence GLSAG into NCAM was insufficient to give CEA-like biological activities, suggesting a requirement for further CEA residues.
Figure 3.
Inserting five–amino acid sequences into NCAM is insufficient to create a protein with CEA-like properties. (A) Five–amino acid CEA sequences were inserted into the corresponding regions of NCAM. FACS means for LR and L6 transfectants are shown. (B) Insolubility as shown by localization to the pellet (P) fraction of all chimeras after cold Triton X-100 lysis demonstrated GPI anchorage (top). Cell surface levels of each protein also decreased after PIPLC treatment (bottom). (C) Adhesion assay demonstrating that these proteins mediated intercellular adhesion. (D) Binding to Fn by LR transfectants. (E) L6 transfectant binding to immobilized Fn. (D and E) Values relative to parental cells represent the mean ± SEM (error bars) from four independent experiments (*, P < 0.002). (F) Effect of chimera expression on L6 morphological differentiation; the fusion index, which is given as a percentage, is also provided in the insets. (G) Immunoblotting on 5 μg of lysate from differentiated cells to examine biochemical differentiation through myosin up-regulation.
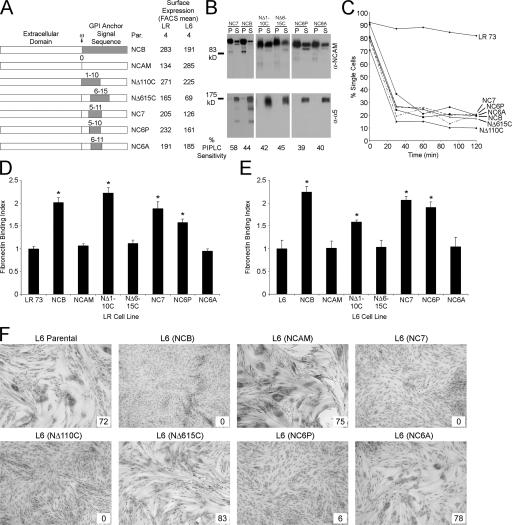
Therefore, larger amounts of the CEA-derived sequence were inserted into NCAM to determine the minimum sequence sufficient for specifying the addition of the CEA GPI anchor (Fig. 4 A). All chimeras contained the GLSAG sequence, with variable amounts of upstream and/or downstream CEA sequence, and the resulting proteins were GPI anchored (Fig. 4 B) and mediated intercellular adhesion (Fig. 4 C). When examined for the effects on LR binding to Fn, adding five upstream CEA amino acids (NΔ110C) but not five downstream amino acids (NΔ615C) resulted in increased binding (P < 0.0001; Fig. 4 D). Simply adding one CEA residue on each side of GLSAG (PGLSAGA; NC7) also produced increased cellular binding, suggesting, along with the NΔ110C result, that the upstream proline was required for CEA anchor addition. This was examined directly by generating constructs containing only the upstream proline (NC6P) or the downstream alanine (NC6A; Fig. 4 A). In LR cells, NC6P expression increased binding to Fn, whereas NC6A had no effect, confirming the importance of the proline (P < 0.0001; Fig. 4 D). These results were recapitulated in L6 cells, where only transfectants of NΔ110C, NC7, and NC6P showed a significant difference in binding to Fn compared with the parental cell line (P < 0.0001; Fig. 4 E).
Figure 4.
The upstream proline is required to confer CEA-like properties. (A) NCAM chimeras that were used to localize the CEA-specific signal. Mean FACS surface expression is indicated for LR and L6 transfectants. (B) The chimeric constructs were GPI anchored, as shown by cold Triton X-100 insolubility (top) and PIPLC sensitivity (bottom). (C) LR transfectants demonstrated that all proteins had the ability to mediate intercellular adhesion. (D) Binding by LR transfectants to immobilized Fn, in which values represent the mean ± SEM (error bars) for at least four independent experiments. (E) L6 cells showed similar effects on Fn binding. Values relative to parental cells represent the mean ± SEM for at least four independent experiments. (D and E) *, P < 0.0001. (F) Morphological differentiation of L6 transfectants. The fusion index, which is provided as a percentage, is given in the insets.
All chimeras containing the sequence PGLSAG blocked differentiation, whereas those lacking the proline fused similarly to NCAM transfectants (Fig. 4 F). Thus, inserting the sequence PGLSAG into the GPI anchor signal sequence of NCAM is sufficient to generate a protein with CEA-like biological properties, demonstrating a requirement for the presence of the proline. In CEA and all of these NCAM chimeras, this proline is a part of a G(X)XP sequence (Fig. 1 A). This consensus sequence has been shown to result in a kink in TM helices (Cordes et al., 2002) and can be suggested to serve a similar function in this GPI anchor signal sequence. The resulting altered structure may be important to determine the addition of a certain functional anchor. However, it should be noted that the lack of the proline can be overcome if a sufficient downstream CEA sequence is included (Fig. 1, NCBΔ5).
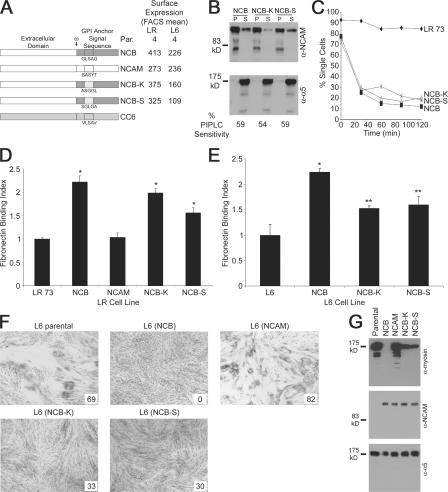
The sequence GLSAG was also randomly scrambled to give sequences of ASGGL (denoted NCB-K) and SGLGA (NCB-S; Fig. 5 A). It was hypothesized that a complete loss of biological function would be observed if there was a requirement for a particular amino acid sequence or a particular amino acid at a given position, whereas at least partial function should be retained if the signal resulted from a general characteristic of this stretch. These proteins were GPI anchored (Fig. 5 B) and promoted intercellular adhesion (Fig. 5 C). LR transfectants of both chimeras significantly increased binding to Fn compared with the NCAM cell line (P < 0.001; Fig. 5 D). However, both scrambled transfectants bound less than NCB, particularly NCB-S (P < 0.002). L6 transfectants showed altered binding compared with the parental cell line (P < 0.003 for NCB and P < 0.05 for NCB-K and NCB-S; Fig. 5 E), although transfectants of NCB-K and NCB-S again bound significantly (P < 0.05) less compared with the NCB transfectants. L6 (NCB-K) and L6 (NCB-S) cells differentiated substantially less than the parental or NCAM controls but did not show completely blocked morphological fusion (Fig. 5 F). Myosin up-regulation was seen in the NCB-K and NCB-S cell lines, which is contrary to NCB, although at lower levels than the NCAM transfectant (Fig. 5 G). Thus, scrambling this region results in an incomplete loss of function, indicating that the primary source of the signal is the overall region's characteristics, although this signal is maximized by the sequence PGLSAG.
Figure 5.
Scrambling the amino acid sequence in the identified key region. (A) Schematic of the constructs containing scrambled amino acids at positions 6–10, with the amino acid sequence shown underneath. Relative cell surface expression is shown. (B) GPI anchorage of these chimeric proteins was demonstrated by insolubility in cold Triton X-100 (top) and sensitivity to PIPLC treatment (bottom). (C) The scrambled constructs mediated intercellular adhesion in LR cells. (D) Binding to Fn by LR transfectants of the scrambled constructs. Values represent the mean ± SEM (error bars) for four independent experiments (*, P < 0.0005). (E) Effect of expression of the scrambled constructs on L6 Fn binding. Values represent the relative mean ± SEM for three independent experiments (*, P < 0.004; **, P < 0.03). (F) Morphological differentiation of L6 transfectants. The fusion index, which is provided as a percentage, is given in the insets. (G) Bio Biochemical differentiation in terms of myosin production as demonstrated by Western blotting on 5 μg of cellular lysate.
The signal for the addition of a GPI anchor consists of a set of small amino acids followed by a spacer and a hydrophobic region (Fig. 1 A; Coyne et al., 1993). Work on the bovine liver 5′-nucleotidase has demonstrated the requirement for particular lengths of both the spacer and the hydrophobic domain for proper processing (Furukawa et al., 1994, 1997). Differences in the efficiency of GPI anchor addition for various signal sequences suggest that these stretches are not processed identically (Chen et al., 2001). However, these previous studies have been concerned with the efficiency of anchor addition; this study is the first to demonstrate that the specificity of anchor addition is the result of a second signal within this sequence. We have previously demonstrated that the CEA GPI anchor signal sequence determines function and localization to a specific membrane raft despite being cleaved in the ER (Screaton et al., 2000; Nicholson and Stanners, 2006). This study was designed to establish the residues that are critical for this specification, with the demonstration that the amino acids PGLSAG in the hydrophilic region of the GPI signal sequence are necessary and sufficient for this effect. The identified sequence from CEA is fairly well conserved in CEACAM6 (PVLSAV) and CEACAM7 (PDLSAG; Fig. 1 A), which are proteins that show similar biological effects to CEA (Rojas et al., 1996; unpublished data), suggesting that it may have a similar role in determining the function of these proteins. Because GPI anchors from five other proteins did not show any of these effects when bound to the CC1 external domain (unpublished data), this amino acid set is quite specific.
This work has identified a novel signal within the GPI anchor signal sequence of CEA, which determines protein functionality. Studies using various GPI-anchored protein comparisons such as CEA and NCAM (Nicholson and Stanners, 2006), Thy-1 and the prion protein (Madore et al., 1999), and folate receptor and placental AP (Wang et al., 2002) have suggested that different GPI-anchored proteins exist in different microdomains on the cell surface. This distribution is important because switching the anchor and the subsequent distribution of NCAM to that of CEA is sufficient to radically alter its function (Screaton et al., 2000; Nicholson and Stanners, 2006). This occurs through association with particular rafts and their specific signaling elements, which determine the downstream tumorigenic effects of CEA (Camacho-Leal et al., 2007). Examining the GPI anchor signal sequences of various other proteins should further elucidate the specificity and importance of this signal in determining specific anchor addition and, ultimately, protein function.
It will also be important to characterize how this region is capable of determining the addition of a particular anchor. It is possible that this stretch of amino acids interacts directly with the GPI anchor precursor before the transamidation binding reaction, and only combinations that match structurally proceed enzymatically. Alternatively, the transamidase complex, which is composed of five different subunits, may play a direct role in this effect. One component, Gaa1p, has been suggested to recognize the signal sequence (Chen et al., 2003), whereas another, Gpi8p, functions as the enzymatic subunit (Ohishi et al., 2000). Either of these or one of the three complex components (PIG-S, PIG-T, and PIG-U) with currently unknown function could serve to bring together specific signal sequences and GPI anchors. This study demonstrates that a specific six–amino acid stretch of the GPI anchor signal sequence determines the addition of a particular functional anchor, which, in turn, can determine the ultimate function of the protein.
Materials and methods
Constructs
All chimeras were generated by PCR overlap extension. The ω site of NCAM remains unknown and was assigned to be A736 on the basis of sequence alignment with chicken NCAM, in which other potential anchor addition sites were not conserved between humans and chickens and, as such, are unlikely to serve as the ω site (Screaton et al., 2000). Constructs were generated using NCAM, NCB, or C1-C cDNA (note that the C1-C used in this study did not contain the I to F point mutation described in the original study [Screaton et al., 2000] and was called 1C to allow for differentiation between the two proteins) and the primers indicated in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200701158/DC1). Initial PCR reactions involved separate extensions using the CC1 or NCAM sense primer with the corresponding antisense chimera primer and the sense chimera primer with the antisense CEA or NCAM primer. These fragments were joined by overlap PCR using the CC1 or NCAM sense primer and the CEA or NCAM antisense primer. The resulting NCAM-like fragments were inserted into the EcoRI sites of NCAM (at positions 1,616 and 2,794) in the p91023b expression vector. The C1-C chimeras replaced the corresponding sequence in C1-C using the internal CC1 BamHI digestion site (at position 971) and the BamHI digestion site located in the polylinker region of the vector pEGFP-C2 (used as a cloning vector; BD Biosciences) and were subcloned into p91023b using flanking EcoRI sites.
Cell culture, FACS analysis, and transfection
CHO-derived LR-73 fibroblasts and rat L6 myoblasts were cultured as previously described (Nicholson and Stanners, 2006). Cell surface protein expression was determined by FACS analysis using the mouse mAbs J22, which recognizes CEA and CC1 (Zhou et al., 1993), and 123C3 (Santa Cruz Biotechnology, Inc.), which recognizes the NCAM external domain. Transfection and sorting for high expression was performed as previously described (Nicholson and Stanners, 2006).
Differentiation assays
L6 myoblasts were seeded at 104 cells/cm2 in 60-mm dishes in medium containing 10% FBS on day 0. 3 d later, the culture medium was changed to 2% horse serum. Myogenic differentiation was assayed 5 d after changing media either by staining with hematoxylin (Sigma-Aldrich) to assess fusion into multinuclear myotubes by light microscopy (Screaton et al., 2000) or by lysing cells and performing Western blots for myosin expression using mouse mAb 47A (De Giovanni et al., 1993). Photomicrographs of representative fields of stained cells were obtained at room temperature using a microscope (Eclipse E800; Nikon) and a 10× NA 0.30 Ph1 ∞/0.17 objective. Images were acquired with a digital camera (DXM1200; Nikon) and ACT-1 image acquisition software (Nikon). The fusion index was determined by counting the number of nuclei present in fused myotubes (taken as cells with three or more nuclei) and comparing this to the total number of nuclei in the field.
Triton X-100 solubility and PIPLC sensitivity assays
Assays were performed essentially as described previously (Screaton et al., 2000). For Triton X-100 solubility, cells were collected with PBSCE and resuspended in cold 1% Triton X-100 with protease inhibitors. Cells were syringed through a 27-gauge needle, incubated on ice for 15 min, and centrifuged at 13,000 g for 15 min. The supernatant fraction was removed, and the pellet was resuspended in the same volume as the supernatant. Partitioning between pellet and soluble fractions was assessed by immunoblotting; integrin α5 should be found in the supernatant (soluble) fraction and was used as a lysis control, with detection by a rabbit polyclonal anti-α5 (H-104; Santa Cruz Biotechnology, Inc.). For PIPLC sensitivity, monolayer cultures were incubated with 0.1 U bacterial PIPLC (Sigma-Aldrich) in a 1:1 solution of DME/PBS containing 0.2% BSA for 45 min at 37°C. Treated and control untreated cultures were then washed with PBS, rendered single-cell suspensions by light (0.063%) trypsin treatment, and were processed for FACS analysis. Percent sensitivity was determined as the percent decrease in mean fluorescence value (relative units) in the treated sample compared with the untreated control.
Adhesion assays
Adhesion assays were performed as previously described (Zhou et al., 1993). Cells were removed from culture flasks by light trypsin treatment (for CC1 external domain chimeras, which are insensitive to trypsin) or PBS citrate + 4 mM EDTA (PBSCE; for NCAM external domain chimeras, which are cleaved by trypsin) and resuspended at a concentration of 106 cells/ml in α-MEM containing 0.8% FBS and 10 μg/ml DNase I (Roche). Single-cell suspensions were obtained by syringing through a 27-gauge needle and were allowed to aggegrate at 37°C with stirring at 100 rpm using a magnetic stirring bar (Spinbar Micro Stir Bar; VWR International). Aliquots were removed at the indicated times, and the percentage of single cells was determined by a hemocytometer (Bright-Line; VWR International).
Fn-binding assay
Assays were performed essentially as previously described (Nicholson and Stanners, 2006). Cells were resuspended at a concentration of 4 × 105 cells/ml for LR or 2 × 105 cells/ml for L6. 100 μl of this suspension was added to Fn-coated plates (Chemicon international) and incubated for 1 h at 37°C. Adherent cells were stained with crystal violet, and the optical density was determined with a plate reader (PowerWave; Bio-Tek Instruments) at 570 nm. Note that in certain cases for L6 cells, depending on the particular experiment, integrin activation resulted in decreased cellular binding to Fn, which is likely the result of the previously described integrin activation-dependent formation of a cocoon of polymerized Fn around the cells (Ordonez et al., 2007). However, the relative difference between parental cells and activated transfectant cells remained in the inverse sense, so data is presented for ease of interpretation as an increase in all cases. Statistical significance was determined using the t-test (http://www.physics.csbsju.edu/stats/t-test_bulk_form.html).
Immunoblotting
Cellular lysates were resolved by SDS-PAGE and transferred to a 0.45-μm polyvinylidene difluoride membrane (Millipore). Antibody binding was detected using the ECL Plus chemiluminescent reagent (GE Healthcare).
Online supplemental material
Table S1 contains the nucleotide sequences of the primers used to generate the chimeras. Fig. S1 demonstrates that replacing less than five CEA amino acids in the region 6–10 is insufficient to cause a complete loss of biological function. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200701158/DC1.
Supplementary Material
Acknowledgments
This work was supported by the Cancer Research Society of Canada and by a research contract from Novothera Biotechnologies. T.B. Nicholson was supported by a scholarship from the Natural Sciences and Engineering Research Council of Canada.
Abbreviations used in this paper: CC1, CEA-related cell adhesion molecule 1; CEA, carcinoembryonic antigen; Fn, fibronectin; GPI, glycophosphatidylinositol; NCAM, neural cell adhesion molecule; PIPLC, phosphatidylinositol PLC; TM, transmembrane.
References
- Benchimol, S., A. Fuks, S. Jothy, N. Beauchemin, K. Shirota, and C.P. Stanners. 1989. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 57:327–334. [DOI] [PubMed] [Google Scholar]
- Brugger, B., C. Graham, I. Leibrecht, E. Mombelli, A. Jen, F. Wieland, and R. Morris. 2004. The membrane domains occupied by glycosylphosphatidylinositol-anchored prion protein and Thy-1 differ in lipid composition. J. Biol. Chem. 279:7530–7536. [DOI] [PubMed] [Google Scholar]
- Camacho-Leal, P., A.B. Zhai, and C.P. Stanners. 2007. A co-clustering model involving alpha5beta1 integrin for the biological effects of GPI-anchored human carcinoembryonic antigen (CEA). J. Cell. Physiol. 211:791–802. [DOI] [PubMed] [Google Scholar]
- Chen, R., J.J. Knez, W.C. Merrick, and M.E. Medof. 2001. Comparative efficiencies of C-terminal signals of native glycophosphatidylinositol (GPI)-anchored proproteins in conferring GPI-anchoring. J. Cell. Biochem. 84:68–83. [DOI] [PubMed] [Google Scholar]
- Chen, R., V. Anderson, Y. Hiroi, and M.E. Medof. 2003. Proprotein interaction with the GPI transamidase. J. Cell. Biochem. 88:1025–1037. [DOI] [PubMed] [Google Scholar]
- Cordes, F.S., J.N. Bright, and M.S. Sansom. 2002. Proline-induced distortions of transmembrane helices. J. Mol. Biol. 323:951–960. [DOI] [PubMed] [Google Scholar]
- Coyne, K.E., A. Crisci, and D.M. Lublin. 1993. Construction of synthetic signals for glycosyl-phosphatidylinositol anchor attachment. Analysis of amino acid sequence requirements for anchoring. J. Biol. Chem. 268:6689–6693. [PubMed] [Google Scholar]
- De Giovanni, C., P.L. Lollini, R. Dolcetti, L. Landuzzi, G. Nicoletti, E. D'Andrea, K. Scotland, and P. Nanni. 1993. Uncoupling of growth inhibition and differentiation in dexamethasone-treated human rhabdomyosarcoma cells. Br. J. Cancer. 67:674–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidelman, F.J., A. Fuks, L. DeMarte, M. Taheri, and C.P. Stanners. 1993. Human carcinoembryonic antigen, an intercellular adhesion molecule, blocks fusion and differentiation of rat myoblasts. J. Cell Biol. 123:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, Y., H. Tamura, and H. Ikezawa. 1994. Mutational analysis of the COOH-terminal hydrophobic domain of bovine liver 5′-nucleotidase as a signal for glycosylphosphatidylinositol (GPI) anchor attachment. Biochim. Biophys. Acta. 1190:273–278. [DOI] [PubMed] [Google Scholar]
- Furukawa, Y., K. Tsukamoto, and H. Ikezawa. 1997. Mutational analysis of the C-terminal signal peptide of bovine liver 5′-nucleotidase for GPI anchoring: a study on the significance of the hydrophilic spacer region. Biochim. Biophys. Acta. 1328:185–196. [DOI] [PubMed] [Google Scholar]
- Homans, S.W., M.A. Ferguson, R.A. Dwek, T.W. Rademacher, R. Anand, and A.F. Williams. 1988. Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature. 333:269–272. [DOI] [PubMed] [Google Scholar]
- Low, M.G. 1989. Glycosyl-phosphatidylinositol: a versatile anchor for cell surface proteins. FASEB J. 3:1600–1608. [DOI] [PubMed] [Google Scholar]
- Madore, N., K.L. Smith, C.H. Graham, A. Jen, K. Brady, S. Hall, and R. Morris. 1999. Functionally different GPI proteins are organized in different domains on the neuronal surface. EMBO J. 18:6917–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghibalhossaini, F., and C.P. Stanners. 2004. Minimal mutations are required to effect a radical change in function in CEA family members of the Ig superfamily. J. Cell Sci. 117:761–769. [DOI] [PubMed] [Google Scholar]
- Nicholson, T.B., and C.P. Stanners. 2006. Specific inhibition of GPI-anchored protein function by homing and self-association of specific GPI anchors. J. Cell Biol. 175:647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi, K., N. Inoue, Y. Maeda, J. Takeda, H. Riezman, and T. Kinoshita. 2000. Gaa1p and gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell. 11:1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez, C., R.A. Screaton, C. Ilantzis, and C.P. Stanners. 2000. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res. 60:3419–3424. [PubMed] [Google Scholar]
- Ordonez, C., A.B. Zhai, P. Camacho-Leal, L. Demarte, M.M. Fan, and C.P. Stanners. 2007. GPI-anchored CEA family glycoproteins CEA and CEACAM6 mediate their biological effects through enhanced integrin alpha5beta1-fibronectin interaction. J. Cell. Physiol. 210:757–765. [DOI] [PubMed] [Google Scholar]
- Rojas, M., A. Fuks, and C.P. Stanners. 1990. Biliary glycoprotein, a member of the immunoglobulin supergene family, functions in vitro as a Ca2(+)-dependent intercellular adhesion molecule. Cell Growth Differ. 1:527–533. [PubMed] [Google Scholar]
- Rojas, M., L. DeMarte, R.A. Screaton, and C.P. Stanners. 1996. Radical differences in functions of closely related members of the human carcinoembryonic antigen gene family. Cell Growth Differ. 7:655–662. [PubMed] [Google Scholar]
- Screaton, R.A., L. DeMarte, P. Draber, and C.P. Stanners. 2000. The specificity for the differentiation blocking activity of carcinoembryonic antigen resides in its glycophosphatidyl-inositol anchor. J. Cell Biol. 150:613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeth, E., T. Wirth, H.J. List, S. Kumbhani, A. Petersen, M. Neumaier, F. Czubayko, and H. Juhl. 2001. Controlled ribozyme targeting demonstrates an antiapoptotic effect of carcinoembryonic antigen in HT29 colon cancer cells. Clin. Cancer Res. 7:2022–2030. [PubMed] [Google Scholar]
- Taheri, M., H.U. Saragovi, and C.P. Stanners. 2003. The adhesion and differentiation-inhibitory activities of the immunoglobulin superfamily member, carcinoembryonic antigen, can be independently blocked. J. Biol. Chem. 278:14632–14639. [DOI] [PubMed] [Google Scholar]
- Wang, J., W. Gunning, K.M. Kelley, and M. Ratnam. 2002. Evidence for segregation of heterologous GPI-anchored proteins into separate lipid rafts within the plasma membrane. J. Membr. Biol. 189:35–43. [DOI] [PubMed] [Google Scholar]
- Zhou, H., A. Fuks, G. Alcaraz, T.J. Bolling, and C.P. Stanners. 1993. Homophilic adhesion between Ig superfamily carcinoembryonic antigen molecules involves double reciprocal bonds. J. Cell Biol. 122:951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.