Abstract
Postnatal growth and regeneration of skeletal muscle requires a population of resident myogenic precursors named satellite cells. The transcription factor Pax7 is critical for satellite cell biogenesis and survival and has been also implicated in satellite cell self-renewal; however, the underlying molecular mechanisms remain unclear. Previously, we showed that Pax7 overexpression in adult primary myoblasts down-regulates MyoD and prevents myogenin induction, inhibiting myogenesis. We show that Pax7 prevents muscle differentiation independently of its transcriptional activity, affecting MyoD function. Conversely, myogenin directly affects Pax7 expression and may be critical for Pax7 down-regulation in differentiating cells. Our results provide evidence for a cross-inhibitory interaction between Pax7 and members of the muscle regulatory factor family. This could represent an additional mechanism for the control of satellite cell fate decisions resulting in proliferation, differentiation, and self-renewal, necessary for skeletal muscle maintenance and repair.
Introduction
The Pax gene family defines an evolutionary conserved group of transcription factors that play critical roles during organogenesis and tissue homeostasis (Chi and Epstein, 2002; Robson et al., 2006). Nine Pax proteins have been described in mammals, where the presence of the paired box DNA binding domain is a common feature. The family is further subgrouped by the presence of an octapeptide motif and the presence, absence, or truncation of a homeodomain region.
Pax3 and Pax7 are two closely related family members (Bober et al., 1994; Goulding et al., 1994; Tajbakhsh et al., 1997; Chi and Epstein, 2002; Robson et al., 2006) that are involved in the specification and maintenance of skeletal muscle progenitors. Genetic analyses in mice showed that Pax3 is critical for delamination and migration of muscle precursors from the somites to the limbs (Bober et al., 1994; Goulding et al., 1994; Tajbakhsh et al., 1997). Pax7 −/− mice have no gross defects in muscle formation. However, in the absence of Pax7, adult skeletal muscles are completely devoid of satellite cells (Seale et al., 2000; Oustanina et al., 2004), which are thought to represent the stem cell compartment responsible for postnatal muscle growth and regeneration. Accordingly, Pax7-null mice exhibit reduced muscle growth, marked muscle wasting, and an extreme deficit in muscle regeneration after acute injury (Seale et al., 2000; Kuang et al., 2006). Despite these differences, both Pax3 and Pax7 appear to mark a population of muscle progenitors (Pax3+/Pax7+ cells) in the dermomyotome of embryonic somites (Ben-Yair and Kalcheim, 2005; Gros et al., 2005; Kassar-Duchossoy et al., 2005; Relaix et al., 2005). Pax3+/Pax7+ cells proliferate and persist throughout embryonic and fetal development and are proposed to be the cellular origin for satellite cells. Pax3 expression is down-regulated in satellite cells before birth and appears to be confined to a subpopulation of satellite cells in specific muscle groups (Kassar-Duchossoy et al., 2005; Relaix et al., 2006). Thus, cumulative evidence supports distinct roles for Pax3 and Pax7 during myogenesis and a critical requirement for Pax7 in satellite cell specification, survival, and potentially, self-renewal (Seale et al., 2000; Olguin and Olwin, 2004; Oustanina et al., 2004; Zammit et al., 2004; Kuang et al., 2006; Shinin et al., 2006).
In adult muscle, quiescent satellite cells express Pax7, whereas expression of Myf5 and MyoD is low or nondetectable (Yablonka-Reuveni and Rivera, 1994; Cornelison and Wold, 1997; Seale et al., 2000). Pax7 persists at lower levels in recently activated, proliferating satellite cells and is rapidly down-regulated in cells that commit to terminal differentiation (Olguin and Olwin, 2004; Zammit et al., 2004). In culture, Pax7 appears to be up-regulated and persists in a small population of myogenic cells that down-regulate MyoD expression. This subpopulation remains undifferentiated and mitotically inactive, resembling a quiescent satellite cell (Olguin and Olwin, 2004; Zammit et al., 2004). We have previously shown that Pax7 overexpression recapitulates these events in proliferating myogenic cells (Olguin and Olwin, 2004). Moreover, ectopic expression of Pax7 can efficiently repress the MyoD-dependent conversion of mesenchymal cells to the muscle lineage (Olguin and Olwin, 2004). Although this is evidence for a functional relationship between Pax7 and the MyoD family of transcription factors, the exact nature of this relationship is controversial (Olguin and Olwin, 2004; Oustanina et al., 2004; Seale et al., 2004; Relaix et al., 2006; Zammit et al., 2006).
Here, we attempted to delineate the molecular mechanisms involved in Pax7-mediated repression of MyoD function and myogenic progression. Our data indicate that Pax7 blocks myogenesis independently of its transcriptional activity, by a mechanism involving regulation of MyoD protein stability. Similarly, myogenin, but not MyoD, appears to regulate Pax7 function by affecting Pax7 levels. These results provide evidence supporting the existence of a reciprocal inhibition between Pax7 and the muscle regulatory factors (MRFs). Our data suggest that this mechanism may function to regulate the decision of an activated satellite cell to proliferate, commit to terminal differentiation, or reacquire a quiescent state.
Results
Pax7 represses myogenesis via inhibition of MyoD activity
During myogenic differentiation, Pax7 up-regulation is observed in cells that remain undifferentiated and down-regulate MyoD expression (Olguin and Olwin, 2004; Zammit et al., 2004), which is reminiscent of the reserve cell phenotype (Yoshida et al., 1998). Furthermore, overexpression of Pax7 down-regulates MyoD in satellite cells and myogenic cells lines, preventing terminal differentiation and cell cycle progression (Olguin and Olwin, 2004). Pax7 also inhibits MyoD-induced myogenic conversion of C3H10T1/2 cells (Olguin and Olwin, 2004), suggesting that Pax7-mediated inhibition of muscle differentiation occurs before induction of myogenin expression. To determine whether these effects are regulated at the transcriptional level, we asked if Pax7 differentially affects MyoD and myogenin transcriptional activity. We first assessed the ability of MyoD and myogenin to activate transcription from a luciferase reporter driven by the proximal regulatory region of the myogenin gene (myogenin-Luc; Fig. 1 A), in the presence or the absence of Pax7. Ectopically expressed MyoD activates the myogenin-Luc reporter gene >5,000-fold during the myogenic conversion of C3H10T1/2 cells, whereas ectopically expressed myogenin activates the reporter gene >700 fold (Fig. 1 A). Cotransfection of Pax7 represses MyoD transcriptional activity up to 90% in a dose-dependent manner (Fig. 1 B). However, myogenin activity was substantially less affected by Pax7 coexpression (approximately threefold repression at the highest Pax7 dose) than MyoD (Fig. 1 B). These data suggest that Pax7-dependent repression of myogenesis is specific for MyoD.
Figure 1.
Differential effects of Pax7 on MyoD and myogenin activity. (A, top). Schematic representation of the myogenin-Luc reporter (see Materials and methods). (bottom) Myogenin-Luc reporter gene is robustly activated by both MyoD (>5,000-fold) and myogenin (>700-fold). Pax7 has no effect on basal activity. Basal reporter activity was normalized to 1. (B) Pax7 coexpression differentially affects MyoD (4.8- ± 0.17- and 16.4- ± 1.9-fold repression at 1:1 and 1:2 molar ratio, respectively; black bars) versus myogenin (1.9- ± 0.15- and 2.8- ± 0.5-fold repression, respectively; white bars) transcriptional activity. (C) Transcriptional activity of a Gal-MyoD fusion protein (activation of the Gal4-luc reporter gene; schematic) is inhibited by Pax7 coexpression (13.6- ± 2.4-fold repression at 1:2 Gal4-MyoD/Pax7 molar ratio). Gal-VP16 transcriptional activity is considerably less sensitive to Pax7 coexpression (3.5- ± 0.06-fold repression). In B and C, maximum reporter activity was normalized to 1. Asterisks indicate that mean values are representative of at least three independent experiments. Error bars indicate standard deviation. (D) Binding of purified MyoD and E47 (E47N) to a DNA target is not disrupted by in vitro translated Pax7 protein (right). MCK-REbox indicates right E-Box of the muscle creatine kinase promoter ◂, E47N–MyoD–DNA complex; ♦, MyoD–DNA complex; ○, E47N–DNA complex. RRL, rabbit reticulocyte lysate. Arrowheads indicate the expected Pax7, MyoD, and E47 bands according to molecular weight. (left) Control in vitro translation for Pax7 expression.
We hypothesized that inhibition of MyoD function could arise via competition of Pax7 and MyoD for binding to common DNA targets. Thus, MyoD transcriptional activity on a noncanonical regulatory element should be insensitive to Pax7 repression. We tested this possibility by changing the DNA binding specificity of MyoD using a Gal4-MyoD fusion protein and determining the activation of a Gal4-Luc reporter gene (Fig. 1 C). Surprisingly, Pax7 was able to repress the activity of the fusion protein (Fig. 1 C). The inhibition of the Gal4-MyoD activity was quantitatively equivalent to that observed for wild-type MyoD (Fig. 1 C). This effect is specific for MyoD because a constitutive activator (Gal4-VP16) shows a greatly reduced sensitivity to cotransfection of Pax7 (Fig. 1 C), suggesting that the ability of Pax7 to repress MyoD transcriptional activity is unlikely to reflect a competitive binding to a common DNA target. This is further supported by the inability of Pax7 to either bind directly to a MyoD target sequence (MCK-REbox) or disrupt the binding of MyoD, E47, or MyoD-E47 dimers to DNA in electrophoretic mobility shift assays (EMSAs; Fig. 1 D). Consequently, we envision at least two mechanisms whereby Pax7 could inhibit MyoD activity: (1) regulating transcription of additional genes required for MyoD function or (2) a nontranscriptional mechanism, such as competition for a common interaction partner. To determine the contribution of Pax7 transcriptional activity to the inhibition of myogenesis, we performed deletion analysis of domains required for this function in Pax7 and tested the ability of the mutant proteins to repress MyoD activity during myogenic conversion of C3H10T1/2 cells.
The Pax7 homeodomain is critical for the repression of MyoD function
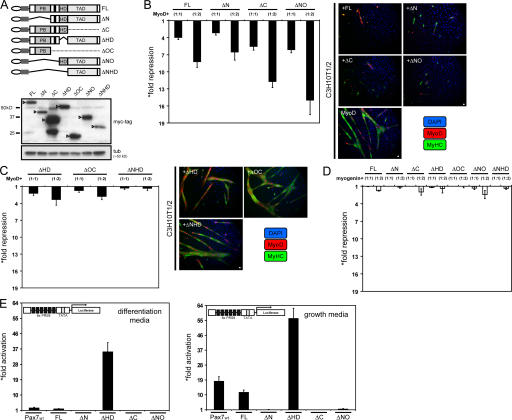
A series of Pax7-deletion mutants were generated (see Materials and methods) containing a myc-tag epitope followed by an NLS inserted at the N terminus of each mutant construct (Fig. 2 A, top). A prior set of mutants lacking the exogenous NLS exhibited cytoplasmic mislocalization and high variability in protein expression, suggesting major differences in protein stability (unpublished data). The mutant proteins used in subsequent assays (myc-NLS) were expressed at relatively similar levels (Fig. 2 A, bottom), with the exception of the ΔC mutant, which showed higher levels of protein expression at equivalent amounts of transfected expression vector (Fig. 2 A, bottom). This difference appears to be related to enhanced protein stability compared with other mutant products (unpublished data). The ability of each Pax7-deletion mutant to repress myogenic conversion of C3H10T1/2 cells induced by ectopic expression of MyoD was then evaluated. Pax7 mutants lacking either the paired-box or the transactivation domains repressed MyoD activity (Fig. 2 B, left), resembling the effect of the full-length Pax7. These findings correlated with a severe reduction in both myotube formation and expression of myosin heavy chain (MyHC), a marker of terminal differentiation (Fig. 2 B, right). In contrast, deletion of the homeodomain region abolished the effect of Pax7 on MyoD activity (Fig. 2 C, left) and failed to block myogenic differentiation (Fig. 2 C, right). Expression of a deletion mutant containing only the homeodomain and transactivation domain is sufficient to repress myogenic conversion of C3H10T1/2 cells, preventing terminal differentiation (Fig. 2 B). Interestingly, this mutant appeared more potent than the full-length Pax7 protein (Fig. 2 B). Ectopic expression of the N terminus plus the paired-box domain or the transactivation domain alone had no considerable effect on MyoD activity (Fig. 2 C). Together, these data suggest a critical role for the Pax7 homeodomain in repressing myogenesis and inhibiting MyoD function. This effect appears specific for MyoD, as neither wild-type Pax7 protein nor the deletion mutants had substantial effects on myogenin transcriptional activity (Fig. 2 D). To further determine whether inhibition of MyoD activity requires Pax7- dependent transcription, we analyzed the transcriptional activity of Pax7 and Pax7 mutants on a reporter gene driven by a regulatory sequence derived from the Drosophila even-skipped gene (Chalepakis et al., 1991; Bennicelli et al., 1999) containing both paired-box and homeodomain binding sites (6xPRS9-Luc). Unexpectedly, we detected only weak Pax7-dependent activation of the reporter gene under conditions that repressed MyoD activity (Fig. 2 E, left). However, the Pax-dependent reporter gene can be activated by full-length Pax7 under proliferation conditions (Fig. 2 E, right). As expected, deletion of either the paired-box or the transactivation domain abolished Pax7 transcriptional activity (Fig. 2 E). Interestingly, deletion of the homeodomain region, required for repression of MyoD activity, increased Pax7 transcriptional activity (Fig. 2 E; both under proliferation and differentiation conditions). This is in agreement with previous studies showing a cis-acting transcription repression activity for this domain in Pax7 (Bennicelli et al., 1999). These observations indicate that the ability to repress MyoD activity does not correlate with active Pax7-dependent transcription, suggesting that MyoD protein could be regulated by Pax7 protein interactions.
Figure 2.
The Pax7 homeodomain regulates MyoD activity. (A, top) Schematic representation of Pax7 functional domains and deletion mutants. FL, full length; PB, paired-box domain; HD, Hox/Homeodomain; TAD, transactivation domain. Black bars indicate octapeptide, ovals indicate myc-tag epitope, and gray boxes indicate SV40 T-antigen NLS. (bottom) Expression of Pax7 mutants (arrowheads) was analyzed by Western blots in C3H10T1/2 cells. Tubulin was used as loading control. (B and C) Deletion of the Pax7 paired-box domain (ΔN; B) or transactivation domain (ΔC; B) has no significant effects on repression of MyoD transcriptional activity (left) or on myogenic differentiation of C3H10T1/2 cells (MyHC expression; right) mediated by Pax7. Deletion of the homeodomain (ΔHD; C) impairs Pax7-mediated inhibition of MyoD transcriptional activity (left) and myogenic differentiation in C3H10T1/2 cells (right). Expression of either PB (ΔOC) or TAD (ΔNHD) domains alone have no significant effects on MyoD activity (C). (D) Effects of Pax7-deletion mutants on myogenin transcriptional activity. Maximum reporter activity was normalized to 1 in B and D. (E) Analysis of the transcriptional activity of Pax7 and Pax7-deletion mutants (on the 6xPRS9-luc reporter gene) during myogenic conversion of C3H10T1/2 cells. Basal reporter activity was normalized to 1. Asterisks indicate that data are representative of at least two independent experiments. Error bars indicate standard deviation. Bars, 12 μm.
Pax7 expression affects MyoD protein stability
Prompted by these observations, we asked if MyoD protein levels were affected by ectopic expression of Pax7. Western blot analyses of myogenic-converted C3H10T1/2 cell lysates revealed that inhibition of MyoD transcriptional activity and terminal differentiation correlated with changes in the levels of MyoD protein (Fig. 3 A, top). Full-length Pax7 (FL) or a Pax7 mutant that represses myogenesis (ΔN) reduced MyoD protein levels upon cotransfection, whereas a Pax7 mutant that does not repress myogenesis (ΔHD) had no effect on MyoD levels (Fig. 3 A, top). These changes appear specific for MyoD, as the levels of Pax7 protein were consistent with the amount of expression plasmid added to the cells (Fig. 3 A, bottom). Thus, MyoD protein stability appears specifically affected by coexpression with Pax7 and Pax7 mutants that inhibit myogenic differentiation.
Figure 3.
MyoD protein stability is affected by Pax7. (A) Western blots from C3H10T1/2 cell lysates normalized to equivalent levels of protein (lysate aliquots from a representative experiment equivalent to those used in Fig. 2, B and C) were probed for MyoD upon coexpression with Pax7, ΔN, and ΔHD Pax7-deletion mutants (top). Relative levels of Pax7 and Pax7 mutants were monitored (using anti-Pax7 and anti–myc-tag antibodies, respectively) in the same extracts (bottom). (B, left) MyoD protein is recovered to control levels (lane 1) after incubation with MG132 (lane 2) even in the presence of Pax7 (compare lanes 2 and 3). Incubation with DMSO has no effect on MyoD levels in the presence of Pax7 (lane 3, control). (bottom) Quantification of lanes 1–3. (right) MyoD levels are unaffected by Pax7 coexpression under proliferation conditions (compare lanes 4 and 5 to 6 and 7, respectively). (C) MG132 treatment results in partial rescue of MyoD protein (yellow arrowheads) in Pax7-overexpressing adult primary myoblasts (compare to DMSO control; white arrowheads). Bar, 10 μm. (D) MyoD transcriptional activity in C3H10T1/2 cells is not recovered by proteasome inhibition (tested as in Fig. 2 A). Error bars indicate standard deviation. Basal reporter activity was normalized to 1. (E) MG132 treatment results in high levels of MyoD and Pax7 coexpression in the majority of transfected C3H10T1/2 cells (white arrows). In control (DMSO) treated cells, only high levels of Pax7 (arrowheads) or MyoD (asterisks) can be detected in single cells. Bar, 12 μm.
MyoD is subject to regulation through proteasome- dependent degradation (Abu Hatoum et al., 1998; Song et al., 1998; Tintignac et al., 2000; Floyd et al., 2001; Lingbeck et al., 2003; Lingbeck et al., 2005; Sun et al., 2005); thus, we asked if this pathway was involved in down-regulating MyoD protein upon Pax7 coexpression. Loss of MyoD protein after the switch to differentiation media in C3H10T1/2 cells expressing both MyoD and Pax7 can be detected clearly by 24 h (Fig. 3 B, lane 3). Treatment with the proteasome inhibitor MG132 prevented loss of MyoD protein and rescues MyoD to control levels (Fig. 3 B, lanes 2 and 1, respectively). Interestingly, MyoD stability is affected by Pax7 coexpression in C3H10T1/2 only upon a switch to differentiation conditions, as we detected no difference in MyoD levels in the presence or absence of Pax7 when cultures were maintained in proliferation media (Fig. 3 B, lanes 4–7). This observation indicates that MyoD degradation does not occur via nonspecific effects derived from Pax7 overexpression. Most important, proteasome-mediated protein degradation also contributes to MyoD down-regulation induced by Pax7 overexpression in adult primary myoblasts (Olguin and Olwin, 2004), as MG132 treatment partially rescues MyoD expression under these conditions (Fig. 3 C). The inability of MG132 to fully rescue MyoD protein levels in adult myoblasts could be due to a decrease in transcription of the endogenous MyoD gene caused by down-regulation of MyoD protein.
We expected that rescuing MyoD protein levels would rescue its transcriptional activity. Interestingly, MyoD function was not restored upon proteasome inhibition, as MG132 treatment did not rescue MyoD-dependent activation of the myogenin-luc reporter in the presence of Pax7 (Fig. 3 D), even when robust nuclear coexpression of both transcription factors was observed under these conditions (Fig. 3 E). This finding suggests that additional events are involved in Pax7-dependent regulation of MyoD activity.
Myogenin can negatively regulate Pax7 expression
Initial events in myoblast differentiation include permanent withdrawal from the cell cycle and induction of myogenin followed by induction of muscle-specific genes. Along with others, we have shown that myogenin and Pax7 expression is mutually exclusive, whereas Pax7 is retained (and up-regulated) only in a small population of cells that escape differentiation and down-regulate MyoD expression (Olguin and Olwin, 2004; Zammit et al., 2004). In light of our new observations, we asked if up-regulation of myogenin controls Pax7 protein levels. Western blot analysis of C3H10T1/2 cell lysates cotransfected with myogenin and Pax7 revealed a reduction in Pax7 protein when compared with Pax7 levels upon cotransfection with MyoD (Fig. 4 A, left; compare lanes 3 and 4). Interestingly, myogenin is also considerably reduced upon Pax7 coexpression (Fig. 4 A, left), suggesting a reciprocal effect on relative protein levels. As observed previously for MyoD, myogenin reduction under these conditions involves proteasome-dependent protein degradation, as treatment with MG132 blocks myogenin loss (Fig. 4 A, right). Interestingly, MG132 treatment also blocks Pax7 reduction when myogenin is coexpressed (Fig. 4 A, right). Although the levels of Pax7 and myogenin appear to be reciprocally affected, Pax7 and myogenin are not coexpressed in adult myoblasts (in mice and humans), indicating that these observations may reflect complex population dynamics inherent in an asynchronous population of cells undergoing terminal differentiation. To definitively determine whether Pax7 and myogenin are coexpressed during the early stages of muscle differentiation, we used the MM14 satellite cell line, where cells can be synchronized at M/G1 by mitotic shake-off (Clegg et al., 1987; Kudla et al., 1998; Jones et al., 2005). When induced to differentiate, synchronized MM14 cells express muscle-specific genes within 6–12 h and begin fusion into multinucleated myotubes by 12–15 h, providing a useful assay for cell cycle–specific events associated with terminal differentiation (Clegg et al., 1987; Kudla et al., 1998; Jones et al., 2005). Synchronized MM14 cells were allowed to adhere for 8–10 h in the presence of growth medium and then cultured in differentiation medium for various periods of time (Fig. 4 B). We observed that Pax7 expression persists in a large fraction of the cell population until 12 h after differentiation induction (Fig. 4 B, left). As expected, myogenin protein was detectable by 8 h after induction of differentiation, reaching a maximum at 21 h (Fig. 4 B, middle). Between 8 and 12 h of differentiation, Pax7 and myogenin proteins were largely coexpressed within the same cell population, as 85.5 ± 1.2% (8 h) and 82.2 ± 5.2% (12 h) of the myogenin+ cells showed robust expression of both markers, indicating that myogenin protein accumulates in Pax7+ cells (Fig. 4 B, right). Coexpression of Pax7 and myogenin is transient because 9 h later (21 h in differentiation medium) the percentage of myogenin+ cells reached a maximum, whereas the percentage of Pax7+ cells dropped to a minimum (Fig. 4 B, middle and left, respectively). At this time point, expression of both Pax7 and myogenin becomes mutually exclusive, as the percentage of Pax7+/myogenin+ cells decreases to 7 ± 1.9%. By 30 h, percentages of myogenin+ and Pax7+ cells have not changed substantially (>80 and <17%, respectively), but we could no longer detect cells that were positive for both Pax7 and myogenin.
Figure 4.
Myogenin negatively regulates Pax7 expression. (A, left) Western blots from C3H10T1/2 cell lysates expressing MyoD, myogenin, and Pax7 reveal reduction of both myogenin and Pax7 levels upon coexpression. (middle) Quantification of lanes 2–4 for myogenin (top) and Pax7 (bottom) abundance, respectively. (right) Proteasome inhibition increases levels of myogenin and Pax7 affected by coexpression. (B) Analysis of Pax7 and myogenin expression in mitotically synchronized MM14 myoblasts during commitment to terminal differentiation (schematic). At the indicated times, cells expressing either Pax7 (white bars) or myogenin (black bars) were independently scored and plotted as a percentage of the total population. Cells coexpressing both Pax7 and myogenin (gray bars) are plotted as the percentage of Pax7+ cells in the myogenin+ subpopulation. (C) Ectopic expression of myogenin down-regulates Pax7 (96.6 ± 1.3% of transfected cells are Pax7−; yellow arrowheads) in MM14 myoblasts under growth conditions. Data are representative of three experiments. (D, left) Endogenous myogenin induction in C3H10T1/2 cells (by MyoD forced expression) is efficiently down-regulated by RNAi, as monitored by Western blot. (right) Quantification of myogenin abundance in the presence of specific or control (ctrl) siRNAs. Inset shows that MyoD protein levels are unaffected. (E) RNAi-mediated down-regulation of myogenin (no detectable myogenin in 60 ± 12.7% of transfected cells; middle and bottom) results in retention of Pax7 expression in differentiating cells (high Pax7 expression in 64.3 ± 18.7% of total transfected cells; bottom, white arrowheads). Control siRNA has no effect on myogenin (high myogenin expression in >80% of transfected cells; top, white arrowheads) or Pax7 expression (not depicted). β-Gal expression was used to identify transfected cells. Bar, 10 μm. (F) Pax7 protein stability is regulated during commitment to terminal differentiation. Mitotically synchronized MM14 cells were induced to differentiate for 15 h and treated with MG132 for 6 h before fixation. In control conditions (DMSO), myogenin (88.9 ± 5.8%) and Pax7 (16.1 ± 6%) are expressed in a mutually exclusive pattern (top and middle, white arrows). Upon MG132 treatment, 77.5 ± 10.1% of the cells are myogenin+, yet 57.2 ± 13% of the cells are also Pax7+ (bottom, white arrows). Bar, 10 μm.
The change in the Pax7/myogenin ratio during myogenic differentiation suggests that accumulation of myogenin protein could down-regulate Pax7. Indeed, we observed a reduction in Pax7 protein levels upon ectopic myogenin expression in MM14 myoblasts, even under proliferation conditions (Fig. 4 C; nondetectable Pax7 in >96% of transfected cells). If myogenin expression is responsible for reducing Pax7 protein, forced loss of myogenin under differentiation conditions should result in the persistence of Pax7 expression. To test this idea, we attempted to knock down myogenin through RNAi. Because siRNA transfection in MM14 myoblasts is inefficient and thus cannot provide a quantitative assessment for the extent of myogenin reduction, we initially tested the efficacy of myogenin-specific siRNAs in C3H10T1/2 cells ectopically expressing MyoD. As determined by Western blot analysis, maximum myogenin knockdown (>120-fold) was obtained at all doses tested (Fig. 4 D). This effect appears to be specific because MyoD expression was not affected under the same conditions (Fig. 4 D, right) and myogenin protein remained unaffected in the presence of a nonspecific control siRNA (Fig. 4 D). RNAi-mediated down-regulation of myogenin prevented the loss of Pax7 protein (high Pax7 signal in ≥60% of total transfected cells) in MM14 myoblasts (Fig. 4 E). As shown previously, control siRNA had no significant effect on myogenin (Fig. 4 E) or Pax7 protein (not depicted). These data support the hypothesis that Pax7 levels are negatively regulated by myogenin in cells undergoing commitment to terminal differentiation.
We then asked whether the rapid loss of Pax7 during commitment to differentiation in MM14 cells involved proteasome activity. Mitotically synchronized MM14 myoblasts were induced to differentiate for 15 h and treated with DMSO (control) or the proteasome inhibitor MG132 for additional 6 h (Fig. 4 F). At this time point (21 h after differentiation induction), >85% of the control cells expressed myogenin, whereas ∼15% of the cells expressed Pax7 in a mutually exclusive pattern (Fig. 4 F). After MG132 treatment, the percentage of Pax7+ cells increased to ∼60%, whereas the percentage of myogenin+ cells remained at ∼80% (Fig. 4 F). Under these conditions, myogenin and Pax7 were coexpressed in ∼50% of the cells analyzed (Fig. 4 F). Together, these results indicate that proteasome-dependent degradation appears to play an important role in the loss of Pax7 during myoblast commitment to terminal differentiation, correlating with the expression and accumulation of myogenin.
Indirect protein–protein interaction between Pax7 and MyoD
Our findings suggest a reciprocal regulation between Pax7/MyoD and Pax7/myogenin during the progression of cell differentiation. We asked whether these observations reflected interactions at the protein level by attempting copurification of Pax7–MyoD complexes or Pax7–myogenin complexes from nuclear extracts. Preliminary data indicated that putative Pax7–MyoD (and Pax7–myogenin) interaction was transient and/or unstable in adult primary myoblasts cultures and in MM14 cells (unpublished data). Thus, we asked whether these complexes could be detected in C3H10T1/2 cells coexpressing myc-tagged Pax7 and MyoD. Under control differentiation conditions, little if any detectable MyoD coimmunoprecipitated with Pax7 (Fig. 5 A, lane 2), yet MyoD was readily detectable in immunoprecipitates from MG132-treated cells (Fig. 5 A, lane 3). We could not detect any significant copurification of MyoD and Pax7 under proliferation conditions (unpublished data). We were unable to detect any specific Pax7–myogenin interactions using the same copurification strategy as for MyoD and Pax7 complexes (unpublished data). This could be explained by the strong effect on protein stability observed when both myogenin and Pax7 are coexpressed and thus may reflect a transient interaction disrupted during isolation.
Figure 5.
Indirect interaction between Pax7 and MRF proteins. (A) Nuclear fractions isolated from C3H10T1/2 cells transfected with MyoD alone or MyoD/myc–NLS-Pax7 (1:1 ratio) and induced to differentiate were immunoprecipitated with an anti-myc antibody and further analyzed by Western blot for MyoD in the eluted fractions. There is a minimal increase in MyoD signal upon Pax7 coexpression (compare lanes 1 and 2) that is substantially increased (more than sevenfold) in the presence of MG132 (compare lanes 1 and 3). (B) 35S-labeled MyoD, myc–NLS-Pax7 (FL-Pax7), and myogenin (left) were combined as indicated and subjected to in vitro coimmunoprecipitation (see Materials and methods) using anti–myc tag antibody. No MyoD–Pax7 or myogenin–Pax7 interactions were detected (lanes 1 and 2, respectively). Arrowheads indicate expected Pax7, MyoD, and E47 bands in the TNT assay, according to molecular weight.
Our previous results (Fig. 1, C and D) and the apparently weak Pax7–MyoD physical interaction, suggest that Pax7 and MyoD coexist in protein complexes through indirect interactions. This idea is further supported by the observation that these proteins do not interact directly during in vitro coimmunoprecipitation assays (Fig. 5 B). Similarly, we cannot detect a direct interaction between Pax7 and myogenin (Fig. 5 B). Together, these data suggest that upon external stimuli, Pax7 and members of the MRF family (i.e., MyoD and myogenin) can interact with common elements in a protein complex, leading to functional inhibition and changes in protein stability perhaps by altering interactions within the protein complexes.
Discussion
The transcription factor Pax7 has been implicated in satellite cell specification, survival, and self-renewal (Seale et al., 2000; Olguin and Olwin, 2004; Oustanina et al., 2004; Zammit et al., 2004; Kuang et al., 2006; Shinin et al., 2006). Although expression profiles and genetic evidence suggest a functional interaction between Pax7 and the MRFs, this interaction has not been defined at the molecular level. We have previously shown that Pax7 overexpression represses myogenesis (Olguin and Olwin, 2004). Here, we attempted to delineate some of the molecular mechanisms responsible for these effects and the functional interactions between Pax7 and the MRFs.
A Pax7–MRF mutually inhibitory circuit for satellite cell fate regulation
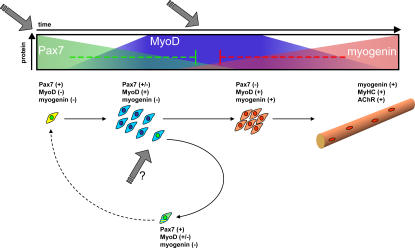
Muscle satellite cells, normally residing in a quiescent state, must be activated, proliferate, differentiate, and self-renew to maintain and repair adult skeletal muscle tissue. The mechanisms involved in regulating these decisions are not well understood. Here, we show evidence for an inhibitory regulatory relationship between Pax7, MyoD, and myogenin that may play a role in determining the cell fate decisions of activated satellite cells (Fig. 6). We propose a working model where, upon satellite cell activation, MyoD is induced and the Pax7/MyoD ratio plays a critical role in cell fate determination. At low Pax7/MyoD ratios, cells commit to terminal differentiation and induce myogenin, causing a rapid loss of Pax7. Intermediate Pax7/MyoD ratios prevent myogenin induction and may favor proliferation/survival of committed cells. A small population of muscle progenitors acquires or maintains a higher Pax7/MyoD ratio, causing a loss of MyoD protein, and may renew the quiescent satellite cell. In our model, the Pax7/MRF expression ratio is likely regulated via extracellular signaling and could integrate with additional external cues to promote commitment to each of these different cell fates (Fig. 6).
Figure 6.
Working model: reciprocal regulation of Pax7 and MRFs during myogenic cell fate commitment. Satellite cells (Pax7+/MyoD−/myogenin−) must commit to proliferate, differentiate, or renew the progenitor population to maintain muscle function. We propose that commitment to proliferate requires environmental cues (gray arrows) that activate satellite cells and up-regulate MyoD (blue) with a concomitant decline in Pax7 expression (green). Upon commitment to terminal differentiation, up-regulation of myogenin (red) down-regulates Pax7. In a small cell population, up- regulation of myogenin is prevented; Pax7 is up-regulated by unknown mechanisms, resulting in MyoD down-regulation (green nucleus and dashed cytoplasm cell) leading to the commitment to a quiescent, undifferentiated phenotype. In this model, the Pax7/MRF expression ratio is critical and integrates with environmental signals (gray arrows) to regulate cell fate commitment.
MyoD as a nodal point for Pax7 regulation of myogenesis
We initially hypothesized that Pax7 would inhibit myogenesis via a transcriptional mechanism that regulated MyoD activity to prevent myogenin induction. Thus, we evaluated the inhibitory effects of Pax7 on MyoD- and myogenin-dependent transcriptional activities in the context of a common target promoter. We found that MyoD activity was inhibited by Pax7 but that myogenin-dependent transcription was only marginally affected. Moreover, we have shown that under conditions where MyoD activity is enhanced, myogenin is up-regulated, promoting terminal differentiation even in the presence of ectopic Pax7 (Olguin and Olwin, 2004). These observations support the idea that once myogenin accumulates, Pax7 is incapable of preventing muscle differentiation, in agreement with our working model (Fig. 6).
In a recent report, Zammit et al. (2006) showed that retrovirus-mediated delivery of Pax7 does not prevent progression through myogenesis in myoblasts cultures. Because we postulate that timing, as well as the level of Pax7 expression, is critical for satellite cell fate decisions, these observations do not necessarily disagree with our own. If Pax7 is expressed after myogenin induction, myoblasts will commit to terminal differentiation (Fig. 6). In agreement with our data on C3H10T1/2 cells, which lack endogenous Pax7 expression, Zammit et al. (2006) show that ectopic expression of Pax7 in a Pax7-null subclone of C2C12 cells perturbs myogenic differentiation.
Genetic interactions indicate that Pax7 could participate in induction of the myogenic program during development (Ben-Yair and Kalcheim, 2005; Gros et al., 2005; Kassar-Duchossoy et al., 2005; Relaix et al., 2005). Moreover, ectopic expression of dominant-repressor Pax7 constructs (Pax7-EnR) suggests that Pax7-dependent transcription could induce MyoD expression (Chen et al., 2006; Relaix et al., 2006). In this context, our observations suggest that Pax7 may have a dual role where it activates the myogenic program by regulating MyoD transcription and prevents commitment to differentiation by regulating MyoD function, similar to what has been shown for Pax3 function in melanocyte development (Lang et al., 2005). Analysis of gene expression profiles from rhabdomyosarcoma cell lines overexpressing Pax-FKHR proteins (both Pax7- or Pax3-FKHR) shows that MyoD expression is twofold higher than in controls; however, several genes related to muscle differentiation (including myogenin) are specifically down-regulated (Davicioni et al., 2006). These data further support the idea that by differentially regulating MyoD expression and function, Pax7 (or Pax-FKHR) may promote retention of muscle progenitor characteristics.
Transcriptional versus nontranscriptional regulation of MyoD activity by Pax7
Intriguingly, we showed that altering MyoD DNA binding specificity did not affect the ability of Pax7 to inhibit MyoD- dependent transcription. Moreover, the binding of MyoD to DNA is not affected by Pax7 protein in vitro. Therefore, inhibition of MyoD activity by Pax7 does not appear to require competitive binding to common DNA targets, suggesting that Pax7 could function to either regulate transcription of additional genes required for MyoD function or via a nontranscriptional mechanism, such as posttranslational control of MyoD and/or MyoD protein interactions.
Pax7 and Pax3 contain all the major functional domains described for the Pax family, and this sequence/structural complexity is thought to be reflected in an increased repertoire of targets and mechanisms for their own regulation (Chi and Epstein, 2002; Robson et al., 2006). Unlike Pax3, Pax7 is a poor transcriptional activator, containing two cis-acting repressor domains (at the N terminus and the homeodomain, respectively) (Bennicelli et al., 1999). Hence, we addressed the contribution of Pax7-dependent transcription on MyoD inhibition by disrupting Pax7 domains thought to be critical for its transcriptional activity. Our data show that Pax7 represses myogenesis in the absence of either its paired-box or the transactivation domains, whereas deletion of the homeodomain abrogates the ability of Pax7 to inhibit myogenesis. Moreover, expression of the C-terminal Pax7 region, including the homeodomain and the transactivation domain, is sufficient to repress MyoD activity in C3H10T1/2 cells. As expected, deletion of either the paired-box domain or the transactivation domain abolishes Pax7-dependent transcription of a Pax3/Pax7-specific reporter gene.
Remarkably, Pax7-dependent transcriptional activation appears to be highly dependent on the cellular context. Although we detected activity from the full-length Pax7 in differentiation media, the activation was modest and only twofold above background. In contrast, we observed robust activation of the Pax7 reporter in cells maintained in proliferation media. Moreover, the ΔHD mutant was transcriptionally active under both conditions, yet this mutant fails to repress MyoD activity. These results contrast directly with recent observations where ectopically expressed Pax7 sustains transcription during myoblast differentiation (Zammit et al., 2006). In this study, the construct used for generation of a reporter mouse strain contained only binding sites for the paired-box domain (derived from the Trp-1 gene) to drive the expression of β-galactosidase. The reporter gene used in the present study contains binding sites for both the paired-box domain and the homeodomain (derived from the e5 sequence in the Drosophila even-skipped promoter) to drive the expression of luciferase. Thus, differences in Pax7-dependent transcription appear to be cell type, cell context, and reporter context dependent. Nevertheless, our findings strongly suggest that Pax7 transcriptional activity is not directly involved in the inhibition of MyoD. We envision that Pax7 acts in part via protein–protein interactions that may disrupt functional MyoD-containing transcriptional complexes, resulting in loss of specific MyoD functions and inhibition of myogenesis. In agreement with this idea, we showed that MyoD protein levels are reduced upon ectopic expression of Pax7. Importantly, the loss of MyoD protein requires the Pax7 homeodomain and can be reverted by inhibition of proteasome activity.
Proteasome-dependent MyoD degradation is inhibited by MyoD binding to DNA in vitro (Abu Hatoum et al., 1998). Interestingly, we showed that although proteasome inhibition rescued MyoD protein levels, its myogenic function was not restored, further supporting the idea that MyoD-containing complexes may be disrupted by Pax7. Although we can detect complexes containing both MyoD and Pax7 consistently in the presence of proteasome inhibitors, we cannot detect interactions between Pax7 and MyoD when purified proteins are used in gel shift assays or after in vitro coimmunoprecipitation. Thus, our data suggest that Pax7 and MyoD coexist in a protein complex through indirect interactions. Pax7 has also been found in a complex with MyoD by mass spectrometry, using alternative cellular sources (unpublished data), supporting the existence of a protein complex containing Pax7 and MyoD. In addition, our results do not rule out an effect of Pax7 on transcription of cofactors required for MyoD function, via inhibitory protein–protein interactions at specific promoters. Current efforts are directed to the development of tools for the unbiased identification of Pax7-interacting partners in myogenic cells.
Commitment to terminal muscle differentiation and the regulation of Pax7 expression
Pax7 and myogenin expression in individual cells occurs in a mutually exclusive pattern (Olguin and Olwin, 2004; Zammit et al., 2004). Prompted by the observation that ectopic coexpression of Pax7 and myogenin results in decreased levels of both proteins in C3H10T1/2 cells, we analyzed the expression of myogenin and Pax7 during commitment to differentiation in mitotically synchronized MM14 myoblasts. During differentiation, Pax7 and myogenin transiently coexist in the same cell population, but as differentiation progresses and myogenin levels increase, Pax7 levels decline, exhibiting the mutually exclusive expression pattern described previously. Interestingly, the time at which Pax7 levels decline correlates with the irreversible commitment of MM14 cells to terminal differentiation (Clegg et al., 1987). At this time point, it is already possible to identify a minor population of Pax7+/myogenin− cells that remains throughout differentiation, reminiscent of the reserve population phenotype (Yoshida et al., 1998; Olguin and Olwin, 2004). Conversely, the loss of Pax7 expression correlates with the loss of the Pax7+/myogenin+ phenotype, suggesting that during this period, myogenin expression down-regulates Pax7. Using a similar strategy used to detect Pax7- and MyoD-containing protein complexes, we were unable to detect Pax7–myogenin interactions. If a Pax7–myogenin complex exists, it may be too weak to be detected by these methods. Alternatively, the lack of a detectable interaction could be due to the observations that Pax7 and myogenin appear unstable when both proteins are present. In summary, these findings are compatible with a model whereby myogenin up-regulation results in the rapid loss of Pax7 during myoblast differentiation, and inhibition of myogenin expression may be necessary for maintenance and up-regulation of Pax7 in activated satellite cells that escape differentiation, eventually contributing to satellite cell self-renewal.
Materials and methods
Cell lines
C3H10T1/2 cells were cultured in DME and 10% fetal bovine serum at 37°C and 5% CO2. For myogenic conversion assays, cultures were induced to differentiate in DME and 2% fetal bovine serum for 48 h or as specified. MM14 cells were cultured in F12-C, 15% horse serum, and 500 pM FGF-2 at 37°C and 5% CO2. Differentiation was induced by culture in F12-C and 10% horse serum. When specified, cells were treated with 20–25 μM MG132 (Calbiochem) for 6–8 h before harvesting or fixation.
Pax7-deletion mutants
Pax7 deletions were constructed via PCR mutagenesis using pcDNA-Pax7d vector (Olguin and Olwin, 2004) as a template. Appropriate restriction sites were included at the 5′-end of forward and reverse primers (Table I). Pax7 (and mutant) cDNAs were subcloned into pcDNA3-myc-NLS expression vector (BamHI and XhoI sites; a gift from J. Lykke-Andersen and G. Singh, University of Colorado, Boulder, CO). In frame cloning introduces a single copy of a myc-tag epitope followed by the SV40 T-antigen NLS, to the 5′-end of each cloned cDNA.
Table I.
Primers and cDNA products for construction of Pax7-deletion mutants
| cDNA | Forward primer (5′→3′)/target location (nt) | Reverse primer (5′→3′)/target location (nt) |
|---|---|---|
| FL | GGATCCATGGCGGCCCTTCCC/1–15 | CTCGAGCTAGTAGGCTTGTCCCGTTTCCAC/1489–1512 |
| ΔN | GGATCCGGGAAGAAAGAGGACGACGAG/487–507 | CTCGAGCTAGTAGGCTTGTCCCGTTTCCAC/1489–1512 |
| ΔC | GGATCCATGGCGGCCCTTCCC/1–15 | CTCGAGCTATCCTGCCTGCTTGCGCCA/811–828 |
| ΔHD | ||
| N-Pax7a | GGATCCATGGCGGCCCTTCCC/1–15 | CTGGTTAGCGCGCTGCTTGCGCTTCAG/631–648 |
| C-Pax7b | AAGCAGCGCGCTAACCAGCTGGCCGCC/826–843 | CTCGAGCTAGTAGGCTTGTCCCGTTTCCAC/1489–1512 |
| ΔHDc | GGATCCATGGCGGCCCTTCCC/1–15 | CTCGAGCTAGTAGGCTTGTCCCGTTTCCAC/1489–1512 |
| ΔNO | GGATCCCAGCGCCGCAGTCGGACC/643–660 | CTCGAGCTAGTAGGCTTGTCCCGTTTCCAC/1489–1512 |
| ΔNHD | GGATCCGCTAACCAGCTGGCCGCC/826–843 | CTCGAGCTAGTAGGCTTGTCCCGTTTCCAC/1489–1512 |
GGATCC, BamHI recognition site; CTCGAG, XhoI recognition site.
N terminus/paired box/octapeptide.
Transactivation domain.
N-Pax7/C-Pax7 template.
Myogenic conversion and reporter assays
Myogenic conversion of C3H10T1/2 cells was induced by transfecting (Superfect; QIAGEN) 1 μg/well (12-well plate) of the pRSV-MyoD vector. Differentiation was induced 24 h after transfection for 24 or 48 h as indicated. When required, pcDNA-Pax7 vector was cotransfected along with pRSV-MyoD or pEMS-ratmyogenin vectors at the indicated molar ratios. pcDNA3 was used as control DNA. To evaluate MyoD transcriptional activity, 1 μg of the myogenin-luc reporter gene was transfected in the absence or the presence of 0.4 μg pRSV-MyoD and in the absence or presence of pcDNA-Pax7 (at the specified molar ratios), in triplicate for each condition. 0.025 μg of the CMV-LacZ expression vector was cotransfected as a marker for transfection efficiency, and pcDNA3 was used as control DNA. After differentiation induction, whole cell lysates were collected and luciferase and β-galactosidase activities were determined using the Dual-Light System (Applied Biosystems) as reported previously (Olguin and Olwin, 2004). Total protein content was estimated (micro BCA; Pierce Chemical Co.) for subsequent analyses. Where indicated, the fold difference between maximum activation (reporter plus MyoD or myogenin expression vector) and the activation in different experimental conditions was represented as fold repression.
C3H10T1/2 cells were cotransfected with Gal4-luc reporter gene and either Gal4-MyoD or Gal4-VP16 fusion proteins in the presence or the absence of pCDNA3-Pax7 at the indicated molar ratios. Pax7 and Pax7-deletion mutants were tested for transcriptional activation in C3H10T1/2 cells as described above by cotransfection with the 6xPRS9-Luc reporter gene (provided by F. Barr, University of Pennsylvania, Philadelphia, PA), in the presence or absence of MyoD.
In vivo and in vitro coimmunoprecipitation
For in vivo coimmunoprecipitation experiments, C3H10T1/2 cells were transiently transfected with 1:1 molar ratio (MyoD/myc-Pax7), as described previously. When indicated, cells were incubated with 20 μM MG132 for 6 h before harvest. Cells were washed twice and harvested in ice-cold PBS using a cell scraper. Cell pellet was recovered by centrifugation and resuspended in 1 ml buffer A (10 mM Hepes, pH 7.6, 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT). After a 10-min incubation in ice, cell pellet was recovered, resuspended in 400 μl buffer A, and disrupted in ice using a Dounce tissue grinder. Cell nuclei were recovered by centrifugation and resuspended in 200 μl buffer B (20 mM Hepes, pH 7.6, 0.5 mM EDTA, 100 mM KCl, 10% glycerol, 2 mM DTT, 3 mM CaCl2, 1.5 mM MgCl2, 0.25 mM Na3VO4, 1 mM NaF, 50 mM β-glycerophosphate, and protease inhibitor cocktail). Nuclear fraction was treated with nuclease S7 (Roche; 6 mU/μg of total DNA) for 10 min at 37°C. Nuclease activity was stopped by addition of EDTA (20 mM final concentration), and nuclear fraction was incubated for 2 h at 4°C with gentle rotation. Extracts were recovered by centrifugation. For immunoprecipitation, total protein was equalized (∼200 μl at 1 mg/ml), precleared with 20 μl of agarose–protein G (50% slurry; Pierce Chemical Co.), and incubated in the presence or absence of anti–myc tag antibody (clone 9B11 at a dilution of 1:1,000; Cell Signaling) at 4°C overnight. Immunocomplexes were captured by incubation with agarose–protein G for 3 h at 4°C, washed five times for 5 min each in buffer B, and eluted by resuspending beads in 50 μl 2× SDS-PAGE loading buffer and boiling for 5 min.
For in vitro coimmunoprecipitation experiments, 35S-labeled proteins were obtained by coupled transcription and translation in rabbit reticulocyte lysate (Promega). Protein interaction and immunopurification (using equivalent protein concentration estimated by autoradiography) was performed as described by Davis et al. (1990) using anti–myc tag antibody. Proteins were visualized by SDS-PAGE and autoradiography (Storm 860 Scanner [Molecular Dynamics]; control software version 5.03).
EMSAs
Gel mobility shift assays were performed from rabbit reticulocyte translated proteins (Davis et al., 1990) or purified proteins (Thayer and Weintraub, 1993) as required. Approximately equal amounts of each factor were added to the binding reactions (estimated by 35S-methionine incorporation in a translation reaction performed in parallel).
Myogenin overexpression and knockdown
pEMS-ratmyogenin (1.5 μg/well; 6-well plate) was used to ectopically express myogenin in MM14 cells and adult primary myoblasts (Lipofectamine 2000; Invitrogen). Cells were fixed and subjected to immunofluorescence staining 24 h after transfection. For myogenin expression knockdown, 200 nM SMARTpool siRNA duplexes (Dharmacon) were transfected in MM14 cells (Transmessenger; QIAGEN). siCONTROL RISC-free siRNA (Dharmacon) was used as a negative control. Cells were fixed 48–72 h after transfection. Specific and control siRNA duplexes were provided by Y. Fedorov (Dharmacon, Lafayette, CO).
Western blotting
Whole C3H10T1/2 cell extracts were obtained by disruption in modified RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% IGEPAL, 1 mM NaFl, 1 mM Na3Vo4, and 1× Complete anti-protease cocktail [Roche]), and incubating for 10 min at 4°C. Lysates were cleared by centrifugation. 30–50 μg total protein were loaded onto 10% SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes (Millipore). Primary antibodies and dilutions used were as follows: mouse monoclonal anti-MyoD1 (clone 5.8A; Vector Laboratories) at 1:100; mouse monoclonal anti-Pax7 (Developmental Studies Hybridoma Bank) at 1:10 (cell culture supernatant); mouse monoclonal anti-myogenin (F5D; Developmental Studies Hybridoma Bank) at 1:10 (cell culture supernatant); mouse monoclonal anti–α-tubulin (DM1A; Sigma-Aldrich) at 1:100; mouse monoclonal anti–myc tag (9B11; Cell Signaling) at 1:1,000. Anti-mouse HRP-conjugated secondary antibodies (Promega) were used at 1:5,000, and HRP activity was visualized using the ECL Plus Western Blotting Detection System (GE Healthcare). When required, x-ray films were scanned (Powerlook 1120 scanner; UMAX), digitalized (VueScan 7.6.8; Hamrick Software), and analyzed (ImageJ; NIH) for figure preparation.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 20 min. Primary antibodies and dilutions used were as follows: mouse monoclonal anti-Pax7 (Developmental Studies Hybridoma Bank) at 1:5 (cell culture supernatant); rabbit polyclonal anti-MyoD (Santa Cruz Biotechnology, Inc.) at 1:30; rabbit polyclonal anti-myogenin (Santa Cruz Biotechnology, Inc.) at 1:30; mouse monoclonal anti-MyHC (MF20; Developmental Studies Hybridoma Bank) at 1:5 (cell culture supernatant). Secondary antibodies conjugated to Alexa 594 or Alexa 488 were obtained from Invitrogen. Vectashield (Vector Laboratories) was used as mounting media. Micrographs were taken from an epifluorescence microscope (Eclipse E800 [Nikon] using 20×/0.50 and 40×/0.75 objectives [Nikon]) at RT, using Slidebook v3.0 acquisition software (Intelligent Imaging Innovations, Inc.) coupled to a digital camera (Sensicam; Cooke). Digital deconvolution for single plane images (no neighbors) was applied (when required) to acquired images (Slidebook v3.0).
Image processing and figure preparation
For figure preparation, images were exported into Photoshop (Adobe). If necessary, the brightness and contrast were adjusted to the entire image, the image was cropped, and individual color channels were extracted (when required) without color correction adjustments or γ adjustments. Final figures were prepared in PowerPoint (Microsoft) and Illustrator (Adobe).
Acknowledgments
The authors acknowledge Dr. Frederick Barr for the 6xPRS9-luc reporter gene and Dr. Jens Lykke-Andersen and Guramrit Singh for the pcDNA3-myc-NLS expression vector. We thank Karen Seaver and Lauren Snider for technical assistance with the EMSAs and Dr. Yuri Fedorov (Dharmacon) for control and myogenin siRNAs. We also thank Dr. Cecilia Riquelme and Melissa Hausburg for critical review of the manuscript.
This work was supported by grants from the Muscular Dystrophy Association (MDA3928) to H.C. Olguin and the National Institutes of Health (AR39467 and AR49446) to B.B. Olwin. Z. Yang is supported by the Fred Hutchinson Cancer Research Center interdisciplinary training grant and National Institutes of Health grant F32, and S.J. Tapscott is supported by National Institutes of Health grant AR45113. The authors have no commercial affiliations or conflicts of interest.
Abbreviations used in this paper: EMSA, electrophoretic mobility shift assay; MRF, muscle regulatory factor; MyHC, myosin heavy chain.
References
- Abu Hatoum, O., S. Gross-Mesilaty, K. Breitschopf, A. Hoffman, H. Gonen, A. Ciechanover, and E. Bengal. 1998. Degradation of myogenic transcription factor MyoD by the ubiquitin pathway in vivo and in vitro: regulation by specific DNA binding. Mol. Cell. Biol. 18:5670–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennicelli, J.L., S. Advani, B.W. Schafer, and F.G. Barr. 1999. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene. 18:4348–4356. [DOI] [PubMed] [Google Scholar]
- Ben-Yair, R., and C. Kalcheim. 2005. Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development. 132:689–701. [DOI] [PubMed] [Google Scholar]
- Bober, E., T. Franz, H.H. Arnold, P. Gruss, and P. Tremblay. 1994. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 120:603–612. [DOI] [PubMed] [Google Scholar]
- Chalepakis, G., R. Fritsch, H. Fickenscher, U. Deutsch, M. Goulding, and P. Gruss. 1991. The molecular basis of the undulated/Pax-1 mutation. Cell. 66:873–884. [DOI] [PubMed] [Google Scholar]
- Chen, Y., G. Lin, and J.M. Slack. 2006. Control of muscle regeneration in the Xenopus tadpole tail by Pax7. Development. 133:2303–2313. [DOI] [PubMed] [Google Scholar]
- Chi, N., and J.A. Epstein. 2002. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 18:41–47. [DOI] [PubMed] [Google Scholar]
- Clegg, C.H., T.A. Linkhart, B.B. Olwin, and S.D. Hauschka. 1987. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J. Cell Biol. 105:949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelison, D.D.W., and B.J. Wold. 1997. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 191:270–283. [DOI] [PubMed] [Google Scholar]
- Davicioni, E., F.G. Finckenstein, V. Shahbazian, J.D. Buckley, T.J. Triche, and M.J. Anderson. 2006. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 66:6936–6946. [DOI] [PubMed] [Google Scholar]
- Davis, R.L., P. Cheng, A.B. Lassar, and H. Weintraub. 1990. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 60:733–746. [DOI] [PubMed] [Google Scholar]
- Floyd, Z.E., J.S. Trausch-Azar, E. Reinstein, A. Ciechanover, and A.L. Schwartz. 2001. The nuclear ubiquitin-proteasome system degrades MyoD. J. Biol. Chem. 276:22468–22475. [DOI] [PubMed] [Google Scholar]
- Goulding, M., A. Lumsden, and A.J. Paquette. 1994. Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development. 120:957–971. [DOI] [PubMed] [Google Scholar]
- Gros, J., M. Manceau, V. Thome, and C. Marcelle. 2005. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 435:954–958. [DOI] [PubMed] [Google Scholar]
- Jones, N.C., K.J. Tyner, L. Nibarger, H.M. Stanley, D.D. Cornelison, Y.V. Fedorov, and B.B. Olwin. 2005. The p38α/β MAPK functions as a molecular switch to activate the quiescent satellite cell. J. Cell Biol. 169:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassar-Duchossoy, L., E. Giacone, B. Gayraud-Morel, A. Jory, D. Gomes, and S. Tajbakhsh. 2005. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 19:1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang, S., S.B. Charge, P. Seale, M. Huh, and M.A. Rudnicki. 2006. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 172:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla, A.J., N.C. Jones, R.S. Rosenthal, K. Arthur, K.L. Clase, and B.B. Olwin. 1998. The FGF receptor-1 tyrosine kinase domain regulates myogenesis but is not sufficient to stimulate proliferation. J. Cell Biol. 142:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, D., M.M. Lu, L. Huang, K.A. Engleka, M. Zhang, E.Y. Chu, S. Lipner, A. Skoultchi, S.E. Millar, and J.A. Epstein. 2005. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 433:884–887. [DOI] [PubMed] [Google Scholar]
- Lingbeck, J.M., J.S. Trausch-Azar, A. Ciechanover, and A.L. Schwartz. 2003. Determinants of nuclear and cytoplasmic ubiquitin-mediated degradation of MyoD. J. Biol. Chem. 278:1817–1823. [DOI] [PubMed] [Google Scholar]
- Lingbeck, J.M., J.S. Trausch-Azar, A. Ciechanover, and A.L. Schwartz. 2005. E12 and E47 modulate cellular localization and proteasome-mediated degradation of MyoD and Id1. Oncogene. 24:6376–6384. [DOI] [PubMed] [Google Scholar]
- Olguin, H.C., and B.B. Olwin. 2004. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev. Biol. 275:375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina, S., G. Hause, and T. Braun. 2004. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 23:3430–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix, F., D. Rocancourt, A. Mansouri, and M. Buckingham. 2005. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 435:948–953. [DOI] [PubMed] [Google Scholar]
- Relaix, F., D. Montarras, S. Zaffran, B. Gayraud-Morel, D. Rocancourt, S. Tajbakhsh, A. Mansouri, A. Cumano, and M. Buckingham. 2006. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 172:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson, E.J., S.J. He, and M.R. Eccles. 2006. A PANorama of PAX genes in cancer and development. Nat. Rev. Cancer. 6:52–62. [DOI] [PubMed] [Google Scholar]
- Seale, P., L.A. Sabourin, A. Girgis-Gabardo, A. Mansouri, P. Gruss, and M.A. Rudnicki. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell. 102:777–786. [DOI] [PubMed] [Google Scholar]
- Seale, P., J. Ishibashi, A. Scime, and M.A. Rudnicki. 2004. Pax7 is necessary and sufficient for the myogenic specification of CD45+:Sca1+ stem cells from injured muscle. PLoS Biol. 2:E130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinin, V., B. Gayraud-Morel, D. Gomes, and S. Tajbakhsh. 2006. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat. Cell Biol. 8:677–682. [DOI] [PubMed] [Google Scholar]
- Song, A., Q. Wang, M.G. Goebl, and M.A. Harrington. 1998. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol. Cell. Biol. 18:4994–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., J.S. Trausch-Azar, A. Ciechanover, and A.L. Schwartz. 2005. Ubiquitin-proteasome-mediated degradation, intracellular localization, and protein synthesis of MyoD and Id1 during muscle differentiation. J. Biol. Chem. 280:26448–26456. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh, S., D. Rocancourt, G. Cossu, and M. Buckingham. 1997. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 89:127–138. [DOI] [PubMed] [Google Scholar]
- Thayer, M.J., and H. Weintraub. 1993. A cellular factor stimulates the DNA-binding of MyoD and E47. Proc. Natl. Acad. Sci. USA. 90:6483–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintignac, L.A., M.P. Leibovitch, M. Kitzmann, A. Fernandez, B. Ducommun, L. Meijer, and S.A. Leibovitch. 2000. Cyclin E-cdk2 phosphorylation promotes late G1-phase degradation of MyoD in muscle cells. Exp. Cell Res. 259:300–307. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni, Z., and A.J. Rivera. 1994. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev. Biol. 164:588–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, N., S. Yoshida, K. Koishi, K. Masuda, and Y. Nabeshima. 1998. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J. Cell Sci. 111:769–779. [DOI] [PubMed] [Google Scholar]
- Zammit, P.S., J.P. Golding, Y. Nagata, V. Hudon, T.A. Partridge, and J.R. Beauchamp. 2004. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 166:347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit, P.S., F. Relaix, Y. Nagata, A.P. Ruiz, C.A. Collins, T.A. Partridge, and J.R. Beauchamp. 2006. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 119:1824–1832. [DOI] [PubMed] [Google Scholar]