Abstract
The yeast chitin synthase Chs3 provides a well-studied paradigm for polytopic membrane protein trafficking. In this study, high-throughput analysis of the yeast deletion collection identifies a requirement for Pfa4, which is an uncharacterized protein with protein acyl transferase (PAT) homology, in Chs3 transport. PATs, which are the enzymatic mediators of protein palmitoylation, have only recently been discovered, and few substrates have been identified. We find that Chs3 is palmitoylated and that this modification is Pfa4-dependent, indicating that Pfa4 is indeed a PAT. Chs3 palmitoylation is required for ER export, but not for interaction with its dedicated ER chaperone, Chs7. Nonetheless, both palmitoylation and chaperone association are required to prevent the accumulation of Chs3 in high–molecular mass aggregates at the ER. Our data indicate that palmitoylation is necessary for Chs3 to attain an export-competent conformation, and suggest the possibility of a more general role for palmitoylation in the ER quality control of polytopic membrane proteins.
Introduction
The ER is an important quality control site within the cell, where proteins destined for secretion or for sorting to post-Golgi organelles are monitored for proper folding and oligomeric assembly. Proteins that fail to fold or assemble are typically retained in the ER and, in some cases, retrotranslocated to the cytoplasm for proteosomal degradation (Meusser et al., 2005). Polytopic membrane proteins receive particular scrutiny in this regard. Indeed, many diseases are attributable to the failed ER export of mutant transmembrane proteins (Schulein, 2004).
Yeast genetic analyses have identified several ER resident proteins that mediate the ER export of specific polytopic membrane proteins. For instance, Shr3 is required for the ER export of the yeast amino acid permeases. In shr3Δ mutants, these permeases are retained in the ER, whereas the transit of other polytopic proteins is unimpaired (Gilstring et al., 1999). Similarly, Gsf2, Pho86, and Chs7, which are unrelated to Shr3 at the sequence level, are specifically required for the ER export of the hexose transporter Hxt1, the phosphate transporter Pho84, and the chitin synthase Chs3, respectively (Kota and Ljungdahl, 2005). These export factors have been suggested either to direct the segregation of their target proteins into budding COPII vesicles for anterograde transport or, alternatively, to act as dedicated chaperones, regulating proper protein folding before transport.
The yeast chitin synthase Chs3, which is a polytopic protein with six to eight predicted transmembrane domains, provides a genetic model for understanding mechanisms of transport through the secretory pathway. Chs3-mediated chitin deposition at the plasma membrane is highly regulated at the level of intracellular trafficking. Chs3 is maintained at steady state in an intracellular pool that may correspond to the TGN or endosomes (Ziman et al., 1996), and it is transported to the plasma membrane upon activation of the BCK1-SLT2 cell-integrity signaling pathway (Valdivia and Schekman, 2003). Mutants that impair cell wall chitin deposition have been found to block the plasma membrane delivery of Chs3 at different intracellular transport steps, whereas Chs7 mediates the ER export of Chs3, Chs5 and Chs6 direct its transport from the TGN to the plasma membrane, and Chs4 is required both for Chs3 activity at the cell surface and for its localization at the bud neck (for review see Roncero, 2002). We describe a genomic analysis of factors that regulate the transport of Chs3 to the cell surface, and identify an unexpected role for protein palmitoylation in the ER export of Chs3.
Palmitoylation, which is the thioester linkage of palmitate to selected cysteine residues, is one of several lipid modifications used for tethering proteins to membranes (Bijlmakers and Marsh, 2003). For transmembrane proteins, which are already embedded in the bilayer, the functional consequences of palmitoylation are not clear, though a role in directing segregation to membrane microdomains (lipid rafts) is often invoked. Enzymes for protein palmitoylation, which are called protein acyl transferases (PATs), were identified only recently by work in yeast (Lobo et al., 2002; Roth et al., 2002). The first two PATs to be characterized, Akr1 and Erf2, were found to contain a conserved zinc finger–like Asp-His-His-Cys (DHHC) domain, suggesting that this motif defines a larger PAT family. Yeast has seven DHHC proteins, whereas 23 are identifiable from the human genome. More recent reports have linked additional DHHC proteins to the palmitoylation of various substrates in both yeast and mammalian cells (for review see Mitchell et al., 2006). In this study, we find the uncharacterized yeast DHHC protein Pfa4 to be required for ER export by acting as the dedicated PAT for Chs3 palmitoylation.
Results and discussion
Chs3 cell surface activity requires the DHHC PAT Pfa4
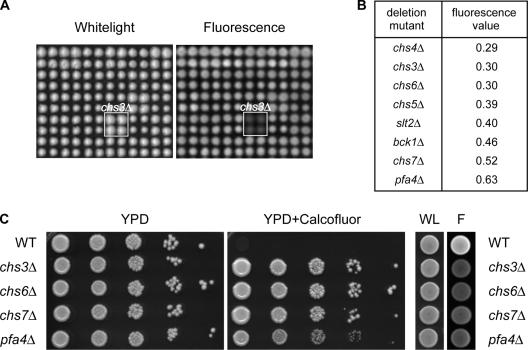
To identify additional genes required for Chs3 trafficking, we developed a fluorescence assay suitable for the large-scale screening of yeast deletion arrays, based on the binding of the fluorescent antifungal drug Calcofluor white (CW) to cell wall chitin (Fig. 1 A; Roncero and Duran, 1985). Mutants that are defective for the transport of Chs3 to the cell surface produce little chitin, and thus exhibit low levels of fluorescence. We screened three independent gene-deletion collections in duplicate and calculated the median fluorescence intensity for each strain. As expected, the mutants with the lowest fluorescence values included chs3Δ cells, which lack chitin synthase III, and strains deleted for the known Chs3 transport factors CHS4–7 (Fig. 1 B). In addition, the screen identified slt2Δ and bck1Δ, which are components of the cell-integrity pathway that stimulates the cell surface transport of Chs3, indicating that colony fluorescence values correlate well with independent measures of cell wall chitin (Lesage et al., 2004). Unexpectedly, cells deleted for the uncharacterized ORF YOL003c (PFA4) consistently displayed fluorescence values comparable to chs3–7 mutants (Fig. 1, B and C). Pfa4 is predicted to be a 45-kD protein containing the signature DHHC cysteine-rich domain that has been linked to protein palmitoylation (Bijlmakers and Marsh, 2003). Like chs mutants, pfa4Δ cells not only bind less CW but are also strikingly resistant to CW toxicity (Fig. 1 C).
Figure 1.
Pfa4 mutants show reduced fluorescence on CW media. (A) Representative white light and fluorescence images showing part of a MATα yeast deletion array grown on 50 μg/ml CW. (B) Ranked list of mutants with the lowest median fluorescence values. (C) Growth and fluorescence phenotypes of wild-type (BY4741) and congenic chs3Δ, chs6Δ, chs7Δ, and pfa4Δ cells on 75 μg/ml CW. F, fluorescence; WL, white light.
Pfa4 is required for the ER exit of Chs3
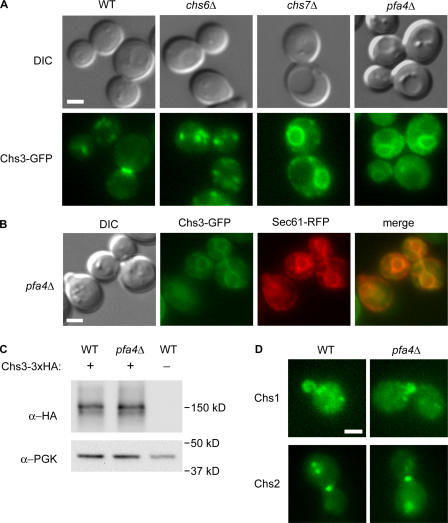
To determine if the low levels of cell wall chitin in pfa4Δ mutants result from alterations in the intracellular transport of Chs3, we examined the subcellular localization of Chs3-GFP in pfa4Δ strains and in mutants with known Chs3-trafficking defects. In wild-type cells, Chs3-GFP is present at the bud neck, bud tip, and intracellular compartments, whereas it is completely restricted to this latter compartment in chs6Δ mutants, which is consistent with a mislocalization to the TGN or endosomes (Fig. 2 A; Ziman et al., 1998). In contrast, Chs3-GFP localized to intracellular rings in pfa4Δ mutants, which are similar to those seen in chs7Δ mutants, where ER exit of Chs3 is blocked (Trilla et al., 1999). Colocalization with the ER marker Sec61 confirmed that Chs3-GFP resided primarily in the ER of pfa4Δ cells (Fig. 2 B); however, unlike chs7Δ cells, a small proportion of cells also showed some Chs3-GFP at the bud neck or bud tip. The ER-localized pool of Chs3 is not unstable or targeted for degradation, as Chs3 is present at wild-type levels in pfa4Δ mutants (Fig. 2 C). These results indicate that loss of cell surface Chs3 activity in pfa4Δ mutants results from a defect in transport at the ER. Moreover, this transport defect is specific to Chs3, as localization of other yeast chitin synthases, Chs1 and Chs2, are unaltered in pfa4Δ cells (Fig. 2 D).
Figure 2.
Chs3-GFP mislocalizes to the ER in pfa4Δ cells. (A) Wild-type (KLY9), chs6Δ (KLY1), chs7Δ (KLY3), and pfa4Δ (KLY5) cells expressing Chs3-GFP were observed by differential interference contrast and fluorescence microscopy. (B) Chs3-GFP colocalizes with the ER-marker Sec61-RFP in pfa4Δ (KLY55) cells. (C) Chs3-3xHA levels in wild-type (KLY41) and pfa4Δ (KLY43) cells, as shown by Western blotting with α-HA mAb. α-PGK1 was used as a loading control. (D) Localization of Chs1- and Chs2-GFP in wild-type and pfa4Δ cells. Bars, 2 μm.
Chs3 is palmitoylated in a Pfa4-dependent manner
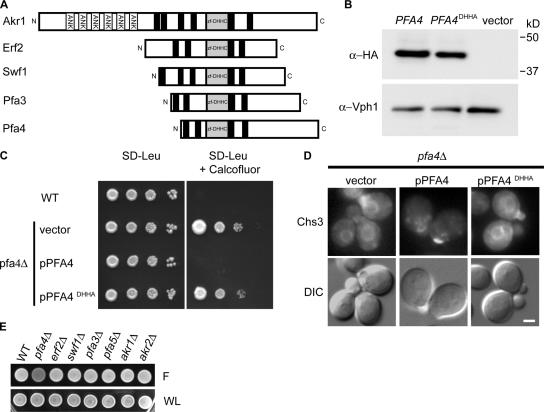
Because Pfa4 is a predicted PAT, we considered the possibility that ER export of Chs3 requires a Pfa4-mediated palmitoylation event. For the few DHHC PATs examined to date, mutation of the cysteine within the core DHHC tetrapeptide element has been found to abolish PAT activity both in vivo and in vitro (Fig. 3 A; Lobo et al., 2002; Roth et al., 2002; Smotrys et al., 2005; Valdez-Taubas and Pelham, 2005). Therefore, cysteine108 within the Pfa4 DHHC sequence was mutated to alanine. The Pfa4DHHA mutant protein accumulated to wild-type levels (Fig. 3 B), indicating that the substitution does not destabilize Pfa4. Nevertheless, plasmid-borne pfa4DHHA failed to complement CW resistance and Chs3-GFP mislocalization in pfa4Δ mutants (Fig. 3, C and D), suggesting these phenotypes are caused by a lack of Pfa4 enzymatic activity. Deletion of other DHHC proteins did not alter CW fluorescence (Fig. 3 E).
Figure 3.
The DHHC domain of Pfa4 is required for Chs3 localization. (A) Topology of the four characterized yeast DHHC proteins (Akr1, Erf2, Swf1, and Pfa3) and Pfa4. ANK, ankyrin repeats; black boxes, hypothetical transmembrane domains; Zf-DHHC, zinc finger–like DHHC cysteine-rich domain. (B) Western blots of lysates prepared from pfa4Δ cells carrying plasmids expressing Pfa4-3xHA, Pfa4DHHA-3xHA, or vector alone indicate the DHHA mutant is stably expressed in vivo. α-Vph1 mAb was used as a loading control. (C) Serial dilutions of strains from B on 100 μg/ml CW. (D) pfa4Δ strains expressing Chs3-GFP (KLY5) were transformed with empty vector, pPFA4, or pPFA4 DHHA, and observed by microscopy. Bar, 2 μm. (E) Fluorescence phenotype of mutants lacking other DHHC proteins on 75 μg/ml CW. F, fluorescence; WL, white light.
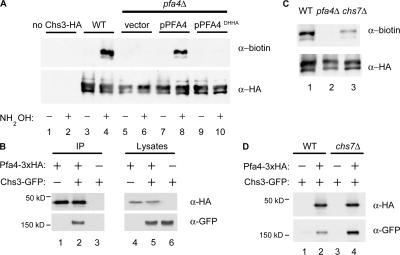
To determine if Chs3 is the direct target of Pfa4, we tested Chs3 for palmitoylation using a modified acyl-biotin exchange assay (Drisdel and Green, 2004; Politis et al., 2005). This three-step protocol involves the following: (1) blockade of free thiols with N-ethylmaleimide, (2) hydroxylamine cleavage of palmitoylation thioester linkages, and (3) thiol-specific biotinylation of the newly exposed cysteinyl thiols. To control for acyl-biotin exchange specificity, samples omitting the key hydroxylamine cleavage step were processed in parallel. We found that Chs3 is palmitoylated, and this modification is Pfa4-dependent, being abolished in pfa4Δ and pfa4DHHA mutants (Fig. 4 A). In contrast, palmitoylation of the other chitin synthases, Chs1 and Chs2, or the ER export factor Chs7, could not be detected (unpublished data). Other PATs have been shown to copurify with their substrates (Keller et al., 2004). Pfa4–Chs3 complexes could be detected by coimmunoprecipitation (Fig. 4 B), suggesting that Pfa4 interacts directly with Chs3 to mediate its palmitoylation. The observation that Chs3 is ER-localized and palmitoylated in chs7Δ cells (Fig. 4 C) is consistent with Chs3 palmitoylation being an early, ER-localized event.
Figure 4.
Pfa4 is required for palmitoylation of Chs3. (A) Lysates from cells expressing C-terminally 3xHA-FLAG epitope-tagged Chs3 from the GAL1 promoter were subjected to the acyl-biotin exchange reactions. Protein extracts were treated with (+) or without (−) hydroxylamine (NH2OH). α-FLAG immunoprecipitates were blotted with α-biotin and α-HA to detect modified and total Chs3, respectively. (B) Cells expressing Chs3-GFP and Pfa4-3xHA were immunoprecipitated with α-HA antiserum and analyzed by Western blotting with α-HA and α-GFP mAbs. (C) Chs3 palmitoylation was assessed in wild-type, pfa4Δ, and chs7Δ cells by acyl exchange, as in A. (D) Coimmunoprecipitation of Chs3-GFP and Pfa4-3xHA in wild-type and chs7Δ cells as performed in B.
To date, four of the seven yeast DHHC proteins have been shown to mediate protein palmitoylation in various cellular locations (Lobo et al., 2002; Roth et al., 2002; Hou et al., 2005; Smotrys et al., 2005; Valdez-Taubas and Pelham, 2005). Our finding of Pfa4-dependent palmitoylation for Chs3 adds a fifth DHHC protein to this list. Two other yeast PATs, Erf2/4 and Swf1, are also known to function at the ER, but recognize distinct substrates. The emerging picture is, thus, of a family of PATs that are distinguished from one another both by intracellular localization and by substrate specificity.
A requirement for palmitoylation and chaperone association in Chs3 ER export
As both Chs7 and Pfa4 are required for Chs3 ER export, we considered the possibility that these two proteins act together. Heterooligomeric PATs have been identified that require binding partners for activity and stability (Lobo et al., 2002). Although Chs3 palmitoylation was reduced in chs7Δ cells, it clearly was not abolished (Fig. 4 C). Furthermore, Chs3 copurifies with its PAT even in the absence of Chs7 (Fig. 4 D), whereas Pfa4–Chs7 interactions could not be detected under similar conditions (not depicted). Therefore, Chs7 does not appear to be required for substrate recognition by Pfa4 and is unlikely to be a subunit of a dimeric PAT.
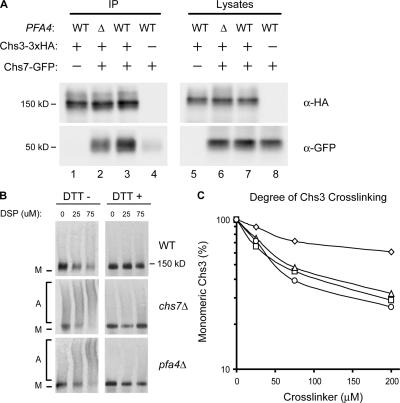
We tested an alternative model, in which Chs7 preferentially interacts with lipid-modified Chs3 to promote its interaction with the COPII vesicle-budding machinery (Gilstring et al., 1999). Although a Chs3–Chs7 physical interaction has not been yet reported, other polytopic yeast proteins, including Gap1 and the vacuolar H-ATPase, have been shown to interact with dedicated accessory factors at the ER during biosynthesis (Gilstring et al., 1999; Malkus et al., 2004). Using coimmunoprecipitation, we demonstrated that Chs3 and Chs7 do interact (Fig. 5 A, lane 3). In pfa4Δ cells, the Chs3–Chs7 interaction was subtly reduced, but not eliminated (Fig. 5 A, lane 2), indicating that Chs3 palmitoylation is not required for recognition by Chs7.
Figure 5.
Loss of PFA4 causes Chs3 aggregation in the ER. (A) Cells expressing Chs3-3xHA and Chs7-GFP were subjected to immunoprecipitation with α-HA antiserum and analyzed by Western blotting with α-HA and α-GFP mAbs. Strains used were KLY46 (lanes 1 and 5), KLY45 + pTM15 (lanes 2 and 6), KLY46 + pTM15 (lanes 3 and 7), and KLY14 + pTM15 (lanes 4 and 8). (B) Lysates of wild-type, chs7Δ, and pfa4Δ cells transformed with plasmid-borne Chs3-3xHA (pHV7) were cross-linked using increasing concentrations of DSP; DTT was added to duplicate samples to reverse cross-links. Chs3 was detected by Western blotting with α-HA mAb. M, monomer; A, aggregates. (C) Cross-linking of chromosomally encoded Chs3-3xHA in wild-type (KLY41; ⋄), chs7Δ (KLY46; ▵), pfa4Δ (KLY43; □), and chs7Δ pfa4Δ (KLY45; ○) strains was performed as for B. Disappearance of monomeric Chs3 as a function of cross-linker concentration was quantified by densitometry.
Chs3 aggregates in the absence of Pfa4
Recent work has suggested a chaperone function for Chs7 and the ER accessory proteins Shr3, Gsf2, and Pho86 (Kota and Ljungdahl, 2005). When their cognate chaperones were absent, the substrate polytopic proteins were found to accumulate in the ER in high molecular mass aggregates, which were visualized by cross-linking with dithiobissuccinimidyl propionate (DSP). Using the DSP cross-linking protocol of Kota and Ljungdahl (2005), we examined Chs3 aggregation in pfa4Δ cells (Fig. 5 B). Chs3-HA was readily cross-linked into high molecular mass forms in chs7Δ, pfa4Δ, and chs7Δ pfa4Δ mutants (Fig. 5, B and C), indicating that Chs7 chaperone function and Pfa4-mediated palmitoylation are both required to circumvent Chs3 aggregation.
Despite the similarity of the chs7Δ and pfa4Δ phenotypes, our data do not support models that Chs7 and Pfa4 act together as part of a complex or linear pathway. Instead, our results are best accommodated by models where Chs7 and Pfa4 act in parallel, mediating separate events that are both required for ER export. Nonetheless, defects in one pathway do appear to affect the other; Chs3 palmitoylation is reproducibly reduced in chs7Δ cells, and the Chs3–Chs7 interaction is decreased in pfa4Δ cells.
As both Chs7 and Pfa4 are required to circumvent Chs3 accumulation in high molecular mass aggregates, both appear to participate in the prerequisite folding of Chs3 that precedes ER export. Hydrophobic mismatch, resulting from an incompatibility between long transmembrane domains of polytopic proteins and the thinner ER bilayer, could explain why a membrane protein such as Chs3 requires both palmitoylation and chaperone association for export. ER chaperones have been hypothesized to shield hydrophobic regions of transmembrane domains to prevent protein aggregation (Levine et al., 2000). Palmitoylation may also promote hydrophobic matching by targeting proteins to cholesterol-rich membrane microdomains, which provide a local region of higher bilayer thickness. It is interesting that Chs1 and Chs2, which are also polytopic proteins, require neither Chs7 nor Pfa4 for ER export. This suggests a requirement for chaperone association and palmitoylation for only a subset of membrane proteins.
Several recent results suggest that the palmitoylation requirement for ER export may not be unique to Chs3. Our concurrent proteomic analysis of yeast protein palmitoylation indicates that several amino acid permeases are palmitoylated in a Pfa4-dependent manner (Roth et al., 2006). Intriguingly, these polytopic proteins also require dedicated accessory proteins for their ER export (Kota and Ljungdahl, 2005). It will be interesting to see if, as for Chs3, palmitoylation plays a role in permease trafficking. The link between palmitoylation and ER exit may hold true for at least some polytopic proteins in higher cells, as it was recently reported that functional cell surface expression of the nicotinic acetylcholine receptor requires an ER palmitoylation event (Drisdel et al., 2004). Thus, palmitoylation may participate more generally in ER quality control mechanisms, particularly for newly synthesized polytopic integral membrane proteins.
Materials and methods
General molecular biology methods were as previously described (Conibear and Stevens, 2000, 2002). Primer sequences are available on request. Protein topologies were predicted by the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/).
CW genomic screen
Three yeast knockout collections (in strain backgrounds BY4741, BY4742, and BY4743) were obtained from Open Biosystems and pinned four times in 1,536-array format onto YPD plates containing 50 μg/ml CW (Sigma-Aldrich), using a Virtek automated colony arrayer (Bio-Rad Laboratories). After incubation at 30°C for 3 d, white-light images were acquired using a flat-bed scanner (model 2400; Epson), and fluorescent-light images were captured with a Fluor S Max MultiImager (Bio-Rad Laboratories) using the 530DF60 filter and Quantity One software (version 4.2.1; Bio-Rad Laboratories). The open-source spot-finding program GridGrinder (http://gridgrinder.sourceforge.net) was used for the densitometry of digital images. Average growth and fluorescence values from two independent screens were calculated for each strain using Excel (Microsoft); slow-growing strains were removed from the analysis.
Strain construction
The BY4741 (MATa his3Δ 1 leu2Δ 0 met15Δ 0 ura3Δ 0) knockout strains akr1Δ, akr2Δ, chs3Δ, chs6Δ, chs7Δ, erf2Δ, pfa3Δ, pfa4Δ, pfa5Δ, and swf1Δ and the BY4742 (MATα his3Δ 1 leu2Δ 0 lys2Δ 0 ura3Δ 0) knockout strains chs7Δ and pfa4Δ used in this study were obtained from Open Biosystems. Deletion mutants used for palmitoylation assays were in the BY4742 background. Other strains are listed in Table I.
Table I.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| KLY1 | BY4741 chs6∷kanR CHS3-GFP∷HIS3 |
| KLY3 | BY4741 chs7∷kanR CHS3-GFP∷HIS3 |
| KLY5 | BY4741 pfa4∷kanR CHS3-GFP∷HIS3 |
| KLY9 | BY4741 CHS3-GFP∷HIS3 |
| KLY14 | BY4741 chs7∷HIS3 |
| KLY41 | BY4741 CHS3-3xHA∷kanR |
| KLY43 | BY4741 pfa4∷natR CHS3-3xHA∷kanR |
| KLY45 | BY4741 chs7∷HIS3 pfa4∷natR CHS3-3xHA∷kanR |
| KLY46 | BY4741 chs7∷HIS3 CHS3-3xHA∷kanR |
| KLY55 | BY4741 pfa4∷kanR CHS3-GFP∷HIS3 SEC61-RFP∷URA3 |
| KLY60 | BY4741 CHS1-GFP∷HIS3 |
| KLY62 | BY4741 CHS2-GFP∷HIS3 |
| KLY65 | BY4741 CHS2-GFP∷HIS3 pfa4∷natR |
| KLY66 | BY4741 CHS1-GFP∷HIS3 pfa4∷natR |
| KLY67 | BY4741 chs7∷kanR pfa4∷natR CHS3-GFP∷HIS3 |
KLY14 was created by transforming a 3.5-kb EcoRV fragment from pTM10 (Cos et al., 1998) into wild-type BY4741. To create KLY45, a natR cassette amplified from p4339 (Tong et al., 2001) using primers with flanking homology to 5′ and 3′ PFA4 sequences was transformed into KLY14. C-terminal tags (GFP and 3xHA) were integrated at the CHS3 locus as previously described (Longtine et al., 1998). C-terminal tagging of SEC61 with RFP was performed as previously described (Sheff and Thorn, 2004).
Plasmid construction
A PCR fragment containing PFA4-3xHA was created using a two-step PCR method (Conibear and Stevens, 2000) and cotransformed into pfa4Δ yeast together with XhoI-cut pRS415 or XhoI-cut pRS426, thereby creating pKL1 and pKL3, respectively. Plasmids were rescued and tested for complementation of pfa4Δ CW resistance phenotypes and expression of Pfa4-3xHA by Western blotting. pKL6 was created by QuikChange (Stratagene) mutagenesis of pKL1, using primers designed to change Cys108 to alanine. The resulting mutant was confirmed by sequencing. For construction of pND2115, which is GAL1-CHS3-3xHA-FLAG-His carried on pRS316, the CHS3 ORF was PCR amplified from yeast genomic DNA and was used to replace the AKR1 ORF of a GAL1-AKR1-3xHA-FLAG-His plasmid construct (Roth et al., 2002). pHV7 (CHS3-3xHA), pTM10 (chs7∷HIS3; Cos et al., 1998), and pTM15 (CHS7-GFP; Trilla et al., 1999) were gifts from C. Roncero (Universidad de Salamanca, Salamanca, Spain).
Microscopy
For fluorescence microscopy of GFP- or RFP-tagged strains, log-phase cells grown in minimal media were observed directly. Indirect immunofluorescence microscopy was performed as previously described (Conibear and Stevens, 2000, 2002). For DAPI staining, cells were mounted in buffered glycerol containing 1 mg/ml p-phenylenediamine and 0.05 μg/ml DAPI. Cells were viewed using a 100× oil-immersion objective on a fluorescence microscope (Axioplan2; Carl Zeiss MicroImaging, Inc.), and images were captured with a camera (CoolSNAP; Roper Scientific) using MetaMorph 6.2r6 software (Molecular Devices). Images were adjusted for brightness with Photoshop CS2 (Adobe).
Cross-linking
Cross-linking of yeast cell lysates with DSP was performed as previously described (Kota and Ljungdahl, 2005) with slight modifications. DSP was added to 10 μg of protein in 40 μL PBS. Reactions were quenched with 40 mM Tris HCl, pH 7.5, at 25°C for 30 min. Parallel samples were treated with 40 mM DTT at 37°C for 30 min.
Palmitoylation assay
Chs3 palmitoylation was assessed by acyl-biotinyl exchange (Politis et al., 2005), using a modified version of the Drisdel and Green method (Drisdel and Green, 2004). Denatured protein extracts, which were prepared from Chs3-3xHA-FLAG-His–expressing cells (2-h galactose-induced expression period) by glass-bead lysis, were subjected to the three steps of acyl-biotinyl exchange protocol, as previously described (Politis et al., 2005). The epitope-tagged Chs3 was immunoprecipitated with M2 anti-FLAG-agarose, and then Western blotted with either anti–biotin-HRP or anti–HA-HRP.
Coimmunoprecipitations
Coprecipitation was performed as previously described (Conibear and Stevens, 2000). 20 OD600 of spheroplasts were resuspended in 1 mL lysis buffer (1% CHAPSO, 50 mM KPO4, pH7.5, 50 mM NaCl, and protease inhibitors) and incubated with 3 μL of rabbit α-HA antiserum for 1 h at 4°C. 30 μL of protein G–Sepharose (GE Healthcare) was added for 1 h at 4°C. Beads were washed twice in lysis buffer and subjected to SDS-PAGE. Coimmunoprecipitated proteins were analyzed by Western blotting with antibodies to HA or GFP.
Acknowledgments
We would like to thank members of the Conibear and Davis laboratories for comments on the manuscript, and Cesar Roncero for providing plasmids. The contributions of Drew Lett and Jenny Bryan to the analysis of high throughput data are gratefully acknowledged.
Funding was provided by grants from the Canadian Institute for Health Research, the Michael Smith Foundation for Health Research (MSFHR), the Canada Foundation for Innovation, and the BC Research Institute for Children's and Women's Health (to E. Conibear), and by National Institutes of Health grant GM65525 (to N.G. Davis). E. Conibear is also the recipient of an MSFHR Scholar Award.
Abbreviations used in this paper: CW, Calcofluor white; DSP, dithiobissuccinimidyl propionate; PAT, protein acyl transferase.
References
- Bijlmakers, M.J., and M. Marsh. 2003. The on-off story of protein palmitoylation. Trends Cell Biol. 13:32–42. [DOI] [PubMed] [Google Scholar]
- Conibear, E., and T.H. Stevens. 2000. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell. 11:305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear, E., and T.H. Stevens. 2002. Studying yeast vacuoles. Methods Enzymol. 351:408–432. [DOI] [PubMed] [Google Scholar]
- Cos, T., R.A. Ford, J.A. Trilla, A. Duran, E. Cabib, and C. Roncero. 1998. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur. J. Biochem. 256:419–426. [DOI] [PubMed] [Google Scholar]
- Drisdel, R.C., and W.N. Green. 2004. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 36:276–285. [DOI] [PubMed] [Google Scholar]
- Drisdel, R.C., E. Manzana, and W.N. Green. 2004. The role of palmitoylation in functional expression of nicotinic alpha7 receptors. J. Neurosci. 24:10502–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilstring, C.F., M. Melin-Larsson, and P.O. Ljungdahl. 1999. Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles. Mol. Biol. Cell. 10:3549–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, H., K. Subramanian, T.J. LaGrassa, D. Markgraf, L.E. Dietrich, J. Urban, N. Decker, and C. Ungermann. 2005. The DHHC protein Pfa3 affects vacuole-associated palmitoylation of the fusion factor Vac8. Proc. Natl. Acad. Sci. USA. 102:17366–17371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, C.A., X. Yuan, P. Panzanelli, M.L. Martin, M. Alldred, M. Sassoe-Pognetto, and B. Luscher. 2004. The gamma2 subunit of GABA(A) receptors is a substrate for palmitoylation by GODZ. J. Neurosci. 24:5881–5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota, J., and P.O. Ljungdahl. 2005. Specialized membrane-localized chaperones prevent aggregation of polytopic proteins in the ER. J. Cell Biol. 168:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage, G., A.M. Sdicu, P. Menard, J. Shapiro, S. Hussein, and H. Bussey. 2004. Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics. 167:35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, T.P., C.A. Wiggins, and S. Munro. 2000. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol. Biol. Cell. 11:2267–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo, S., W.K. Greentree, M.E. Linder, and R.J. Deschenes. 2002. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277:41268–41273. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Malkus, P., L.A. Graham, T.H. Stevens, and R. Schekman. 2004. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol. Biol. Cell. 15:5075–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meusser, B., C. Hirsch, E. Jarosch, and T. Sommer. 2005. ERAD: the long road to destruction. Nat. Cell Biol. 7:766–772. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.A., A. Vasudevan, M.E. Linder, and R.J. Deschenes. 2006. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J. Lipid Res. 47:1118–1127. [DOI] [PubMed] [Google Scholar]
- Politis, E.G., A.F. Roth, and N.G. Davis. 2005. Transmembrane topology of the protein palmitoyl transferase Akr1. J. Biol. Chem. 280:10156–10163. [DOI] [PubMed] [Google Scholar]
- Roncero, C. 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41:367–378. [DOI] [PubMed] [Google Scholar]
- Roncero, C., and A. Duran. 1985. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: in vivo activation of chitin polymerization. J. Bacteriol. 163:1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A.F., Y. Feng, L. Chen, and N.G. Davis. 2002. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, A.F., J. Wan, A.O. Bailey, B. Sun, J.A. Kuchar, W.N. Green, B.S. Phinney, J.R. Yates III, and N.G. Davis. 2006. Global analysis of protein palmitoylation in yeast. Cell. 125:1003–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulein, R. 2004. The early stages of the intracellular transport of membrane proteins: clinical and pharmacological implications. Rev. Physiol. Biochem. Pharmacol. 151:45–91. [DOI] [PubMed] [Google Scholar]
- Sheff, M.A., and K.S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. [DOI] [PubMed] [Google Scholar]
- Smotrys, J.E., M.J. Schoenfish, M.A. Stutz, and M.E. Linder. 2005. The vacuolar DHHC-CRD protein Pfa3p is a protein acyltransferase for Vac8p. J. Cell Biol. 170:1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H., M. Evangelista, A.B. Parsons, H. Xu, G.D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C.W. Hogue, H. Bussey, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368. [DOI] [PubMed] [Google Scholar]
- Trilla, J.A., A. Duran, and C. Roncero. 1999. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 145:1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Taubas, J., and H. Pelham. 2005. Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J. 24:2524–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia, R.H., and R. Schekman. 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA. 100:10287–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman, M., J.S. Chuang, and R.W. Schekman. 1996. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell. 7:1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman, M., J.S. Chuang, M. Tsung, S. Hamamoto, and R. Schekman. 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell. 9:1565–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]