Abstract
From a differential display designed to isolate genes that are down-regulated upon differentiation of the central nervous system in Danio rerio embryos, we isolated d-asb11 (ankyrin repeat and suppressor of cytokine signaling box–containing protein 11). Knockdown of the d-Asb11 protein altered the expression of neural precursor genes sox2 and sox3 and resulted in an initial relative increase in proneural cell numbers. This was reflected by neurogenin1 expansion followed by premature neuronal differentiation, as demonstrated by HuC labeling and resulting in reduced size of the definitive neuronal compartment. Forced misexpression of d-asb11 was capable of ectopically inducing sox2 while it diminished or entirely abolished neurogenesis. Overexpression of d-Asb11 in both a pluripotent and a neural-committed progenitor cell line resulted in the stimulus-induced inhibition of terminal neuronal differentiation and enhanced proliferation. We conclude that d-Asb11 is a novel regulator of the neuronal progenitor compartment size by maintaining the neural precursors in the proliferating undifferentiated state possibly through the control of SoxB1 transcription factors.
Introduction
Precursor cells that generate the various differentiated cell types of the central nervous system (CNS) are generally located in defined areas of the developing embryo, which frequently correspond to the ventricular zones of the developing nervous system. During a large part of the development of the embryo, neural progenitors proliferate to renew the precursor pool and, in parallel, give rise to postmitotic cells that move out of the progenitor areas (Temple, 2001). The balance between the generation of precursor and postmitotic cells shifts during development but has to be tightly controlled to guarantee the formation of the appropriate neuronal cell numbers and the size of the tissue domains. Nervous tissue in the vertebrate embryo arises through complex processes of neural induction of the ectoderm, the delineation of neurogenesis-competent domains within the neural plate, and differentiation of neural precursors within these regions (Harland, 2000; Bally-Cuif and Hammerschmidt, 2003; Hammerschmidt et al., 2003). Simultaneously, the expansion of each neuronal compartment takes place until its definitive size and form are reached. At this point in time, the neurons undergo terminal differentiation, thereby acquiring the characteristics necessary for participation in nervous signal propagation while simultaneously losing their capacity for further proliferation.
As the mechanisms controlling the spatio-temporal timing of the sequence of proliferation and differentiation of neuronal-committed cells are still only partly understood, we performed a differential display of mRNAs designed to isolate genes that are differently expressed during neuronal differentiation as well as genes responsible for the proliferation of the stem cell compartment in vivo.
Results
Identification of d-Asb11 as a differentiation down-regulated gene
We used a differential display screen to discover genes that are involved in control of neural stem cell compartment proliferation. To this end, zebrafish embryos were either left untreated or were pulse treated with 0.5 μM all-trans retinoic acid (RA) between 50 and 60% epiboly when neural induction and patterning are initiated. Subsequently, mRNA was isolated from wild-type and treated embryos at 90% epiboly, which, after conversion into cDNA, was used for a differential display using the transcripts from the untreated embryos as driver cDNA and the transcripts from the treated embryos as tester cDNA. The resulting differential fragments were tested using whole-mount in situ hybridization at different developmental stages. One fragment was singled out for detailed characterization based on its restricted spatio-temporal expression pattern during late gastrulation and early somitogenesis. Obtaining a full-length sequence of the down-regulated fragment revealed that the gene was most homologous to mammalian ASB11 (ankyrin repeat and suppressor of cytokine signaling [SOCS] box–containing protein 11), thus identifying this gene as d-asb11 (sequence is deposited at the National Center for Biotechnology Information under GenBank/EMBL/DDBJ accession no. AJ510191; Fig. 1 A, supplemental material, and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200601081/DC1).
Figure 1.
Discovery, characterization, and expression of d-asb11. (A) Amino acid sequence of the d-Asb11 protein. The detection of a down-regulated 0.34-kb fragment in RA-treated embryos in the differential display screen. (B) Expression of d-asb11 in zebrafish embryos. (top) At 50% epiboly (5 hpf), d-asb11 transcripts are present throughout the blastoderm (animal pole up). (bottom) At 95% epiboly (9.5 hpf), d-asb11 transcripts are at a high level localized to the polster (p; lateral view, anterior up). (C) At the tailbud stage (10 hpf), the expression is strongest in the polster and along the margins of the neural plate. Weaker expression is present in two stripes extending toward the midline (asterisks) and in paraxial domains (a compilation of images; dorsal view anterior to the top). (D) Double label in situ hybridization at approximately three somites; d-asb11 is in blue, and deltaA is in red. The expression of deltaA in longitudinal proneuronal domains is complemented by the expression of d-asb11 in the neural plate margins. (E) Asterisks mark regions of the prospective mid/hindbrain boundary. (left) Expression pattern of d-asb11 in zebrafish embryos at 90% epiboly. (right) Embryos treated with RA in which d-asb11 expression is absent in the anterior and diminished in the posterior part of the embryo (anterior to the left). The asterisks represent similar positions of the embryo. Bars, 100 μm.
Expression of d-Asb11 suggests a role in the developing nervous system
ASB family members contain a relatively divergent N-terminal domain followed by a varying number of ankyrin repeats and a C-terminal SOCS box acting as a part of a ubiquitin ligase complex (Kile et al., 2002; Wilcox et al., 2004; Chung et al., 2005; Heuze et al., 2005) and are expressed in members throughout the chordate phylum (Kile et al., 2000; Liu et al., 2003; Guo et al., 2004). d-asb11 was not maternally expressed because the transcripts were not detected by in situ hybridization or Northern blotting in 2.5 h postfertilization (hpf) embryos (unpublished data). The expression of d-asb11 was by in situ hybridization first detected at 4 hpf and was ubiquitous throughout the blastoderm (Fig. 1 B), and such uniform expression was maintained until late gastrula, when d-asb11 transcripts were restricted to the polster (Fig. 1 B). At the tailbud, neuroectodermal expression extends as two stripes along the margins of the neural plate (Fig. 1 C), most likely overlapping with the posterior lateral neural plate expression of notch 1b and the lateral neural plate expression of notch 3 (Figs. 2 and 3 in Westin and Lardelli, 1997) and being complementary to proneural domains expressing deltaA (Fig. 1 D). At the level of the prospective mid/hindbrain boundary, two bilateral domains of expression are detected (Fig. 1, C and E). The expression is transient, as this pattern is maintained until 12 somites. From 12 somites to at least 3 d after fertilization, there was no expression detectable by in situ hybridization, although low levels of mRNA were still detected in 24 hpf embryos by Northern blotting (unpublished data). In RA-treated embryos, the expression of d-asb11 is absent in the anterior half and down-regulated in the posterior half of the embryo, which is consistent with its isolation as a down-regulated fragment from the differential display screen (Fig. 1 E). Thus, the expression of d-asb11 may indicate its possible function in the spatio-temporal timing of proliferation and differentiation in the developing nervous system.
Knockdown of d-Asb11 causes premature neuronal commitment
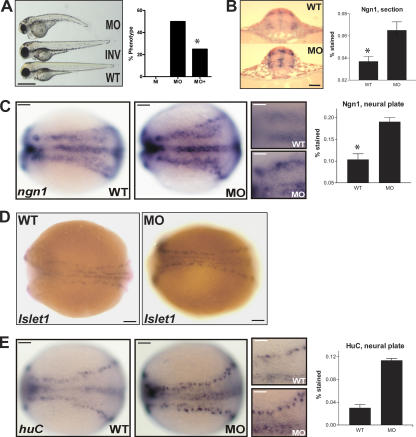
Direct in vivo evidence for a role of d-asb11 in zebrafish development was obtained from d-Asb11 knockdown by the injection of morpholinos (MOs) designed to inactivate d-asb11 (d-Asb11–MO; Nasevicius and Ekker, 2000). We generated an antibody against d-Asb11, tested it on transfected cells, and confirmed it by Western blotting of embryo extracts upon MO injection of zygotes, which resulted in embryos devoid of d-Asb11 protein (supplemental material and Fig. S2 C, available at http://www.jcb.org/cgi/content/full/jcb.200601081/DC1). We assessed the effects of two different d-asb11 ATG MOs on development, and both gave similar phenotypes (see Materials and methods section MOs). At 3 d after fertilization, morphants were typically smaller with a shortened trunk, sometimes accompanied by a downward-directed curly tail and a hyperpericardium (Fig. 2 A). Rescue experiments were performed in which d-Asb11 knockdown by MO that targets sequences upstream of ATG was rescued with the overexpression of myc tag (MT)–d-asb11 mRNA lacking these sequences. Based on the morphology, the overexpression of MT–d-asb11 (MO+) rescued 50% of the d-asb11 phenotype (MO), as shown by a decrease in phenotype from 50 to 25% (Fig. 2 A).
Figure 2.
Modulation of the neural precursor compartment upon knockdown of d-Asb11. (A) Phenotypes of a wild-type (WT), d-Asb11 morpholino (MO)-injected, and inverted MO (INV)-injected embryo at 3 d after fertilization. (right) Rescue of d-asb11 MO-injected embryos with MT–d-asb11 mRNA (see Materials and methods). NI, not injected; MO, MO-injected embryos; MO+, MO and MT–d-asb11–injected embryos. The percentage of altered phenotype was scored for the different groups, and the data shown are representative of four independent experiments. (B) The level and surface of ngn1 expression in the neural tube of 20 hpf embryos were enhanced in morphants (MO) as compared with the control (embryos injected with inverted MO), and quantification of ngn1 induction shows its significant increase in the morphants (n = 5). (C and E) Increase in ngn1 (C) and huC (E) mRNA-expressing cells in the d-asb11 morphant. Left, control embryos; middle, morphant at approximately three somites; right, magnification of wild-type and MO regional expression of ngn1 (C) and huC (E). (far right) Quantification of the amount of staining to calculate the amount of stained cells (C, n = 5; E, n = 6). (A–C) The asterisks mark significant differences using a chi-squared test (P < 0.05). Error bars represent SEM. (D) Increase in islet1 primary neurons in the morphants at the approximately five-somite stage (right) as compared with embryos injected with inverted MO (for photography improvement, polster was removed from the specimens). Bar (A), 200 μm; (B) 20 μm; (C–E) 100 μm; (C and E, right) 20 μm.
Next, d-Asb11 knockdown experiments were performed to determine changes in the expression of the proneural basic helix loop helix gene neurogenin1 (ngn1) that actively promotes neuronal differentiation, inhibits gliogenesis, and is also expressed in still-dividing neuronal precursors (Blader et al., 1997; Morrison, 2001). Strikingly, d-Asb11 knockdown led to the expansion of ngn1 that was first observed at the three-somite stage, when in the morphants, ngn1 was expressed at a higher level and more contiguously in an otherwise unaffected pattern (Fig. 2 C). In the neural tube of d-asb11 morphants, lateral and dorsal expression domains of ngn1 were substantially expanded, as assessed by image quantification of the ngn1-positive surface in transversal sections of representative embryos (Fig. 2 B). Furthermore, strong ectopic ngn1 expression appeared at the center of the neural tube in the proliferative zone (Fig. 2 B). These data show that knockdown of d-Asb11 results in a premature commitment to the neuronal cell lineage.
d-Asb11 knockdown leads to premature postmitotic neuronal differentiation and reduced compartment size
Next, we investigated whether the inhibition of d-asb11 translation affects the generation of terminally differentiated neurons by analyzing the expression of huC, a marker for postmitotic neurons. Again at the three-somite stage, huC expression was increased in the morphants (Fig. 2 E) as compared with embryos injected with inverted MO. A comparable, albeit more subtle, phenotype was obtained by labeling of a subpopulation of the primary neurons by islet1. As in morphants, islet1 neurons were slightly more numerous with what appeared to be higher levels of transcript than in the controls (Fig. 2 D). The increase of HuC-positive cells in the neural tube as evaluated by antibody to HuC protein is maintained at 24, 48, and 72 hpf (Fig. 3, A and B). The proportion of HuC+ cells in the neural tube was at all times higher in morphants as compared with wild-type embryos and, at 72 hpf, occupied even most of the ventricular zone known to consist of the proliferating cells (Fig. 3, A and B). However, low numbers of HuC-negative cells were at 72 hpf still present in the ventricular zone, suggesting that the proliferating compartment was not entirely depleted by loss of function d-Asb11.
Figure 3.
Modulation of differentiation, proliferation, or apoptosis in the neural tube upon the knockdown of d-Asb11. (A) HuC antibody labeling of postmitotic neurons in the spinal cord at 24, 48, and 72 hpf embryos of wild type (WT) and morphants (MO). (B) 3D reconstruction of a representative HuC-labeled 24 hpf morphant and wild-type embryo to calculate the proportion of postmitotic neurons (red, HuC-positive neurons; green, unlabeled cells; yellow, notochord). (C–E) Double immunofluorescence labeling of a mitotic cell with anti-PH3 antibody (green) and terminally differentiated neurons with anti-HuC antibody (red) at 24 (C) and 48 hpf (D) in morphants and wild-type neural tubes (anterior to the left) showing a transient, slight increase in proliferation at 24 hpf followed by a strong decrease in proliferation by 48 hpf and relative increase of HuC+ neurons (red). (E) Quantification of PH3+ mitotic cells. Error bars represent SEM. *, P < 0.05; **, P < 0.005. (F, left) Acridine orange staining at 24 hpf showing enhanced apoptosis in the neural tube of morphants (arrowhead) and few apoptotic cells in wild-type embryos (lateral view, rostral to the left). (right) TUNEL assay shows increased apoptosis (blue cells) throughout in the neural tube of 24 hpf morphants versus only Rohon-Beard apoptosis in the dorsal-most neural tube of the control embryo. Transverse sections through the neural tube are shown. Bars (A), 20 μm; (B–D) 100 μm; (F, left) 200 μm; (F, right) 20 μm.
To quantify the proportion of postmitotic neurons relative to presumably proliferating unlabeled precursors that reside in the ventricular zone of the CNS, a 3D reconstruction of serial sections from a 24 hpf wild-type embryo and a representative 24 hpf morphant was made (Verbeek and Boon, 2002). The measurements show that HuC+ cells contribute to 28% of the total volume of the neural tube in the wild-type embryo and 36% in the morphant, confirming that the proportion of postmitotic neurons is increased in the morphant embryo (Fig. 3, A and B). These data suggest that the targeted knockdown of d-Asb11 results in premature postmitotic neuronal commitment of a subpopulation of neural precursors.
Knockdown of d-Asb11 impairs cell division in the developing nervous system
At 24 hpf, the expanded ngn1 compartment in the morphants could hold both terminally differentiated neurons as well as still-dividing ngn1+ precursors. To verify whether the HuC+ cells were indeed nondividing terminally differentiated neurons, we performed double fluorescent whole-mount antibody labeling with HuC and a mitotic marker anti–phosphohistone-3 (PH3) antibody. Examination of nine morphants and seven wild-type embryos revealed that there was no colocalization of the HuC+ and PH3+ signals (see Whole-mount immunolabeling). The data show that the knockdown of d-Asb11 resulted in a relative and premature increase of terminally differentiated nondividing neurons already at 24 hpf (Fig. 3 C). It is very well possible that a fraction of the supernumerary ngn1+ cells in the morphants was still proliferating and, therefore, was not expressing HuC. The comparison of numbers of mitotic PH3+ cells between morphants and controls at 24 hpf showed that in the morphants, there was a slight but significant (P < 0.03) increase in proliferation that was most likely caused by ectopic proliferating ngn1+ neurons (Fig. 3 E). These data suggest that d-Asb11 functions to prevent neuronal commitment possibly by sustaining proliferation. The reduction of the definitive neuronal compartment may be the result of the depletion of neural progenitors through decline in proliferation. This is supported by a highly significant (P = 0.008) decrease in PH3+ mitotic cells at 48 hpf (Fig. 3, D and E).
Effects of d-Asb11 on compartment size through apoptosis
Concurrent to the decrease in proliferation, the apoptosis of prematurely committed precursors could also contribute to a reduction in compartment size. To study this possibility, we analyzed whether the increase of HuC+ cells in the morphants was accompanied by apoptosis in the neural tube. We found slightly enhanced apoptosis at 24 hpf morphants using acridine orange as an indicator of apoptosis (Fig. 3 F). Analysis of apoptotic cells was extended using TUNEL assay (Fig. 3 F). Transversal sections of the neural tube of wild-type embryos showed apoptotic Rohon-Beard neurons, whereas throughout the entire neural tube of the morphants, there was an increase in apoptosis (Fig. 3 F). This suggests that cell survival of the prematurely committed neuronal precursors may be compromised as a consequence of d-Asb11 knockdown.
Effects of d-Asb11 knockdown on neuronal development are upstream of the SoxB1 HMG group of transcription factors
Because d-Asb11 appears to function in preventing neuronal commitment of the progenitors, we studied whether knockdown of d-Asb11 altered the expression of neural progenitor genes sox2 or sox3 of the SoxB1 high mobility group (HMG) box transcription factors. The expression of sox2 and sox3 through evolution correlates directly with ectodermal cells that are competent to acquire neural fate and subsequently may correlate with the commitment of cells to a neuronal fate (Pevny and Placzek, 2005; Wegner and Stolt, 2005). The expression of sox2 and sox3 in the zebrafish developing CNS is found in the regions containing neural precursors as well as those containing neurons (Okuda et al., 2006). The situation is complex, as neither sox2 nor sox3 is uniformly expressed in all neural-competent cells of the neural plate. This is because there are other SoxB1 genes expressed in the developing CNS of zebrafish and because the kinetics of expression during the transition between the neural precursor and neuron has thus far not been elucidated.
Strikingly, the knockdown of d-Asb11 affected the expression of both sox2 and sox3, albeit differently. At 20 hpf, sox3 expression in the neural tube was reduced in 50% of the morphants (Fig. 4 B), suggesting that d-Asb11 may function to maintain specific levels of sox3 transcripts that may be required for precursor cell maintenance. If this is the case, d-Asb11 does so, most likely, in conjunction with other factors because the expression of sox3 was weaker but still present. This difference in the intensity of sox3 expression between morphants and controls is already clear in whole-mount in situ hybridizations (Fig. 4 B). At 20 hpf in the neural tube of controls, sox2 transcripts were cellularly condensed and appeared to be localized in specific compartments of the cell (Fig. 4 A). In contrast, in 40% of the morphants, sox2 transcripts were diffuse and appeared scattered evenly throughout the cell (Fig. 4 A). The data suggest that d-Asb11 may be required for subcellular condensation of sox2 transcripts. We speculate that such condensed transcripts may be representative of a specific localization, and, in this form, sox2 may be required for maintenance of the precursor cell fate. Such a mechanism of segregating and subcellularly localizing molecular determinants in precursor cells through, for instance, interactions with the cytoskeleton has extensively been studied in neuroblast development in Drosophila melanogaster (Wang et al., 2005). These data show that the loss of d-Asb11 may mediate the reduction of a population of neural precursors through affecting neural competence factors of SoxB1 HMG group transcription factors.
Figure 4.
Effect of d-Asb11 knockdown and misexpression on Sox2 and Sox3 expression in embryos. (A and B) Altered mRNA expression of neural precursor genes sox2 and sox3 upon the knockdown of d-Asb11 in transversal sections of neural tubes and whole-mount in situ hybridizations (insets) showing at 20 hpf the lower expression of sox3 in the morphants and diffuse as opposed to condensed (in the wild types) expression of sox2 in the morphants. (C) Double whole-mount in situ hybridization for sox2 and anti-MT antibody labeling showing that MT–d-Asb11 is present in the cells ectopically expressing sox2 (four-somite stage embryo; anterior to the left). Bars, 100 μm.
Forced expression of d-Asb11 maintains progenitor expansion and impairs neurogenesis
Confirmation for d-Asb11 function in vivo and the possibility of its involvement in the transition of neural precursor to neuron was obtained in experiments in which we investigated whether the misexpression of d-asb11 mRNA would interfere with primary neurogenesis by evaluating islet1 expression, a marker for primary neurons, as well as the expression of neuronal markers ngn1 and huC (Fig. 5, A and B). Indeed, in 70% of the embryos, islet1 expression was affected (Fig. 5 A). Most prominent was a reduction or total absence of islet1 expression in the lateral sensory Rohon-Beard neurons. Occasionally, defects in the ventromedial motor neurons or the cranial ganglia were observed. Immunolabeling demonstrated that MT–d-Asb11 was present in the domains that did not contain islet1 labeling. Comparable results were obtained with ngn1 and huC (Fig. 5 B).
Figure 5.
Misexpression of d-Asb11 causes altered expression of different markers. (A) Quantitative analysis of defects in islet1 expression upon the misexpression of MT–d-asb11, MT–d-asb11–ΔC, or MT–d-asb11–ΔSOCS. Data are pooled from four independent experiments, and the SEM (error bars) is shown. (right images) Repression of islet1 expression by the misexpression of d-asb11 mRNA. (top left) Wild-type (WT) islet1. (top right) Absence of Rohon-Beard neurons at the injected side. (middle) Absence of the primary motor neurons and displacement of the cranial ganglia. (bottom) Immunolabeling of MT–d-Asb11 in islet1-labeled injected embryos. Immunolabeling of MT–d-asb11–injected embryos with the anti-myc antibody (9E10) demonstrates that one half is positive for MT–d-Asb11 translation and corresponds to defects in islet1 labeling. (far right) Misexpression of d-asb11–ΔC or d-asb11–ΔSOCS mRNA induces significantly fewer defects in islet1 expression. (B) Loss and abnormalities in ngn1- and huC-labeled neurons upon the misexpression of MT–d-asb11 mRNA. (C) Misexpression of MT–d-asb11 mRNA induces ectopic expression of her1 (middle) or a subtle defect in her1 expression (bottom) as compared with wild-type her1 expression (top). (far right) Patterning of the neural plate in wild-type embryos and embryos misexpressing MT–d-asb11 mRNA as revealed by the mid/hindbrain marker her5. Misexpression of d-asb11–ΔC or d-asb11–ΔSOCS induces significantly fewer defects. (D) A combination of mid/hindbrain marker pax2.1 and krox20 marker for rhombomeres 3 and 5 reveals the loss of expression in embryos overexpressing d-asb11 mRNA. (E, left) GCNF is in the wild-type embryos expressed in the entire neural plate (middle), whereas GCNF expression is absent in the areas where MT–d-Asb11 is misexpressed (right). Anterior to the top in A–C and to the left in D and E. Asterisks mark the injected side of the embryo (three- to five-somite stage embryos). mRNA was injected into one of the two blastomeres. Bars, 100 μm.
To verify whether aberrant patterning of the neural plate could underlie abnormalities in primary neurogenesis, we evaluated the expression of regional neural plate markers her5, pax2.1, and krox20 upon the misexpression of d-asb11. Next to the wild-type expression pattern, we observed a reduction of marker gene expression in embryos (Fig. 5, C and D). Most interestingly, we observed embryos in which marker expression was indicative of a split or otherwise deformed neural plate (Fig. 5, C and D), suggesting that in regions misexpressing MT–d-Asb11, neural plate formation was disrupted. To investigate whether the loss of islet1-positive neurons and/or abnormalities in her5, pax2.1, and krox20 expression were secondary to neural plate defects, we studied the effect of d-asb11 misexpression on the expression of germ cell nuclear factor (GCNF) as a neural plate marker (Braat et al., 1999; Barreto et al., 2003). The regional overexpression of MT–d-Asb11 as detected by anti-MT antibody labeling at the three-somite stage overlapped with the region of the neural plate where GCNF was absent (Fig. 5 E). This suggests that d-Asb11 negatively interfered with the establishment of the neural plate and that the absence of islet1-, ngn1-, and huC-labeled neurons and aberrant her5, pax2.1, and krox20 expression are the consequence of this event. To exclude the possibility that the misexpression of d-asb11 mediates nonspecific repression of gene expression, we evaluated several markers upon the misexpression of d-asb11. Interestingly, the expression of the Notch target gene her1 was ectopically expanded in embryos misexpressing d-asb11 (Fig. 5 C), whereas the expression of ptc1 was unaffected (not depicted), ruling out the possibility that d-asb11 is a general transcriptional repressor.
Effects of d-Asb11 misexpression on neuronal development are upstream of the SoxB1 HMG group of transcription factors
The aforementioned experiments suggest that d-Asb11 may, when misexpressed, function to block neuralization through forced maintenance of the precursor cell fate. In support of this notion, the phenotype of d-Asb11 morphants is characterized by altered expression of neural competence factors sox2 and sox3. To establish whether d-Asb11 misexpression may mediate its effects in preventing neuronal differentiation through the induction of sox2 expression, we misexpressed MT–d-asb11 mRNA. As a result, we sporadically observed scarce ectopic sox2-positive (sox2+) cells (unpublished data). In an attempt to increase the inefficiency of MT–d-asb11 mRNA misexpression, we microinjected MT–d-Aasb11 DNA in embryos. Although the frequency of ectopic induction of sox2+ cells (∼5%) remained low, the ectopically induced cells were more numerous. The ectopic expression of sox2 coincided with MT–d-Asb11 protein expression (Fig. 4 C). These data show that when misexpressed, d-Asb11 is capable of inducing the neural precursor gene sox2. sox2 is expressed in neural precursors as well as in neurons. As d-Asb11 misexpression inhibited neurogenesis (Fig. 5), we propose that through its positive regulation of the sox2 gene, d-Asb11 may maintain neural-competent cells in their precursor state. The low frequency of this ectopic induction suggests a requirement for other limiting factors in order for d-Asb11 to induce sox2. Alternatively, potential negative regulators of sox2 would have to be excluded from cells to ectopically express sox2.
The SOCS box is required for defects upon the misexpression of d-Asb11
We obtained functional insight into the molecular mechanism by which d-Asb11 acts from experiments in which we investigated the action of mutated versions of d-asb11 with a specific deletion of the SOCS box (d-asb11–ΔSOCS) or of the entire C-terminal part after the last ankyrin repeat (d-asb11–ΔC) using as a read-out their effects on islet1 neurons or on neural plate patterning as revealed by her5. Importantly, both mutant molecules resulted in a marked decrease of an altered expression pattern of islet1 when injected into zebrafish embryos (Fig. 5 A). As we have already shown, the expression of her5 at early somitogenesis was clearly affected by the misexpression of d-asb11, as in ∼50% of the embryos, one half of the V-shaped expression domain at the mid/hindbrain boundary was shifted laterally or was diminished (Fig. 5 C). Likewise, when d-asb11–ΔC or d-asb11–ΔSOCS mRNA was microinjected, the wild-type expression pattern of her5 was maintained in a high percentage of embryos (Fig. 5 C). Thus, the SOCS box of d-Asb11 is required for its biological effects.
The SOCS box of d-Asb11 is a component of the ubiquitin ligase complex
As mentioned in the second paragraph of Results, SOCS boxes frequently act in ubiquitin ligase complexes, and because the SOCS box in d-Asb11 appears to play an important role during embryogenesis, we determined whether the SOCS box in d-Asb11 is also able to function as an ubiquitin ligase. Indeed, the overexpression of d-Asb11 either in zebrafish embryos or in HeLa cells led to the ubiquitination of a variety of cellular proteins in a SOCS box–dependent fashion (Fig. 6, A and B), demonstrating that d-Asb11 is capable of functionally participating in a ubiquitin ligase complex and again showing that d-Asb11 is a member of the ASB family (Kamura et al., 1998; Chung et al., 2005; Heuze et al., 2005). Importantly, cells transfected with the control expression vector or with SOCS box–deficient d-Asb11 were incapable of ubiquitination (Fig. 6, A and B). Thus, d-Asb11 can act as an ubiquitin ligase in a SOCS box–dependent fashion. Ankyrin repeats are important mediators of protein–protein interaction, and, thus, we reasoned that if d-Asb11 has the capacity to act as an ubiquitin ligase, it is most likely acting on ankyrin repeat–containing proteins. Indeed, we observed that upon coexpression in HeLa cells, d-Asb11 coimmunoprecipitates with various ankyrin repeat–containing proteins, including I-κBα and Notch, but not recombining binding protein-Jκ, which does not contain ankyrin repeats (Fig. 6, C–E).
Figure 6.
d-Asb11 acts as an ubiquitin ligase in vitro. (A) HeLa cells were transfected with either MT alone, MT–d-Asb11, or MT–d-Asb11–ΔC in combination with HA-ubiquitin. Before stimulation, 10 μM of the proteasome inhibitor MG-132 was added, and cells were stimulated for 60 min with 10 ng/ml IL-1β. Equal amounts of protein were immunoprecipitated with anti–IκB-α antibody and subsequently analyzed for the presence of ubiquitinated proteins by immunoblotting with ubiquitin antibody. (B) HeLa cells were transfected with MT-Notch1a intracellular domain and either HA–d-Asb11 or HA–d-Asb11–ΔC in combination with HA-ubiquitin. Before immunoprecipitation, 10 μM of the proteasome inhibitor MG-132 was added, and cells were further processed after 60 min. Equal amounts of protein were immunoprecipitated with anti-MT antibody and subsequently analyzed for the presence of ubiquitinated proteins by immunoblotting with ubiquitin antibody. (C) d-Asb11 can bind to Notch intracellular domain in a SOCS box–independent manner. (D) HEK293 cells were transfected with HA–d-Asb11 and/or IκB-α (middle and bottom). Equal amounts of cell lysates were immunoprecipitated with the anti-HA antibody and analyzed for IκB-α expression. IκB-α was detected in the precipitate of HA-tagged proteins (top). (E) Recombining binding protein-Jκ is not able to bind to d-Asb11.
d-Asb11 overexpression prevents terminal neuronal differention in vitro
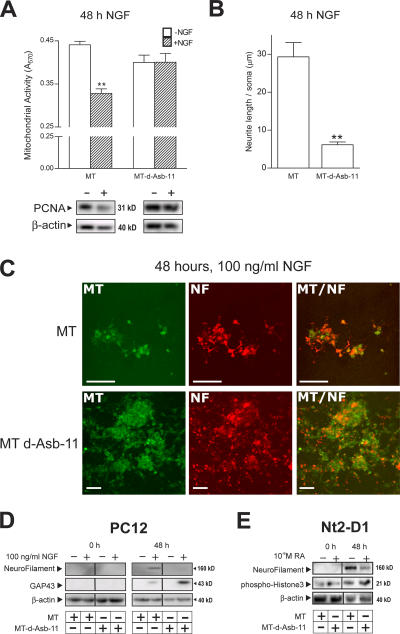
The loss and gain of function experiments in zebrafish embryos suggest that d-Asb11 acts as a molecule that supports the maintenance of neuronal precursors possibly by safeguarding the proper identity and expansion of the precursor stem cell compartment. This notion was further tested using a specific neuronal progenitor cell line (PC12) and a pluripotent embryonic carcinoma cell line (Nt2-D1; Greene and Tischler, 1976; Andrews et al., 1984). Experiments on PC12 cells were performed using an NGF-induced in vitro differentiation of PC12 pheochromocytoma cells according to the standard protocols (Greene and Tischler, 1976; Vaudry et al., 2002). Upon NGF stimulation, the cells transfected with control DNA showed a decrease in proliferation, which was determined by MTT (3-[4,5-dimethyl-2-thiazolyl]-2,5-diphenyl-2H-tetrazolium bromide) activity, and the expression levels of proliferating cell nuclear antigen (PCNA) and neurites were formed (Fig. 7, A and B). Strikingly, when d-Asb11 was overexpressed in PC12 cells, the proliferation was sustained, and the amount of neurites was significantly reduced (Fig. 7, A and B). Importantly, PC12 cells overexpressing d-Asb11 appear to continue proliferating upon NGF-induced differentiation, as they form overlaying colonies in contrast to control cells that remained in a monolayer (Fig. 7 C). The lack of NGF-induced neurite extension in d-Asb11–expressing PC12 cells may be caused by the incapacity of these cells to initiate neuronal differentiation but, alternatively, may reflect a block further down in the execution of the neuronal differentiation program. To distinguish between these possibilities, we investigated the expression of growth cone–associated protein 43 (GAP-43), a marker already expressed in proliferating neuroblasts, and the neurofilament, a marker of terminally differentiated neurons (Kanazir et al., 1996; Mani et al., 2001; Shen et al., 2002). As expected from the neurite extension experiments, transfection of MT–d-asb11 into PC12 cells inhibited neurofilament expression as assayed either by immunoblotting (Fig. 7 D) or immunofluorescence (Fig. 7 C). Importantly, however, d-Asb11 enhanced the NGF-dependent expression of GAP-43 (Fig. 7 D).
Figure 7.
Effect of d-Asb11 on differentiation in PC12 or Nt2-D1 cells. (A) Proliferation of PC12 cells upon NGF stimulation. Relative mitochondrial activity was measured (MTT assay), and PCNA expression levels were determined. The experiments were performed in duplicate, and the values shown are the means and SEM (error bars) of 11 different samples. (B) Neurite length was measured in at least 10 different fields of cells for each condition. **, P < 0.01. (C) Fluorescent double labeling of PC12 cells transfected with MT or MT–d-Asb11. d-Asb11 inhibited neurofilament (NF) expression and allowed continued cell proliferation as compared with MT-transfected cells. (D) The protein expression profile of GAP-43 and neurofilament of whole cell lysates in PC12 cells after NGF stimulation. (E) Expression of neurofilament, PH3 (Ser10), and actin in Nt2-D1 cells after RA addition. Bars, 200 μm.
To examine the effects that d-Asb11 may have upon the differentiation of uncommitted pluripotent embryonic carcinoma cells (Andrews et al., 1984, 1994), we chose Nt2-D1 cells that differentiate primarily into neurons upon RA treatment (Andrews, 1984). When d-Asb11 was overexpressed in the Nt2-D1 cell line, it inhibited the RA-induced differentiation into neurons, as shown by diminished neurofilament expression (Fig. 7 E). Importantly, as in PC12 cells, these cells were still mitotically active, as shown by increased PH3 expression, suggesting that these pluripotent cells are also blocked in a phase before terminal neuronal differentiation. Thus, the overexpression of d-Asb11 in both PC12 and Nt2-D1 cells enhances proliferation of the committed neural precursor in spite of differentiation stimuli. These in vitro data are therefore consistent with the inhibition of neurogenesis upon the misexpression of d-Asb11 in the embryo.
Discussion
We have shown that in the developing zebrafish embryo, d-asb11 is expressed in the neural plate margins and is complementary to and abutting the proneuronal zone, as defined by deltaA staining. Before early somitogenesis, d-asb11 is ubiquitously expressed in the pluripotent cells of the blastoderm (Fig. 1 B). Progressive restriction of d-asb11 to the lateral regions of the neural plate as well as its transient expression at that location suggests that d-Asb11 may indeed support the stem cell fate.
Upon the knockdown or misexpression of d-asb11, the size of the terminally differentiated neuronal compartment increases or decreases, respectively, suggesting that d-Asb11 is involved in regulation of the number of neurons that eventually arise. The relatively subtle minor initial increase in neurogenesis upon d-Asb11 knockdown, as not all dorsal or lateral precursors are induced to differentiate, may implicate redundancy with other ASB family members from this group (Fig. S1). Alternatively, the strictly localized and transient expression of d-asb11 could affect only a small subset of neural precursors, thereby resulting in this subtle phenotype.
We propose that during zebrafish development, d-Asb11 maintains the undifferentiated neural-competent state of a subpopulation of cells in the neural plate until its expression is down-regulated by subsequent differentiation signals (e.g., RA; Sockanathan and Jessell, 1998). Because d-asb11 is only transiently expressed from 4 hpf until the 12-somite stage, upon its disappearance, cells devoid of d-Asb11 would become sensitive to secondary inductive signals and would be able to pursue the appropriate neuronal cell fate. Interestingly, the timing of extinguishing d-asb11 mRNA expression at ∼12 somites roughly corresponds with the final round of DNA synthesis occurring between 9 and 16 hpf in motor neurons (Myers et al., 1986). As first secondary neurons are born ∼6 h after the first primary neurons, it may be that d-Asb11 functions to separate waves of primary and secondary neurogenesis by maintaining a pool of precursors to respond to inductive signals operating later in development.
The loss of function of d-Asb11 causes premature neuronal commitment that is reflected in ectopic ngn1 expression and followed by a relative increase of HuC+ terminally differentiated neurons. In parallel, a progressive loss of ventricular zone cells occurs, resulting in reduction of the definitive neuronal compartment. Importantly, d-Asb11 may mediate its effects through control of SoxB1 neural precursor genes, as the knockdown results in a reduced and aberrant expression of sox3 and sox2, respectively. Concurrently, when misexpressed, d-Asb11 prevents neuronal differentiation possibly through the induction of sox2 or yet another SoxB1-type factor. Both gain of function and loss of function of d-Asb11 phenotypes in zebrafish are in agreement with experiments in the chick in which the overexpression of Sox2 and/or Sox3 also inhibits neuronal differentiation of neural progenitors, which then maintain their undifferentiated state and capacity to proliferate. In contrast, dominant-negative Sox2 and/or Sox3 in neural progenitors results in premature exit from the cell cycle, and neuronal differentiation ensues (Bylund et al., 2003; Graham et al., 2003). Our data are consistent with a model in which d-Asb11 supports the maintenance of a proliferative subpopulation of neural precursors. We further propose that d-Asb11 may control this process through SoxB1 neural precursor genes.
Consistent with such a model, it has been proposed that SoxB1 genes play a role in maintaining neural progenitors in the cell cycle and regulate the timing of their exit from the cell cycle. The findings that the inhibition of SOX2 function in chick neural progenitors (Graham et al., 2003) and rat oligodendrocyte precursor cells (Kondo and Raff, 2004) leads to their premature exit from mitosis support this hypothesis and are in agreement with loss of function d-Asb11, as d-Asb11 knockdown similarly leads to the reduction of mitotic cell numbers after 24 hpf.
The role of d-Asb11 in maintaining the proliferative state of precursors was also illustrated in in vitro differentiation models. When d-Asb11 is overexpressed in a neural-committed progenitor or a pluripotent cell line, these cells are unable to execute the terminal neuronal differentiation program, as shown by the inhibition of neurofilament expression upon exposure to stimuli that cause neuronal differentiation. Consistently, the proliferation was not inhibited in these cells as it was in the controls, indicating that d-Asb11 is able to block terminal neuronal differentiation and allow cells to proliferate.
Because PC12 is already committed to neural crest fate, the overexpression of d-Asb11 cannot prevent the PC12 cell in activating its neuronal differentiation program (enhanced GAP-43 expression in d-Asb11–transfected cells), but it can hold these cells in a state of proliferating neuroblast precursors. Consistent with the proposed role of d-Asb11 in the maintenance of neural precursors, GAP-43 is reported to be expressed in proliferating neuroblasts, and its function has been implicated in the cell cycle, differentiation, and, eventually, also cell survival (Kanazir et al., 1996; Mani et al., 2001; Shen et al., 2002). Importantly, PC12 cells overexpressing d-Asb11 appear to have a block in further execution of the neural differentiation program (as they have diminished neurites and lower neurofilament expression). Our data suggest that d-Asb11 acts to maintain cells in the neuroblast state independently of their primary status, being either pluripotent (Nt2-D1) or neural progenitor (PC12) cells.
Consistently, the misexpression of XSox3 with XBF1 promotes the proliferation of neuroectodermal cells in Xenopus laevis embryos (Hardcastle and Papalopulu, 2000). We speculate that the misexpression of d-Asb11 in zebrafish embryos perhaps through the induction of sox2 or another SoxB1-type factor may similarly affect the fate of zebrafish neuroectodermal cells, thereby abolishing neuronal differentiation. The in vitro data on Nt2-D1 and PC12 cells, a pluripotent and a neuronal-committed cell line, respectively, also suggest that d-Asb11 functions by maintaining the proliferative state of the neural precursors, thereby making them refractory to differentiation signals perhaps through regulating SoxB genes.
Our findings show that d-Asb11 functions as a regulator of progenitor cell maintenance. More specifically, it keeps progenitors in a less differentiated, more proliferative state. Although little is known about the functions of ASB proteins, the fact that ASB5 (which is highly homologous to d-Asb11) has a role in arteriogenesis strengthens the notion that this class of ASB proteins may have a role in cell fate decisions (Boengler et al., 2003).
Interestingly, a possible role for d-Asb11 in the development of the brain has recently indirectly been uncovered by the experiments performed by Nielsen et al. (2005). They compared the genetic mutations between the human and chimpanzee genome, looking for genes with an unusually higher number of functional nucleotide changes between the two species, which would suggest a possible important role for such genes in the definition of being a chimpanzee or a human. Most of these genes were involved in the immune and reproductive systems, but ASB11 was among the highly altered genes between the human and chimpanzee genome despite its generally very high level of conservation in the chordate phylum. It is of course tempting to suggest that the obvious size differences between human and chimpanzee brains are also a result of altered ASB11 function.
Materials and methods
Fish and embryos
Zebrafish were kept at 27.5°C. Embryos were obtained by natural matings, cultured in embryo medium, and staged according to Kimmel et al. (1995; Westerfield, 1994).
Cell cultures
HEK293, HeLa, Nt2-D1, and PC12 cells were maintained in DME containing 10% FCS. The culture medium was supplemented with 5 mM glutamine and antibiotics/antimitotics. Cells were incubated in a 5% CO2 humidified incubator at 37°C.
Plasmid construction
pCS2+MT (MT; myc-tagged expression vector), pCS2+MT–d-Asb11 (MT–d-Asb11), pCS2+MT–d-Asb11–ΔC (MT–d-Asb11–ΔC), and pMT2SM-HA–d-Asb11 (HA-tagged expression vector; HA–d-Asb11) were constructed as follows: an expression vector containing d-asb11, the coding region, was cloned into the BamHI–XhoI sites of the pCS2+ expression vector. To obtain a myc-tagged version of d-asb11 (MT–d-asb11), the coding region was cloned into the NcoI–XhoI sites of pCS2+MT. An MT–d-asb11–ΔC construct encoding a C-terminal deletion (aa 229–293) was generated by digesting MT–d-asb11 with XbaI. All expression constructs were checked for errors by sequencing. For HA-tagged d-asb11 (HA–d-asb11), d-asb11–ΔC (HA–d-asb11–ΔC), and d-asb11–ΔSOCS (HA–d-asb11–ΔSOCS), the coding region was cloned into the XhoI–EcoRI sites of pMT2SM-HA, and the expression constructs were subsequently verified by sequencing. To obtain a construct for RNA synthesis of asb-a to control mRNA injections of MT–d-asb11 mRNA, the 1.6-kb asb-a cDNA was cloned into the BglII and XhoI sites of pT7TS+. A partial cDNA fragment of asb-a in pBluescript was used as a template to generate a riboprobe for in situ hybridizations.
mRNA synthesis, mRNA and DNA microinjections, and rescue of d-Asb11 knockdown with the misexpression of MT–d-asb11 mRNA
Capped mRNAs were synthesized using the mMESSAGE mMACHINE kit (Ambion). In rescue experiments of d-Asb11, knockdown MT–d-asb11 mRNA at concentrations ranging from 2.5 to 50 pg was injected in 1 nl at the yolk/blastoderm interface in the one-cell stage upon microinjection with d-asb11 MO in one-cell stage embryos such that MO-injected only embryos were the control for embryos first injected with the MO and then with d-asb11 mRNA. A chi-squared test was performed to determine the statistical differences in percent morphology in MO and potentially rescued MO-injected embryos The rescue could be performed because the MO-1 sequence was complementary to the 5′ untranslated sequences not incorporating the ATG translation start site. In the MT–d-asb11 construct, myc was fused directly to the ATG, thus lacking the MO-1 target sequences.
To misexpress wild-type or mutant versions of d-asb11 mRNA, zebrafish embryos were injected at the two-cell stage in one of the two blastomeres (300 pg mRNA), resulting in defects in the injected half of the embryo. To misexpress MT–d-asb11 DNA, 10 pg was microinjected into zygotes.
In situ hybridization
Whole-mount in situ hybridizations were performed according to Joore et al. (1994) and for sox2 were performed according to Cunliffe and Casaccia-Bonnefil (2006). Double label whole-mount in situ hybridization was performed according to Hauptmann and Gerster (2000). For histological analysis, upon in situ hybridization, embryos were overstained for 48 h, subsequently fixed for 1 h in 4% PFA in phosphate buffer, dehydrated, embedded in plastic, and sectioned. deltaA and huC probes were provided by B. Appel (Vanderbilt University, Nashville, TN). Probes for her1 and her5 were provided by L. Bally-Cuif (Institute of Developmental Genetics, Neuherberg, Germany). U. Strahle (Institute of Toxicology and Genetics, Forschungszentrum Karlsruhe, Karlsruhe, Germany) provided the probes for ngn1 and islet1. V. Cunliffe (Centre for Developmental Genetics, University of Sheffield, Sheffield, United Kingdom) and S. Wilson (University College London, London, United Kingdom) provided the sox2 and sox3 probes, respectively.
Acridine orange staining
Acridine orange staining was performed as described previously (Furutani-Seiki et al., 1996) with modification of the acridine orange (Sigma-Aldrich) solution (0.078 μg/ml in embryo medium).
TUNEL assay
TUNEL assay was performed as described previously by Cole and Ross (2001) with modification of the staining reaction. Antidigoxigenin antibody was preabsorbed in blocking buffer (1% DMSO and 2% BSA in PBS) for 1 h at 4°C. Embryos were incubated overnight in blocking buffer with 1:2,000 antidigoxigenin antibody at 4°C, washed eight times with PBS, and stained according to the method used for in situ hybridization.
cDNA library screen
A random primed λZAP neurula cDNA library prepared from 3 to 15 hpf embryos was used to screen 1.5 × 106 phages to obtain a full-length clone. The probe was labeled to high specific activity with α-[32P]dCTP (GE Healthcare) using a Rediprime labeling kit (GE Healthcare). After in vivo excision, the longest positive clone was subcloned and sequenced.
MOs
Antisense MOs (Gene Tools, LLC) were designed to complement the 5′ untranslated sequences of d-asb11 (d-asb11–MO-1; 5′-AGAAACCTCGCAGACAGCAACGGTC-3′) or sequences upstream and including the ATG start site (d-asb11–MO-2; 5′-CCATCTCTAAACTAAAACACAGCCA-3′), and, as a control, an inverted MO (d-asb11–MO; 5′-CTGGCAACGACAGACGCTCCAAGA-3′) was used. Approximately 9 ng/1 nl was injected into one-cell stage embryos, whereas as a control, we injected ∼15 ng/1 nl of inverted MO.
Whole-mount immunolabeling
For whole-mount immunohistochemistry, fixed embryos were rinsed in PBS and incubated in blocking solution for 1 h (PBS + 0.1% Tween 20 containing 1% BSA and 2% normal lamb serum). The primary antibody (anti-HuC; Sigma-Aldrich) was added, and embryos were incubated overnight at 4°C. After washing in PBS + 0.1% Tween 20, embryos were incubated overnight at 4°C in goat anti–mouse peroxidase-conjugated secondary antibody. After extensive washing, embryos were incubated in 0.5 mg/ml DAB and reacted in 0.003% H2O2. For the double fluorescent whole-mount antibody staining with anti-HuC and anti-PH3 antibody (Upstate Biotechnology), fixed embryos were permeabilized for 4 min in 2.5 mg/ml ice-cold trypsin in PBT (PBS + 0.1% BSA + 0.1% Tween 20; Worthington Biochemical Corp.) on ice, subsequently washed three times for 5 min each in PBT, and incubated for at least 1 h in blocking buffer (2% lamb serum, 0.1% BSA, 0.1% DMSO, and 1% Triton X-100 in PBS). To detect HuC and PH3, respectively, secondary antibodies were Cy3 and Cy5 conjugated. Immunofluorescently labeled embryos were analyzed in 100% glycerol using confocal laser scanning microscopy at ambient temperature.
Analysis of confocal images
Potential cellular colocalization of HuC and PH3 signal in double immunofluorescence experiments was analyzed using Volocity 3 software (Improvision). To this end, all 50 optical z slices per embryo were scored (in total seven wild type and nine morphants at 24 hpf and seven wild types and eight morphants at 48 hpf).
To determine numbers of PH3-positive cells, they were scored manually on the cumulative z series for each embryo (for n see Results). The cells were counted at 30,000× magnification in a standardized square of 7-cm length and depth covering the extent of the neural tube defined by HuC antibody labeling. Comparable regions were chosen in morphants and wild types with reference to the yolk-tube extension.
3D reconstruction and calculation
Volumes were calculated from a 3D reconstruction from 7-μm serial sections with an acquisition station (Verbeek and Boon, 2002). This resulted in 123 section images for the control and 149 section images for the treated fish. The notochord and otic vesicle were included to have a frame of reference.
Microscopy and image quantification
Pictures were obtained using a microscope (Axioplan; Carl Zeiss MicroImaging, Inc.) with a 10× NA 0.30 plan Neofluar or a 40× NA 0.65 Achroplan lens on a Leica microscope (TCS NT). Images were digitized with a camera (DFC480; Leica) and processed with the IM500 Image Manager (Leica). A stereomicroscope (MZ FLIII; Leica) with a camera (DC 300F; Leica) was used for Figs. 2 A and 7 B. Digital pictures of transversal sections of ngn1 in situ hybridizations were then quantified with Image Analysis software (EFM Software) by calculating the total signal intensity. The pictures of sections were corrected from embryo size by dividing the amount of staining by the size of the embryo surface.
Digital pictures of whole-mount in situ hybridizations were obtained using a digital camera (DXM1200; Nikon). Some in situ images were used to determine the size of the stained compartments by counting the amount of positive pixels on the embryo and dividing this by the total size of the whole embryo. This number represents the relative size of positive cells in the whole embryo. All images were only adjusted for brightness, contrast, or color balance (autoequalize option).
MTT assay
PC12 cells were transfected with MT alone or MT–d-Asb11 plus neomycin resistance plasmid (see supplemental material for plasmid construction). After 5 d, polyclonal neomycin-resistant PC12 cells were reseeded in 96-well plates and stimulated with 100 ng/ml NGF-2.5S (Invitrogen). After 3 or 6 d, 0.1 mg MTT (M5655; Sigma-Aldrich) was added to the medium. The cells were lysed by the addition of 50 μl MTT lysis buffer (20% SDS and 10% dimethylformamide) to the medium, and absorbance was measured at 570 nm. The results are the mean and standard error of 11 independent cultures obtained in two different experiments.
Measurement of the neurite length
The mean neurite length in Fig. 7 B was measured by blinded counting of the total amount of neurites divided by the amount of counted cells using the Image Analysis software package (EFM Software).
Western blotting and immunofluorescence
PC12 cells were transfected with MT alone or MT–d-Asb11 plus a neomycin resistance plasmid. After 5 d, the polyclonal neomycin-resistant PC12 cells were reseeded in six-well plates for neurofilament and GAP-43 expression and on glass coverslips for immunofluorescence. Cells were subsequently stimulated with 100 ng/ml NGF-2.5S (Invitrogen). After stimulation, the cells were lysed in cell lysis buffer (Cell Signaling) supplemented with phosphatase and protease inhibitors or fixed in 4% PFA for 15 min and kept in PBS at 4°C.
Nt2-D1 cells were seeded in a 24-well plate 1 d before transfection using the MATra and IBAFect (IBA GmbH) transfection technique according to the standard supplier's protocol. After 24 h, the medium was refreshed and 10−6 M RA was added, and, after 48 h, the cells were lysed (Cell Signaling) and supplemented with phosphatase and protease inhibitors.
After cell lysis, the samples were loaded on SDS-polyacrylamide gels. The material was transferred to Immobilon (Millipore) by wet blotting in transfer buffer (50 mM Tris, pH 8.0, 40 mM glycine, 0.0375% SDS, and 20% methanol), and the blots were incubated for 1 h in blocking buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Tween 20, and 5% nonfat milk). Blots were incubated overnight at 4°C in TBS-T (50 mM Tris, pH 8.0, 150 mM NaCl, and 0.05% Tween 20) containing 10% blocking buffer and 1:1,000 PCNA (Sigma-Aldrich), GAP-43 (Sigma-Aldrich), neurofilament (for PC12; 2H3; Developmental Studies Hybridoma Bank), neurofilament-M (for Nt2-D1; Cell Signaling), or β-actin (SC-1615; Santa Cruz Biotechnology, Inc.). The neurofilament antibody 2H3 was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences at the University of Iowa. The blots were washed in TBS-T. After incubation with the appropriate secondary antibody and extensive washing in TBS-T, the blots were developed by ECL.
After fixation, the cells were treated with PBS + 0.1% Triton X-100 for 5 min and subsequently blocked with PBS containing 5% normal goat serum. After blocking, the cells were incubated with 1:100 MT antibody (Cell Signaling) in PBS + 3% BSA and afterward incubated with 1:5,000 anti–rabbit Cy3. After the first staining, the cells were stained for neurofilament (2H3; Developmental Studies Hybridoma Bank) identical to the MT staining but with anti–mouse Cy5 as a secondary antibody. The cells were washed three times in PBS between every step. After staining, the glass coverslips were mounted in moviol. Immunofluorescently labeled cells were analyzed using confocal laser scanning microscopy at ambient temperature.
Rabbit immunization and serum preparation
GST-tagged d-Asb11 was purified from Escherichia coli BL21 bacteria using glutathione–Sepharose beads. Rabbits were immunized with GST-tagged d-Asb11. Subsequently, the serum containing the polyclonal antibody directed against d-Asb11 was used in immunoblotting analysis.
Zebrafish cell lysates for immunoblotting
Embryos were lysed in cell lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, and 1% Triton X-100) at 4°C (7.5–12.5 μl/embryo). Loading buffer was added to the cell lysis buffer, and samples were boiled for 3 min. A total equivalent of five embryos was loaded per lane.
Online supplemental material
Supplemental material provides information about the identification of d-Asb11 and testing of d-asb11 morphants. Fig. S1 presents the characterization of d-Asb11 and the Asb subfamily of proteins. Fig. S2 shows the generation of a d-Asb11 antibody and validation of d-Asb11 expression during embryogenesis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200601081/DC1.
Supplementary Material
Acknowledgments
We thank Drs. Bruce Appel, Laure Bally-Cuif, Vincent Cunliffe, Uwe Strahle, and Steve Wilson for providing the in situ probes, Drs. Ajay Chitnis and Laure Bally-Cuif for stimulating discussions, and Dr. Vincent Cunliffe for help with specific experimental protocols. We thank J. Korving for histology and Dr. L. Tertoolen for initiation in confocal microscopy. We are grateful to A. Ensing, J. Kloots, M. Melis, and M. Phernambucq for technical assistance. We thank the animal care unit at the Hubrecht Laboratory for services rendered.
We acknowledge financial support from the NWO Genomics (to D. Zivkovic) and NWO Casimir (to S.H. Diks) programs.
S.H. Diks and R.J. Bink contributed equally to this paper.
Abbreviations used in this paper: ASB, ankyrin repeat and SOCS box–containing protein; CNS, central nervous system; GAP-43, growth cone–associated protein 43; GCNF, germ cell nuclear factor; HMG, high mobility group; hpf, hours postfertilization; MO, morpholino; MT, myc tag; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide; ngn1, neurogenin1; PCNA, proliferating cell nuclear antigen; PH3, phosphohistone-3; RA, retinoic acid; SOCS, suppressor of cytokine signaling.
References
- Andrews, P.W. 1984. Retinoic acid induces neuronal differentiation of a cloned human embryonal carcinoma cell line in vitro. Dev. Biol. 103:285–293. [DOI] [PubMed] [Google Scholar]
- Andrews, P.W., I. Damjanov, D. Simon, G.S. Banting, C. Carlin, N.C. Dracopoli, and J. Fogh. 1984. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab. Invest. 50:147–162. [PubMed] [Google Scholar]
- Andrews, P.W., I. Damjanov, J. Berends, S. Kumpf, V. Zappavigna, F. Mavilio, and K. Sampath. 1994. Inhibition of proliferation and induction of differentiation of pluripotent human embryonal carcinoma cells by osteogenic protein-1 (or bone morphogenetic protein-7). Lab. Invest. 71:243–251. [PubMed] [Google Scholar]
- Bally-Cuif, L., and M. Hammerschmidt. 2003. Induction and patterning of neuronal development, and its connection to cell cycle control. Curr. Opin. Neurobiol. 13:16–25. [DOI] [PubMed] [Google Scholar]
- Barreto, G., U. Borgmeyer, and C. Dreyer. 2003. The germ cell nuclear factor is required for retinoic acid signaling during Xenopus development. Mech. Dev. 120:415–428. [DOI] [PubMed] [Google Scholar]
- Blader, P., N. Fischer, G. Gradwohl, F. Guillemot, and U. Strahle. 1997. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development. 124:4557–4569. [DOI] [PubMed] [Google Scholar]
- Boengler, K., F. Pipp, B. Fernandez, A. Richter, W. Schaper, and E. Deindl. 2003. The ankyrin repeat containing SOCS box protein 5: a novel protein associated with arteriogenesis. Biochem. Biophys. Res. Commun. 302:17–22. [DOI] [PubMed] [Google Scholar]
- Braat, A.K., M.A. Zandbergen, E. De Vries, B. Van der Burg, J. Bogerd, and H.J.T. Goos. 1999. Cloning and expression of the zebrafish germ cell nuclear factor. Mol. Reprod. Dev. 53:369–375. [DOI] [PubMed] [Google Scholar]
- Bylund, M., E. Andersson, B.G. Novitch, and J. Muhr. 2003. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 6:1162–1168. [DOI] [PubMed] [Google Scholar]
- Chung, A.S., Y.J. Guan, Z.L. Yuan, J.E. Albina, and Y.E. Chin. 2005. Ankyrin repeat and SOCS box 3 (ASB3) mediates ubiquitination and degradation of tumor necrosis factor receptor II. Mol. Cell. Biol. 25:4716–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, L.K., and L.S. Ross. 2001. Apoptosis in the developing zebrafish embryo. Dev. Biol. 240:123–142. [DOI] [PubMed] [Google Scholar]
- Cunliffe, V.T., and P. Casaccia-Bonnefil. 2006. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech. Dev. 123:24–30. [DOI] [PubMed] [Google Scholar]
- Furutani-Seiki, M., Y.J. Jiang, M. Brand, C.P. Heisenberg, C. Houart, D. Beuchle, F.J. van Eeden, M. Granato, P. Haffter, M. Hammerschmidt, et al. 1996. Neural degeneration mutants in the zebrafish, Danio rerio. Development. 123:229–239. [DOI] [PubMed] [Google Scholar]
- Graham, V., J. Khudyakov, P. Ellis, and L. Pevny. 2003. SOX2 functions to maintain neural progenitor identity. Neuron. 39:749–765. [DOI] [PubMed] [Google Scholar]
- Greene, L.A., and A.S. Tischler. 1976. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth-factor. Proc. Natl. Acad. Sci. USA. 73:2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J.H., H. Saiyin, Y.H. Wei, S. Chen, L. Chen, G. Bi, L.J. Ma, G.J. Zhou, C.Q. Huang, L. Yu, and L. Dai. 2004. Expression of testis specific ankyrin repeat and SOCS box-containing 17 gene. Arch. Androl. 50:155–161. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt, M., C. Kramer, M. Nowak, W. Herzog, and J. Wittbrodt. 2003. Loss of maternal smad5 in zebrafish embryos affects patterning and morphogenesis of optic primordia. Dev. Dyn. 227:128–133. [DOI] [PubMed] [Google Scholar]
- Hardcastle, Z., and N. Papalopulu. 2000. Distinct effects of XBF-1 in regulating the cell cycle inhibitor p27(XIC1) and imparting a neural fate. Development. 127:1303–1314. [DOI] [PubMed] [Google Scholar]
- Harland, R. 2000. Neural induction. Curr. Opin. Genet. Dev. 10:357–362. [DOI] [PubMed] [Google Scholar]
- Hauptmann, G., and T. Gerster. 2000. Multicolor whole-mount in situ hybridization. Methods Mol. Biol. 137:139–148. [DOI] [PubMed] [Google Scholar]
- Heuze, M.L., F.C. Guibal, C.A. Banks, J.W. Conaway, R.C. Conaway, Y.E. Cayre, A. Benecke, and P.G. Lutz. 2005. ASB2 is an elongin BC-interacting protein that can assemble with cullin 5 and Rbx1 to reconstitute an E3 ubiquitin ligase complex. J. Biol. Chem. 280:5468–5474. [DOI] [PubMed] [Google Scholar]
- Joore, J., G.B. van der Lans, P.H. Lanser, J.M.A. Vervaart, D. Zivkovic, J.E. Speksnijder, and W. Kruijer. 1994. Effects of retinoic acid on the expression of retinoic acid receptors during zebrafish embryogenesis. Mech. Dev. 46:137–150. [DOI] [PubMed] [Google Scholar]
- Kamura, T., S. Sato, D. Haque, L. Liu, W.G. Kaelin, R.C. Conaway, and J.W. Conaway. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazir, S., S. Ruzdijic, S. Vukosavic, S. Ivkovic, A. Milosevic, N. Zecevic, and L. Rakic. 1996. GAP-43 mRNA expression in early development of human nervous system. Brain Res. Mol. Brain Res. 38:145–155. [DOI] [PubMed] [Google Scholar]
- Kile, B.T., E.M. Viney, T.A. Willson, T.C. Brodnicki, M.R. Cancilla, A.S. Herlihy, B.A. Croker, M. Baca, N.A. Nicola, D.J. Hilton, and W.S. Alexander. 2000. Cloning and characterization of the genes encoding the ankyrin repeat and SOCS box-containing proteins Asb-1, Asb-2, Asb-3 and Asb-4. Gene. 258:31–41. [DOI] [PubMed] [Google Scholar]
- Kile, B.T., B.A. Schulman, W.S. Alexander, N.A. Nicola, H.M.E. Martin, and D.J. Hilton. 2002. The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 27:235–241. [DOI] [PubMed] [Google Scholar]
- Kimmel, C.B., W.W. Ballard, S.R. Kimmel, B. Ullmann, and T.F. Schilling. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310. [DOI] [PubMed] [Google Scholar]
- Kondo, T., and M. Raff. 2004. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 18:2963–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., J. Li, F. Zhang, W. Qin, G. Yao, X. He, P. Xue, C. Ge, D. Wan, and J. Gu. 2003. Molecular cloning and characterization of the human ASB-8 gene encoding a novel member of ankyrin repeat and SOCS box containing protein family. Biochem. Biophys. Res. Commun. 300:972–979. [DOI] [PubMed] [Google Scholar]
- Mani, S., Y. Shen, J. Schaefer, and K.F. Meiri. 2001. Failure to express GAP-43 during neurogenesis affects cell cycle regulation and differentiation of neural precursors and stimulates apoptosis of neurons. Mol. Cell Neurosci. 17:54–66. [DOI] [PubMed] [Google Scholar]
- Morrison, S.J. 2001. Neuronal potential and lineage determination by neural stem cells. Curr. Opin. Cell Biol. 13:666–672. [DOI] [PubMed] [Google Scholar]
- Myers, P.Z., J.S. Eisen, and M. Westerfield. 1986. Development and axonal outgrowth of identified motoneurons in the zebrafish. J. Neurosci. 6:2278–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius, A., and S.C. Ekker. 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26:216–220. [DOI] [PubMed] [Google Scholar]
- Nielsen, R., C. Bustamante, A.G. Clark, S. Glanowski, T.B. Sackton, M.J. Hubisz, A. Fledel-Alon, D.M. Tanenbaum, D. Civello, T.J. White, et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed]
- Okuda, Y., H. Yoda, M. Uchikawa, M. Furutani-Seiki, H. Takeda, H. Kondoh, and Y. Kamachi. 2006. Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 235:811–825. [DOI] [PubMed] [Google Scholar]
- Pevny, L., and M. Placzek. 2005. SOX genes and neural progenitor identity. Curr. Opin. Neurobiol. 15:7–13. [DOI] [PubMed] [Google Scholar]
- Shen, Y., S. Mani, S.L. Donovan, J.E. Schwob, and K.F. Meiri. 2002. Growth-associated protein-43 is required for commissural axon guidance in the developing vertebrate nervous system. J. Neurosci. 22:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockanathan, S., and T.M. Jessell. 1998. Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell. 94:503–514. [DOI] [PubMed] [Google Scholar]
- Temple, S. 2001. The development of neural stem cells. Nature. 414:112–117. [DOI] [PubMed] [Google Scholar]
- Vaudry, D., P.J.S. Stork, P. Lazarovici, and L.E. Eiden. 2002. Signaling pathways for PC12 cell differentiation: making the right connections. Science. 296:1648–1649. [DOI] [PubMed] [Google Scholar]
- Verbeek, F.J., and P.J. Boon. 2002. High-resolution 3D reconstruction from serial sections: microscope instrumentation, software design, and its implementations. In Three-Dimensional and Multidimensional Microscopy: Image Acquisition and Processing IX. J.-A. Conchello, C.J. Cogswell, and T. Wilson, editors. SPIE–The International Society for Optical Engineering, Bellingham, WA. 65–76.
- Wang, H., K.H. Ng, H. Qian, D.P. Siderovski, W. Chia, and F. Yu. 2005. Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat. Cell Biol. 7:1091–1098. [DOI] [PubMed] [Google Scholar]
- Wegner, M., and C.C. Stolt. 2005. From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 28:583–588. [DOI] [PubMed] [Google Scholar]
- Westerfield, M. 1994. The Zebrafish Book: a Guide for the Laboratory Use of Zebrafish (Brachydanio rerio). University of Oregon Press, Eugene, OR.
- Westin, J., and M. Lardelli. 1997. Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Dev. Genes Evol. 207:51–63. [DOI] [PubMed] [Google Scholar]
- Wilcox, A., K.D. Katsanakis, F. Bheda, and T.S. Pillay. 2004. Asb6, an adipocyte-specific ankyrin and SOCS box protein, interacts with APS to enable recruitment of elongins B and C to the insulin receptor signaling complex. J. Biol. Chem. 279:38881–38888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.