Abstract
The meiotic cohesin Rec8 is required for the stepwise segregation of chromosomes during the two rounds of meiotic division. By directly measuring chromosome compaction in living cells of the fission yeast Schizosaccharomyces pombe, we found an additional role for the meiotic cohesin in the compaction of chromosomes during meiotic prophase. In the absence of Rec8, chromosomes were decompacted relative to those of wild-type cells. Conversely, loss of the cohesin-associated protein Pds5 resulted in hypercompaction. Although this hypercompaction requires Rec8, binding of Rec8 to chromatin was reduced in the absence of Pds5, indicating that Pds5 promotes chromosome association of Rec8. To explain these observations, we propose that meiotic prophase chromosomes are organized as chromatin loops emanating from a Rec8-containing axis: the absence of Rec8 disrupts the axis, resulting in disorganized chromosomes, whereas reduced Rec8 loading results in a longitudinally compacted axis with fewer attachment points and longer chromatin loops.
Introduction
During the mitotic cell cycle, the regulated segregation of sister chromatids is achieved by the mitotic cohesin complex, and upon entering meiosis, additional meiosis-specific cohesin variants appear to choreograph meiosis-specific chromosomal events (Nasmyth, 2001; Revenkova and Jessberger, 2006). The meiotic cohesin complex not only mediates sister chromatid cohesion but also plays a critical role in assembling a proteinaceous chromosome axis as its major component (Revenkova and Jessberger, 2006). This chromosome axis later forms the axial/lateral element of the synaptonemal complex (SC; Klein et al., 1999; Pasierbek et al., 2001; Pelttari et al., 2001; Eijpe et al., 2003). In many organisms, longitudinal compaction of the chromosome is observed along with chromosome axis formation during meiotic prophase (Heng et al., 1996; Moens et al., 1998; Zickler and Kleckner, 1999; Kleckner et al., 2003). However, the molecular mechanisms that underlie meiotic prophase chromosome compaction remain unclear.
In addition to the core cohesin complex, a conserved protein known as Pds5, Spo76, or BimD is associated with cohesin and is implicated in sister chromatid cohesion and chromosome structure maintenance. This protein has been studied in Sordaria macrospora (called Spo76), Aspergillus nidulans (called BimD), Saccharomyces cerevisiae, and Caenorhabditis elegans (van Heemst et al., 1999, 2001; Hartman et al., 2000; Wang et al., 2003; Zhang et al., 2005). Two Pds5s, Pds5A and Pds5B, have been identified in vertebrate cells (Sumara et al., 2000; Losada et al., 2005). A role for Mcd1/Scc1 (the Rad21 homologue) and Pds5 in mitotic chromosome condensation has been suggested in S. cerevisiae: FISH analysis demonstrated that these molecules are required for ribosomal DNA (rDNA) condensation at metaphase of mitosis (Guacci et al., 1997; Hartman et al., 2000). It has also been shown that Pds5/Spo76 has a role in meiosis because its loss results in defects in spore formation and SC integrity (van Heemst et al., 1999; Zhang et al., 2005).
In the fission yeast Schizosaccharomyces pombe, the mitotic cohesin complex is composed of two structure maintenance of chromosome (SMC) subunits, Psm1 and -3, and two non-SMC subunits, Rad21 and Psc3 (Tomonaga et al., 2000). In meiosis, Rad21 is largely replaced by a meiosis-specific cohesin, Rec8, and this exchange is essential for reductional segregation of chromosomes in the first meiotic division (meiosis I; Molnar et al., 1995; Parisi et al., 1999; Watanabe and Nurse, 1999; Yokobayashi et al., 2003). Rec11 is the meiotic Psc3 counterpart and acts at the chromosome arms (Kitajima et al., 2003). In the absence of Rec11, arm cohesion is reduced to the level seen in rec8 − mutants (Molnar et al., 2003). Unlike many other organisms, S. pombe shows no obvious chromosome condensation in meiotic prophase. In addition, this organism does not assemble canonical SC structures but forms so-called linear elements (LEs), which are evolutionally related to the axial/lateral elements of the SC (Bahler et al., 1993; Lorenz et al., 2004; Loidl, 2006). It is known that the meiotic cohesin is required for LE formation because aberrant LE structures form in both rec8 − and rec11 − mutants (Molnar et al., 1995, 2003). On the other hand, S. pombe Pds5 is nonessential for mitotic growth, but its loss reduces viability in G2-arrested cells (Tanaka et al., 2001; Wang et al., 2002), suggesting a role for Pds5 in maintenance of sister chromatid cohesion. Pds5 is also required for spore formation in S. pombe (Wang et al., 2002), suggesting that it plays a role in meiosis.
In S. pombe, meiotic prophase is characterized by an elongated nucleus, which is generally called a “horsetail” nucleus. The horsetail nucleus moves back and forth between the cell ends during meiotic prophase, and telomeres remain clustered at the leading edge of the moving nucleus (Chikashige et al., 1994; Ding et al., 1998). Observation of homologous pairing in living meiotic cells has demonstrated that telomere clustering and oscillatory chromosome movements spatially align homologous chromosomes in the early stages of meiotic prophase to promote their contact, which is stabilized later by homologous recombination (Ding et al., 2004). The telomere-clustered movement aligns chromosomes along the direction of the movements, providing a unique opportunity to examine chromatin structures within a defined orientation of the chromosome. In this study, we have identified a role for Rec8 and Pds5 in chromosome compaction by directly measuring chromosome compaction in living cells: Rec8 modulates chromosome compaction during meiotic prophase, and Pds5 is required for stable binding of Rec8 to the chromosome. Our results demonstrate that meiotic cohesins are essential for compaction of chromosomes in meiotic prophase.
Results
Aberrant chromatin structure during meiotic prophase in the absence of Rec8 and Pds5
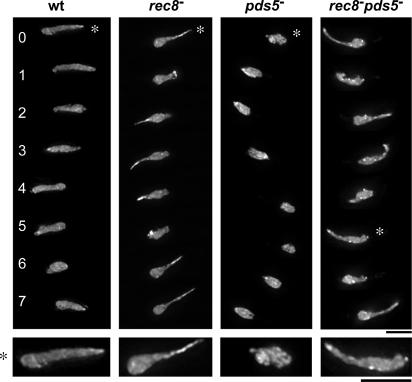
In a screen of meiotic mutants, we have observed nuclear movement during meiotic prophase in living cells of S. pombe (Hiraoka et al., 2000). In such a screen, we observed aberrant nuclear movements in the rec8 − mutant: although the telomeres repeatedly traversed the cell, the bulk of the chromosomes did not follow (Fig. 1 and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200605074/DC1; Molnar et al., 2001). This suggests that chromatin architecture is altered in this mutant. In contrast to the elongated morphology of the nucleus in the rec8 − mutant, we found that pds5 − cells had a shorter nucleus. However, the rec8 − pds5 − double mutant showed an elongated morphology similar to that observed in the rec8 − single mutant (Fig. 1), indicating that the shorter nucleus in pds5 − cells is dependent on Rec8. These results suggest that Rec8 and Pds5 are required for proper chromosome structure in meiotic prophase and that the function of Pds5 requires the presence of Rec8.
Figure 1.
Morphology of the horsetail nucleus in meiotic cohesin mutants. Time-lapse images of nuclear movements in wild-type, rec8 −, pds5 −, and rec8 − pds5 − cells are shown. Chromosomes were stained with histone H3-GFP. The bottom panels are twofold-enlarged images of the nuclei indicated by asterisks. Bars, 5 μm.
Measurement of longitudinal compaction ratio of chromatin in living cells
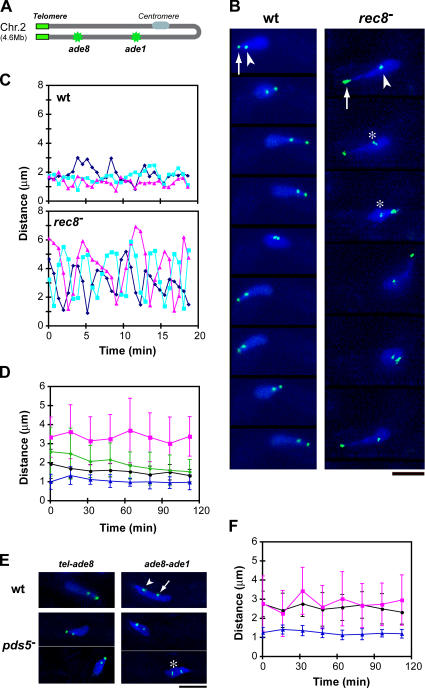
To further examine chromosome structures, we took advantage of the polarized orientation of chromosomes in the telomere-led nuclear movement to directly measure chromosome compaction in living cells. We measured the distance between the telomere and the ade8 locus, using a GFP-tagged telomere protein (Taz1) and a lacO/lacI-GFP tag at the ade8 locus (Fig. 2, A and B). The distance measured was relatively constant in wild-type cells but was greater and varied in rec8 − cells (Fig. 2 C), indicating that the chromosomes were more extended or more flexible in the absence of Rec8. We then examined the distance between the telomere and ade8 locus during the entire meiotic prophase and found that the distance was always significantly larger in rec8 − cells than in wild-type cells (Fig. 2 D). Extension of chromatin was more significant at the moving edge (telomere to ade8) than at the trailing region (ade8 to ade1) in rec8 − as well as wild-type cells (Fig. 2, D and F). Thus, chromatin structure is altered in the absence of Rec8, being more flexible to allow for the pulling forces of nuclear movements.
Figure 2.
Meiotic prophase chromosome compaction. (A) Schematic drawing showing the position of lacO inserts (green) in chromosome II with the telomeres clustered. (B) Time-lapse images of living wild-type and rec8 − cells. DNA was stained with Hoechst 33342 (blue). Green spots represent telomeres stained with Taz1-GFP (arrows) and the ade8 locus stained with lacO/lacI-GFP (arrowheads). Slight separation of sister chromatid loci is seen in the rec8 − cell (asterisks). (C) Distance between the telomere and the ade8 locus in examples of three individual cells. Time 0 represents the start of observations. (D) Changes in the telomere to ade8 distance during meiotic prophase in wild-type (black circle), rec8 − cells (magenta square), rec11 − cells (green diamond), and pds5 − cells (blue triangle). The distance was measured during a 2-min period every 15 min: values were obtained only when the nucleus was moving straight in either direction (not making a turn) and averaged from 10–20 measurements from 10 cells at each time point. The error bars indicate standard deviations. Time 0 represents the start of meiotic prophase, when karyogamy has just finished. (E) Chromosome compaction in pds5 − mutants. DNA was stained with Hoechst 33342 (blue). The green spots in left panels are telomeres and the ade8 locus, as in B. The green spots in the right panels are the ade8 locus (arrowhead) and the ade1 locus (arrow). Slight separation of sister chromatid loci in a pds5 − cell is indicated by an asterisk. (F) Changes in the ade8 to ade1 distance during meiotic prophase in wild-type (black circle), rec8 − cells (magenta square), and pds5 − cells (blue triangle). Bars, 5 μm.
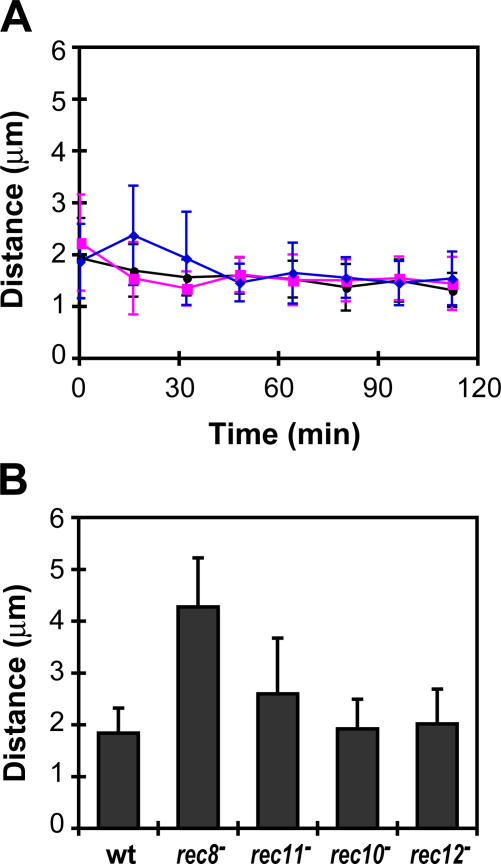
To determine whether meiotic prophase chromosome structure depends on the presence of other cohesin subunits, we examined chromosome compaction in the absence of another meiotic cohesin component, Rec11. In rec11-156 mutant cells (Li et al., 1997), chromosome compaction was also relaxed but to a lesser extent than in the rec8 − mutant (Fig. 2 D and see Fig. 4 B). In the rec11 − mutant, although localization of Rec8 and Psm3 was similar to that of wild-type cells (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200605074/DC1), the mitotic counterpart of Rec11, Psc3, which concentrates at the centromere in wild-type cells (Kitajima et al., 2003), was relocalized along the entire length of the chromosomes (Fig. S1 B). Thus, Psc3 could partially supplement the usual roles of Rec11, thereby resulting in a milder phenotypic alteration in chromosome compaction.
Figure 4.
Chromosome compaction in recombination mutants during meiotic prophase. (A) Changes in the telomere to ade8 distance during meiotic prophase in wild-type (black circle), rec10 − (magenta square), and rec12 − cells (blue diamond). (B) The mean telomere to ade8 distances in wild-type cells and mutants during meiotic prophase. The means were calculated from 140 to 190 measurements from time-lapse series of 15–20 cells per strain; the error bars represent the standard deviation. The difference between the wild-type and rec8 − or rec11 − mutants is significant (P < 0.0001), as detected by a t test.
In contrast to the rec8 − and rec11 − mutants, we found that in pds5 − cells, the distance from the telomere to ade8 or from ade8 to ade1 loci was about one half of that seen in wild-type cells (Fig. 2 E). These distances were always shorter in pds5 − cells than in wild-type cells throughout meiotic prophase (Fig. 2, D and F). Thus, the chromosome was hypercompacted in the absence of Pds5.
We then calculated the apparent longitudinal DNA compaction of meiotic prophase chromosomes using the measured distance data. In wild-type cells, the DNA compaction ratio was 80–110, approximately two times more compact than the 30-nm chromatin fiber (Sedat and Manuelidis, 1978; Table I). However, in the absence of Rec8, the apparent DNA compaction ratio decreased to 32 at the moving edge, similar to the 30-nm chromatin level. In the pds5 − mutant, the apparent compaction ratio significantly increased to 140–290 (Table I). Extension of chromatin was more significant in the telomere-proximal region (telomere to ade8) than in the telomere-distal region (ade8 to ade1) in all cases of rec8 −, pds5 −, and wild-type cells (Table I). Thus, chromatin can be extended by the pulling forces of nuclear movements, but its flexibility differs in rec8 −, pds5 −, and wild-type cells.
Table I.
Spatial distance between two chromosome loci and the apparent chromosome compaction ratio in meiotic prophase
| Distance | Apparent compaction ratio |
|
|---|---|---|
| μm | ||
| Wild type | ||
| Telomere to ade8 a | 1.8 ± 0.5 (161) | 77 |
| ade8 to ade1 b | 3.1 ± 0.5 (117) | 111 |
| rec8 − | ||
| Telomere to ade8 | 4.3 ± 1.0 (144) | 32 |
| ade8 to ade1 | 3.2 ± 1.4 (90) | 106 |
| pds5 − | ||
| Telomere to ade8 | 1.0 ± 0.4 (249) | 137 |
| ade8 to ade1 | 1.2 ± 0.2 (189) | 287 |
| 30-nm fiber | 40 | |
| Telomere to ade8 | 3.7 | |
| ade8 to ade1 | 9.6 |
Distance is represented as mean ± SD. The number of cells examined is indicated in parentheses. Distances were obtained only when the nucleus was moving straight in either direction (not making a turn). The compaction ratio was estimated from B-type DNA as 0.34 μm/kb.
0.4 Mb.
1.0 Mb.
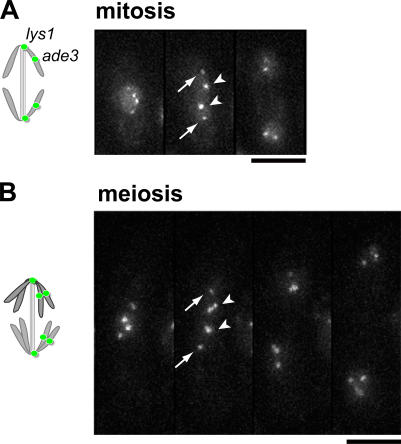
To address whether Rec8 and Pds5 are also involved in chromosome architecture in both mitosis and meiosis I, we evaluated chromosome compaction at early anaphase by labeling two loci on the same arm of chromosome I: the lys1 locus and ade3 locus (Fig. 3). We found a DNA compaction ratio of ∼600–700 in wild-type cells, and similar chromosome compaction was calculated for both the rec8 − and pds5 − mutants (Table II). These results suggest that Rec8 and Pds5 do not significantly contribute to the chromosome architecture, at least not at anaphase.
Figure 3.
Chromosome compaction at anaphase. Anaphase chromosomes in mitosis (A) and meiosis I (B). Two lacO arrays were inserted on one of the arms of chromosome I at lys1 (arrows) and ade3 (arrowheads) loci and stained with lacI-GFP (schematic drawing). Data in Table II were collected only from cells in early anaphase: when the spindle elongation was ongoing and nuclear separation was not complete (second panel). Bars, 5 μm.
Table II.
Spatial distance between two chromosome loci and the apparent chromosome compaction ratio in early anaphase in mitosis and meiosis I
| Distance from ade3 to lys1 a |
Apparent compaction ratio |
|
|---|---|---|
| μm | ||
| Wild type | ||
| Mitosis | 1.26 ± 0.20 (130) | 683 |
| Meiosis I | 1.37 ± 0.33 (72) | 628 |
| rec8 − | ||
| Mitosis | 1.35 ± 0.25 (80) | 638 |
| Meiosis I | 1.39 ± 0.33 (65) | 619 |
| pds5 − | ||
| Mitosis | 1.26 ± 0.25 (111) | 683 |
| Meiosis I | 1.47 ± 0.54 (81) | 585 |
| 30-nm fiber | 24 | 40 |
Distance is represented as mean ± SD. The number of cells examined is indicated in parentheses. A t test shows that the distance between ade3 and lys1 in rec8 − or pds5 − mutant cells is not significantly different from that of wild-type cells (P > 0.004).
2.5 Mb.
LE and double-strand break are not required for meiotic prophase chromatin compaction
In S. pombe, Rec8 is required for LE formation and homologous recombination (DeVeaux and Smith, 1994; Molnar et al., 1995; Watanabe and Nurse, 1999). To examine whether these processes are required for chromosome compaction, we measured chromosome compaction in rec10 − and rec12 − mutants. Rec10 is an essential component for LE formation because, in the rec10 − mutant, no LE structures form (Molnar et al., 2003; Lorenz et al., 2004). Rec12, a homologue of S. cerevisiae Spo11, generates double-strand breaks and is required for homologous recombination (Cervantes et al., 2000). We found that chromosome compaction was similar in the rec10 −, rec12 −, and wild-type cells (Fig. 4). In addition, in a strain of rec10 − pds5 − double mutant, the hypercompaction caused by Pds5 loss was also observed (unpublished data). Thus, the meiotic cohesins and Pds5 support chromosome compaction during meiotic prophase in a manner independent of LE formation or recombination.
Defective localization of cohesins in the absence of Rec8 and Pds5
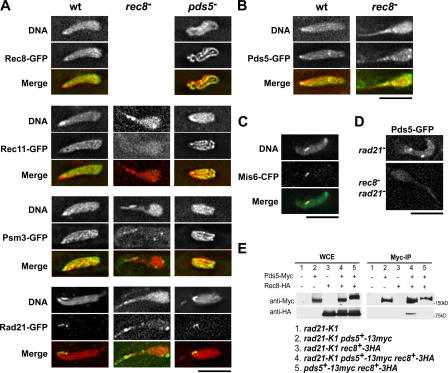
To evaluate the contribution of cohesins to meiotic chromosome structures, we examined the interdependency of cohesins and Pds5 in their localization. In wild-type meiotic prophase nuclei, Rec8-, Rec11-, and Psm3-GFP formed thin filaments (Fig. 5 A, Fig. S2, and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200605074/DC1), which probably represent the axis of the chromosome. Pds5-GFP also formed thin filaments, with additional foci at the centromeres (Fig. 5 B), as confirmed by its colocalization with the centromere protein Mis6 (Fig. 5 C). In rec8 − cells, however, Rec11- and Psm3-GFP did not concentrate at the chromosome axis; only a residual punctate Psm3-GFP signal was found at the leading edge and putative centromere regions of the nucleus in such cells (Fig. 5 A and Fig. S3). Thus, localization of Rec11 and Psm3 on the chromosome axis is dependent on Rec8.
Figure 5.
Localization and interaction of cohesins and Pds5 in the meiotic prophase nucleus. (A) Localization of Rec8-, Rec11-, Psm3-, and Rad21-GFP in rec8 − and pds5 − mutants. The rec8 − has no Rec8-GFP. (B) Localization of Pds5-GFP in wild-type and rec8 − cells. In both A and B, DNA was stained with Hoechst 33342; red represents DNA and green represents GFP in the merged images. (C) Double staining of Pds5-GFP (green in merged image) and Mis6-CFP (red in merged image) in a wild-type cell. (D) Localization of Pds5-GFP in the rad21-K1 mutant and the rec8 − rad21-K1 double mutant at 31°C. (E) Immunoprecipitation of Pds5-13Myc and Rec8-3HA in mitotic cells. Immunoprecipitation was performed with anti-Myc mouse antibody. Proteins from whole cell extracts and from precipitates were electrophoresed and immunoblotted with anti-HA and anti-myc antibodies. Bars, 5 μm.
On the other hand, localization of Pds5 was not greatly affected by the loss of Rec8 or Rad21 (Fig. 5, B and D). In mitotic cells, Pds5 binds chromosomes in a Rad21-dependent manner (Hartman et al., 2000; Panizza et al., 2000; Tanaka et al., 2001; Wang et al., 2002). Whereas Rad21 binds along with Rec8 on the chromosome axis in meiotic cells of many other eukaryotes (Klein et al., 1999; Prieto et al., 2002), Rad21-GFP is confined to the rDNA region in S. pombe, which is located at the leading edge of the horsetail nucleus (because rDNA locates next to the telomeres of chromosome III; Yokobayashi et al., 2003; Fig. 5 A). In rec8 − cells, some Rad21 extends to the chromosome arm (Yokobayashi et al., 2003; Fig. 5 A). Localization of Pds5 on the chromosome axis decreased when Rad21 was inactivated in the temperature-sensitive mutant rad21-K1 in the rec8-deletion background (Fig. 5 D), indicating that Pds5 localization depends on Rec8 and, in the absence of Rec8, on Rad21. These localization results are consistent with the results of immunoprecipitation experiments. Pds5 is known to coprecipitate with Rad21 (Tanaka et al., 2001), and we found that mitotically expressed Rec8-HA could also coprecipitate with Pds5-Myc, but only when Rad21 was absent (Fig. 5 E). These results suggest that Pds5 associates with either cohesin, predominantly Rad21 in mitosis, and could associate with Rec8 in the absence of Rad21. It should be noted that rec8 − cells had loose chromosomes despite the presence of Rad21 on the chromosome; thus, the relocated Rad21 is not sufficient for replacing the function of Rec8 in compaction of chromosome arms. Collectively, the results show that Rec8 recruits cohesins to the chromosome axis and plays a key role in forming meiotic prophase chromosome structure.
In the absence of Pds5, Rec8-GFP showed distinct staining along the compacted chromosomal axis (Fig. 5 A and Fig. S2 A), and Rec11-GFP and Psm3-GFP showed a similar pattern to Rec8-GFP (Fig. 5 A, Fig. S2, and Fig. S3). Therefore, lack of Pds5 causes aberrant localization of Rec8 and other cohesins along the chromosome, which may reflect the hypercompaction of meiotic prophase chromosomes in the pds5 − mutant. In addition, Rad21-GFP was retained in the rDNA region as in wild-type cells (Fig. 5 A), suggesting that Rad21 is not involved in the regulation of chromosome compaction in the pds5 − mutant.
Decreased Rec8 binding on chromosomes in the absence of Pds5
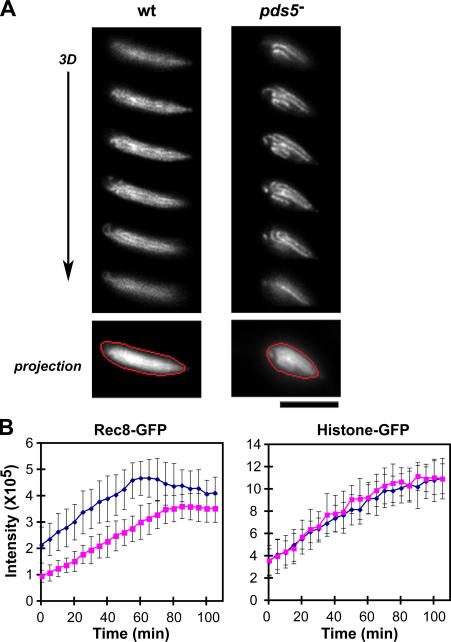
Because Rec8 is a key molecule in meiotic prophase chromosomes, the binding of Rec8 to chromosomes must be precisely controlled. We therefore examined the amount of Rec8-GFP in meiotic cells. Because it is difficult to follow temporal changes of the amount of Rec8 binding on chromosomes by detergent extraction or chromatin spreads, we determined the amount of Rec8 by measuring GFP fluorescence intensity in the nucleus of individual living cells from karyogamy to meiosis I. Rec8-GFP signal could be detected in both wild-type and pds5 − mutants before karyogamy, with its intensity reaching a peak during the horsetail stage (∼60 min in Fig. 6 B and Fig. S2). Total amounts of Rec8 were similar in both wild-type and pds5 − mutants, as determined by Western blot analyses (unpublished data). However, quantitative analysis of fluorescence intensity in the nucleus of living cells clearly detected a decrease in the total intensity of Rec8-GFP signals in the pds5 − mutants when compared with wild-type cells (Fig. 6). The total intensity of the GFP signal in wild-type cells was, on average, ∼1.5 times higher than in pds5 − mutants (Fig. 6 B, left), even though the cohesin axis was more clearly observed in pds5 − mutants. As a control, the intensity of histone-GFP signals was examined and found to be the same in both wild-type and pds5 − mutant cells (Fig. 6 B, right). As chromatin unbound cohesin subunits increased in the cytoplasm (Fig. S3), their retention within the nucleus may be mediated through binding to chromatin. Thus, this decrease in nuclear Rec8 fluorescence may reflect a reduction in the amount of chromatin bound Rec8 in the absence of Pds5, raising the possibility that Rec8 binding to the chromosome is reduced in the absence of Pds5.
Figure 6.
Quantitative analysis of the fluorescent intensity of Rec8- and histone-GFP in the nucleus in meiotic prophase. (A) Images of Rec8-GFP are shown in 3D focal planes, with an additive projection image (bottom). The red polygons represent the satisfactory polygon defined by the 2D Polygon Finder of the SoftWoRx imaging analysis software. (B) Changes in intensity of Rec8- and histone H3-GFP signals in nuclei of wild-type (blue) and pds5 − mutant (magenta) cells during meiotic prophase. The data presented are means from 19 cells for Rec8-GFP and 14 cells for histone H3-GFP. The error bars represent the standard deviation. Time 0 represents the start of meiotic prophase, when karyogamy has just finished. Bar, 5 μm.
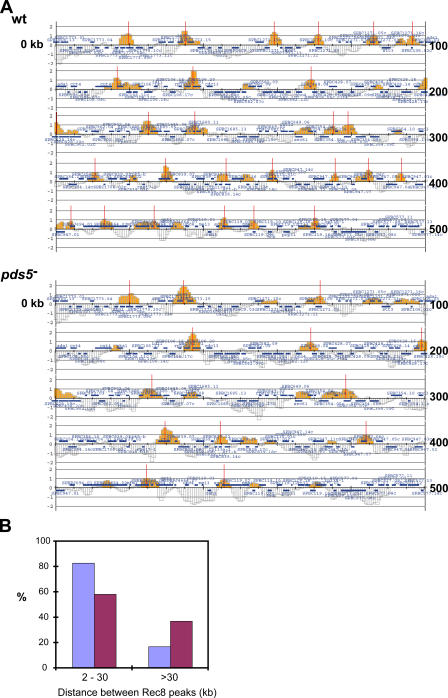
To further explore this possibility, we performed chromatin immunoprecipitation (ChIP) of Rec8 in synchronized meiosis induced by inactivation of pat1 (Yamamoto and Hiraoka, 2003), followed by hybridization to a high-density oligonucleotide array (ChIP-chip analysis; Katou et al., 2003; Lengronne et al., 2004). The association of Rec8 with chromosomes II and III was examined. In wild-type cells, Rec8 bound to ∼300 distinct sites, each spanning 2–5 kb, along the arms of the 5.5 Mb of chromosomes II and III (complete DNA array data in Gene Expression Omnibus under accession no. GSE5284). We repeated the experiment twice. Of the Rec8 binding sites, ∼200 sites were detected in both of the two independent experiments. In the absence of Pds5, ∼100 of the Rec8 binding sites were lost in each set of experiments (see Fig. 7 A for a portion of the array data). Thus, these Rec8 binding sites depend on Pds5 for stable binding to chromatin. In wild-type cells, 83% of the neighboring Rec8 binding sites were within a distance of 2–30 kb (Fig. 7 B), whereas in the absence of Pds5, this population decreased to 58%, and those at a distance of >30 kb increased (Fig. 7 B). Accordingly, the mean distance between the Rec8 sites increased from 18 kb in wild type to 28 kb in pds5 − mutants. These data suggest that loss of Pds5 reduced Rec8 binding to the meiotic prophase chromosome. These results are consistent with the reduction in Rec8-GFP signal that was observed in the live cell analysis of the pds5 − mutants (Fig. 6).
Figure 7.
Rec8 binding on chromosomes in wild-type and pds5− mutant cells. ChIP was performed against Rec8-HA in pat1-114 synchronized meiosis after the temperature was shifted up to 34°C for 3 h. The premeiotic DNA synthesis was complete at this time (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200605074/DC1). (A) Enrichment in the immunoprecipitated fraction relative to a whole genome DNA sample is shown along a 500-kb region at left arm of chromosome II. Each bar represents the mean of 11 oligonucleotide probes within adjacent 250-bp windows. Orange bars represent significant binding. Blue bars above and below the midline indicate open reading frames transcribed from left to right and from right to left, respectively. The y-axis scale is log2. The Rec8 binding sites (red lines) were estimated using a threshold of above 0.8 for peak height and 2 kb for width. (B) Distribution of distance between neighboring Rec8 binding sites on chromosome II and III. Centromere regions were excluded from this analysis. Data are means of two independent experiments.
Role of Pds5 in sister chromatid cohesion in meiotic prophase
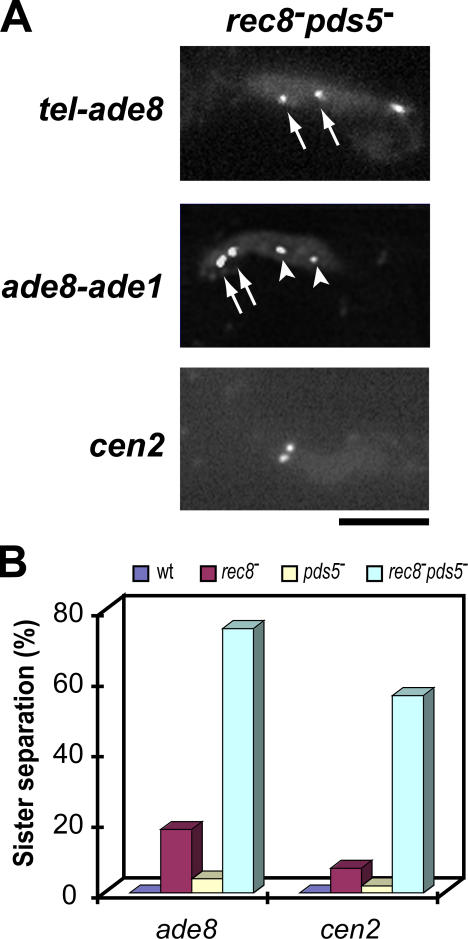
As it is known that Rec8 plays a central role in sister chromatid cohesion (Watanabe and Nurse, 1999; Kitajima et al., 2003), we examined the role of Pds5 in this Rec8-mediated meiotic event. Precocious separation of sister chromatids in meiotic prophase was occasionally observed in both rec8 − and pds5 − mutants (Fig. 2, B and E, respectively), but the frequency of such events was significantly higher in the rec8 − pds5 − double mutant than in either single mutant (Fig. 8). As this phenotype resembles that of the rec8 − rad21-K1 double mutant (Yokobayashi et al., 2003), we propose that in the absence of Rec8, Rad21 cooperates with Pds5 to assist sister chromatid cohesion, although Rec8 normally promotes cohesion independently of Pds5.
Figure 8.
Sister chromatid cohesion in rec8− and pds5− mutants. (A) Precocious separation of sister chromatids in the rec8 − pds5 − double mutant. A pair of sister chromatid loci on one of the homologous chromosomes was marked with lacO/lacI-GFP: (top) Taz1-GFP and ade8 locus (arrows); (middle) ade1 (arrowheads) and ade8 loci (arrows); (bottom) cen2 locus. (B) Frequency of sister chromatid separation during meiotic prophase. A pair of sister chromatid loci on one of the homologous chromosomes was marked with lacO/lacI-GFP at the ade8 or cen2 locus. Bar, 5 μm.
Discussion
We have shown that meiotic prophase chromosome compaction is reduced when meiotic cohesins Rec8 or -11 are absent. Furthermore, the hypercompaction caused by loss of Pds5 requires the presence of Rec8, and the number of Rec8 binding sites on the chromosome is reduced in the absence of Pds5. In addition, we found that the LE component Rec10 is not required for meiotic prophase chromosome compaction. These results indicate that meiotic cohesins have a critical role in meiotic prophase chromosome compaction, which is independent of LE formation, in S. pombe.
A role for meiotic cohesins in chromosome compaction in meiotic prophase
Many studies have suggested that chromosomes in meiotic prophase are packed along an axis with chromatin loops (Zickler and Kleckner, 1999). In this chromosome architecture, when the density of binding sites of chromatin along the axis is reduced, a shorter axis with larger chromatin loops would arise (Revenkova and Jessberger, 2006). This model explains our observation that the reduction in Rec8 binding on chromosomes in the pds5 − mutant results in higher longitudinal chromosome compaction. Further reduction in cohesin levels decreases chromosome compaction because of the resulting loss of the axis required for the loops and explains the results observed in the rec8 − mutants. Similar models that predict the link between cohesion sites and chromosome condensation in mitotic cells have also been proposed in S. cerevisiae (Guacci et al., 1997; Losada and Hirano, 2001).
Our observations show that longitudinal chromatin length remains relatively constant throughout the horsetail stage in rec8 − and pds5 − cells as well as in wild-type cells (Fig. 2, D and F). In addition, Rec8 and other cohesin subunits could be clearly detected at the beginning of the horsetail stage (Figs. S2 and S3). Because premeiotic DNA replication occurs during the horsetail stage and can be observed as an increase in the intensity of the histone-GFP signal (Chikashige et al., 2004; Ding et al., 2004), the Rec8-dependent chromosome architecture may form before premeiotic DNA replication.
In S. pombe, chromosome compaction was normal in a rec10 − mutant, in which the formation of LEs was shown to be abolished completely (Molnar et al., 2003). Although Rec8 and -11 are required for the formation of LEs, their role in compaction of meiotic prophase chromosomes is not mediated through LE formation. Furthermore, no significant increase in compaction was observed during the progression of meiotic prophase in S. pombe, and the compaction in recombination-deficient cells was similar to that in wild-type cells. This indicates that meiotic prophase chromatin is already formed at the onset of meiosis and, thus, independently of subsequent meiotic events such as LE formation or recombination. In contrast, in mouse spermatocytes, the cohesin core is extended in length when the SC protein SYCP3 is deficient (Kolas et al., 2004). Thus, additional chromosome compaction may be mediated by SC proteins or by the formation of SC (Revenkova and Jessberger, 2006).
Another interesting question is whether condensin, the protein complex essential for mitotic chromosome condensation, is involved in meiotic prophase chromosome compaction. In S. cerevisiae, condensin localizes on pachytene chromosomes and is necessary for pachytene chromosome compaction and SC assembly (Yu and Koshland, 2003). In C. elegans, condensin localizes to sister chromatids during diplotene-diakinesis of meiotic prophase I and is necessary for diplotene chromosome condensation (Chan et al., 2004). In S. pombe meiosis, however, the SMC subunits of the condensin complex Cut3 and -14 (Sutani et al., 1999) were localized in the cytoplasm for the entire horsetail stage and localized to the nucleus only after the horsetail movement stopped and meiotic division I was about to begin (unpublished data). Thus, condensin may primarily contribute to metaphase/anaphase chromosome events, or low levels of condensin may be sufficient for meiotic prophase chromosome compaction. The function of condensin in S. pombe in meiotic prophase remains to be elucidated.
The role of Pds5 in the functions of meiotic cohesin on the chromosome
Our results suggest that Pds5 has roles in meiotic chromosome compaction and on sister chromatid cohesion. The chromosome localization of Pds5 is mediated by Rec8, and in the absence of Rad8, by Rad21. On the other hand, we detected a decrease in Rec8 binding to the chromosome in the absence of Pds5 by both live cell imaging and ChIP-chip analysis, indicating that Pds5 plays a role in promoting the binding of Rec8 to meiotic chromosomes. Thus, Pds5 and Rec8 are interdependent in their functions.
In mitotic cells of S. cerevisiae, reduced chromosome binding of Scc1 in a pds5-99 mutant was reported (Panizza et al., 2000). Therefore, the role of Pds5 in maintenance of cohesins on mitotic and meiotic chromosomes seems to be conserved. In addition, we found fragmentation of the cohesin axis in the absence of Pds5 (unpublished data). Similarly, in S. macrospora, the axial elements are split and discontinuous in the Pds5 homologue spo76-1 mutant (van Heemst et al., 1999); in S. cerevisiae, the pds5-1 mutation causes fragmentation of the SC (Zhang et al., 2005). These observations suggest a common role for Pds5 in forming the integral axis of chromosomes. However, in a C. elegans Pds5 mutant, evl-14, apparently normal SC formation and Rec8 localization was found (Wang et al., 2003). In S. cerevisiae, Rec8 binding to chromosomes is slightly decreased in pds5-1 mutant cells (Zhang et al., 2005). Given the fact that S. pombe Pds5 plays a dual role in both establishment and maintenance of cohesion (Tanaka et al., 2001), the difference in phenotypes may result from allele specificity or may reflect differences in the contributions of Pds5 in this organism.
Diverse phenotypes out of the conserved cohesins in meiotic chromosome architecture
The role of cohesins in meiotic chromatin structures has also been identified in other organisms, although different phenotypes have been observed in each organism. In S. cerevisiae, lack of Rec8 resulted in undercompacted chromosomes and deficient axial element and SC formation (Klein et al., 1999). In Arabidopsis thaliana, AtRec8, but not AtScc1, is required for an intact chromosome axis (Chelysheva et al., 2005). In C. elegans, depletion of REC-8 as well as SCC-3 disrupts SC formation (Pasierbek et al., 2001, 2003; Wang et al., 2003). In mice, lack of Rec8 or Smc1β resulted in intact but shorter axial elements (Bannister et al., 2004; Revenkova et al., 2004; Xu et al., 2005) and larger loops of chromatin (Revenkova et al., 2004). Unlike in S. pombe, Rad21 also localizes along the SC in mouse meiotic cells (Prieto et al., 2002), suggesting a potential role in meiotic chromosome compaction. These diverse phenotypes may be attributed to the variety and redundancy of cohesin components in different organisms. Considering that the meiotic-specific cohesin complexes appear upon entering meiosis from yeasts to humans, the underlying function of the meiotic cohesin complex in constructing fundamental chromosome architecture is likely to be conserved, but subtle evolutionary changes in their regulation may lead to the diverse phenotypes among organisms.
Materials and methods
Strains
The fission yeast strains used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200605074/DC1). Strains bearing Rec8- and Psc3-GFP were gifts from Y. Watanabe (University of Tokyo, Tokyo, Japan). Disruption of the pds5 gene and construction of the Pds5-GFP fusion gene were performed as previously described (Wang et al., 2002). Psm3-GFP, Rec11-GFP, and Rad21-GFP fusions were constructed using the PCR-based gene targeting method (Bahler et al., 1998), where the ORF of GFP was integrated at the C-terminal end of the endogenous gene locus in the genome. All the GFP fusions showed vegetative growth rates indistinguishable from that of wild-type cells at normal (33°C), lower (20°C), and higher (36°C) temperatures. Spore formation and viability were also comparable with that of wild-type cells.
Image acquisition and manipulation
A computer-controlled fluorescence microscope system (DeltaVision [Applied Precision]; Haraguchi et al., 1999) was used for imaging of live cells. This microscope system is based on an inverted fluorescence microscope (IX70; Olympus) equipped with a charge-coupled device (CoolSNAP HQ; Photometrics). The objective lens used was an oil-immersion lens (Plan Apo 60×; NA = 1.4; Olympus). For time-lapse observation, living fission yeast cells were mounted in a 35-mm glass-bottomed culture dish (MatTek) coated with concanavalin A and observed in EMM2 medium at 26°C. A set of images at 10 focal planes at 0.3-μm intervals was taken at each time point. Image deconvolution was performed using an imaging workstation (SoftWoRx; Applied Precision). For quantitative analysis of GFP signals, a set of images at 10 focal planes at 0.4-μm intervals was taken at each time point. Quantitative projections were generated using an additive image projecting method. On the projected images, 2D polygons (Fig. 6 A) were drawn with an automatically set threshold value using the 2D Polygon Finder in the software, and the sums of the fluorescence intensities in the polygons were obtained. To minimize any error resulting from the progressive decline of the mercury-arc output, we collected datasets from wild-type and mutant cells using the same lamp and within 8 h on the same day. Photoshop 6.0 (Adobe) was used to adjust the linear image intensity (brightness and contrast) for figure production.
Chromosomal loci were visualized by the use of a lac repressor (lacI)/lac operator (lacO) recognition system; i.e., repeats of the lacO sequence were integrated at a chromosome locus and detected by the GFP-lacI fusion protein (Robinett et al., 1996; Straight et al., 1996). Original constructions for each chromosomal loci used in this work are cited in the strain list (Table S1). For observation of meiosis, haploid cells of the opposite mating type were conjugated on a plate to form a diploid zygote. GFP-labeled chromosomal loci or proteins were observed in living zygotes at 26°C, as described previously (Ding et al., 2004).
Immunoprecipitation
To express Rec8 in mitotic cells, the rec8 + -3HA gene was cloned and inserted at the lys1 locus under the control of an inducible nmt1 promoter, and the cells were cultured in medium without thiamine. Cells carrying the rad21-K1 mutation were precultured at 26 or 30°C overnight and then cultured at 36°C for 5 h. The 36°C step was used to inactivate the temperature-sensitive rad21-K1 protein. Protein extracts were prepared as previously described (Tanaka et al., 2001). To liberate chromosome bound proteins, cell extracts were treated with DNase I at 25°C for 5 min. Immunoprecipitation was performed with anti-Myc mouse antibody (9E10; Santa Cruz Biotechnology, Inc.). Proteins from whole cell extracts and from precipitates were electrophoresed in SDS-polyacrylamide gels and immunoblotted with anti-HA (3F10; Roche) and anti-Myc antibodies.
ChIP-chip assay
Haploid h − pat1 − strains carrying the rec8 + -3HA gene were synchronized to meiosis by overnight nitrogen starvation at 26°C, and the culture temperature was shifted to 34°C. After 3 h at 34°C, the cells were fixed with 1% formaldehyde for 30 min at room temperature. ChIP and DNA chip analyses were performed as previously described (Katou et al., 2003; Lengronne et al., 2004). In brief, 5 × 108 cells were disrupted using a multibeads shocker. Whole cell extracts were sonicated (250D; Branson) to obtain 400–600-bp genomic DNA fragments. Anti-HA mouse monoclonal antibody (16B12; Babco) coupled to protein A Dynabeads (Dynal) were used for ChIP. The immunoprecipitates were eluted and incubated overnight at 65°C to reverse the cross-linking. The genomic DNA was precipitated, purified, and amplified by PCR using random primers. For ChIP-chip analyses of Rec8-3HA, S. pombe chromosome II and III tiling array (part 520106; Affymetrix, Inc.) was used. Chip data presented in this paper can be obtained from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession no. GSE5284).
Online supplemental material
Table S1 shows the strain list used in this study and references about the source of the strains. Fig. S1 shows the localization of cohesins Rec8, Psm3, Pds5, and Psc3 in rec11 − mutant cells. Fig. S2 shows the dynamics of Rec8 and Pds5 in the entire meiosis process in wild-type and rec8 − or pds5 − mutant cells. Fig. S3 shows the dynamics of Rec11 and Psm3 in the entire meiosis process in wild-type, rec8 −, and pds5 − mutant cells. Fig. S4 shows the synchronization of pat1-induced meiosis in the ChIP-chip analysis. Video 1 shows the nuclear movement in meiotic prophase in wild- type and rec8 − mutant cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200605074/DC1.
Supplementary Material
Acknowledgments
We thank Monika Molnar, Ayumu Yamamoto, Aki Hayashi, and Yuji Chikashige for sharing strains and GFP constructs; Douglas Koshland, Tatsuya Hirano, Julia Cooper, Ayumu Yamamoto, and Hirohisa Masuda for critical reading of earlier versions of the manuscript; and Abby Dernburg for editorial comments.
This work was supported by grants from the Ministry of Education, Science, and Culture of Japan (D.-Q. Ding) and the Japan Science and Technology Agency (T. Haraguchi and Y. Hiraoka).
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; LE, linear element; rDNA, ribosomal DNA; SC, synaptonemal complex; SMC, structure maintenance of chromosome.
References
- Bahler, J., T. Wyler, J. Loidl, and J. Kohli. 1993. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol. 121:241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler, J., J.Q. Wu, M.S. Longtine, N.G. Shah, A. McKenzie III, A.B. Steever, A. Wach, P. Philippsen, and J.R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [DOI] [PubMed] [Google Scholar]
- Bannister, L.A., L.G. Reinholdt, R.J. Munroe, and J.C. Schimenti. 2004. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 40:184–194. [DOI] [PubMed] [Google Scholar]
- Cervantes, M.D., J.A. Farah, and G.R. Smith. 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell. 5:883–888. [DOI] [PubMed] [Google Scholar]
- Chan, R.C., A.F. Severson, and B.J. Meyer. 2004. Condensin restructures chromosomes in preparation for meiotic divisions. J. Cell Biol. 167:613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva, L., S. Diallo, D. Vezon, G. Gendrot, N. Vrielynck, K. Belcram, N. Rocques, A. Marquez-Lema, A.M. Bhatt, C. Horlow, et al. 2005. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 118:4621–4632. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., D.Q. Ding, H. Funabiki, T. Haraguchi, S. Mashiko, M. Yanagida, and Y. Hiraoka. 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science. 264:270–273. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., R. Kurokawa, T. Haraguchi, and Y. Hiraoka. 2004. Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe. Genes Cells. 9:671–684. [DOI] [PubMed] [Google Scholar]
- DeVeaux, L.C., and G.R. Smith. 1994. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 8:203–210. [DOI] [PubMed] [Google Scholar]
- Ding, D.Q., Y. Chikashige, T. Haraguchi, and Y. Hiraoka. 1998. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111:701–712. [DOI] [PubMed] [Google Scholar]
- Ding, D.Q., A. Yamamoto, T. Haraguchi, and Y. Hiraoka. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 6:329–341. [DOI] [PubMed] [Google Scholar]
- Eijpe, M., H. Offenberg, R. Jessberger, E. Revenkova, and C. Heyting. 2003. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1beta and SMC3. J. Cell Biol. 160:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 91:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi, T., D.Q. Ding, A. Yamamoto, T. Kaneda, T. Koujin, and Y. Hiraoka. 1999. Multiple-color fluorescence imaging of chromosomes and microtubules in living cells. Cell Struct. Funct. 24:291–298. [DOI] [PubMed] [Google Scholar]
- Hartman, T., K. Stead, D. Koshland, and V. Guacci. 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151:613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng, H.H., J.W. Chamberlain, X.M. Shi, B. Spyropoulos, L.C. Tsui, and P.B. Moens. 1996. Regulation of meiotic chromatin loop size by chromosomal position. Proc. Natl. Acad. Sci. USA. 93:2795–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka, Y., D.Q. Ding, A. Yamamoto, C. Tsutsumi, and Y. Chikashige. 2000. Characterization of fission yeast meiotic mutants based on live observation of meiotic prophase nuclear movement. Chromosoma. 109:103–109. [DOI] [PubMed] [Google Scholar]
- Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 424:1078–1083. [DOI] [PubMed] [Google Scholar]
- Kitajima, T.S., S. Yokobayashi, M. Yamamoto, and Y. Watanabe. 2003. Distinct cohesin complexes organize meiotic chromosome domains. Science. 300:1152–1155. [DOI] [PubMed] [Google Scholar]
- Kleckner, N., A. Storlazzi, and D. Zickler. 2003. Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet. 19:623–628. [DOI] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S.B. Buonomo, C. Michaelis, K. Nairz, and K. Nasmyth. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 98:91–103. [DOI] [PubMed] [Google Scholar]
- Kolas, N.K., L. Yuan, C. Hoog, H.H. Heng, E. Marcon, and P.B. Moens. 2004. Male mouse meiotic chromosome cores deficient in structural proteins SYCP3 and SYCP2 align by homology but fail to synapse and have possible impaired specificity of chromatin loop attachment. Cytogenet. Genome Res. 105:182–188. [DOI] [PubMed] [Google Scholar]
- Lengronne, A., Y. Katou, S. Mori, S. Yokobayashi, G.P. Kelly, T. Itoh, Y. Watanabe, K. Shirahige, and F. Uhlmann. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 430:573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y.F., M. Numata, W.P. Wahls, and G.R. Smith. 1997. Region-specific meiotic recombination in Schizosaccharomyces pombe: the rec11 gene. Mol. Microbiol. 23:869–878. [DOI] [PubMed] [Google Scholar]
- Loidl, J. 2006. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 115:260–271. [DOI] [PubMed] [Google Scholar]
- Lorenz, A., J.L. Wells, D.W. Pryce, M. Novatchkova, F. Eisenhaber, R.J. McFarlane, and J. Loidl. 2004. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 117:3343–3351. [DOI] [PubMed] [Google Scholar]
- Losada, A., and T. Hirano. 2001. Shaping the metaphase chromosome: coordination of cohesion and condensation. Bioessays. 23:924–935. [DOI] [PubMed] [Google Scholar]
- Losada, A., T. Yokochi, and T. Hirano. 2005. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J. Cell Sci. 118:2133–2141. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., R.E. Pearlman, H.H. Heng, and W. Traut. 1998. Chromosome cores and chromatin at meiotic prophase. Curr. Top. Dev. Biol. 37:241–262. [DOI] [PubMed] [Google Scholar]
- Molnar, M., J. Bahler, M. Sipiczki, and J. Kohli. 1995. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 141:61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, M., J. Bahler, J. Kohli, and Y. Hiraoka. 2001. Live observation of fission yeast meiosis in recombination-deficient mutants: a study on achiasmate chromosome segregation. J. Cell Sci. 114:2843–2853. [DOI] [PubMed] [Google Scholar]
- Molnar, M., E. Doll, A. Yamamoto, Y. Hiraoka, and J. Kohli. 2003. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J. Cell Sci. 116:1719–1731. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673–745. [DOI] [PubMed] [Google Scholar]
- Panizza, S., T. Tanaka, A. Hochwagen, F. Eisenhaber, and K. Nasmyth. 2000. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 10:1557–1564. [DOI] [PubMed] [Google Scholar]
- Parisi, S., M.J. McKay, M. Molnar, M.A. Thompson, P.J. van der Spek, E. van Drunen-Schoenmaker, R. Kanaar, E. Lehmann, J.H. Hoeijmakers, and J. Kohli. 1999. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell. Biol. 19:3515–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek, P., M. Jantsch, M. Melcher, A. Schleiffer, D. Schweizer, and J. Loidl. 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15:1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek, P., M. Fodermayr, V. Jantsch, M. Jantsch, D. Schweizer, and J. Loidl. 2003. The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp. Cell Res. 289:245–255. [DOI] [PubMed] [Google Scholar]
- Pelttari, J., M.R. Hoja, L. Yuan, J.G. Liu, E. Brundell, P. Moens, S. Santucci-Darmanin, R. Jessberger, J.L. Barbero, C. Heyting, and C. Hoog. 2001. A meiotic chromosomal core consisting of cohesin complex proteins recruits DNA recombination proteins and promotes synapsis in the absence of an axial element in mammalian meiotic cells. Mol. Cell. Biol. 21:5667–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto, I., N. Pezzi, J.M. Buesa, L. Kremer, I. Barthelemy, C. Carreiro, F. Roncal, A. Martinez, L. Gomez, R. Fernandez, et al. 2002. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 3:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova, E., and R. Jessberger. 2006. Shaping meiotic prophase chromosomes: cohesins and synaptonemal complex proteins. Chromosoma. 115:235–240. [DOI] [PubMed] [Google Scholar]
- Revenkova, E., M. Eijpe, C. Heyting, C.A. Hodges, P.A. Hunt, B. Liebe, H. Scherthan, and R. Jessberger. 2004. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6:555–562. [DOI] [PubMed] [Google Scholar]
- Robinett, C.C., A. Straight, G. Li, C. Willhelm, G. Sudlow, A. Murray, and A.S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedat, J., and L. Manuelidis. 1978. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harb. Symp. Quant. Biol. 42:331–350. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., A.S. Belmont, C.C. Robinett, and A.W. Murray. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608. [DOI] [PubMed] [Google Scholar]
- Sumara, I., E. Vorlaufer, C. Gieffers, B.H. Peters, and J.M. Peters. 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J. Cell Biol. 151:749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani, T., T. Yuasa, T. Tomonaga, N. Dohmae, K. Takio, and M. Yanagida. 1999. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 13:2271–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Z. Hao, M. Kai, and H. Okayama. 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20:5779–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga, T., K. Nagao, Y. Kawasaki, K. Furuya, A. Murakami, J. Morishita, T. Yuasa, T. Sutani, S.E. Kearsey, F. Uhlmann, et al. 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14:2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst, D., F. James, S. Poggeler, V. Berteaux-Lecellier, and D. Zickler. 1999. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 98:261–271. [DOI] [PubMed] [Google Scholar]
- van Heemst, D., E. Kafer, T. John, C. Heyting, M. van Aalderen, and D. Zickler. 2001. BimD/SPO76 is at the interface of cell cycle progression, chromosome morphogenesis, and recombination. Proc. Natl. Acad. Sci. USA. 98:6267–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., J. Yoder, I. Antoshechkin, and M. Han. 2003. Caenorhabditis elegans EVL-14/PDS-5 and SCC-3 are essential for sister chromatid cohesion in meiosis and mitosis. Mol. Cell. Biol. 23:7698–7707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.W., R.L. Read, and C.J. Norbury. 2002. Fission yeast Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J. Cell Sci. 115:587–598. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., and P. Nurse. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 400:461–464. [DOI] [PubMed] [Google Scholar]
- Xu, H., M.D. Beasley, W.D. Warren, G.T. van der Horst, and M.J. McKay. 2005. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell. 8:949–961. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., and Y. Hiraoka. 2003. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 22:2284–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi, S., M. Yamamoto, and Y. Watanabe. 2003. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol. Cell. Biol. 23:3965–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.G., and D.E. Koshland. 2003. Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J. Cell Biol. 163:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Q. Ren, H. Yang, M.N. Conrad, V. Guacci, A. Kateneva, and M.E. Dresser. 2005. Budding yeast PDS5 plays an important role in meiosis and is required for sister chromatid cohesion. Mol. Microbiol. 56:670–680. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner. 1999. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 33:603–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.