Abstract
Satellite cells play a critical role in skeletal muscle regeneration in response to injury. Notch signaling is vital for satellite cell activation and myogenic precursor cell expansion but inhibits myogenic differentiation. Thus, precise spatial and temporal regulation of Notch activity is necessary for efficient muscle regeneration. We report that the basic helix-loop-helix transcription factor Stra13 modulates Notch signaling in regenerating muscle. Upon injury, Stra13−/− mice exhibit increased cellular proliferation, elevated Notch signaling, a striking regeneration defect characterized by degenerated myotubes, increased mononuclear cells, and fibrosis. Stra13−/− primary myoblasts also exhibit enhanced Notch activity, increased proliferation, and defective differentiation. Inhibition of Notch signaling ex vivo and in vivo ameliorates the phenotype of Stra13−/− mutants. We demonstrate in vitro that Stra13 antagonizes Notch activity and reverses the Notch-imposed inhibition of myogenesis. Thus, Stra13 plays an important role in postnatal myogenesis by attenuating Notch signaling to reduce myoblast proliferation and promote myogenic differentiation.
Introduction
Skeletal muscle exhibits a tremendous capacity for regeneration in response to muscle injury induced by disease, trauma, or intensive exercise. The process of muscle regeneration involves two distinct phases: a degenerative phase, in which inflammatory cells infiltrate the site of injury and play a critical role in the phagocytosis of necrotic myofibers, and a regenerative phase, in which muscle stem cells are activated, proliferate, differentiate, and fuse to form new myofibers (Hawke and Garry, 2001; Chen and Goldhammer, 2003; Charge and Rudnicki, 2004; for review see Tidball, 2005). Crushed muscle fibers produce mitogenic factors, and, in addition, activated macrophages also secrete various cytokines and growth factors that are important for the activation and proliferation of satellite cells. Normally, the regenerated muscle is functionally identical to uninjured muscle, but if the regeneration process is compromised, muscle tissue is replaced by scar tissue. Defects in the immune response (Lescaudron et al., 1999; Warren et al., 2005), altered production or signaling by growth factors (Floss et al., 1997; McCroskery et al., 2003; Cornelison et al., 2004), or disruption of molecules that regulate cellular proliferation (Megeney et al., 1996; Garry et al., 2000; Hawke et al., 2003) result in impaired muscle regeneration.
Satellite cells (the skeletal muscle stem cells) are primarily responsible for regeneration (Hawke and Garry, 2001; Chen and Goldhammer, 2003; Charge and Rudnicki, 2004). Although normally quiescent, satellite cells are activated upon muscle damage and reenter the cell cycle, providing a pool of proliferating myoblasts that differentiate and fuse to form new myofibers, leading to the complete regeneration of damaged muscles. Quiescent satellite cells do not express the myogenic basic helix-loop-helix (bHLH) factor MyoD (Cornelison and Wold, 1997) but are positive for Pax7 (Seale et al., 2000) and CD34 (Beauchamp et al., 2000). Upon activation by damage, satellite cells proliferate and rapidly up-regulate the expression of myogenic factors MyoD and Myf5, whereas myogenin and MRF4 are up-regulated later during differentiation (Charge and Rudnicki, 2004). Several pathways, including hepatocyte growth factor (Tatsumi et al., 1998; Miller et al., 2000), myostatin (McCroskery et al., 2003), Notch (Conboy and Rando 2002), and p38α/β MAPK (Jones et al., 2005), regulate the transition of satellite cells from quiescence to an activated state. In addition, various cytokines and growth factors such as interleukin-4, transforming growth factor-β, and insulin-like growth factor (Allen and Boxhorn, 1989; Horsley et al., 2003; Li et al., 2004) regulate proliferation and differentiation of myogenic precursor cells. Among the various pathways that regulate distinct steps of satellite cell activation, Notch signaling regulates not only the transition of satellite cells from quiescence to actively proliferating myoblasts but also plays a role in subsequent myogenic differentiation (Kopan et al., 1994; Conlon et al., 1995; Nofziger et al., 1999; Conboy and Rando 2002; Conboy et al., 2003).
The Notch signaling pathway plays an important role in cellular proliferation, differentiation, and apoptosis (Artavanis-Tsakonas and Lake, 1999; Kadesch, 2004; Lai, 2004). Notch signaling is triggered by the interaction between Notch ligands and receptors. This interaction results in the proteolytic cleavage of Notch, which allows the release and translocation of Notch intracellular domain (NICD) into the nucleus, where it interacts with the DNA-bound transcription factor CBF-1/RBPJk (recombination signal sequence–binding protein for Jκ). This association results in the recruitment of coactivators and the activation of downstream target genes Hes1, Hes5, Hey1, Hey2, and HeyL (Jarriault et al., 1995; Ohtsuka et al., 1999; Iso et al., 2003), which are members of the bHLH family of transcription factors. Because Notch signaling regulates different biological processes, it is not surprising that the pathway is tightly regulated at different levels, including expression of the ligands, translocation of Notch into the nucleus, and regulation of NICD activity in the nucleus (Artavanis-Tsakonas and Lake, 1999; Kadesch, 2004; Lai, 2004).
Stra13/DEC1/SHARP-2 is a bHLH transcription factor that is expressed in a number of cell types during mouse embryogenesis (Boudjelal et al., 1997). Similar to Hes and Hey proteins, Stra13 contains an orange (O) domain, which is characteristically seen in all members of the bHLH-O transcriptional repressor subfamily (Davis and Turner, 2001). However, Stra13 differs from the Hes and Hey families in having distinct DNA-binding properties as well as transcriptional repression mechanisms (Boudjelal et al., 1997; Sun and Taneja, 2000). Stra13 expression is inducible by several stimuli in different cell types, and gain of function in cultured cells has demonstrated its role in the regulation of cellular differentiation, cell cycle arrest, stress response, and apoptosis. For instance, the overexpression of Stra13 in many cell types causes cell cycle arrest (Sun and Taneja, 2000; Beadling et al., 2001; Li et al., 2002; Seimiya et al., 2002) and promotes neurogenic and chondrogenic differentiation (Boudjelal et al., 1997; Shen et al., 2002) but inhibits adipogenesis (Yun et al., 2002). We have previously shown that Stra13-null mice exhibit defects in T cell activation, which leads to the development of a lupus-like autoimmune disorder (H. Sun et al., 2001) as well as defects in radiation-induced apoptosis (Thin et al., 2007). However, the role of Stra13 in the development or function of other cell types has not been determined in vivo.
Because Stra13 is expressed in skeletal muscle (Boudjelal et al., 1997), in this study, we investigated its function in myogenesis and muscle regeneration in vivo. We found that although Stra13 is expressed in embryonic and adult muscles, it is dispensable for skeletal muscle development. However, analysis of Stra13−/− muscle upon injury revealed a striking defect in muscle regeneration characterized by degenerated myotubes, increased mononuclear cells, and fibrosis. Interestingly, Notch signaling is elevated in Stra13−/− primary myoblasts as well as in regenerating muscle. Strikingly, blocking Notch signaling ex vivo in Stra13−/− myoblasts reduces cellular proliferation and enhances myogenic differentiation. Moreover, the inhibition of Notch signaling at late stages subsequent to satellite cell activation in vivo results in improved regeneration with a concomitant reduction in cellular proliferation in Stra13−/− mice. We demonstrate that Stra13 antagonizes the Notch-imposed inhibition of myogenic differentiation and inhibits Notch activity in vitro by interaction with the intracellular domain of Notch1, resulting in its reduced association with CBF-1. We conclude that Stra13 plays an important role in the regulation of postnatal myogenesis by antagonizing Notch signaling.
Results
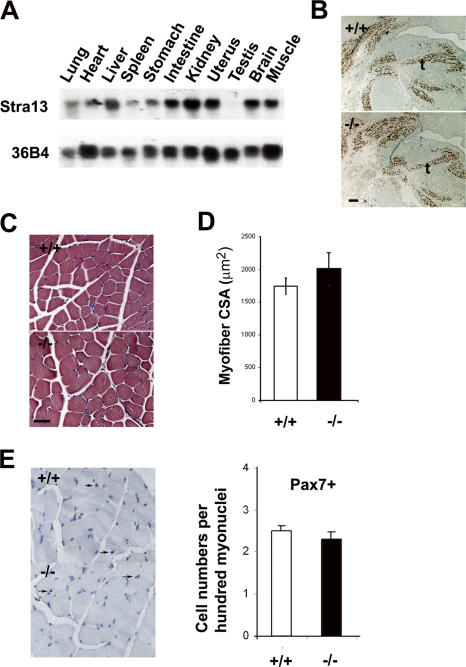
We have previously demonstrated that Stra13 is expressed in skeletal muscle during embryonic development (Boudjelal et al., 1997), and Northern blot analysis of various adult tissues further confirmed that Stra13 was expressed in differentiated adult muscles (Fig. 1 A). To evaluate myogenic differentiation in Stra13−/− mutants, embryonic day (E) 13.5 embryo sections were stained with myogenin (Fig. 1 B) or myosin heavy chain (MHC) antibodies (not depicted), which did not reveal any substantial difference between wild-type (WT) and Stra13−/− embryos. Moreover, histological analysis of adult muscle sections from WT and Stra13−/− mice did not reveal any overt differences in fiber morphology (Fig. 1 C) and fiber size (Fig. 1 D). The number of Pax7+ satellite cells in tissue sections (Fig. 1 E) was also similar in both genotypes, indicating that Stra13 is dispensable for skeletal muscle development.
Figure 1.
Normal skeletal muscle development in Stra13−/− mice. (A) RNA from adult mouse tissues analyzed by Northern blotting revealed Stra13 expression in skeletal muscle. 36B4 was used as an internal control. (B) E13.5 sagittal sections through the developing tongue of WT (+/+) and Stra13−/− (−/−) embryos stained with anti-myogenin antibody revealed comparable expression, indicating normal muscle development. (C) HE staining of tibialis anterior muscles from WT (+/+) and Stra13−/− (−/−) mice showed normal histology of Stra13−/− muscle. (D) Measurement of the cross section area (CSA) of quadriceps did not reveal any overt difference in myofiber size between WT and Stra13−/− mice (n = 4). The cross section area of at least 1000 individual muscle fibers was analyzed, and the mean areas were compared. (E) Tibialis anterior muscle from WT (+/+) and Stra13−/− (−/−) mice (n = 3) stained with anti-Pax7 antibody indicated a comparable number of satellite cells (left). Arrows indicate satellite cells expressing Pax7. Quantification of Pax7+ cells per 100 myonuclei is shown on the right. At least 1,000 nuclei were counted for each animal, and the data presented are a mean of three animals. Data are means ± SEM (error bars). t, tongue. Bars, 100 μm.
Impaired muscle regeneration upon injury in Stra13−/− mice
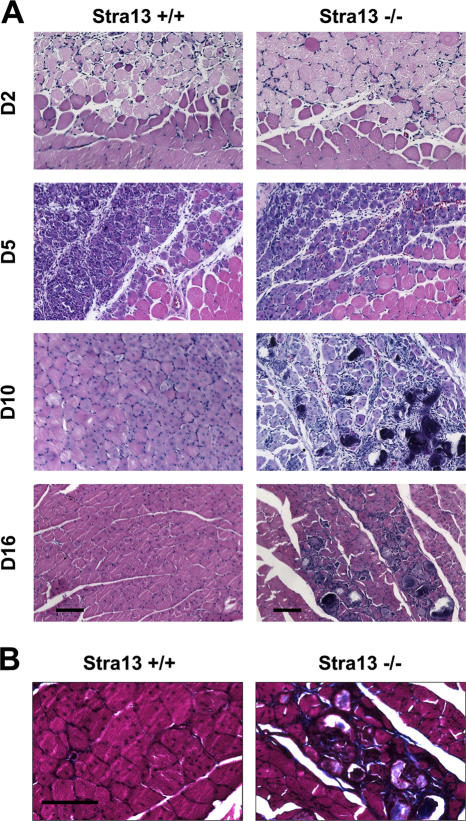
We next investigated whether Stra13 plays a role in postnatal myogenesis by inducing muscle regeneration in Stra13−/− mice. WT and Stra13−/− muscle were subjected to freeze injury (Conboy and Rando, 2002) and analyzed histologically at days 2, 5, 10, and 16 after injury. 2 d after injury, cross sections of quadriceps muscles showed substantial fiber damage and infiltration of immune cells in both WT and Stra13−/− mice (Fig. 2 A), and no phenotypic differences were apparent. 5 d after injury, the damaged area exhibited a mixed population of cells, which included necrotic fibers, activated satellite cells, proliferating myoblasts, differentiating myocytes, and newly formed myotubes (Hawke and Garry, 2001) in both WT and Stra13−/− mice. By days 10 and 16 after injury, damage was no longer visible in WT muscle, and the regenerated muscle contained newly formed myofibers. In contrast, Stra13−/− muscle exhibited a regeneration defect characterized by degenerated myotubes and the presence of many mononuclear cells. Moreover, Masson's trichrome staining revealed collagen deposition in the interstitium of injured muscles in Stra13−/− mice (Fig. 2 B). These results showed that upon muscle injury, Stra13−/− muscle exhibited impaired regeneration and enhanced fibrosis.
Figure 2.
Impaired muscle regeneration in Stra13−/− mice. (A) Skeletal muscle regeneration was induced by freeze injury in WT and Stra13−/− mice, and muscle was collected on days 2 (D2), 5, 10, and 16 after injury. A clear boundary between the injured and uninjured tissue is visible. At days 10 and 16 after injury, myonecrosis and mononuclear cells are apparent in Stra13−/− muscle. (B) Masson's trichrome staining revealed collagen deposition (blue staining) in Stra13−/− (−/−) muscle at day 16 after injury. Bars, 100 μm.
Enhanced satellite cell activation in injured Stra13−/− muscle
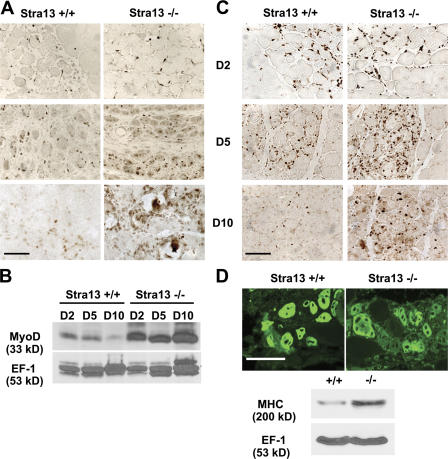
Because satellite cell activation is crucial for efficient regeneration, we examined whether satellite cell activation is altered in Stra13−/− muscle. MyoD, an early marker of satellite cell activation, and Ki67, a marker for proliferating cells, were used to analyze the presence of activated myogenic precursor cells. MyoD+ cells were detected in the injured tissue in both WT and Stra13−/− muscle 2 d after injury (Fig. 3 A). At day 5, MyoD+ cells increased in both genotypes as a result of the expansion of activated satellite cells, although a higher number of MyoD+ cells were seen in Stra13−/− muscle. 10 d after injury, MyoD expression dropped dramatically with myofiber maturation, which left few MyoD+ cells in WT muscle. In contrast, a substantially higher number of MyoD+ cells were apparent in the damaged area in Stra13−/− muscle by immunostaining, and Western blot analysis confirmed increased MyoD levels in the mutant tissue (Fig. 3 B). Similar results were seen with Ki67 antibody (Fig. 3 C). These results indicated a sustained satellite cell activation leading to an increased number of myogenic precursor cells in Stra13−/− regenerating muscle. To examine the differentiation status in Stra13−/− regenerating muscle, muscle sections were stained on day 5 after injury with an antibody against embryonic MHC (eMHC), which is expressed in newly formed myotubes. Consistent with the histological analysis (Fig. 2 A), eMHC was expressed in Stra13−/− myotubes (Fig. 3 D), and Western blot analysis revealed increased MHC levels at day 5 after injury (Fig. 3 D), likely as a result of an increase in the total number of newly formed myotubes. These results suggested that although the number of myogenic precursor cells is increased, myogenic differentiation during muscle regeneration was not affected in the absence of Stra13.
Figure 3.
Enhanced satellite cell activation in Stra13−/− regenerating muscle. (A) Sections of injured muscle from WT and Stra13−/− mice were immunostained on days 2 (D2), 5, and 10 after injury with an antibody against MyoD. (B) Protein extracts from WT and Stra13−/− regenerating muscle on days 2, 5, and 10 after injury were analyzed using anti-MyoD antibody. (C) Sections of injured muscle from WT and Stra13−/− mice were immunostained on days 2, 5, and 10 after injury with antibody against Ki67. (D) Muscle sections on day 5 after injury were stained with monoclonal antibody against eMHC (top), and protein extracts from WT and Stra13−/− regenerating muscle on day 5 after injury were analyzed using anti-MHC antibody (bottom). Bars, 100 μm.
Increased proliferation and impaired differentiation of Stra13−/− myoblasts
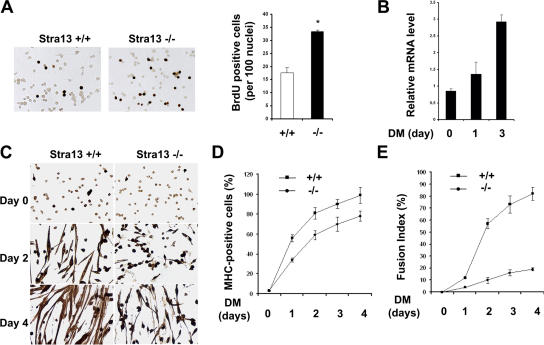
To examine whether defective regeneration in Stra13−/− mutants occurs as a result of a cell-autonomous defect in the proliferation and differentiation of myogenic precursor cells, primary myoblasts were isolated from WT and Stra13−/− mice and analyzed for their ability to proliferate and differentiate in vitro. In growth medium, 20% of WT myoblasts exhibited BrdU incorporation, whereas 33% of Stra13−/− myoblasts were labeled (Fig. 4 A). When cultured in differentiation medium, myoblasts exit the cell cycle, differentiate, and fuse to form myotubes. Stra13 mRNA (Fig. 4 B) was expressed at low levels in proliferating WT myoblasts and was up-regulated upon the induction of differentiation. After 4 d in differentiation medium, almost 90% of WT cells were MHC+, whereas 70% of Stra13−/− cells turned on MHC expression, suggesting defective differentiation (Fig. 4, C and D). Calculation of the fusion index revealed that >80% of WT myocytes contained more than two myonuclei in contrast to 20% for mutant myocytes (Fig. 4 E). Thus, although satellite cells from Stra13−/− mutants exhibit an increased proliferation capacity both in vivo and in vitro, their inability to undergo myogenic differentiation and fusion was apparent only in vitro.
Figure 4.
Increased proliferation and impaired differentiation of Stra13−/− primary myoblasts. (A) WT and Stra13−/− cells cultured in growth medium were pulsed with BrdU and immunostained with anti-BrdU antibody (left). The percentage of BrdU-positive cells was significantly higher in Stra13−/− cells compared with WT cells (right; *, P < 0.05). At least 500 cells were counted for each experiment, and three independent primary myoblast isolates for each genotype were assayed. Data are means ± SEM (error bars). (B) WT primary myoblasts were cultured in differentiation medium and collected on days 0, 1, and 3. Stra13 mRNA level was measured by quantitative PCR. The results were normalized with glyceraldehyde-3-phosphate dehydrogenase and are presented as the mean of triplicates. Data are means ± SD. The experiment was repeated with three independent myoblast isolates, which gave similar results. (C) Primary myoblast cells were differentiated and stained with antibody against MHC. Stra13−/− myoblasts exhibited impaired differentiation and myotube formation. (D) The percentage of differentiated cells was calculated by counting the number of MHC-stained myocytes/total number of nuclei, which revealed lower myogenic differentiation in Stra13−/− cells. (E) Calculation of fusion indices as the percentage of cells containing more than or equal to two nuclei within a differentiated myotube revealed the impaired fusion capacity of Stra13−/− cells. (D and E) Data are means ± SEM (n = 3).
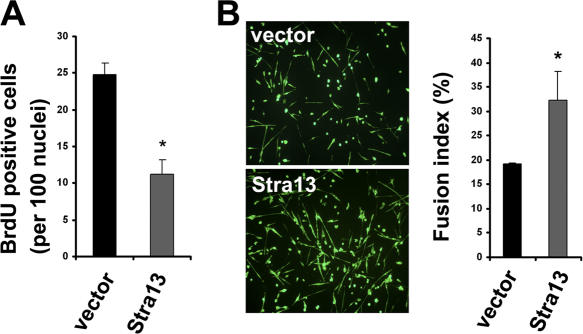
To confirm that increased myoblast proliferation and defective differentiation is a direct effect of Stra13 disruption, Stra13−/− myoblasts were infected with a retrovirus expressing Stra13 or, as a control, with the retroviral vector alone. Stra13- expressing cells exhibited decreased BrdU incorporation in growth medium (Fig. 5 A). In addition, exogenously expressed Stra13 partially rescued the fusion index in differentiation medium (Fig. 5 B), indicating that the differentiation-defective phenotype may be partly noncell autonomous.
Figure 5.
Reexpression of Stra13 in Stra13−/− myoblasts rescues the proliferation and differentiation defect. Stra13−/− myoblasts were transduced with retrovirus expressing Stra13 (pBabe-Stra13) or with vector alone (pBabe). After selection, infected cells were either pulsed with BrdU and analyzed for cell proliferation (A) or changed to differentiation medium for 4 d and analyzed for myotube formation by MHC staining (B). The fusion index was calculated as described in Materials and methods. Data are means ± SEM (error bars). *, P < 0.05.
Enhanced Notch signaling in Stra13−/− mutants
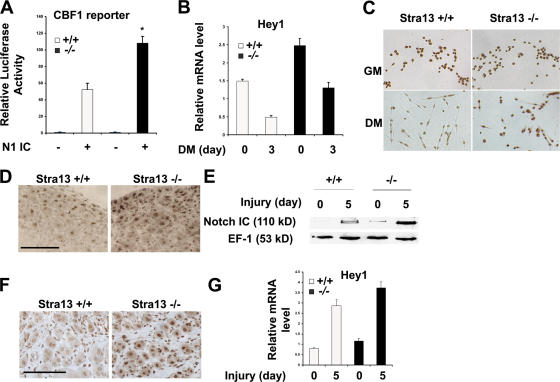
Because Stra13−/− myoblasts exhibit a phenotype reminiscent of constitutively active Notch signaling (Conboy and Rando, 2002), we hypothesized that increased Notch activity may underlie the altered proliferation and differentiation capacity of Stra13−/− cells. To test this hypothesis, we examined Notch activity in WT and Stra13−/− myoblasts using a CBF-1 reporter. N1IC (intracellular domain of Notch1)-induced CBF-1 reporter activity was twofold higher in Stra13−/− myoblasts compared with WT myoblasts (Fig. 6 A), indicating that N1IC is more active in the absence of Stra13. Among the Notch target genes, Hey1 has been shown to be a regulator of myogenic differentiation (J. Sun et al., 2001). As predicted with increased Notch activity, Hey1 RNA was expressed at higher levels in both proliferating and differentiating Stra13−/− myoblasts (Fig. 6 B). Moreover, unlike WT cells, in which Hey1 expression was down-regulated upon differentiation, Stra13−/− cells retained high levels of Hey1 (Fig. 6, B and C) similar to undifferentiated cells.
Figure 6.
Increased Notch activity in Stra13−/− myoblasts. (A) A CBF-1 reporter was transfected in WT and Stra13−/− myoblasts in the absence or presence of N1IC. A higher level of Notch-induced reporter activity was detected in Stra13−/− myoblasts. Data are means ± SEM (error bars; n = 3). *, P < 0.05. (B and C) Primary myoblasts from WT and Stra13−/− mice were induced to differentiate. Hey1 mRNA levels (B) were analyzed at days 0 and 3 by quantitative PCR, and Hey1 protein levels (C) were detected by immunostaining with anti-Hey1 antibody. GM, growth medium; DM, differentiation medium. (D and E) Immunostaining with anti-Notch1 antibody recognizing the activated form of Notch1 (D), and Western blot analysis (E) of WT and Stra13−/− muscle analyzed on day 5 after injury exhibited enhanced Notch activation in Stra13−/− regenerating muscle (n = 2). (F and G) Immunohistochemical detection (F) and quantitative PCR analysis (G) of regenerating muscle 5 d after injury showed increased Hey1 protein and mRNA level in Stra13−/− regenerating muscle (n = 2). (B, C, F, and G) Data indicate means ± SD. Bars, 100 μm.
Because Notch signaling is essential for progenitor cell expansion during muscle regeneration (Conboy and Rando, 2002), we also examined Notch activity in regenerating muscle using antibodies directed against the activated form of Notch1 and its target Hey1. A higher level of activated Notch1 was seen in Stra13−/− muscle by immunostaining of regenerating muscle (Fig. 6 D) as well as by Western blot analysis (Fig. 6 E). Consistent with elevated Notch activity, increased Hey1 expression was also evident in regenerating Stra13−/− muscle by immunostaining in myofibers and, to a lesser degree, in mononuclear cells (Fig. 6 F). Quantitative PCR analysis confirmed increased Hey1 expression in the mutant tissue (Fig. 6 G), although changes in Hey1 RNA levels were less dramatic than changes in protein levels likely because of the contribution by both myofibers and mononuclear cells, which already express Hey1.
Inhibition of Notch signaling reduces proliferation, enhances differentiation, and improves regeneration in Stra13−/− mice
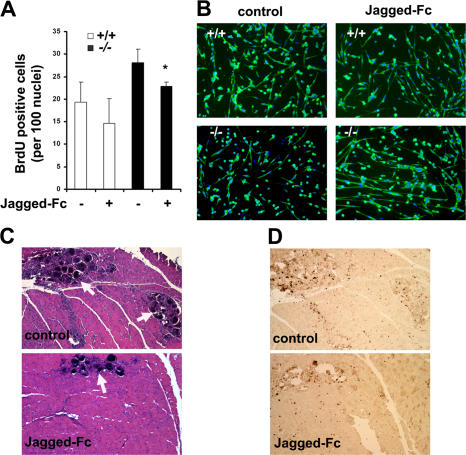
To test whether enhanced Notch activity was causal to the increased proliferation and reduced differentiation of Stra13−/− myoblasts, we treated WT and Stra13−/− cells with the soluble Notch ligand Jagged-Fc fusion protein, which has previously been shown to antagonize Notch activity (Conboy et al., 2003). After treatment with Jagged-Fc, the proliferation of both WT and Stra13−/− myoblasts was reduced, as indicated by decreased BrdU incorporation (Fig. 7 A). Moreover, when induced to differentiate in the presence of Jagged-Fc, myotube formation was increased in Stra13−/− myoblasts (Fig. 7 B). The fusion index of Stra13−/− cells in the presence of Jagged-Fc also increased considerably from <20 to >30% compared with untreated cells (not depicted), which is comparable with the effect seen with the reexpression of Stra13 in the mutant myoblasts (Fig. 5). Similar results were obtained using the γ-secretase inhibitor L685,458 (unpublished data).
Figure 7.
Modulation of Notch signaling attenuates the proliferation and differentiation defect of Stra13−/− cells and improves muscle regeneration. (A) WT and Stra13−/− myoblasts were cultured in growth medium in the absence and presence of Jagged-Fc. Cells were pulsed with BrdU and stained with anti-BrdU antibody. The percentage of BrdU-positive cells was significantly lower in Jagged-Fc–treated Stra13−/− cells compared with untreated cells. Data are means ± SEM (error bars; n = 4). *, P < 0.05. (B) WT and Stra13−/− myoblasts were cultured in the absence and presence of Jagged-Fc in differentiation medium for 4 d. Myogenic differentiation was visualized by MHC staining. Note that most nuclei are located within MHC+ cells in the presence of Jagged-Fc. (C) Stra13−/− mice were injured and, 7 d later, injected with Jagged-Fc or the control vehicle. 3 d after injection, muscle was analyzed histologically by HE staining. Using ImageJ, the area covered by necrotic fibers within the total damaged area was analyzed for each animal. Mean areas were compared and quantified. Jagged-Fc–injected muscle shows reduced necrosis (arrows) compared with the control uninjected muscle (P < 0.05). (D) Control and Jagged-Fc–injected sections were stained with anti-Ki67 antibody. A reduction in Ki67+ cells in the Jagged-Fc–injected muscles was seen by immunostaining. The results are representative of three independent animals.
To determine whether sustained Notch signaling is responsible for increased cellular proliferation and defective regeneration in Stra13−/− mice, we tested whether the inhibition of Notch signaling at late stages could improve the regeneration status of Stra13−/− mice. Injection of Jagged-Fc fusion protein directly into muscle 48 h after injury has previously been shown to inhibit Notch signaling and muscle regeneration by inhibiting satellite cell activation (Conboy et al., 2003). To circumvent such effects of Jagged-Fc on satellite cell activation at early stages, we injected Jagged-Fc into the injured muscle of Stra13−/− mice 7 d after injury. 3 d later, mice were killed, and muscles were analyzed histologically for regeneration. Strikingly, the injection of Jagged-Fc fusion protein resulted in improved regeneration as assessed histologically by reduced necrosis (Fig. 7 C). Analysis of degenerated myotubes within the total injured region indicated that necrosis was seen in only 8–10% of the injured tissue in the presence of the inhibitor. In contrast, the contralateral muscle, which was not injected with the soluble ligand, exhibited impaired regeneration, and 25–30% of the injured tissue contained degenerated myotubes and necrosis. All three mice responded similarly to the Notch inhibitor and exhibited virtually identical effects. The number of proliferating Ki67+ cells was also reduced by 50% in Jagged-Fc–injected muscle relative to untreated controls (Fig. 7 D). Therefore, our in vitro and in vivo data demonstrate that inhibition of Notch signaling can partially reverse the mutant phenotype, indicating that sustained Notch signaling underlies, at least in part, the impaired muscle regeneration of Stra13−/− mutants.
Stra13 suppresses Notch signaling by interaction with NICD
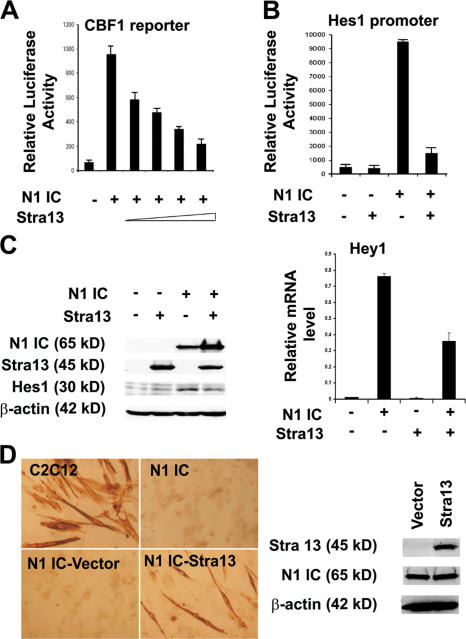
Enhanced Notch activity in Stra13−/− cells suggested that Stra13 inhibits Notch signaling. To examine this possibility, a CBF-1 reporter was transfected in 10T1/2 cells along with N1IC in the absence or presence of Stra13. The overexpression of Stra13 repressed N1IC-induced reporter activity in a dose-dependent manner (Fig. 8 A). Consistently, N1IC-induced activation of the Hes1 promoter (Takebayashi et al., 1994) was also inhibited by Stra13 (Fig. 8 B). We then examined whether Stra13 exerted a similar effect on the endogenous expression of Notch target genes. 10T1/2 cells were transfected with expression vectors for Stra13, N1IC, or both N1IC and Stra13 and were analyzed by Western blotting and quantitative PCR for the endogenous expression of Hes1 and Hey1. Although Stra13 expression alone had no considerable effect, both genes were up-regulated by N1IC (Fig. 8 C). However, the coexpression of Stra13 along with N1IC suppressed Notch-induced Hes1 (Fig. 8 C, left) and Hey1 expression (Fig. 8 C, right), indicating that Stra13 inhibits the transcriptional activity of N1IC. To directly test the possibility that Stra13 inhibits Notch signaling in myogenic cells, we established stable C2C12 cell lines expressing N1IC, which is constitutively active. Consistent with previous studies (Kopan et al., 1994; Nofziger et al., 1999), unlike control C2C12 cells that efficiently differentiate into myotubes, N1IC-expressing cells failed to differentiate (Fig. 8 D). Interestingly, however, the retroviral expression of Stra13 in N1IC-expressing cells rescued 20% of myogenic differentiation as assessed by quantifying the percentage of multinucleated myotubes within MHC+ cells, indicating that Stra13 regulates myogenic differentiation through the inhibition of Notch signaling.
Figure 8.
Stra13 inhibits Notch signaling. (A) 100 ng of a CBF-1 reporter construct was transfected in 10T1/2 cells along with 10 ng N1IC in the absence or presence of increasing amounts of Stra13 (10, 25, 50, and 100 ng). (B) 100 ng Hes-1 promoter was transfected with 10 ng N1IC in the absence or presence of 100 ng Stra13. Reporter assays were repeated at least three times, each with duplicates. Data are means ± SEM (error bars). (C) 10T1/2 cells were transfected with vector (pCS2), Stra13, N1IC, or both N1IC and Stra13. The expression of Stra13, N1IC, and Hes1 was detected by Western blotting (left). Hey1 mRNA level was analyzed by quantitative PCR (right). (D) Control C2C12 cells or stable cell lines expressing Notch (C2C12-N1IC) were stained for MHC 4 d after culturing in differentiation medium. C2C12-N1IC cells were blocked in myogenic differentiation. C2C12-N1IC cells were transduced with an empty retroviral vector (Babe) or one expressing Stra13 and stained for MHC after 4 d in differentiation medium (left). The expression of Notch and Stra13 was detected in N1IC-vector and N1IC-Stra13 cell lines by Western blot analysis (right).
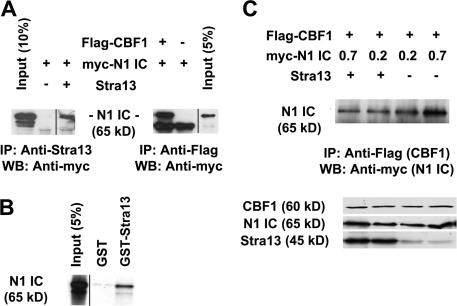
To examine the mechanism by which Stra13 inhibits Notch signaling, we tested whether Stra13 interacts with N1IC by coimmunoprecipitation assays. Myc-tagged N1IC was cotransfected with Stra13 or with Flag-tagged CBF-1 in 293T cells. Lysates were immunoprecipitated with anti-Stra13 antibody or with anti-Flag antibody and subjected to SDS-PAGE. Western blot analysis with anti-myc antibody revealed that Stra13 interacts with N1IC (Fig. 9 A), and, as expected, an interaction between N1IC and CBF-1 was also seen. GST pull-down assays confirmed a direct interaction between Stra13 and the Notch1 intracellular domain (Fig. 9 B). Because Stra13 interacts with N1IC, we considered the possibility that this interaction may reduce the association of N1IC with CBF-1. Therefore, we examined the interaction of CBF-1 and N1IC in the absence or presence of Stra13 by coimmunoprecipitation assays. Interestingly, the amount of CBF-1 that is associated with N1IC was reduced in the presence of Stra13 (Fig. 9 C). Thus, we conclude that the interaction of Stra13 with N1IC reduces the association of N1IC with CBF-1, resulting in inhibition of the Notch-dependent activation of target genes.
Figure 9.
Stra13 physically interacts with Notch1 intercellular domain and prevents it from binding to CBF-1. (A) 293T cells were transfected with myc-tagged N1IC and Stra13 or Flag–CBF-1 as indicated. Lysates were immunoprecipitated with anti-Stra13 or anti-Flag (CBF-1) antibodies and analyzed by Western blotting with anti-myc (N1IC) antibody, which shows the interactions of Stra13 and N1IC (left) and N1IC with CBF-1 (right). (B) GST-Stra13 was tested for interaction with in vitro translated N1IC. GST was used as a control. 5% of input was run on the gel. (C) 293T cells were transfected with 2 μg Flag–CBF-1, and 0.2 and 0.7 μg of different amounts of myc-N1IC in the presence (2 μg) or absence of Stra13. Cell lysates were immunoprecipitated with anti-Flag (CBF-1) antibody and analyzed by Western blotting with anti-myc (N1IC) antibody (top). The bottom panel shows Western blot analysis of CBF-1, N1IC, and Stra13 expression.
Discussion
Notch signaling plays a prominent role in the regeneration of skeletal muscle (Conboy and Rando, 2002), and insufficient Notch activity in ageing satellite cells leads to reduced regenerative potential (Conboy et al., 2003). However, sustained Notch activity is incompatible with myogenic differentiation (Kopan et al., 1994; Nofziger et al., 1999; Conboy and Rando, 2002). Subsequent to satellite cell activation, Notch signaling is attenuated during differentiation by up-regulation and asymmetric localization of its antagonist Numb (Conboy and Rando, 2002). Because alterations in Notch activity can have profound effects on the regenerative response, it is likely that Notch signaling is controlled at multiple steps. The results of this study reveal an unanticipated function for Stra13 as an inhibitor of Notch transcriptional activity. We demonstrate that Stra13 regulates myoblast proliferation and differentiation by modulating Notch signaling and is required for efficient muscle regeneration.
Several lines of evidence suggest that Stra13, like Numb, is critical for the attenuation of Notch signaling in satellite cells. First, Stra13−/− primary myoblasts exhibit enhanced proliferation and defective differentiation, a phenotype that is remarkably similar to the expression of constitutively active Notch in myoblasts (Nofziger et al., 1999; Conboy and Rando, 2002). Second, Notch reporter activity and the expression of its downstream target Hey1 are increased in Stra13−/− myoblasts. The increased Notch activity in proliferating Stra13−/− myoblasts suggest that persistent Notch activity is not likely to be secondary to defective differentiation but rather causal to it. Note that Stra13 is expressed at low levels in proliferating satellite cells when Notch activity is high, and its expression is up-regulated during terminal differentiation concomitant with reduced Notch signaling. Third, treatment with the Notch antagonist Jagged-Fc resulted in reduced proliferation and enhanced myotube formation in Stra13−/− myoblasts. Fourth, Notch-induced inhibition of myogenic differentiation in C2C12 cells can be partially overcome by the forced expression of Stra13. Finally, Stra13 interacts with NICD and inhibits its transcriptional activity.
Consistent with these observations in primary myoblasts, increased levels of activated Notch and Hey1 are also evident in regenerating Stra13−/− muscle. As predicted with elevated Notch signaling, the number of myogenic precursor cells is increased, indicating elevated satellite cell activation and myogenic precursor cell expansion during regeneration in Stra13−/− muscle. However, in contrast to the differentiation defect of Stra13−/− myoblasts in vitro, myogenic differentiation is not perturbed in Stra13−/− regenerating muscle. These observations suggest that in vivo, compensatory mechanisms allow myogenic differentiation to proceed despite elevated Notch activity. Several possibilities may account for this paradox. During regeneration, both MyoD and activated Notch levels are elevated in the mutant tissue. It is possible that the increase in Notch activity is insufficient to completely block MyoD levels (Nofziger et al., 1999; Conboy and Rando, 2002), and, thus, despite elevated Notch activity, myogenic differentiation in vivo proceeds normally. Moreover, elevated Notch1 and Hey1 are apparent in newly formed myofibers in the mutant tissue. Although the role of Notch signaling in the proliferation and differentiation of progenitor cells is well established, its potential function in differentiated cells is unclear. Because Notch activity is high in the mutants in newly formed myofibers that have already differentiated, it is possible that under these conditions, Notch is unable to inhibit MyoD levels and block the differentiation of progenitor cells. However, sustained Notch activity in newly formed myofibers may result in their degeneration, leading to defective regeneration. In support of this possibility, the injection of Jagged-Fc into injured muscles of Stra13−/− mice at day 7 results in improved histopathology, as seen by reduced necrosis and Ki67+ cells, suggesting that deregulated Notch signaling in vivo is indeed one underlying basis of the defective regeneration of Stra13−/− mice.
An alternative possibility for the relatively normal differentiation during regeneration could be through the altered production of growth factors and cytokines by injured muscle and immune cells that regulate satellite cell activation, proliferation, and differentiation. For instance, we have previously demonstrated the aberrant CD4+ T cell activation (H. Sun et al., 2001) and reduced production of several cytokines, including interleukin-4 in Stra13−/− mice, which is required for effective regeneration (Horsley et al., 2003). Activation of peritoneal macrophages is also impaired in Stra13−/− mice, resulting in the reduced production of cytokines interleukin-1β and TNF-α (unpublished data). Macrophages play a critical role in muscle regeneration not only in the clearance of necrotic debris but also at late stages in the fusion of myofibers, and TNF-α receptor mutants are impaired in their regenerative response (Warren et al., 2005). It is conceivable that the inflammatory reaction and expression of various cytokines after muscle injury may be altered in Stra13−/− mice. The reduced production of inhibitory cytokines that normally block myogenesis may allow myotube formation in vivo but would obviously have no impact on the differentiation of cultured myoblasts in vitro. Therefore, it is likely that Stra13 regulates postnatal myogenesis through cross talk with distinct pathways, and the cumulative effects of muscle–cell intrinsic and extrinsic changes lead to diminished regenerative capacity in vivo.
Few inhibitors of Notch signaling have been identified, which include the cytoplasmic regulator Numb as well as the nuclear antagonists Msx2-interacting nuclear target protein (MINT), Hairless, and Nubbin (Kopan, 1999; Kadesch, 2004). Unlike Numb, which attenuates Notch signaling by interacting with NICD and regulating the endocytosis and ubiquitination of Notch, Stra13 represses Notch signaling in the nucleus by reducing its interaction with CBF-1. Because Notch activity needs to be precisely regulated spatially and temporally during muscle regeneration, it is likely controlled at multiple steps. Whether Numb and Stra13 function at distinct temporal phases or, alternatively, function at the same time but at distinct locations to ensure the complete inhibition of Notch activity remains to be determined. The intracellular domain of Notch is constitutively active in the nucleus and bypasses ligand requirement. Because Stra13 antagonizes the activity of NICD both in vitro as well as functionally in myogenic C2C12 cells, our results suggest that Stra13 may directly repress Notch signaling independently of the effects on ligand expression. Nevertheless, we cannot exclude the possibility that in vivo, Stra13 may regulate Notch signaling at additional levels, including the transcriptional regulation of Delta, regulation of Numb expression or localization, or by dimerization with Hes or Hey proteins. It will be of interest to precisely define these changes in order to fully understand the molecular mechanisms by which Stra13 regulates skeletal muscle regeneration.
Materials and methods
Regeneration studies
Stra13−/− mice have been previously described (H. Sun et al., 2001). 3-mo-old littermate WT and Stra13−/− mice were injured by applying a metal probe precooled in dry ice for 10 s. At least two mice were analyzed per time point (2, 5, 10, and 16 d after injury) in each experiment, and regeneration studies were performed four times. All animal protocols followed institutional guidelines. To inhibit Notch signaling, the quadriceps muscles were subjected to freeze injury. 7 d after injury, 15 μl (1.5 μg) Jagged-Fc fusion protein (R&D Systems) was injected into six sites of injured muscles. PBS (in 0.1% BSA) was injected in the contralateral injured muscle. 3 d after injection, muscles were collected and analyzed histologically.
Antibodies
Anti-Stra13 (Dec1) antibody was provided by B. Yan (University of Rhode Island, Kingston, RI). Anti-MyoD, anti-myogenin, and anti-Notch1 (mN1A and C-20) were obtained from Santa Cruz Biotechnology, Inc. Antibody against activated Notch1 (Val1744) was purchased from Cell Signaling, anti-Ki67 was purchased from Novocastra, and anti-Hey1 was obtained from Chemicon. Anti-BrdU, anti-MHC, and anti–β-actin were purchased from Sigma-Aldrich, anti-Hes1 was a gift from Y.N. Jan (University of California, San Francisco, San Francisco, CA), anti-EF1α was purchased from Upstate Biotechnology, and anti-Pax7 and anti-eMHC were obtained from Developmental Studies Hybridoma Bank.
Histology, immunohistochemistry, and microscopy
Serial cross sections were collected along the length of muscle, and one in every six slides was stained with hematoxylin and eosin (HE). Sections from both genotypes with comparable damaged areas were used for histological and immunohistochemical analysis. Immunohistochemistry was performed as described previously (H. Sun et al., 2001). In brief, paraffin-embedded sections were incubated with primary antibody, and staining without primary antibody served as a negative control. After washes, slides were incubated with biotinylated secondary antibodies and developed using VECTASTAIN Elite ABC kit (Vector Laboratories). Masson's trichrome staining was performed using a kit from Diagnostic Biosystems. Phase-contrast microscopy was performed using a microscope (Eclipse TS100; Nikon) with plan Fluor 10× NA 0.3 and 20× NA 0.45 objectives (Nikon) at room temperature, and images were captured using a camera (2.2.1 Spot RT Color; Diagnostic Instruments) and Spot software (version 3.5.9; Diagnostic Instruments). All figures were prepared using Photoshop version 7.0 (Adobe).
Fiber size
Quadriceps muscles from four mice of each genotype were embedded in paraffin. Sections from the center of the muscle were stained with HE, and the cross section area of individual myofibers was measured using ImageJ software (version 1.36b; National Institutes of Health).
Statistical analysis
For all analysis, at least three to four mice per time point or treatment were used. An unpaired t test was used for all statistical calculations, and P-values of <0.05 were considered to be statistically significant.
Satellite cell number
Frozen sections of tibialis anterior muscle from 1-mo-old mice were stained with anti-Pax7 antibody and counterstained with hematoxylin. Both Pax7+ nuclei and myonuclei were counted from several random fields for each animal, and the percentage of satellite cells was calculated as the number of Pax7+ cells per 100 myonuclei. Myonuclei were identified based on their location being closely associated with myofibers, and interstitial nuclei were excluded from the counting.
Cell culture, transfection, and luciferase assays
C3H10T1/2 and 293T were maintained in DME containing 10% FBS. Transient transfections were performed using LipofectAMINE Plus (Invitrogen). Empty expression vectors were added to normalize the amount of DNA in each well. Luciferase assays were performed using the dual luciferase system (Promega). C2C12 cells were maintained in DME containing 20% FBS. To establish stable cell lines, C2C12 cells were transfected with a vector expressing N1IC. Cells were selected with G418, and several independent colonies were tested for N1IC expression. For rescue experiments, two independent N1IC-expressing cell lines were transduced with a vector alone or retroviral vector expressing Stra13 (pBabe-Stra13) and selected with puromycin. After selection, the bulk population of infected cells was switched to differentiation medium (DME with 2% horse serum) for 4 d and immunostained with anti-MHC antibody.
Primary myoblasts, proliferation, and differentiation
Primary myoblasts were isolated as previously described (Rando and Blau, 1994). In brief, hindlimb muscles were dissected from 3-wk-old mice and digested with collagenase. Single-cell suspensions were resuspended in growth medium (20% FBS in F-10 medium supplemented with 5 ng/ml basic FGF) and plated on collagen-coated cell culture dishes. After two to three passages, >95% of cells were myogenic, as seen by staining with Pax7, c-met, and desmin antibodies. All assays were performed with primary myoblasts within passages four to seven, and three to four independent isolates were used for each assay. For differentiation assays, myoblasts were cultured in differentiation medium (DME with 5% horse serum) and harvested at the times indicated. The percentage of differentiated cells was calculated by counting the number of MHC-stained myocytes/total number of nuclei. The fusion index was calculated as the percentage of cells containing more than two nuclei within MHC+ cells. For proliferation assays, myoblasts in growth medium were incubated with 20 μM BrdU for 3.5 h and immunostained with an antibody against BrdU. For rescue experiments, Stra13−/− myoblasts were infected with pBabe or pBabe-Stra13. Infected cells were selected with 1 μg/ml puromycin for 2 d, expanded, and subjected to proliferation and differentiation assays. To inhibit Notch signaling, WT and Stra13−/− myoblasts were cultured in the presence of 1 μg/ml Jagged-Fc (R&D Systems) and subjected to proliferation and differentiation assays.
Coimmunoprecipitation and GST pull-down assays
Coimmunoprecipitation and GST pull-down assays were performed as described previously (Sun and Taneja, 2000). For coimmunoprecipitation assays with transfected plasmids, 400 μg of cell lysate was incubated with 2 μg of antibodies followed by the addition of protein A/G plus agarose beads. Proteins were eluted and subjected to SDS-PAGE. Western blotting was performed with anti-myc, anti-Flag, or anti-Stra13 antibodies. For GST pull- down assays, N1IC was translated in vitro and labeled with [35S]methionine using the rabbit reticulocyte in vitro transcription-translation system (TNT; Promega). 35S-labeled N1IC was incubated with purified GST-Stra13 (Sun and Taneja, 2000) or GST in binding buffer. Samples were run on SDS gels and detected by autoradiography.
Quantitative real-time PCR
Total RNA was extracted using TRIzol (Invitrogen) or with an RNeasy kit (QIAGEN) and treated with Rnase-free DNaseI (TURBO DNA-free kit; Ambion). Quantitative real-time PCR was performed using the Quantitect SYBR green PCR kit (QIAGEN) with 10 ng cDNA as a template. Each sample was amplified in triplicate in a thermocycler (ABI Prism 7900HT; Applied Biosystems). PCR cycling conditions were as follows: 95°C for 15 min and 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. Amplicon size was confirmed by agarose gel electrophoresis. Quantitative PCR standard curves were constructed by using serial dilutions of total RNA isolated from day 5 regenerating muscle. All data were normalized to glyceraldehyde-3-phosphate dehydrogenase. The primer pairs used were as follows: Hey1 (CTTGAGTTCGGCTCTGTGTTCC and GATGCCTCTCCGTCTTTTCCT), Stra13 (TACAAGCTGGTGATTTGTCGG and CTGGGAAGATTTCAGGTCCCG), and glyceraldehyde-3-phosphate dehydrogenase (AGGAGCGAGACCCCACTAACAT and GTGAAGACACCAGTAGACTCCACG).
Northern blot analysis
20 μg of total RNA was run on formaldehyde-agarose gels and hybridized with 32P-labeled cDNA probes for Stra13 and 36B4.
Acknowledgments
We sincerely thank G. Marazzi, D. Sassoon, and M. Arufe for discussions and invaluable help with regeneration experiments. We are grateful to D. Hayward, T. Honjo, R. Kageyama, J.S. Kang, R. Kopan, A. Ozog, L. Vales, and B. Yan for various reagents.
This work was supported by funds from the Muscular Dystrophy Association and a Scholar Award from the Leukemia and Lymphoma Society to R. Taneja. T.H. Thin was supported by a National Cancer Institue postdoctoral training grant (CA88796).
L. Li and C. Vercherat contributed equally to this paper.
Abbreviations used in this paper: bHLH, basic helix-loop-helix; eMHC, embryonic MHC; HE, hematoxylin and eosin; MHC, myosin heavy chain; NICD, Notch intracellular domain; WT, wild type.
References
- Allen, R.E., and L.K. Boxhorn. 1989. Regulation of skeletal muscle satellite cell proliferation and differentiation by transforming growth factor-beta, insulin-like growth factor I, and fibroblast growth factor. J. Cell. Physiol. 138:311–315. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. [DOI] [PubMed] [Google Scholar]
- Beadling, C., A. Cereseto, W. Fan, M. Naramura, and K.A. Smith. 2001. Cytokine response gene 8 (CR8) regulates the cell cycle G1-S phase transition and promotes cellular survival. Oncogene. 20:1771–1783. [DOI] [PubMed] [Google Scholar]
- Beauchamp, J.R., L. Heslop, D.S. Yu, S. Tajbakhsh, R.G. Kelly, A. Wernig, M.E. Buckingham, T.A. Partridge, and P.S. Zammit. 2000. Expression of CD34 and myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 151:1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudjelal, M., R. Taneja, S. Matsubara, P. Bouillet, P. Dollé, and P. Chambon. 1997. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 11:2052–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charge, S.B., and M.A. Rudnicki. 2004. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84:209–238. [DOI] [PubMed] [Google Scholar]
- Chen, J.C., and D.J. Goldhammer. 2003. Skeletal muscle stem cells. Reprod. Biol. Endocrinol. 1:101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy, I.M., and T.A. Rando. 2002. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 3:397–409. [DOI] [PubMed] [Google Scholar]
- Conboy, I.M., M.J. Conboy, G.M. Smythe, and T.A. Rando. 2003. Notch- mediated restoration of regenerative potential to aged muscle. Science. 302:1575–1577. [DOI] [PubMed] [Google Scholar]
- Conlon, R.A., A.G. Reaume, and J. Rossant. 1995. Notch1 is required for the coordinate segmentation of somites. Development. 121:1533–1545. [DOI] [PubMed] [Google Scholar]
- Cornelison, D.D., and B.J. Wold. 1997. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 191:270–283. [DOI] [PubMed] [Google Scholar]
- Cornelison, D.D., S.A. Wilcox-Adelman, P.F. Goetinck, H. Rauvala, A.C. Rapraeger, and B.B. Olwin. 2004. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 18:2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R.L., and D.L. Turner. 2001. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 20:8342–8357. [DOI] [PubMed] [Google Scholar]
- Floss, T., H.H. Arnold, and T.A. Braun. 1997. Role for FGF-6 in skeletal muscle regeneration. Genes Dev. 11:2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garry, D.J., A. Meeson, J. Elterman, Y. Zhao, P. Yang, R. Bassel-Duby, and R.S. Williams. 2000. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. USA. 97:5416–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawke, T.J., and D.J. Garry. 2001. Myogenic satellite cells: physiology to molecular biology. J. Appl. Physiol. 91:534–551. [DOI] [PubMed] [Google Scholar]
- Hawke, T.J., A.P. Meeson, N. Jiang, S. Graham, K. Hutcheson, J.M. DiMaio, and D.J. Garry. 2003. p21 is essential for normal myogenic progenitor cell function in regenerating skeletal muscle. Am. J. Physiol. Cell. Physiol. 285:C1019–C1027. [DOI] [PubMed] [Google Scholar]
- Horsley, V., K.M. Jansen, S.T. Mills, and G.K. Pavlath. 2003. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 113:483–494. [DOI] [PubMed] [Google Scholar]
- Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237–255. [DOI] [PubMed] [Google Scholar]
- Jarriault, S., C. Brou, F. Logeat, E.H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature. 377:355–358. [DOI] [PubMed] [Google Scholar]
- Jones, N.C., K.J. Tyner, L. Nibarger, H.M. Stanley, D.D. Cornelison, Y.V. Fedorov, and B.B. Olwin. 2005. The p38α/β MAPK functions as a molecular switch to activate the quiescent satellite cell. J. Cell Biol. 169:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadesch, T. 2004. Notch signaling: the demise of elegant simplicity. Curr. Opin. Genet. Dev. 14:506–512. [DOI] [PubMed] [Google Scholar]
- Kopan, R. 1999. All good things must come to an end: how is Notch signaling turned off? Sci. STKE. 10.1126/stke.1999.9.pe1. [DOI] [PubMed]
- Kopan, R., J.S. Nye, and H. Weintraub. 1994. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 120:2385–2396. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. 2004. Notch signaling: control of cell communication and cell fate. Development. 131:965–973. [DOI] [PubMed] [Google Scholar]
- Lescaudron, L., E. Peltekian, J. Fontaine-Perus, D. Paulin, M. Zampieri, L. Garcia, and E. Parrish. 1999. Blood borne macrophages are essential for the triggering of muscle regeneration following muscle transplant. Neuromuscul. Disord. 9:72–80. [DOI] [PubMed] [Google Scholar]
- Li, Y., H. Zhang, M. Xie, M. Hu, S. Ge, D. Yang, Y. Wan, and B. Yan. 2002. Abundant expression of Dec1/stra13/sharp2 in colon carcinoma: its antagonizing role in serum deprivation-induced apoptosis and selective inhibition of procaspase activation. Biochem. J. 367:413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., W. Foster, B.M. Deasy, Y. Chan, V. Prisk, Y. Tang, J. Cummins, and J. Huard. 2004. Transforming growth factor-beta1 induces the differentiation of myogenic cells into fibrotic cells in injured skeletal muscle: a key event in muscle fibrogenesis. Am. J. Pathol. 164:1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCroskery, S., M. Thomas, L. Maxwell, M. Sharma, and R. Kambadur. 2003. Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 162:1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeney, L.A., B. Kablar, K. Garrett, J.E. Anderson, and M.A. Rudnicki. 1996. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 10:1173–1183. [DOI] [PubMed] [Google Scholar]
- Miller, K.J., D. Thaloor, S. Matteson, and G.K. Pavlath. 2000. Hepatocyte growth factor affects satellite cell activation and differentiation in regenerating skeletal muscle. Am. J. Physiol. Cell Physiol. 278:C174–C181. [DOI] [PubMed] [Google Scholar]
- Nofziger, D., A. Miyamoto, K.M. Lyons, and G. Weinmaster. 1999. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 126:1689–1702. [DOI] [PubMed] [Google Scholar]
- Ohtsuka, T., M. Ishibashi, G. Gradwohl, S. Nakanishi, F. Guillemot, and R. Kageyama. 1999. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18:2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando, T.A., and H.M. Blau. 1994. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125:1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale, P., L.A. Sabourin, A. Girgis-Gabardo, A. Mansouri, P. Gruss, and M.A. Rudnicki. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell. 102:777–786. [DOI] [PubMed] [Google Scholar]
- Seimiya, M., R. Bahar, Y. Wang, K. Kawamura, Y. Tada, S. Okada, M. Hatano, T. Tokuhisa, H. Saisho, T. Watanabe, M. Tagawa, and J. O-Wang. 2002. Clast5/Stra13 is a negative regulator of B lymphocyte activation. Biochem. Biophys. Res. Commun. 292:121–127. [DOI] [PubMed] [Google Scholar]
- Shen, M., E. Yoshida, W. Yan, T. Kawamoto, K. Suardita, Y. Koyano, K. Fujimoto, M. Noshiro, and Y. Kato. 2002. Basic helix-loop-helix protein DEC1 promotes chondrocyte differentiation at the early and terminal stages. J. Biol. Chem. 277:50112–50120. [DOI] [PubMed] [Google Scholar]
- Sun, H., and R. Taneja. 2000. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA. 97:4058–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., B. Lu, R.Q. Li, R.A. Flavell, and R. Taneja. 2001. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat. Immunol. 2:1040–1047. [DOI] [PubMed] [Google Scholar]
- Sun, J., C.N. Kamei, M.D. Layne, M.K. Jain, J.K. Liao, M.E. Lee, and M.T. Chin. 2001. Regulation of myogenic terminal differentiation by the hairy-related transcription factor CHF2. J. Biol. Chem. 276:18591–18596. [DOI] [PubMed] [Google Scholar]
- Takebayashi, K., Y. Sasai, Y. Sakai, T. Watanabe, S. Nakanishi, and R. Kageyama. 1994. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J. Biol. Chem. 269:5150–5156. [PubMed] [Google Scholar]
- Tatsumi, R., J.E. Anderson, C.J. Nevoret, O. Halevy, and R.E. Allan. 1998. HGF/SF is present in normal adult skeletal muscle and is capable of activating satellite cells. Dev. Biol. 194:114–128. [DOI] [PubMed] [Google Scholar]
- Thin, T.H., L. Li, T.-K. Chung, H. Sun, and R. Taneja. 2007. Stra13 is induced by genotoxic stress and regulates ionizing-radiation-induced apoptosis. EMBO Rep. 8:401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball, J.G. 2005. Inflammatory processes in muscle injury and repair. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288:R345–R353. [DOI] [PubMed] [Google Scholar]
- Warren, G.L., T. Hulderman, D. Mishra, X. Gao, L. Millecchia, L. O'Farrell, W.A. Kuziel, and P.P. Simeonova. 2005. Chemokine receptor CCR2 involvement in skeletal muscle regeneration. FASEB J. 19:413–435. [DOI] [PubMed] [Google Scholar]
- Yun, Z., H.L. Maecker, R.S. Johnson, and A.J. Giaccia. 2002. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell. 2:331–341. [DOI] [PubMed] [Google Scholar]