Abstract
Quiescence is the most common and, arguably, most poorly understood cell cycle state. This is in part because pure populations of quiescent cells are typically difficult to isolate. We report the isolation and characterization of quiescent and nonquiescent cells from stationary-phase (SP) yeast cultures by density-gradient centrifugation. Quiescent cells are dense, unbudded daughter cells formed after glucose exhaustion. They synchronously reenter the mitotic cell cycle, suggesting that they are in a G0 state. Nonquiescent cells are less dense, heterogeneous, and composed of replicatively older, asynchronous cells that rapidly lose the ability to reproduce. Microscopic and flow cytometric analysis revealed that nonquiescent cells accumulate more reactive oxygen species than quiescent cells, and over 21 d, about half exhibit signs of apoptosis and necrosis. The ability to isolate both quiescent and nonquiescent yeast cells from SP cultures provides a novel, tractable experimental system for studies of quiescence, chronological and replicative aging, apoptosis, and the cell cycle.
Introduction
In addition to being the most common cell state on earth (Lewis and Gattie, 1991), quiescence, or G0, is critically important in the development and survival of all organisms and plays a significant role in disease, such as cancer (Gray et al., 2004). Quiescent cells are directly involved in the establishment and persistence of microbial infectious diseases such as tuberculosis (Parrish et al., 1998) and cryptosporosis (Alexander and Perfect, 1997). In complex eukaryotes, quiescence is essential for stem-cell maintenance (Suda et al., 2005) as well as for activating cells for wound healing (Chang et al., 2002) and sexual reproduction. Both wound healing and sexual reproduction require regulated exit from G0 (Gouge et al., 1998).
Difficulties in isolating populations of quiescent cells have made a clear understanding of quiescence almost impossible. For example, in metazoans, quiescent cells usually closely associate with nonquiescent cells; thus, the isolation of pure populations of quiescent cells in quantities large enough for systems-level analysis has not usually been feasible (Gray et al., 2004). In addition, stationary-phase (SP) cultures of the unicellular yeast Saccharomyces cerevisiae include both quiescent and nonquiescent cells, but this heterogeneity was not recognized so that all cells in an SP culture were sometimes mistakenly referred to as quiescent (Werner-Washburne et al., 2002; Radonjic et al., 2005). This heterogeneity has led to serious confusion about the relationship between SP and quiescence in yeast. This confusion has been compounded by the lack of a concise definition of quiescence; quiescent yeast cells have been defined more by what they do not do than by what they do (Coller et al., 2006).
Nonetheless, a comparison of yeast cells in SP cultures with those in exponentially growing cultures has identified several characteristics associated with quiescence, including a thickened cell wall, decreased metabolic rate, and accumulation of a variety of storage molecules, such as trehalose and glycogen (Werner-Washburne et al., 1993). In addition, quiescent cells exhibit decreased transcription (Choder, 1991) and translation (Fuge et al., 1994), have characteristically condensed chromosomes (Pinon, 1978), and show increased resistance to a variety of stresses (Werner-Washburne et al., 1993). Because cells under other conditions can exhibit these characteristics, the hallmark of quiescent cells was proposed to be their ability to retain viability without added nutrients, including the ability to reproduce.
We report here the separation of quiescent and nonquiescent cells from S. cerevisiae SP cultures by density-gradient centrifugation. Cells in the lower, denser fraction have characteristics of quiescent cells, suggesting that they are bona fide G0 cells. They are mostly unbudded daughter cells that retain both viability and the ability to reproduce. Furthermore, they synchronously reenter mitosis when inoculated into fresh medium. Additionally, they are very thermotolerant and highly refractive by phase-contrast microscopy. Cells in the less dense, upper fraction are heterogeneous. They include both budded and unbudded cells and do not synchronously reenter mitosis, but they all rapidly lose the ability to reproduce over time. They are less resistant to heat stress than quiescent cells and exhibit signs of oxidative stress and apoptosis.
Microarray analysis also revealed distinct differences between the cell fractions. Transcripts encoding proteins required for response to water stress, fatty acid oxidation, and some metabolic pathways, including coenzyme metabolism, were more abundant in quiescent cells. Transcripts encoding genes involved in DNA recombination and transposition were more abundant in nonquiescent cells, consistent with a high percentage of these cells being apoptotic. Interestingly, the transcripts that differentiate quiescent and nonquiescent cells are not those that have previously been identified as SP transcripts (Gasch et al., 2000; Martinez et al., 2004).
The significance of this work is that this represents the first time quiescent, putative G0 cells have been isolated and studied from yeast SP cultures. It was unexpected that the quiescent cell fraction would consist mostly of daughter cells and that, although viable, nonquiescent cells would so rapidly lose their ability to reproduce. Based on these observations, we present models for the development of quiescent and nonquiescent cells during the growth of cultures to SP and their relationship to cells in the mitotic cell cycle.
Results
Two distinct cell fractions are present in yeast cultures during growth to SP
Previously, cells in yeast SP cultures were observed to be asynchronous and contain a mixture of unbudded and budded cells (Werner-Washburne et al., 2002; Gendron et al., 2003). To further characterize different cell types within an SP culture, we sought a method that would separate the cell types. Separation of cells from SP yeast cultures by equilibrium density-gradient centrifugation revealed the presence of two fractions with different buoyant densities: ∼1.10 g/ml (the upper fraction) and 1.14 g/ml (the lower fraction; Fig. 1 a). In contrast, cells from exponentially growing cultures formed a single band with a buoyant density of 1.12 g/ml. Two fractions were also observed for SP cultures of other strains, including W303, BY4742, and BY4743 (unpublished data). Additionally, several (atp1, qcr7, rip1, sdh4, kgd1, and cor1) of >50 mitochondrial mutants evaluated (Martinez et al., 2004) were unable to form a lower band at 7 d (unpublished data), whereas other, functionally related mutants, such as atp2, atp3, sdh2, and cox6, were able to do so. These differences in ability of mitochondrial mutants to form a lower band suggest that there are some as-yet-uncharacterized functional differences between these mitochondrial genes.
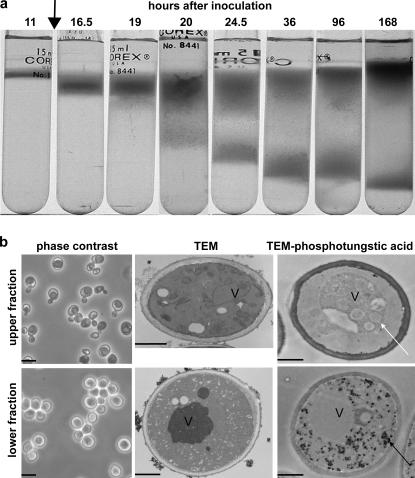
Figure 1.
Two distinct cell populations are formed in yeast cultures entering SP. (a) Density-gradient separation of two distinct cell fractions in S288c cultures as a function of time after inoculation. Glucose exhaustion (arrow) occurred 12 h after inoculation. (b) Phase contrast, transmission EM (TEM), and phosphotungstic acid–stained transmission EM micrographs of upper- and lower-fraction cells from SP cultures (7 d after inoculation). The white arrow indicates vacuolar vesicles, and the black arrow indicates accumulated glycogen. V, vacuole. Bars: 10 μm (left); 1 μm (middle and right).
We examined cultures to determine when the two fractions began to differentiate in the culture over time (Fig. 1 a). A single band of cells was detected 11 and 12 h after inoculation. Notably, glucose exhaustion also occurred at 12 h after inoculation (Fig. 1 a, arrow). A distinct broadening of the band of cells was detected by 19 h after inoculation, and two bands were observed by 20–24.5 h. The lower cell fraction continued to increase in density for at least 7 d, whereas the upper cell fraction exhibited a slight decrease in density between 3 and 7 d after inoculation. We conclude that the lower-fraction cells form after glucose exhaustion during the transition to growth on nonfermentable carbon sources.
Morphological characterization of the two cell fractions
To determine whether there were morphological differences between the fractions, they were examined by light and electron microscopy. The more dense, lower-fraction cells were unbudded and highly refractive by phase-contrast microscopy (Fig. 1 b) as well as uniform in size (4.6 ± 0.33 μm). These cells contained nuclei and electron-dense vacuoles but, strikingly, other internal organelles, including mitochondria and endoplasmic reticulum, were not detectable (Fig. 1 b, lower fraction, TEM). They also contained significant concentrations of glycogen (Fig. 1 b, lower fraction, TEM–phosphotungstic acid), which is known to increase after the diauxic shift (Lillie and Pringle, 1980).
The less dense, upper-fraction cells exhibited some distinct characteristics. They contained budded and unbudded cells and were dark by phase-contrast microscopy (Fig. 1 b, upper fraction, phase contrast) but had mean diameters similar to the lower-fraction cells (4.1 ± 0.34 μm). These cells had large vacuoles and numerous mitochondria and ER profiles but only trace amounts of glycogen (Fig. 1 b, upper fraction, TEM and TEM–phosphotungstic acid). In addition, their vacuoles contained numerous vesicles (Fig. 1 b, TEM–phosphotungstic acid, white arrow), suggesting enhanced autophagy (Abeliovich and Klionsky, 2001). We conclude that the two cell fractions are morphologically distinct, with the more uniform, lower-fraction cells exhibiting characteristics associated with quiescent cells.
Physiological characterization of the two cell types
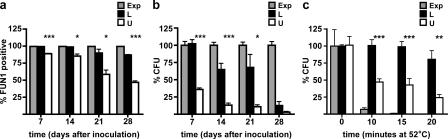
To determine whether these cell fractions were physiologically different, we tested for long-term viability, colony-forming capacity, and thermotolerance. Lower-fraction cells exhibited >87% viability over 28 d in culture, as detected by FUN-1 (2-chloro-4-[2,3-dihydro-3-methyl-{benzo-1,3-thiazol-2-yl}-methylidene]-1-phenylquinolinium iodide) uptake and flow cytometry (Fig. 2 a). Upper-fraction cells (>85%) were viable at 7 and 14 d but decreased in viability (to 47%) by 28 d. The two-cell fractions exhibited even larger differences in colony-forming capacity (Fig. 2 b). Essentially all viable lower-fraction cells could produce colonies at 7 d. This decreased to ∼65% at both 14 and 21 d and to 12% by 28 d.
Figure 2.
FUN-1 viability, colony-forming capacity, and thermotolerance of cells from upper and lower fractions from SP cultures. (a) Viability of cells from lower (L) and upper (U) fractions from 7-, 14-, 21-, and 28-d-old S288c SP cultures as determined by FUN-1 uptake measured by flow cytometry. Exponentially growing cells (Exp) were positive controls. (b) Colony-forming capacity of cells from S288c SP cultures determined by plating assay. Values are expressed as the percentage of colony-forming units (CFU) of exponentially growing cultures plated in parallel on each day. (c) Constitutive thermotolerance of cells from S288c SP cultures and fractions as well as exponentially growing cultures as a function of time at 52°C. At T = 0, the number of colonies produced by each population was normalized to 100%. Error bars indicate SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In contrast, upper-fraction cells exhibited a much more rapid decline in reproductive capacity. Only 36% were able to form colonies at 7 d, ∼10% at 14 and 21 d, and only 3% by day 28. A two-way analysis of variance (ANOVA) confirmed significant differences between lower and upper fractions with respect to the ability to reproliferate (P < 0.0001) and with respect to time (P = 0.0016). This is the first time, to our knowledge, that such a dramatic difference between FUN-1 viability and the ability to form colonies has been observed in yeast. We conclude that during differentiation of the two cell populations, upper-fraction cells can retain viability but experience some physiological change that makes them unable to reproduce.
Because cells in SP cultures are known to become thermotolerant (Gray et al., 2004), we wanted to determine whether these cell fractions differed in their response to heat shock. Cells from both fractions and exponential cultures were tested for their ability to survive a heat shock at 52°C (Fig. 2 c). Lower-fraction cells were the most heat resistant, retaining essentially 100% viability over 15 min. As expected, cells from exponentially growing cultures rapidly lost the ability to form colonies after heat shock. Upper-fraction cells were more resistant than cells from exponentially growing cultures but significantly less resistant than lower-fraction cells. Approximately 50% of the upper-fraction cells were unable to form colonies after 10–15 min at 52°C, decreasing to 25% after 20 min. We conclude that the lower fraction is uniformly thermotolerant, whereas the upper fraction is heterogeneous and contains a small percentage of thermotolerant cells.
Based on all of the aforementioned experiments, we conclude that the cells in the lower fraction are quiescent. As an operative term, we will refer to the upper-fraction cells as nonquiescent, recognizing that this fraction is heterogeneous and may include a very small population of quiescent cells.
Synchronous reentry into mitosis by quiescent cells
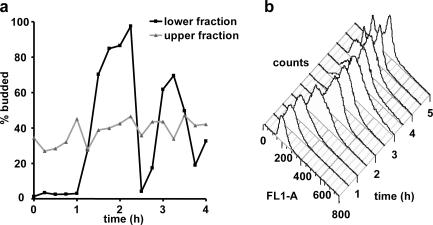
In eukaryotes, the hallmark of G0 cells obtained by serum starvation is their ability to synchronously reenter the mitotic cell cycle (Pardee, 1974). Microscopic analysis revealed that lower-fraction, quiescent cells budded synchronously after slightly more than 1 h (Fig. 3 a, black line). In contrast, nonquiescent, upper-fraction cells were not synchronous (Fig. 3 a, gray line). Cell counts (unpublished data), using a particle counter, demonstrated that the lag phase for both fractions was ∼4 h, consistent with previous studies (Bonini et al., 2000; Brejning et al., 2003; Radonjic et al., 2005). For both the lower- and upper-fraction cells, we observed microscopically that the daughter cells did not separate from the mothers until the second cell division (4.5 h after refeeding). This could account for the time difference between budding and the increase in cell number.
Figure 3.
Lower-fraction, quiescent cells synchronously reenter the mitotic cell cycle. (a) Budding index as a function of time after inoculation into fresh, glucose-rich medium for nonquiescent, upper-fraction and quiescent, lower-fraction cells from S288c SP cultures. (b) DNA content as measured by SYBR Green I fluorescence (FL1-A) and flow cytometry of S288c quiescent, lower-fraction cells after inoculation into fresh, glucose-based media.
DNA-content analysis further demonstrated the synchrony of the quiescent cells (Fig. 3 b). The first cell cycle contains a single 1N peak at T0 that broadens and becomes an intermediate S-phase peak between 1 and 1.5 h. A 2N shoulder is evident by 2 h. The cells rapidly lost synchrony during the second cell cycle, with both 1N and 2N peaks present by 3.5 h (Fig. 3 b). Because the quiescent cells, like mammalian G0 cells, are obtained as a result of starvation and are synchronous upon reentry into the cell cycle, we conclude that they are also likely to be in a G0 state. Further work is needed to determine cell cycle–specific differences between cells in this state and those in the G1 phase of the mitotic cell cycle.
Replicative age of quiescent and nonquiescent cells
Because lower-fraction, quiescent cells retained their reproductive capacity markedly better than the upper-fraction cells, we were interested in determining whether the quiescent cells were all of the same replicative age as assayed by their number of bud scars. Bud scars were measured by calcofluor white staining and flow cytometry. Flow cytometry gates were set to enrich for populations of cells with 0, 1, 2, or 3 or more bud scars (Fig. 4 a), and the number of bud scars on separated cells were verified microscopically (see the supplemental text, available at http://www.jcb.org/cgi/content/full/jcb.200604072/DC1). Microscopic evaluation of FACS-sorted cells revealed that 95% of cells sorted using the first gate (0 bud scar group) had 0 bud scars and 5% had 1 or 2 bud scars (n = 61). Of cells sorted using the second gate (1 bud scar group), 90% had 1 bud scar and 10% had 2 bud scars (n = 40). Finally, cells sorted using the third gate (2 bud scars), 81% had 2 bud scars, and 19% had 3 or more bud scars (n = 53). Therefore, the flow cytometry gates were most accurate for unbudded daughter cells.
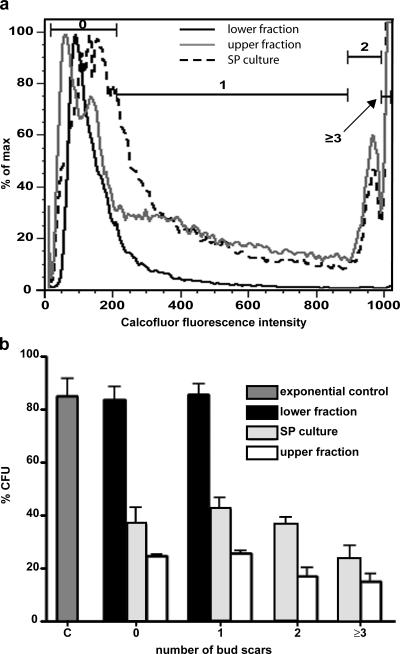
Figure 4.
Replicative age and colony-forming capacity as a function of replicative age. (a) The number of bud scars as an indicator of replicative age was assayed by calcofluor white M2R staining and flow cytometry. Shown is the fluorescence-intensity histogram with gates set to sort cells by number of bud scars. Gates used to sort cells by bud scar number are indicated by the bracketed lines. (b) Colony-forming capacity of cells separated by number of bud scars from SP cultures, quiescent, lower fractions, and nonquiescent, upper fractions in panel a. Cells from unfractionated exponential cultures (C) were used as a positive control for colony-forming capacity. Error bars indicate SD.
Flow cytometry of calcofluor-stained cells from unfractionated SP cultures revealed a broad peak of fluorescence that included cells with 0 bud scars, an extended region that included cells with 1 bud scar, and two additional peaks representing cells with 2 and 3 or more bud scars, respectively (Fig. 4 a). If all mother cells divided once after glucose exhaustion, 50% of cells in SP cultures would be expected to be unbudded daughter cells. A slightly higher than expected percentage of unbudded daughter cells (55%) was observed in cells from SP cultures (Table I). Upper-fraction, nonquiescent cells exhibited a profile similar to that observed with SP cultures (Fig. 4 a, gray line; and Table I). In contrast, lower-fraction, quiescent cells, which were purified twice by density-gradient centrifugation, were predominantly daughter cells (91% had no detectable bud scars and only 9% had one bud scar; Table I). We conclude from these results that the quiescent fraction is predominantly composed of daughter cells produced in the first round of budding after glucose exhaustion, consistent with the appearance of the lower band (Fig. 1 a) and that these cells comprised <50% of the cells in SP cultures in this separation. We also conclude that nonquiescent mother cells continue to bud during the postdiauxic phase, producing additional unbudded, nonquiescent daughters. This is consistent with the observation of budded cells in upper fraction 7 d after inoculation (Fig. 1 b).
Table I.
Percentage of bud scars in nonseparated SP cultures and from upper and lower cell fractions
| Number of bud scars |
0 | 1 | 2 | 3 or more |
|---|---|---|---|---|
| SP cultures | 55 ± 1.5 | 26 ± 1.4 | 6 ± 0.4 | 11 ± 0.7 |
| Upper fraction | 49 ± 4.9 | 29 ± 2.4 | 7 ± 0.7 | 13 ± 1.9 |
| Lower fraction | 91 ± 2.2 | 9 ± 2.0 | 0.2 ± 0.02 | 0.1 ± 0.04 |
Cell percentages do not add up to 100% because a small percentage of cells detected by the flow cytometer are not captured within the established gates.
The colony-forming capacity of cells with different numbers of bud scars was also evaluated (Fig. 4 b). Quiescent, lower-fraction cells, which have either 0 or 1 bud scars were similar to cells from nonseparated, exponentially growing cultures in their ability to form colonies. There were no cells with >1 bud scar in this fraction. For nonquiescent, upper-fraction cells, all cells exhibited reduced colony-forming capacity; however, cells with 2 or more bud scars showed a slightly greater reduction. The overall loss of replicative capacity as compared with cells with 0 or 1 bud scars in nonquiescent cells suggests that the switch that limits the ability of these cells to reproduce occurs in most, if not all, upper-fraction cells and is relatively independent of replicative age. As expected, cells from SP cultures exhibited intermediate colony-forming capacities. Furthermore, the unbudded and singly budded cells in the lower, quiescent fraction are indistinguishable with respect to colony-forming capacity, suggesting that quiescence is not simply a property unique to virgin cells but, under some set of circumstances, singly budded, new mother cells can also acquire quiescent characteristics.
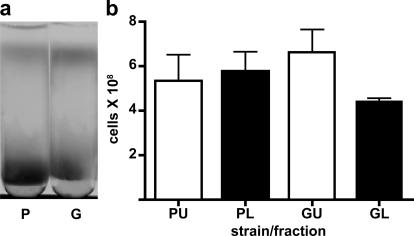
Glycogen accumulation has little effect on the density of quiescent cells
Because glycogen accumulation occurred predominantly in lower-fraction, quiescent cells (Fig. 1 b, TEM– phosphotungstic acid), we tested a glc3 mutant that fails to accumulate glycogen because it lacks a glycogen-branching enzyme, to determine whether glycogen was responsible for the increase in density. There were no significant differences in cell number between upper fractions from parental or glc3 mutant strains (Fig. 5). However, the lower cell fraction from the glc3 mutant exhibited a small but significant decrease in cell number (Fig. 5 b). glc3 mutant strains did exhibit a slight decrease in density (Fig. 5 a), suggesting that glycogen accumulation does not contribute substantially to the increased density in these cells. These results are consistent with our finding that some mitochondrial mutants, such as cox6 and atp2, which are also defective for glycogen accumulation (Saccharomyces Genome Database) are able to form lower bands (unpublished data).
Figure 5.
Glycogen accumulation has only a slight effect on the density and number of cells in the quiescent, lower fraction. (a) Parental (P) and glc3 mutant (G) cells from SP cultures separated by density-gradient centrifugation. (b) Number of cells in nonquiescent, upper (U) and quiescent, lower (L) fractions in the parental and glc3 mutant strain. The results in panel b are the mean and SD of three biological replicates.
Reactive oxygen species (ROS), apoptosis, and necrosis in nonquiescent cells
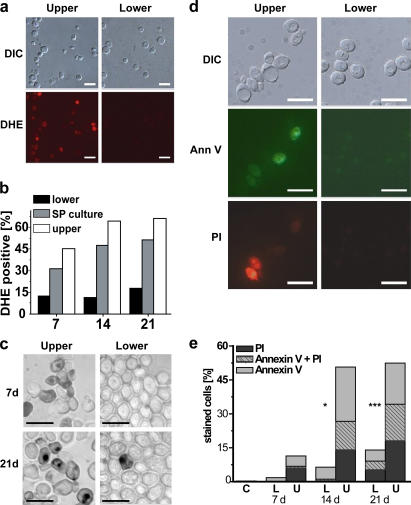
Chronologically aged SP cultures have been shown to display markers of apoptosis (Fabrizio et al., 2004; Herker et al., 2004). Aging cultures have also been demonstrated to accumulate ROS, which is typically a precursor to age-induced apoptosis (Herker et al., 2004). We examined 7-, 14-, and 21-d-old cells to determine whether quiescent and nonquiescent cell fractions differed with respect to ROS (detected by dihydroethidium [DHE] staining) and three other markers for apoptosis (Madeo et al., 1997).
ROS-positive cells were predominantly in the nonquiescent fraction. Lower-fraction, quiescent cells showed very low percentages of DHE-positive cells at all time points, reaching only slightly more than 15% by day 21 (Fig. 6, a and b). In contrast, 45% of the upper-fraction, nonquiescent cells were DHE-positive cells by day 7, increasing to >60% by 21 d (Fig. 6 b). As expected, cells from unfractionated SP cultures were intermediate between the quiescent and nonquiescent fractions (Fig. 6 b).
Figure 6.
Evaluation of nonquiescent, upper-fraction and quiescent, lower-fraction cells for ROS, apoptosis, and necrosis. (a) DHE staining to detect ROS in cells from upper and lower fractions from 7-d-old S288c SP cultures. DIC, differential interference contrast. (b) Flow cytometric quantification of DHE-positive, fractionated 7-, 14-, and 21-d-old S288c cells. (c) TUNEL staining to detect DNA fragmentation in 7- and 21-d-old S289 nonquiescent, upper- and quiescent, lower-fraction cells. (d) Ann V and PI costaining of 14-d-old S289 cells. (e) Flow cytometric quantification of 7-, 14-, and 21-d-old S289 lower (L) and upper (U) fraction cells or exponentially growing (C) cells costained with AnnV and PI. Bars, 10 μm.
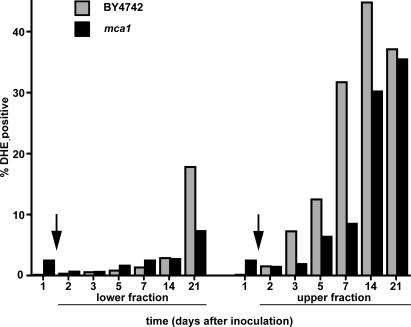
ROSs have been shown to cause cell death during SP (Fabrizio et al., 2004; Herker et al., 2004), and it has been suggested that yeast caspase is required for generation of oxygen radicals during induction of apoptosis (Fahrenkrog et al., 2004; Flower et al., 2005). We were interested in determining whether ROS accumulation was an immediate response to glucose exhaustion at the diauxic shift and dependent on caspase encoded by MCA1. For this experiment, we tested both fractions from parental (BY4742) and mca1 mutant strains (Madeo et al., 2002) over a 21-d period.
Quiescent cells, as expected, exhibited very low ROS accumulation at all time points (Fig. 7). For the first 14 d, very few quiescent cells from either the parental or mca1 mutant strain were ROS positive and, by day 21, only 18% of parental cells and <10% of mca1 mutants exhibited ROS accumulation, indicating a slight dependency of ROS accumulation on Mca1p. As expected, upper-fraction, nonquiescent cells showed much more rapid and extensive ROS accumulation. However, this accumulation occurred 2 d after glucose exhaustion and, thus, is not an immediate response to the change in carbon source. ROS accumulation in the mca1 mutant was delayed significantly (P < 0.05) from that in the parental strain, but by day 21, ROS accumulation in both strains was indistinguishable. These results suggest that the function of Mca1p is transient, consistent with previous evidence for additional sources of caspase-like activity in aging yeast cultures (Herker et al., 2004). Cells from nonseparated, SP cultures exhibited ROS accumulation that was typically intermediate between the two cell fractions (unpublished data). We conclude from these results that Mca1p has a positive but transient effect on ROS accumulation and that ROS accumulation is not fully induced by glucose exhaustion.
Figure 7.
Evaluation of ROS in parental and metacaspase-deficient (mca1) strains. Flow cytometric quantification of DHE-positive parental BY4742 and mca1 mutant cells from upper and lower fractions of 1-, 2-, 3-, 5-, 7-, 14-, and 21-d-old cultures. Arrows indicate the time of glucose exhaustion (diauxic shift).
Quiescent and nonquiescent cell fractions also differed with respect to other apoptosis-related markers: DNA fragmentation as detected by TUNEL staining (Fig. 6 c) and externalization of phosphatidyl serine and loss of membrane integrity by Annexin V (AnnV) and propidium iodide (PI) staining (Fig. 6, d and e). TUNEL staining was more pronounced in upper- fraction, nonquiescent cells than in quiescent cells at both 7 and 21 d after inoculation (Fig. 6 c), indicating apoptotic DNA fragmentation in nonquiescent cells. Flow cytometric analysis of AnnV and PI staining allowed differentiation of nonquiescent cells into four subpopulations: (1) nonstaining, viable cells; (2) early apoptotic cells with intact membranes (AnnV positive); (3) late apoptotic cells (AnnV + PI positive); and (4) necrotic cells (PI positive). 11% of upper-fraction, nonquiescent cells were stained with either AnnV (early apoptosis) or PI (necrosis) by day 7, and >50% of these cells were stained by day 14 (Fig. 6, d and e), including ∼15% that were doubly stained, indicating late-apoptotic cells (Fig. 6 e). In contrast, <2% of quiescent cells were positively stained with AnnV by day 7, which increased to 6 and 14% by days 14 and 21, respectively (Fig. 6, d and e). We conclude that there is a significant difference between quiescent and nonquiescent cells with respect to the timing and extent of induction of apoptosis and necrosis. Furthermore, the stability of AnnV and PI staining at 14 and 21 d suggests that the nonquiescent cell fraction is heterogeneous with respect to the induction of apoptosis.
Transcript abundance differs significantly between lower-fraction, quiescent and upper-fraction, nonquiescent cells
We examined transcript abundance by microarray analysis in quiescent and nonquiescent cell fractions to gain further insight into these cell populations. Statistical ranking analysis of biological replicates revealed 68 and 266 transcripts that distinguished quiescent and nonquiescent fractions, respectively (P < 10−5; Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200604072/DC1). Gene Ontology analysis revealed that 65 of the 266 transcripts in nonquiescent cells are known to be associated with DNA recombination (Kim et al., 1998), including Ty-element transposition.
These cells also accumulate transcripts encoding proteins involved in the resolution of stalled replication forks, including the DNA helicase Rrm3p (Ivessa et al., 2000), the conserved Mus81p-resolvase binding partner Mms4p (Boddy et al., 2001; Mullen et al., 2001; Ogrunc and Sancar, 2003), and the replication and DNA damage checkpoint proteins Mrc1p (Alcasabas et al., 2001) and Rad17p (Lydall et al., 1996). Additionally, nonquiescent cells accumulate transcripts encoding Amn1p, a negative regulator of exit from mitosis that helps reset the cell cycle, and Clb2p, a G2 cyclin that binds to Amn1p (Wang et al., 2003). Based on these observations, we hypothesize that nonquiescent cells are unable to arrest and that the consequences of entering S-phase under carbon starvation may contribute to the development of apoptosis in these cells.
In the lower, quiescent fraction, the most significant Gene Ontology–process categories were response to water stress and energy metabolism (Table II). Transcripts that differentiated quiescent from nonquiescent cells encode proteins involved in sensing and responding to ROS, such as Trr2p, a thioredoxin reductase involved in protection against oxidative stress and essential for survival during respiratory conditions (Trotter and Grant, 2005); Gtt1p, glutathione transferase; and Hyr1p (Gpx3), a glutathione peroxidase–like enzyme that functions as a hydroperoxide receptor involved in transducing a ROS-induced, redox signal to Yap1 (Delaunay et al., 2002); Mcr1p, involved in ergosterol biosynthesis (Lamb et al., 1999) and response to oxidative stress (Lee et al., 2001); and Bcy1p, the regulatory subunit for cAMP-dependent protein kinase, which is essential for entry into SP (Cannon and Tatchell, 1987) and activation of the oxidative stress response (Charizanis et al., 1999). Quiescent cells also accumulate three transcripts involved in high-affinity iron transport (FTR1), siderophore activity (ARN1), and mitochondrial iron ion homeostasis (MMT1), underscoring the importance of iron, which is essential for the survival of mitochondrial function in cells in SP cultures (Longo et al., 1999). We conclude from these results that studies of separated quiescent and nonquiescent cells from SP cultures provides cell type–specific information that cannot be determined from studies of unfractionated cultures alone.
Table II.
Biological processesa for genes strongly associatedb with lower- or upper-fraction cells from SP cultures
| Fraction | Biological processc | Gene (P < 0.01) |
|---|---|---|
| Lower/quiescent | Response to water, water deprivation, and dessication |
SIP18, GRE1 |
| Main pathways of carbohydrate metabolism and fatty acid oxidation |
MDH3, GND2, FBA, ACO1, PEX11 | |
| Generation of precursor metabolites and energy, energy derivation by oxidation of organic compounds, and oxidoreduction coenzyme metabolism |
MDH3, GND2, FBA1, MCR1, PDC5, ACO1, ADH1 | |
| Coenzyme metabolism | MDH3, COQ6, GND2, GTT1, ACO1 | |
| Organic acid metabolism and carboxylic acid metabolism |
MDH3, LYS21, AYR1, ARG3, FBA1, PDC5, ACO1, PEX11 | |
| Response to abiotic stimulus | AZR1, TRR2, BCY1, HYR1, MCR1, ORM2, SIP18, GRE1 | |
| Upper/nonquiescent | Ty element transposition; DNA transposition, recombination and metabolism; nucleobase, nucleoside, nucleotide, and nucleic acid metabolism; biopolymer and macromolecule metabolism |
RAD17, MLH2, RRM3, EXO1, BRR2, MIG2, ISN1, ELG1, SMP1, RPA12, RRP9, SFH1, MED7,PRP39, MMS4, MRC1, SCC2, UTP6, PRI1, PUF3, CFT2, MMS22, NAM2, OGG1, RIT1, HST3, MED4, SPT15, KAR4, ESF1, VPS72, YAP3, POL1, YAP6, IME4, RPO41, TRF5, RFC4, SKS1, HAP4, HNT2, ARP6, YOR356W, YGR161W-B, YGR161W-A, YCL019W, YJR027W, YLR410W-B, YDR210C-D/YDR210W-D YPR137C-B, YFL002W-A, YBL100W-B/YBL101W-B, YDR034C-D, YBR012W-B, YDR210W-B, YDR098C-B, YOR142W-B, YPR158C-D, YER138C, YOR192C-B, YHR214C-B, YML039W, YGR161C-D, YDR365W-B, YDR261C-D, YOR343W-B/YOR343C-B, YBL005W-B, YER160C, YNL054W-B, YMR045C, YMR050C, YLR157C-B, YLR035C-A, YPR158W-B, YAR009C, YDR316W-B, YML045W, YER137C-A, YPL257W-B, YJR029W, YOL103W-B, YGR038C-B, YDR316W-A, YNL284C-B, YGR027W-A, YPR158C-C, YER159C-A, YGR161C-C, YLR227W-B, YNL054W-A, YMR051C, YML045W-A, YHR214C-C, YDR210C-C/YDR210W-C, YGR027W-B, YML040W, YBR012W-A, YDR261W-B, YDR261W-A, YDR098C-A, YPR137C-A, YGR038C-A, YDR261C-C, YLR157C-A |
Gene Ontology, Saccharomyces Genome Database.
Statistical ranking analysis.
Processes are listed in order of decreasing statistical significance for each fraction.
Discussion
Two cell fractions are separable from yeast SP cultures by density-gradient centrifugation. Morphological and physiological characteristics, including DNA content and budding, indicate that cells in the denser, lower-fraction cells are quiescent and likely to be in a G0 state. The less dense, upper-fraction cells are heterogeneous, mostly nonquiescent, and, although viable as analyzed by FUN-1 staining, rapidly lose the ability to reproduce. About half of the nonquiescent cells in 14-d-old cultures appear to be apoptotic or necrotic, but a small percentage (∼10%) are thermotolerant, suggesting that there may be a few cells with quiescent properties. Further work is needed to determine the number of distinct populations present in the nonquiescent fraction and the regulation of reproductive capacity in these cells.
These findings demonstrate, in the face of years of misconceptions about yeast SP cultures, that there is a quiescent and, likely, a G0 state in yeast that is very distinct from other cells in SP cultures. As cultures approach and enter SP, there is a process that leads to morphologically and physiologically unique populations of unbudded, quiescent daughter cells and nonquiescent cells. There is also a switch that significantly changes the reproductive capacity of mother cells in the nonquiescent population. Recent studies of stem cells in tissues and microniches (Suda et al., 2005) and on yeast colonies (Palkova and Forstova, 2000; Minarikova et al., 2001) have demonstrated functional heterogeneity of cells as a function of position within a tissue or colony. Our observation of different life cycle trajectories within SP cultures is consistent with the hypothesis that both unicellular and metazoan species have evolved mechanisms for functional differentiation of cells in close proximity. This differentiation is likely to provide a selective advantage to microbes as well as to metazoans.
More than 260 transcripts were identified that distinguish quiescent from nonquiescent cells. Interestingly, the most abundant transcripts that distinguish quiescent from nonquiescent cell fractions are not the same as those transcripts that distinguish SP cultures from exponentially growing cultures (Gasch et al., 2000; Martinez et al., 2004; Radonjic et al., 2005). This important finding suggests that the transcriptional changes observed as cultures enter SP are likely to be physiological responses occurring in both quiescent and nonquiescent cells. One might consider that this is similar to a tissue-level response in metazoans. This further suggests that studying the development of cellular heterogeneity in living systems and the responses of functionally distinct populations at the genomic level will require examination of homogenous cell subpopulations in isolation.
Models of the development of quiescent and nonquiescent cells in yeast and their relationship to the mitotic cell cycle
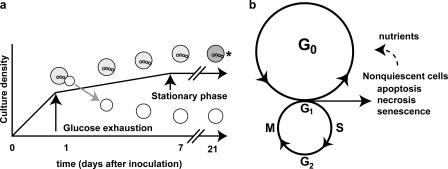
Our data suggest two models: one for the process by which quiescent and nonquiescent cells form in cultures grown to SP in rich, glucose-based medium (Fig. 8 a) and another for the relationship of these cells to the mitotic cell cycle (Fig. 8 b). During growth to SP, most quiescent cells (91% of which are daughter cells) are formed during the transition from fermentation to respiratory growth (the diauxic shift; Fig. 8 a). We hypothesize that there is a cell division during the diauxic shift by which mother cells give rise to quiescent daughter cells. During the postdiauxic phase, daughter cells increase in density, whereas the heterogeneous, nonquiescent cells undergo physiological differentiation, with about half of these cells becoming apoptotic. Most of the nonquiescent cells lose their ability to reproduce, apparently independent of the replicative age of the cell.
Figure 8.
Models for the formation of quiescent and nonquiescent cells in chronologically aged yeast cultures and the relationship between the cells in SP cultures and the mitotic cell cycle. (a) After glucose exhaustion, during the diauxic shift, quiescent daughter cells are formed and can be isolated from nonquiescent cells by density. (b) Daughter cells produced during the diauxic shift enter G0 and reenter the mitotic cell cycle when nutrients become available. Nonquiescent cells are heterogeneous; may continue in the mitotic cell cycle; and can become senescent, apoptotic, or necrotic. As dead cells break down, they release nutrients that can be used by G0 cells (broken arrow to the G0 cycle). A small percentage of nonquiescent cells may retain the ability to reproduce when nutrients become available. The asterisk indicates a nonquiescent fraction that is heterogeneous and contains both budded and unbudded cells.
There are a few caveats to the first model (Fig. 8 a). It does not include singly budded quiescent cells, of which ∼9% are also able to become quiescent, nor does it include the unbudded cells in the nonquiescent cell fraction. It seems likely that once the process by which mother cells give rise to quiescent daughter cells is understood, the factors that affect the ability of unbudded and singly budded mother cells to become quiescent will be easier to address. We have also not examined the cell division leading to the formation of quiescent cells yet. Nevertheless, the cell division that leads to quiescent daughter cells is likely to be asymmetric, as nonquiescent mother cells are very different morphologically and physiologically from quiescent cells in SP cultures.
This model (Fig. 8 a) does, however, lead to several important questions. For example, what are the specific roles of signal transduction pathways known to be required for the diauxic shift, including PKA, TOR, PKC, PHO85, and SNF1, in the differentiation of quiescent and nonquiescent cells (Gray et al., 2004)? It seems likely that at least one of these signaling pathways functions differently in the two cell types because, based on the physiological and transcript abundance differences, cells in the nonquiescent fraction may be unable to down-regulate PKA, a requirement for survival of SP cultures (Cannon and Tatchell, 1987).
The second model diagrams the relationship between cells in SP cultures and the mitotic cell cycle (Fig. 8 b). Normally, mother and daughter cells in exponentially growing cultures with unlimited glucose have identical outcomes: they both reenter the mitotic cell cycle. In this model, if a dividing cell (in the mitotic cell cycle) runs out of glucose, the daughter and mother cells have different outcomes in which daughter, but not mother, cells enter G0. In addition, a switch is activated in mother cells that make them unable to reproduce. The signal that limits reproduction is not absolutely a function of the replicative age of the mother but the physiological status shared by all mother cells.
In conclusion, we have been able to isolate and characterize two previously undiscovered cell types from yeast SP cultures. This characterization lays the ground work for examining the biogenesis of quiescence and the G0 state in an experimentally tractable organism. Notably, it has already produced a fundamental paradigm shift in our understanding of SP in the yeast S. cerevisiae. There are clear differences in response to glucose exhaustion by mother cells, which become nonquiescent, and daughter cells, which become quiescent. The evolutionary conservation of basic processes in eukaryotes would argue that the differentiation of these two cell types is not unique to yeast. Therefore, understanding the regulation of this process will be relevant to quiescence in the life cycle of pathogens, the development of cancer, and the survival and reproduction of all living organisms.
Materials and methods
Growth conditions
S288C (MATα SUC2 mal mel gal2 CUP1 flo1 flo8-1) was obtained from G. Fink (Massachusetts Institute of Technology, Cambridge, MA). S289 (MATa/α; SUC2/SUC2; ma/ mal; me/ mel; gal2/ gal2; CUP/CUP1 flo1/flo1 flo8-1/flo8-1), an S288c diploid derivative, was obtained by mating. BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), BY4743 (MATa/α; his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0), and mca1 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 mca1∷KANMX) were obtained from Open Biosystems. W303 (MATα ade2-1 ade3 trp1-1 leu2-3 leu2-112 his3-11 his3-15 ura3 can1-100) was obtained from L. Breeden (Fred Hutchinson Cancer Research Center, Seattle, WA). JC889-14B (MATα ura3 leu2 can1 his3 trp1 glc3∷TRP1) and the parental strain JC746-9D (MATα leu2-3, 112 ura3-52 can 1–100 trp1 his3-11, 15) were obtained from J. Cannon (University of Missouri, Columbia, MO). Strains were cultured at 30°C with aeration in YPD + A (2% yeast extract, 1% peptone, 2% glucose, 0.04 mg/ml adenine, and 50 μg/ml ampicillin; Rose et al., 1990).
Fractionation
RediGrad or Percoll density gradients (GE Healthcare) were prepared using the manufacturer's preformed gradient protocol with modifications. Percoll was diluted 9:1 (vol/vol) with 1.5 M NaCl, for a final NaCl concentration of 167 mM. To form the gradients, 10 ml of the Percoll solution was put into 15-ml Corex tubes and centrifuged at 13,800 RPM (19,240 g av) for 15 min at 20°C. Approximately 2 × 109 cells (200 OD600) were pelleted, resuspended in 1 ml Tris buffer, overlaid onto the preformed gradient, and centrifuged at 400 g av for 60 min in a tabletop centrifuge equipped with a swinging bucket rotor (Beckman Instruments) at 20°C. Fractions were collected, washed once in 40 ml Tris buffer, pelleted, and resuspended in ddH20 or conditioned medium for subsequent assays. For microarray and bud scar analysis, upper- and lower-fraction cells from the first fractionation were overlaid onto preformed gradients and centrifuged at 400 g av for 60 min, resulting in a two-step purification of the upper- and lower-fraction cells. For the time course separations (Fig. 1 A), glucose concentration was determined by using Precision glucose test strips (Precision Labs).
Cell counts for each fraction were determined using a particle count and size analyzer (Z2; Beckman Instruments). Buoyant densities of the fractions were determined by comparison to density marker beads (GE Healthcare) in gradients processed in parallel with samples. The fraction migrations were measured, as well as the bead migrations. The results for the beads were plotted and the buoyant densities of the samples were determined by spline fitting.
Quantification of fractions from parental and glc3 strains
For fraction quantification, ∼1.0 × 109 cells were pelleted and resuspended in 500 μl of Tris buffer and counted. For both the parental and glycogen mutant strains, 1.73 × 109 cells were overlaid onto three identical preformed gradients and centrifuged at 400 g av for 60 min in a tabletop centrifuge equipped with a swinging bucket rotor (Beckman Instruments) at 20°C. The upper cell fractions were collected into 50-ml conical tubes using a 1,000-ml pipetman. The remaining portion containing Percoll/NaCl/cells was vortexed and collected in separate 50-ml conical tubes. These fractions were washed once in 40 ml of Tris buffer, pelleted, and resuspended in 800 ml ddH2O for quantification. Cell counts for each fraction were determined using the particle count and size analyzer. Values represent the mean (±SD) for three replicates per fraction per strain.
Microscopy
A microscope (Optiphot; Nikon) equipped with a Phase 3 Plan 40 NA 0.70 DL objective was used for phase contrast light microscopy. Images were acquired with a digital camera (Coolpix 995; Nikon) affixed to the microscope with a 0.55× CCTV adaptor (Diagnostic Instruments). Differential interference contrast and fluorescent microscopy were done using an Axioskop 2 mot plus microscope (Carl Zeiss MicroImaging, Inc.) equipped with Plan-Apochromat 63× NA 1.40 oil (Carl Zeiss MicroImaging, Inc.) or Plan-Neofluar 100× NA 1.30 oil (Carl Zeiss MicroImaging, Inc.) objectives using DAPI (excitation, G 365, and emission, 445/50 [Carl Zeiss MicroImaging, Inc.], for Calcofluor white staining), FITC (excitation, BP 485/20, and emission, BP 515–565 [Carl Zeiss MicroImaging, Inc.], for AnnV staining), or Rhodamine (excitation, BP 546/12, and emission, FT 580 LP590 [Carl Zeiss MicroImaging, Inc.], for PI staining) filter sets. Images were acquired using AxioVision 4.4 software (Carl Zeiss MicroImaging, Inc.). Bud scar micrographs are composite images from Z stack acquisitions using AxioVision 4.4 software. Image postprocessing was done using Photoshop CS 8.0 and Illustrator CS 11.0.0 (Adobe). No image manipulations other than contrast, brightness, and color balance adjustments were used.
Transmission EM
For fixation of SP cultures (7 d after inoculation), ∼1 × 109 (100 OD600) cells were pelleted and washed twice in distilled H2O. Washed cells were resuspended in 1 ml of freshly prepared fixation solution (cold 2.5% EM grade glutaraldehyde in 0.1 M cold sodium cacodylate buffer, pH 7.2; Electron Microscopy Sciences) and fixed for 90 min on ice. The cells were pelleted, resuspended in 1 ml of fresh cacodylate buffer, and stored at 4°C until the cells were processed. Cells were embedded in Epon 812 (Shell) and examined in an electron microscope (CM12; Philips) as previously described (Otzen et al., 2004). Phosphotungstic acid staining to detect glycogen was performed as described previously (Farragiana and Marinozzi, 1979).
FUN-1 assay for quantitation of live/dead status
For assays at 7, 14, and 21 d, ∼2 × 107 S288c cells from each fraction of separated cultures, cultures grown overnight, and “killed cells” (ATP1 and ATP2 mutants grown for 14 or more days in YPD; Martinez et al., 2004) were harvested and washed twice in glucose-Hepes buffer (2% [wt/vol] d-[+]-glucose [Sigma-Aldrich] and 10 mM Na-Hepes [Sigma-Aldrich]). Each sample was resuspended in 500 μl of the glucose-Hepes buffer containing 2 μl FUN-1 (Invitrogen) and incubated for 45–60 min at 30°C. Cells were diluted to 1 × 106 cells/ml in Isoton II (Beckman Instruments), and 30,000 cells per sample were analyzed with a flow cytometer (FACSCalibur; Becton Dickinson) using 488 nm excitation and collecting fluorescent emission with filters at 530/30 nm for FL-1 parameter and 585/42 nm for FL-2 parameter. CellQuest software (Becton Dickinson) was used for data collection and analysis.
To establish flow cytometry parameters, 30,000 cells from cultures grown overnight (∼100% live) were assayed. Bright red (CIVS) and green fluorescent emissions were detected and plotted, and quadrants were established (Fig. S1 C, available at http://www.jcb.org/cgi/content/full/jcb.200604072/DC1) such that >99.9% of the fluorescent emissions from these positive control cells were contained in the top right quadrant. 30,000 killed cells stained with FUN-1 (Fig. S1 E), unstained overnight culture cells (Fig. S1 D), and unstained killed cells (Fig S1 F) were assayed and plotted as negative controls. >99% of all the negative control emissions were found in the bottom left quadrants. Error bars in Fig. 2 A represent the SD from the mean for n = 4–5 measurements. Heteroscedastic, two-tailed t tests were performed using Excel 2002 (Microsoft). Two-way ANOVAs were performed using Prism 4.01 (GraphPad Software).
Colony-forming assay
Unstained cells from samples used to assay live/dead status (see the previous section) were diluted, and 400 upper- or 200 lower-fraction cells were spread onto each of two YPD + A plates per sample and incubated at 30°C for 3 d. Colonies were counted, and the percentage of survival was calculated relative to a control sample of exponentially growing cells (overnight culture). Values represent the mean (±SEM) for 4–7 biological replicates. Heteroscedastic, two-tailed t tests were performed using Excel 2002. Two-way ANOVAs were performed using Prism 4.01.
Machine plating for colony-forming units (Fig. 4) was done using a cell sorter (MoFlo; DakoCytomation) equipped with an automated stage. A YPD + A 10-cm plate was placed on the stage, and the sorter was adjusted so that the machine would deposit single cells in a 12 × 12 square grid on the plate. The plates were incubated for 3 d at 30°C. The percentage of colony-forming units was determined using the following equation: (the number of colonies that grew/144) × 100. Values represent the mean (±SD) for three replicate plates per sample.
Thermotolerance assay
To test thermotolerance, 4 × 107 S288c haploid upper- and lower-fraction cells in glass test tubes were incubated for 0, 10, 15, or 20 min in a 52°C water bath. After the heat shock, the sample tubes were placed on ice until all samples were treated. For cell viability analysis, the cells were diluted and 400 cells from upper and 200 cells from lower fractions were spread onto each of two YPD + A plates per sample and incubated for 3 d at 30°C. The percentage of cells able to form colonies was calculated relative to plating identical numbers of untreated cells. 200 cells from S288c haploid cultures grown overnight and thermotolerance induced by incubation at 37°C for 1 h were used as a positive control for heat shock sensitivity. Values represent the mean (±SEM) for 5–6 biological replicates. t tests and ANOVAs were performed as described in the previous sections.
Budding index
S288c haploid lower- and upper-fraction cells were examined microscopically for the presence of new buds. Three fields of ∼50–60 cells were examined per time point, and the budding percentage was calculated by comparing the number of cells with new buds to the total number of cells counted.
DNA content analysis for determination of cell cycle stage
DNA content was analyzed using a SYBR Green I staining protocol (Fortuna et al., 2000) with modifications. For analysis, 600 μl (∼4.5 × 106 cells) of S288c lower-fraction cells from a 7-d-old SP culture were harvested into tubes containing 900 μl of 100% ethanol (for a final ethanol concentration of 70%), fixed overnight at 4°C, washed twice in 1 ml Tris buffer, resuspended in 900 μl Tris buffer and 100 μl of a 10× RNase A solution (10 mg/ml RNase A and 100 mM NaOAc; Sigma-Aldrich), and incubated overnight at 37°C. Samples were pelleted, resuspended in 1 ml of freshly prepared pepsin solution (5 mg/ml pepsin in H2O, pH adjusted, with 55 μl 1N HCl per milliliter of solution), and incubated at room temperature for 5 min. The samples were pelleted, washed twice in TE buffer (10 mM Tris base and 1 mM EDTA, pH 8.0), resuspended in 1 ml of a SYBR Green I staining solution (SYBR Green I stock solution [Invitrogen] diluted 1:10,000 in TE buffer and 0.25% Nonidet P40 [Sigma-Aldrich]), and stained overnight at 4°C in the dark. On the following day, the cells were washed twice in Tris buffer before dilution in Isoton II, and 30,000 cells per sample were analyzed on a FACSCalibur flow cytometer using 488 nm excitation and collecting fluorescent emission with filters at 530/30 nm for FL-1 parameter. CellQuest software was used for data collection and analysis. The voltage was adjusted to center the FL1-A G1 intensity peak at 200.
Calcofluor white M2R bud scar assay for quantitation of replicative age
Calcofluor white M2R 5 mM stock solution (Invitrogen) was diluted 1:200 in ddH2O for a 25-μM working solution. Approximately 2 × 107 S288c cells from SP cultures and the separated upper- and lower-cell fractions were pelleted, resuspended in the working solution, and incubated for 90 min at room temperature in the dark. The samples were washed twice in ddH2O. The cells were diluted to 1 × 106 cells/ml in Isoton II, and ∼30,000 cells per sample were analyzed with a flow cytometer (MoFlo; DakoCytomation) using 351 nm excitation and collecting fluorescent emission with filters at 450/65 nm for FL-6 parameter. Summit software (DakoCytomation) was used for data collection and analysis.
Gates were established such that ∼50% of the cells from a nonseparated SP culture were gated as having 0 bud scars. Gates for 1, 2, and 3 or more bud scars were established by examination of the histogram peaks, and, after sorting into microfuge tubes, by epifluorescent microscopic examination using a DAPI filter (Carl Zeiss MicroImaging, Inc.). The gating was refined through multiple rounds of sorting followed by microscopic examination. We were unable to discriminate between cells with various numbers of multiple bud scars because of lack of sensitivity at the high end of the linear fluorescence intensity scale. Two-way ANOVAs were performed using SAS software.
Apoptosis assays
DHE, TUNEL, AnnV, and PI staining were performed as described previously (Madeo et al., 1997, 1999). Cells were observed microscopically and quantitatively using flow cytometry.
DHE assay for quantitation of ROS
DHE stock solution (Invitrogen) was diluted 1:10 in PBS (Fluka) for a working solution. Approximately 1 × 108 S288c upper- and lower-fraction cells per sample were pelleted and resuspended in 100 μl of the YPD + A supernatant that had been filter sterilized. 1 μl DHE working solution was added to each sample and incubated for 3 min at room temperature in the dark. The samples were washed three times in PBS. The samples were diluted to 1 × 106 cells/ml in Isoton II, and 30,000 cells per sample were analyzed with a FACScan flow cytometer (CLONTECH Laboratories, Inc.) using 488 nm excitation and collecting fluorescent emission with filters at 585/42 nm for FL-1 parameters. CellQuest software was used for data collection and analysis.
AnnV/PI for determination of early- and late-stage apoptosis and necrosis
AnnV and PI costaining were done as described previously (Madeo et al., 1997). After staining, the samples were diluted to 1 × 106 cells/ml in Isoton II and 30,000 cells per sample were analyzed with a FACScan flow cytometer using 488 nm excitation and collecting fluorescent emission with filters at 530/30 nm for FL-1 parameter and 585/42 nm for FL-2 parameter. CellQuest software was used for data collection and analysis. Quadrants were established such that >99.9% of the autofluorescent emissions from unstained cells were contained in the bottom left quadrant.
Microarrays
BY4742 cells were grown for 7 d and separated into upper and lower fractions using the two-step density-gradient protocol. RNA was isolated, and labeled cDNA was prepared and hybridized to 70-mer DNA oligonucleotide microarrays as described previously (Aragon et al., 2006). Transcript abundance was analyzed using GenePix 6.0 as described previously (Aragon et al., 2006). Six replicates from each fraction were used for a total of 12 microarrays.
Statistical ranking analysis
To identify genes strongly associated with either fraction, we calculated for each gene the difference between lower and upper gene expression for all microarrays in the dataset and then calculated the mean of these differences for each. If the mean difference for each gene is positive, this gene will be more highly expressed in the lower fraction. Conversely, if the mean paired difference is negative, this gene will be more highly expressed in the upper fraction. As a result, we transformed the mean difference in expression for each gene to a t distribution (Moore, 2004) with n − 1 degrees of freedom, where n is the number microarrays, and generated a two-tailed p-value using the t cumulative distribution function. We established a significance cutoff level of P = 0.001 to generate gene lists for the upper and lower fractions. The upper- and lower-fraction gene lists (Table S1) were queried, and annotations for gene processes were obtained from the Saccharomyces Genome Database Gene Ontology (http://db.yeastgenome.org/cgi-bin/GO/goTermMapper). A step-by-step description of this method can be found in the supplemental text.
Online supplemental material
Fig. S1 shows flow cytometric scatter plots of FUN-1–stained upper- and lower-fraction cells from SP cultures, controls, and a representative micrographic image of a FUN-1–stained cell. Fig. S2 shows micrographs of bud scars from FACS sorting. The supplemental text gives a step-by-step description of the microarray ranking analysis. Table S1 provides gene lists for genes strongly associated with quiescent and nonquiescent cells from the microarray statistical ranking analysis. Online supplemental material is available at http://jcb.org/cgi/content/full/jcb.200604072/DC1.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratory for discussions, especially Steve Phillips and Mark Carter for their careful reading of the manuscript, and to Linda Breeden and John Cannon for providing yeast strains. Technical and instrument support was provided by the University of New Mexico Shared Flow Cytometry Resource.
This work was supported by grants from the National Institutes of Health (GM-060201 to J.A. Thomas and A.D. Aragon, GM-072351 to C. Allen, and GM-67593 to M. Werner-Washburne), the United States Department of Agriculture (99-38422-8034 to A.D. Aragon), the National Science Foundation (MCB0220425 to S.W. Ruby and MCB-0092364 to M. Werner-Washburne), the Fonds zur Förderung der wissenschaftlichen Forschung (Austria; S-9304-B05 to F. Madeo and S. Buttner), and deutsche Forschungsgemeinschaft (MA2587 to F. Madeo).
Abbreviations used in this paper: AnnV, Annexin V; ANOVA, analysis of variance; DHE, dihydroethidium; FUN-1, 2-chloro-4-(2,3-dihydro-3-methyl-[benzo-1, 3-thiazol-2-yl]-methylidene)-1-phenylquinolinium iodide; PI, propidium iodide; ROS, reactive oxygen species; SP, stationary phase.
References
- Abeliovich, H., and D.J. Klionsky. 2001. Autophagy in yeast: mechanistic insights and physiological function. Microbiol. Mol. Biol. Rev. 65:463–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcasabas, A.A., A.J. Osborn, J. Bachant, F.H. Hu, P.J.H. Werler, K. Bousset, K. Furuya, J.F.X. Diffley, A.M. Carr, and S.J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958–965. [DOI] [PubMed] [Google Scholar]
- Alexander, B.D., and J.R. Perfect. 1997. Antifungal resistance trends towards the year 2000. Implications for therapy and new approaches. Drugs. 54:657–678. [DOI] [PubMed] [Google Scholar]
- Aragon, A.D., G.A. Quiñones, E.V. Thomas, S. Roy, and M. Werner-Washburne. 2006. Release of extraction-resistant mRNA in stationary phase Saccharomyces cerevisiae produces a massive increase in transcript abundance in response to stress. Genome Biol. 7:R9; 10.1186/gb- 2006-7-2-r9. [DOI] [PMC free article] [PubMed]
- Boddy, M.N., P.H.L. Gaillard, W.H. McDonald, P. Shanahan, J.R. Yates, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 107:537–548. [DOI] [PubMed] [Google Scholar]
- Bonini, B.M., C. van Vaeck, C. Larsson, L. Gustafsson, P.S. Ma, J. Winderickx, P. van Dijck, and J.M. Thevelein. 2000. Expression of Escherichia coli otsA in a Saccharomyces cerevisiae tps1 mutant restores trehalose 6-phosphate levels and partly restores growth and fermentation with glucose and control of glucose influx into glycolysis. Biochem. J. 350:261–268. [PMC free article] [PubMed] [Google Scholar]
- Brejning, J., L. Jespersen, and N. Arneborg. 2003. Genome-wide transcriptional changes during the lag phase of Saccharomyces cerevisiae. Arch. Microbiol. 179:278–294. [DOI] [PubMed] [Google Scholar]
- Cannon, J.F., and K. Tatchell. 1987. Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7:2653–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, E., J.W. Yang, U. Nagavarapu, and G.S. Herron. 2002. Aging and survival of cutaneous microvasculature. J. Invest. Dermatol. 118:752–758. [DOI] [PubMed] [Google Scholar]
- Charizanis, C., H. Juhnke, B. Krems, and K.D. Entian. 1999. The oxidative stress response mediated via Pos9/Skn7 is negatively regulated by the Ras PKA pathway in Saccharomyces cerevisiae. Mol. Gen. Genet. 261:740–752. [DOI] [PubMed] [Google Scholar]
- Choder, M. 1991. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 5:2315–2326. [DOI] [PubMed] [Google Scholar]
- Coller, H.A., L. Sang, and J.M. Roberts. 2006. A new description of cellular quiescence. PLoS Biol. 4:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaunay, A., D. Pflieger, M.-B. Barrault, J. Vinh, and M.B. Toledano. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 111:471–481. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., L. Battistella, R. Vardavas, C. Gattazzo, L.-L. Liou, A. Diaspro, J.W. Dossen, E.B. Gralla, and V.D. Longo. 2004. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog, B., U. Sauder, and U. Aebi. 2004. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell Sci. 117:115–126. [DOI] [PubMed] [Google Scholar]
- Farragiana, T., and V. Marinozzi. 1979. Phosphotungstic acid staining of polysaccharides containing structures on epoxy embedded tissues. J. Submicrosc. Cytol. 11:263–265. [Google Scholar]
- Flower, T.R., L.S. Chesnokova, C.A. Froelich, C. Dixon, and S.N. Witt. 2005. Heat shock prevents alpha-synuclein-induced apoptosis in a yeast model of Parkinson's disease. J. Mol. Biol. 351:1081–1100. [DOI] [PubMed] [Google Scholar]
- Fortuna, M., M.S. Joao, M. Corte-Real, C. Leäo, A. Salvador, and F. Sansonetty. 2000. Cell cycle analysis of yeasts. In Current Protocols in Cytometry. Vol. 13. John Wiley & Sons, New York, New York. 11.13.1–11.13.9. [DOI] [PubMed]
- Fuge, E.K., E.L. Braun, and M. Werner-Washburne. 1994. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J. Bacteriol. 176:5802–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch, A., P. Spellman, C. Kao, O. Carmel-Harel, M. Eisen, G. Storz, D. Botstein, and P. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11:4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron, C.M., N. Minois, V.D. Longo, S.D. Pletcher, and J.W. Vaupel. 2003. Biodemographic trajectories of age-specific reproliferation from stationary phase in the yeast Saccharomyces cerevisiae seem multiphasic. Mech. Ageing Dev. 124:1059–1063. [DOI] [PubMed] [Google Scholar]
- Gouge, R.C., P. Marshburn, B.E. Gordan, W. Nunley, and Y.M. Huet-Hudson. 1998. Nitric oxide as a regulator of embryonic development. Biol. Reprod. 58:875–879. [DOI] [PubMed] [Google Scholar]
- Gray, J.V., G.A. Petsko, G.C. Johnston, D. Ringe, R.A. Singer, and M. Werner-Washburne. 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68:187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker, E., H. Jungwirth, K.A. Lehmann, C. Maldener, K.U. Frohlich, S. Wissing, S. Buttner, M. Fehr, S. Sigrist, and F. Madeo. 2004. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa, A.S., J.Q. Zhou, and V.A. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 100:479–489. [DOI] [PubMed] [Google Scholar]
- Kim, J.M., S. Vanguri, J.D. Boeke, A. Gabriel, and D.F. Voytas. 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8:464–478. [DOI] [PubMed] [Google Scholar]
- Lamb, D.C., D.E. Kelly, N.J. Manning, M.A. Kaderbhai, and S.L. Kelly. 1999. Biodiversity of the P450 catalytic cycle: yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction. FEBS Lett. 462:283–288. [DOI] [PubMed] [Google Scholar]
- Lee, J.-S., W.-K. Huh, B.-H. Lee, Y.-U. Baek, C.-S. Hwang, S.-T. Kim, Y.-R. Kim, and S.-O. Kang. 2001. Mitochondrial NADH-cytochrome b5 reductase plays a crucial role in the reduction of d-erythroascorbyl free radical in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1527:31–38. [DOI] [PubMed] [Google Scholar]
- Lewis, D.L., and D.K. Gattie. 1991. The ecology of quiescent microbes. ASM News. 57:27–32. [Google Scholar]
- Lillie, S., and J. Pringle. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143:1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, V.D., L.L. Liou, J.S. Valentine, and E.B. Gralla. 1999. Mitochondrial superoxide decreases yeast survival in stationary phase. Arch. Biochem. Biophys. 365:131–142. [DOI] [PubMed] [Google Scholar]
- Lydall, D., Y. Nikolsky, D.K. Bishop, and T. Weinert. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 383:840–843. [DOI] [PubMed] [Google Scholar]
- Madeo, F., E. Frohlich, and K.-U. Frohlich. 1997. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139:729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, F., E. Frohlich, M. Ligr, M. Grey, S.J. Sigrist, D.H. Wolf, and K.-U. Frohlich. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lachelt, M. Herian, M. Fehr, K. Lauber, S.J. Sigrist, S. Wesselborg, and K.U. Frohlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell. 9:911–917. [DOI] [PubMed] [Google Scholar]
- Martinez, M.J., S. Roy, A.B. Archuletta, P.D. Wentzell, S. Santa Anna-Arriola, A.L. Rodriguez, A.D. Aragon, G.A. Quinones, C. Allen, and M. Werner-Washburne. 2004. Genomic analysis of stationary-phase and exit in Saccharomyces cerevisiae: gene expression and identification of novel essential genes. Mol. Biol. Cell. 15:5295–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minarikova, L., M. Kuthan, M. Ricicova, J. Forstova, and Z. Palkova. 2001. Differentiated gene expression in cells within yeast colonies. Exp. Cell Res. 271:296–304. [DOI] [PubMed] [Google Scholar]
- Moore, D.S. 2004. The Basic Practice of Statistics. W.H Freeman and Company, New York, New York. 691 pp.
- Mullen, J.R., V. Kaliraman, S.S. Ibrahim, and S.J. Brill. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 157:103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrunc, M., and A. Sancar. 2003. Identification and characterization of human MUS81-MMS4 structure-specific endonuclease. J. Biol. Chem. 278:21715–21720. [DOI] [PubMed] [Google Scholar]
- Otzen, M., U. Perband, D.Y. Wang, R.J.S. Baerends, W.H. Kunau, M. Veenhuis, and I.J. Van der Klei. 2004. Hansenula polymorpha Pex19p is essential for the formation of functional peroxisomal membranes. J. Biol. Chem. 279:19181–19190. [DOI] [PubMed] [Google Scholar]
- Palkova, Z., and J. Forstova. 2000. Yeast colonies synchronise their growth and development. J. Cell Sci. 113:1923–1928. [DOI] [PubMed] [Google Scholar]
- Pardee, A. 1974. A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. USA. 71:1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, N.M., J.D. Dick, and W.R. Bishai. 1998. Mechanisms of latency in Mycobacterium tuberculosis. Trends Microbiol. 6:107–112. [DOI] [PubMed] [Google Scholar]
- Pinon, R. 1978. Folded chromosomes in non-cycling yeast cells: evidence for a characteristic G0 form. Chromosoma. 67:263–274. [DOI] [PubMed] [Google Scholar]
- Radonjic, M., J.C. Andrau, P. Lijnzaad, P. Kemmeren, T.T.J.P. Kockelkorn, D. van Leenen, N.L. van Berkum, and F.C.P. Holstege. 2005. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 18:171–183. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 198 pp.
- Suda, T., F. Arai, and A. Hirao. 2005. Hematopoietic stem cells and their niche. Trends Immunol. 26:426–433. [DOI] [PubMed] [Google Scholar]
- Trotter, E.W., and C.M. Grant. 2005. Overlapping roles of the cytoplasmic and mitochondrial redox regulatory systems in the yeast Saccharomyces cerevisiae. Eukaryot. Cell. 4:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., T. Shirogane, D. Liu, J.W. Harper, and S.J. Elledge. 2003. Exit from exit: resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell. 112:697–709. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne, M., E. Braun, G. Johnston, and R. Singer. 1993. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev. 57:383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne, M., B. Wylie, K. Boyack, E. Fuge, J. Galbraith, J. Weber, and G. Davidson. 2002. Comparative analysis of multiple genome-scale data sets. Genome Res. 12:1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.