Abstract
The local accumulation of phosphatidylinositol (3,4,5) trisphosphate (PIP3) and resulting activation of Rac1/Cdc42 play a critical role in nerve growth factor (NGF)–induced neurite outgrowth. To further explore the mechanism, we visualized PIP3, phosphatidylinositol (3,4) bisphosphate, and Rac1/Cdc42 activities by fluorescence resonance energy transfer (FRET) imaging in NGF-stimulated PC12 cells. Based on the obtained FRET images, and with the help of in silico kinetic reaction model, we predicted that PI-5-phosphatase negatively regulates PIP3 upon NGF stimulation. In agreement with this model, depletion of Src homology 2 domain–containing inositol polyphosphate 5-phosphatase 2 (SHIP2) markedly potentiated NGF-induced Rac1/Cdc42 activation and PIP3 accumulation and considerably increased the number and the length of neurites in phosphate and tensin homologue–depleted PC12 cells. Further refinement of the computational model predicted Rac1 regulation of PI3-kinase and SHIP2, which was also validated experimentally. We propose that the SHIP2-mediated negative feedback on PIP3 coordinately works with the PI3-kinase–mediated positive feedback to form an initial protrusive pattern and, later, to punctuate the PIP3 accumulation to maintain proper neurite outgrowth.
Introduction
Neuritogenesis is an essential event in making the complex architecture of neuronal networks. Initially, the original round shape of neurons is broken down to make buds. Thereafter, some protrusions are selected and stabilized into neurites. These morphological changes are accompanied by the cytoskeletal reorganization of actin and microtubules (da Silva and Dotti, 2002). Rho-family GTPases (RhoA, Rac1, and Cdc42) play central roles in cytoskeletal regulation involved in a range of cellular functions (Van Aelst and D'Souza-Schorey, 1997; Hall, 1998). Also, they are committed to neuronal morphogenesis, including axon growth and guidance, dendritic elaboration, and the formation of synapses (Luo, 2000; Govek et al., 2005). Our previous studies using fluorescence resonance energy transfer (FRET)–based probes have shown that Rac1 and Cdc42 are locally and repetitively activated at protruding sites during neurite outgrowth in NGF-stimulated PC12 cells and in sensory neurons (Aoki et al., 2004; Nakamura et al., 2005), suggesting that their local activation is required for neurite outgrowth.
Phosphatidylinositol 3-kinase (PI3-kinase) has been shown to be required for NGF-induced neurite outgrowth of PC12 cells (Kimura et al., 1994) and sufficient to induce neurite extension in PC12 (Kobayashi et al., 1997; Kita et al., 1998) and SH-SY5Y cells (Pan et al., 2005). Although PI3-kinase is a multifunctional signaling molecule having various effectors (Cantley, 2002), a close linkage between PI3-kinase and Rac1/Cdc42 has been reported in a wide range of morphological responses to external stimuli (Bourne and Weiner, 2002; Merlot and Firtel, 2003). Moreover, a local positive feedback loop composed of Rac1/Cdc42 and PI3-kinase has been hypothesized to be responsible for the symmetry breaking and persistent activation required for morphogenesis (Merlot and Firtel, 2003; Shi et al., 2003). We have previously shown that in PC12 cells, NGF/TrkA signaling drives the cycling of a positive feedback loop composed of PI3-kinase, Vav2/Vav3, and Rac1/Cdc42 at neurite tips (Aoki et al., 2005). On the other hand, it is argued that negative regulators define the spatiotemporal window of active signaling. Thus, the local and repetitive activation of PI3-kinase and Rac1/Cdc42 in NGF-treated PC12 cells implies the involvement of negative regulators in this signaling pathway. However, the dynamic and interdependent properties of this signaling network make it difficult to analyze these negative regulators intuitively. We have therefore used in silico simulation of the kinetic reaction (Sasagawa et al., 2005; Fujioka et al., 2006) as a tool to help generate hypotheses and to validate the reliability of experimental results.
In this study, we demonstrate that two phosphatidylinositol (3,4,5) trisphosphate (PIP3) phosphatases, i.e., Src homology 2 domain–containing inositol phosphatase 2 (SHIP2) and phosphate and tensin homologue (PTEN), act as key molecules of negative regulation in the NGF–PIP3–Rac1/Cdc42 signaling network. Depletion of SHIP2 and PTEN by RNA interference markedly potentiated NGF-induced Rac1/Cdc42 activation and PIP3 accumulation and substantially increased the number and the length of neurites in PC12 cells. Moreover, our computational and experimental studies showed the presence of an NGF-dependent negative feedback from Rac1 to SHIP2. This negative feedback loop has previously been unexpected. Finally, direct activation of PI3K and SHIP2 by Rac1 was indicated by acute Rac1 activation using the inducible translocation technique. We propose that SHIP2 and PTEN work coordinately with positive regulators to form the initial protrusive pattern and, after initial neuritogenesis, punctuate the PIP3 accumulation to maintain proper neurite outgrowth in neuronal cells.
Results
NGF-induced negative regulation of PIP3, Rac1, and Cdc42
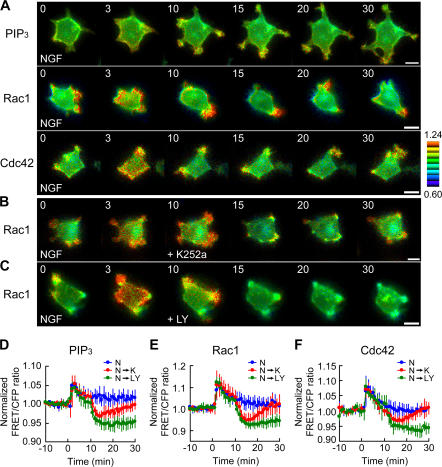
We examined the effect of shutting down TrkA activity on the regulation of PIP3, Rac1, and Cdc42 in NGF-treated PC12 cells. As a control, Fig. 1 A shows the spatiotemporal change of PIP3 level and Rac1/Cdc42 activity after NGF stimulation. Immediately after NGF addition (<5 min), PI3-kinase, Rac1, and Cdc42 were transiently activated in broad areas at the cell periphery, where the lamellipodial extension was concurrently observed. Thereafter, the local accumulation of PIP3 and active Rac1 and Cdc42 was observed at the protruding tips (>10 min). Next, we inhibited TrkA phosphorylation with 10 nM K252a at 10 min after NGF addition (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200609017/DC1). K252a treatment immediately induced a transient decrease in Rac1 activity to below the basal level and the suppression of cell protrusion (Fig. 1, B and E, red line). Rac1 activity and the morphological repression returned to the basal levels within 20 min. A similar change was observed for PIP3 level and Cdc42 activity (Fig. 1, D and F, red lines). Meanwhile, without NGF pretreatment, K252a did not induce any changes in the PIP3 level or Rac1/Cdc42 activity (Fig. S1 D). Therefore, the K252a-induced super-suppression in NGF-treated cells indicated the presence of an NGF-dependent negative regulation of PIP3, Rac1, and Cdc42. We similarly examined the effect of LY294002, a specific PI3-kinase inhibitor. 20 μM LY294002 treatment at 10 min after NGF addition decreased the PIP3 concentration and Rac1/Cdc42 activity to below the basal level for >20 min (Fig. 1, C–F, green lines). The addition of LY294002 without NGF pretreatment also suppressed the PIP3 concentration and Rac1/Cdc42 activity to below the basal level (Fig. S1 E). The kinetics of the LY294002-induced down-regulation of Rac1/Cdc42 activity in unstimulated cells (Fig. S1 E) was comparable to that of NGF-stimulated cells (Fig. 1, E and F). This observation excludes GTPase-activating proteins (GAPs) for Rac1 and Cdc42 from the potential NGF-dependent negative regulator.
Figure 1.
Perturbation of PIP3 production and Rac1/Cdc42 activation with K252a or LY294002 in NGF-stimulated PC12 cells. (A–C) PC12 cells expressing Pippi-PIP3, Raichu-Rac1, or Raichu-Cdc42 were starved for 6 h and then treated with 50 ng/ml NGF. In B and C, the cells were treated with 10 nM K252a (B) or 20 μM LY294002 (C) at 10 min after NGF addition. Images were obtained every 1 min for 30 min after NGF stimulation. Representative ratio images of FRET/CFP at the indicated time points (in minutes) after NGF addition are shown in the intensity-modulated display mode. In the intensity-modulated display mode, eight colors from red to blue are used to represent the FRET/CFP ratio, with the intensity of each color indicating the mean intensity of FRET and CFP. FRET/CFP ratio images were shown after normalization with a reference value acquired from images of unstimulated cells. The upper and lower limits of fold values are shown on the right. Bars, 10 μm. (D–F) The mean FRET/CFP ratios over the whole cell are expressed by measuring the relative increase as compared with the reference value, which was averaged over 10 min before NGF addition. The blue, red, or green line indicates the result of treatment with NGF (N), NGF followed by K252a (N→K), or NGF followed by LY294002 (N→LY), respectively. The number of experiments is as follows: (D) PIP3 (N, n = 6; N→K, n = 11; N→L, n = 11), (E) Rac1 (N, n = 11; N→K, n = 9; N→L, n = 5), (F) Cdc42 (N, n = 9; N→K, n = 5; N→L, n = 5). Error bars show SD.
Monitoring of PI(3,4)P2 levels after NGF stimulation
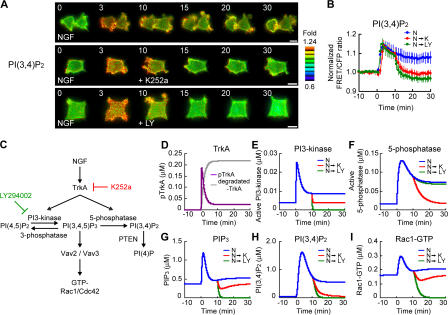
To obtain a clue for identifying the negative regulators of PIP3, Rac1, and Cdc42, we examined the change in phosphatidylinositol (3,4) bisphosphate (PI[3,4]P2) level after NGF stimulation. The basic structure of the FRET probe for PI(3,4)P2 named Pippi-PI(3,4)P2 (Yoshizaki et al., 2007) was identical to that of the PIP3 reporter Pippi-PIP3/fllip-pm (Sato et al., 2003) except that the pleckstrin homology (PH) domain of TAPP1, which binds specifically to PI(3,4)P2 (Dowler et al., 2000), was substituted for the PH domain of general receptor of phosphoinositides (GRP). Upon NGF stimulation of PC12 cells, PI(3,4)P2 was rapidly produced and then gradually decreased (Fig. 2, A [top] and B [blue line]). The spatiotemporal pattern of PI(3,4)P2 production was different from that of PIP3; the increase in PI(3,4)P2 showed a wide distribution throughout the cells and was sustained above the basal level for >30 min. K252a or LY294002 treatment at 10 min after NGF stimulation gradually decreased PI(3,4)P2 to or below the basal level, respectively (Fig. 2, A [middle and bottom] and B [red and green lines]). A PI-5-phosphatase generates PI(3,4)P2 from PIP3. If this 5-phosphatase activity is NGF independent, the time course of PI(3,4)P2 level after K252a treatment should be similar to that of PIP3 level. However, this was not the case. Thus, the 5-phosphatase is a candidate for NGF-dependent negative regulator of PIP3.
Figure 2.
PI(3,4)P2 production during NGF stimulation and in silico analysis of the NGF–PIP3–Rac1 signaling cascade. (A) PC12 cells expressing Pippi-PI(3,4)P2 were stimulated with NGF. Then, the cells were either untreated (top) or treated with K252a (middle) or LY294002 (bottom) at 10 min after NGF addition. Representative images of normalized FRET/CFP ratios of Pippi-PI(3,4)P2 are shown at the indicated time points (in minutes) as described in the legend to Fig. 1 (A–C). Bars, 10 μm. (B) The means of normalized FRET/CFP ratios of Pippi-PI(3,4)P2 (N, n = 10; N→K, n = 9; N→LY, n = 9) are expressed as in the legend to Fig. 1 (D–F). Error bars show SD. (C) Model of the NGF–PIP3–Rac1/Cdc42 signaling cascade. TrkA activates PI3-kinase to generate PIP3, which activates Rac1 and Cdc42. On the other hand, TrkA simultaneously activated PI-5-phosphatase to down-regulate PIP3 levels. In silico, K252a or LY294002 inhibits the binding of PI3-kinase and PI-5-phosphatase to TrkA or PI3-kinase activity, respectively. (D–I) Computer simulations in the model of C are shown for TrkA (D), active PI3-kinase (E), active PI-5-phosphatase (F), PIP3 (G), PI(3,4)P2 (H), and Rac1-GTP (I). TrkA (D) was set to be phosphorylated (purple) upon NGF stimulation at 0 min and then degraded (gray). The blue, red, or green line (E–I) indicates the result of treatment with NGF (N), NGF followed by K252a (N→K), or NGF followed by LY294002 (N→LY), respectively.
Modeling and computational simulation of the NGF–PIP3–Rac1 signaling pathway
We attempted to construct a simple kinetic model (Fig. 2 C and Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200609017/DC1) that could reproduce the behaviors of PIP3, Rac1, Cdc42, and PI(3,4)P2 in NGF-treated PC12 cells. At first, we determined the concentration of TrkA, p85 (a regulatory subunit of PI3-kinase), Vav2, and Rac1 in PC12 cells (Fig. S2, G and H; and Table I). Then, we constructed the kinetic model by assuming (1) that the putative 3-phosphatase activity on PIP3 was constant with and without NGF, (2) that TrkA activated both PI3-kinase and a putative 5-phosphatase for PIP3 upon NGF stimulation (based on the results in Fig. 1 D and Fig. 2 B), and (3) that the kinetics of the 5-phosphatase for PIP3 (activation and inactivation rates) were slower than that of PI3-kinase (Fig. 2, E and F, blue lines). We simulated this minimal model numerically in silico and demonstrated that the model approximately reproduced the results of our experiments (Fig. 2, G–I). This implies that NGF-induced PI-5-phosphatase activity inhibited an excess production of PIP3 by NGF stimulation and that the constant activity of PI-3-phosphatase determined the basal level of PIP3.
Table I.
The number and concentration of TrkA, p85, Vav2, Rac1, SHIP2, and PTEN in PC12 cells
| Protein | Number per cell | Concentrationa |
|---|---|---|
| μM/cell | ||
| TrkA | 3.2 × 105 | 0.24 |
| p85 | 1.2 × 105 | 0.091 |
| Vav2 | 4.1 × 105 | 0.31 |
| Rac1 | 1.1 × 106 | 0.80 |
| SHIP2 | 4.4 × 105 | 0.33 |
| PTEN | 4.6 × 104 | 0.035 |
Concentration was calculated from the number of molecules per cell volume. The diameter of a suspended PC12 cell was 16.1 μm; therefore, the volume was 2.2 pl.
Depletion of SHIP2 and PTEN and validation of kinetic simulation
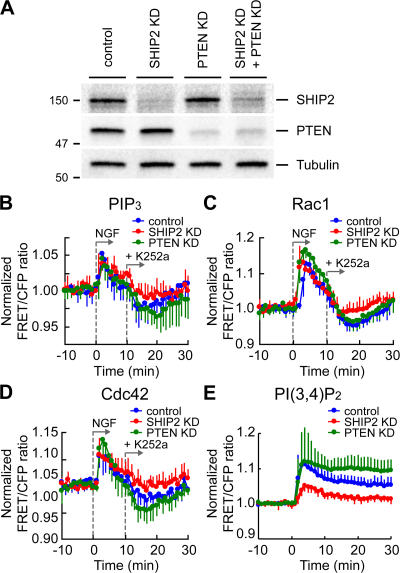
To validate our kinetic model, we used targeted depletion of candidate phosphatases for PIP3 by RNA interference. Candidates for such PI-5- and PI-3-phosphatases include SHIP2 and PTEN, respectively, which are expressed in a wide range of tissues, including neuronal cells. We used a short hairpin RNA (shRNA) expression vector with the pac gene to allow selection of shRNA-expressing cells with puromycin. In SHIP2 or PTEN shRNA-expressing cells, 80–90% of endogenous SHIP2 or PTEN protein was depleted, respectively (Fig. 3 A). In Fig. 3 (B–D), K252a was added to the control or knockdown cells at 10 min after NGF stimulation. In the control and PTEN-depleted cells, K252a treatment induced a transient decrease in the level of PIP3 and in the activity of Rac1/Cdc42 to below the basal level (Fig. 3, B–D, blue and green lines). However, in the SHIP2-knockdown cells, the PIP3 level and Rac1/Cdc42 activity decreased only up to the basal level after K252a addition (Fig. 3, B–D, red lines). Therefore, SHIP2 was responsible for the NGF-dependent negative regulation of PIP3. The data in Fig. 3 E and Fig. S3 (A–C; available at http://www.jcb.org/cgi/content/full/jcb.200609017/DC1) confirmed that SHIP2 and PTEN had a major role in PI(3,4)P2 production and degradation after NGF stimulation of PC12 cells, respectively.
Figure 3.
Depletion of SHIP2 inhibited the K252a-induced decrease in PIP3, Rac1, and Cdc42 to below the basal level. (A) PC12 cells were transfected with an empty pSUPER vector, pSUPER-SHIP2, pSUPER-PTEN, or both pSUPER-SHIP2 and pSUPER-PTEN. After selection with 3 μg/ml puromycin for 2 d, the cells were analyzed by immunoblotting. (B–D) PC12 cells transfected with an empty pSUPER vector (blue line; control), pSUPER-SHIP2 (red line; SHIP2 KD [knockdown]), or pSUPER-PTEN (green line; PTEN KD). After puromycin selection, the cells were further transfected with pPippi-PIP3 (B), pRaichu-Rac1 (C), or pRaichu-Cdc42 (D). After serum starvation for 6 h, the cells were stimulated at 0 min with NGF. Then, K252a was added at 10 min after NGF addition. The means of normalized FRET/CFP ratios are shown as described in the legend to Fig. 1 (D–F). The number of experiments is as follows: (B) PIP3 (control, n = 5; SHIP2 KD, n = 5; PTEN KD, n = 7), (C) Rac1 (control, n = 5; SHIP2 KD, n = 8; PTEN KD, n = 7), (D) Cdc42 (control, n = 5; SHIP2 KD, n = 13; PTEN KD, n = 5). Error bars show SD. (E) In the control (blue), SHIP2-knockdown cells (red), or PTEN-knockdown cells (green), the means of normalized FRET/CFP ratios of Pippi-PI(3,4)P2 after NGF stimulation are shown (control, n = 5; SHIP2 KD, n = 6; PTEN KD, n = 8). Error bars show SD.
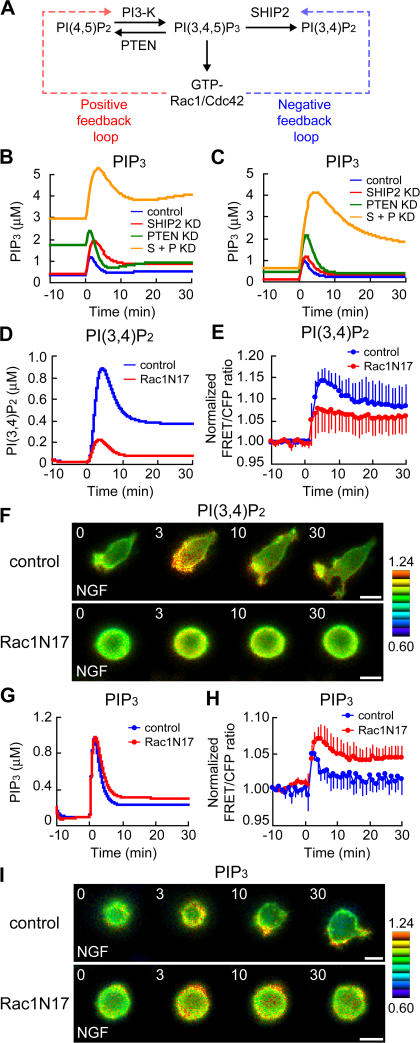
Hyperaccumulation of PIP3 and overactivation of Rac1/Cdc42 in NGF-treated PC12 cells deficient for SHIP2 and PTEN
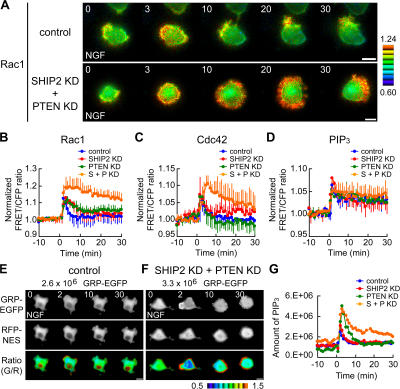
Next, we examined roles of SHIP2 and PTEN in PIP3 production and Rac1/Cdc42 activation after NGF stimulation. In the SHIP2 and PTEN double-knockdown cells, marked and prolonged activation of Rac1 and Cdc42 was observed upon NGF stimulation, although there was no substantial difference amongst the control, SHIP2-knockdown, and PTEN-knockdown cells (Fig. 4, A–C). Furthermore, the SHIP2 and PTEN double-knockdown cells exhibited a sustained lamellipodial structure and delayed localization of Rac1 activity (Fig. 4 A). However, we could not detect a further increase in PIP3 levels, as monitored by Pippi-PIP3, in the knockdown cells as compared with the control cells (Fig. 4 D). This seemed strange considering the results on Rac1/Cdc42 activity in double-knockdown cells (Fig. 4, A–C); thus, we assumed that the PIP3 production in these cells exceeded the upper limit of detection by Pippi-PIP3. To overcome this difficulty, we developed another method to semiquantitatively measure the amount of PIP3 using EGFP-tagged GRP (for detail, see Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200609017/DC1). Fig. 4 E indicates that the control cells showed a transient translocation of GRP-EGFP by 2 min after the addition of NGF. In contrast, the SHIP2 and PTEN double-knockdown cells demonstrated a massive and sustained translocation of GRP-EGFP upon NGF stimulation (Fig. 4 F). Fig. 4 G shows the time course for the amount of PIP3 measured during NGF treatment. This demonstrates that the depletion of both SHIP2 and PTEN caused the sustained overproduction of PIP3 (orange line). The SHIP2-knockdown cells (red line) showed no difference from the control cells (blue line), whereas in the PTEN-knockdown cells (green line), a transient PIP3 overproduction was observed in the early phase after NGF treatment (<10 min).
Figure 4.
Increase in Rac1/Cdc42 activity and PIP3 production by depletion of SHIP2 and PTEN. (A) PC12 cells were transfected with pSUPER vector (top) or both pSUPER-SHIP2 and pSUPER-PTEN (bottom). After selection with puromycin, the cells were further transfected with pRaichu-Rac1. After serum starvation, images were obtained for 30 min after NGF stimulation. Representative images of the normalized FRET/CFP ratios at the indicated time points (in minutes) are shown as described in the legend to Fig. 1 (A–C). Bars, 10 μm. (B–D) In control (blue), SHIP2-knockdown (SHIP2 KD; red), PTEN-knockdown (PTEN KD; green), or SHIP2 and PTEN double-knockdown cells (S + P KD; orange), the means of normalized FRET/CFP ratios of Rac1 (B), Cdc42 (C), and PIP3 (D) are expressed as in the legend to Fig. 1 (D–F). The number of experiments is as follows: (B) control, n = 6; SHIP2 KD, n = 6; PTEN KD, n = 8; S + P KD, n = 5; (C) control, n = 5; SHIP2 KD, n = 5; PTEN KD, n = 5; S + P KD, n = 5; (D) control, n = 6; SHIP2 KD, n = 7; PTEN KD, n = 7; S + P KD, n = 6. Error bars show SD. (E and F) NGF-dependent translocation of GRP-EGFP from the cytosol to the plasma membrane was imaged. In the control (E) or SHIP2 and PTEN double-knockdown cells (F), the localization of GRP-EGFP (top) and RFP-NES (middle) were shown at the indicated time points after NGF stimulation. Total fluorescent intensity of RFP over the whole cell was normalized to that of GFP. These two images were used to obtain the raw number of translocated proteins. Ratio images of GFP/normalized RFP are shown in the intensity-modulated display mode (bottom). Upper and lower limits of ratio images are 1.5 and 0.5, respectively. (G) In control (blue), SHIP2-knockdown (red), PTEN-knockdown (green), or SHIP2 and PTEN double-knockdown cells (orange), the amount of PIP3 is plotted against time. The amount of PIP3 was obtained as an asymptotic value using the raw number of translocated proteins based on the theoretical model.
NGF-dependent negative feedback loop from Rac1 to SHIP2
To reproduce the temporal pattern of PIP3 production in knockdown cells (Fig. 4 G), we improved the initial model by introducing NGF-dependent positive and negative feedback loops (Fig. 5 A). In SHIP2- and PTEN-knockdown cells of Fig. 4 G, the level of PIP3 was transiently increased upon NGF stimulation and returned to the basal levels during the late phase (>10 min). However, this was not reproduced in our initial model (Fig. 5 B). To resolve this contradiction, we added a negative feedback loop from Rac1 to SHIP2 to our model (Fig. 5 A). In this improved model, computer simulations of SHIP2- and PTEN-knockdown cells reproduced a return to the basal level in the late phase (Fig. 5 C). A transient PIP3 overproduction in the early phase of NGF stimulation in PTEN-knockdown cells was also reproduced. Furthermore, the depletion of both SHIP2 and PTEN in the improved model showed an overproduction of PIP3 upon NGF stimulation, as observed in Fig. 4 G.
Figure 5.
NGF-dependent negative feedback loop. (A) Schematic diagram of the NGF-dependent positive and negative feedback loops. (B and C) Results of computer simulation for PIP3 based on the models of Fig. 2 C (B) and Fig. 5 A (C). In knockdown cells, 80% of SHIP2 and/or PTEN was depleted in silico. The blue, red, green, or orange line indicates the control, SHIP2-knockdown (SHIP2 KD), PTEN-knockdown (PTEN KD), or SHIP2 and PTEN double-knockdown cells (S + P KD), respectively. (D–F) In silico (D) or in vivo (E and F) effects of the expression of Rac1N17 on PI(3,4)P2. (E) In control (blue; n = 8) or Rac1N17-expressing cells (red; n = 8), the means of normalized FRET/CFP ratios of Pippi-PI(3,4)P2 are shown as described in the legend to Fig. 1 (D and E). Error bars show SD. (F) Representative images of normalized FRET/CFP ratios of Pippi-PI(3,4)P2 in the control (top) or Rac1N17-expressing cells (bottom) are shown at the indicated time points (in minutes). The Rac1N17-expressing cells were identified by a red fluorescence derived from dsFP593. Bars, 10 μm. (G–I) In silico (G) or in vivo (H and I) effects of the Rac1N17 expression on PIP3. (H) In control (blue; n = 6) or Rac1N17-expressing cells (red; n = 5), the means of normalized FRET/CFP ratios of Pippi-PIP3 are shown. Error bars show SD. (F) Representative images of normalized FRET/CFP ratios of Pippi-PIP3 in the control (top) or Rac1N17-expressing cells (bottom) are shown at the indicated time points (in minutes). Bars, 10 μm.
Next, we attempted to provide evidence of this negative feedback from Rac1 to SHIP2. The production of PI(3,4)P2 and PIP3 in PC12 cells expressing the dominant-negative mutant of Rac1 (Rac1N17) was examined. Our improved model predicted that the expression of Rac1N17 would inhibit the up-regulation of SHIP2 by Rac1, leading to a decrease in PI(3,4)P2 production and an increase in PIP3 production (Fig. 5, D and G). We monitored the change of PI(3,4)P2 levels in PC12 cells expressing Rac1N17 and demonstrated that PI(3,4)P2 production induced by NGF treatment was decreased as compared with the control cells (Fig. 5, E and F). Further, NGF-induced PIP3 production was increased in the Rac1N17-expressing cells (Fig. 5, H and I). Interestingly, the local accumulation of PIP3 was not observed in Rac1N17-expressing cells (Fig. 5 I), suggesting the involvement of SHIP2 in localizing the PIP3 production during NGF treatment.
Excess number and length of neurites in PC12 cells deficient for SHIP2 and PTEN
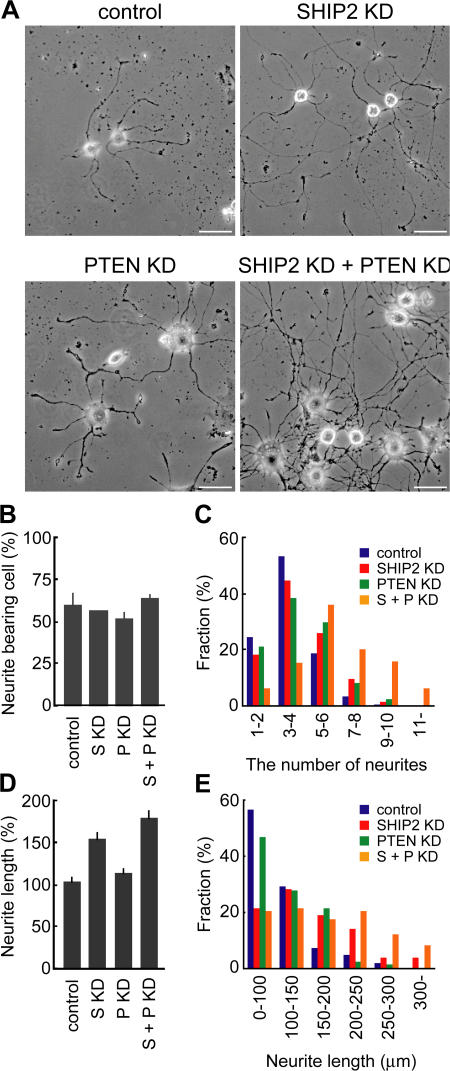
We examined the effect of depletion of SHIP2 and PTEN on NGF-induced neurite outgrowth in PC12 cells. In the absence of NGF, depletion of SHIP2 and/or PTEN did not induce any neurites in PC12 cells (Fig. S3 D). Within 60 h of treatment with NGF, the control PC12 cells transfected with an empty pSUPER vector developed neurites (Fig. 6 A, top left). It is remarkable that depletion of both SHIP2 and PTEN caused a considerable increase in the number and the length of neurites extending from cell bodies (Fig. 6 A, bottom right). The proportions of neurite-bearing cells showed no clear difference among the control and single- and double-knockdown cells (Fig. 6 B). However, the depletion of both SHIP2 and PTEN obviously increased the fraction of cells having more than five neurites, although there was no clear difference between the control and single-knockdown cells (Fig. 6 C). This increase in the neurite number of double-knockdown cells seemed consistent with the sustained overproduction of PIP3 and the overactivation of Rac1 and Cdc42 during NGF treatment in these cells. We noted that differentiated PC12 cells deficient for SHIP2 had longer neurites as compared with controls (Fig. 6, D and E). The length of neurites in double-knockdown cells was even longer than that in the SHIP2-knockdown cells.
Figure 6.
Increase in the number and the length of neurites by depletion of SHIP2 and PTEN. PC12 cells were transfected with the indicated pSUPER constructs and incubated with puromycin for 2 d. The selected cells were then cultured with NGF for 60 h and fixed for microscopy. (A) Representative phase-contrast images of control (top left), SHIP2-knockdown (top right), PTEN-knockdown (bottom left), and SHIP2 and PTEN double-knockdown cells (bottom right). Bars, 50 μm. (B) Cells having neurites whose lengths were twofold longer than their cell body lengths were scored as neurite bearing. At least 50 cells were counted in each experiment, and the experiments were repeated three times. The results are expressed as the mean percentage of neurite-bearing cells with SD. (C) The number of neurites in neurite-bearing cells was divided into six segments, as indicated under the horizontal axis. A histogram plotting the percentage of cell number in each segment is presented for control (blue; n = 173), SHIP2-knockdown (red; n = 143), PTEN-knockdown (green; n = 135), and double-knockdown (orange; n = 182) cells. (D) A bar graph shows the mean length of the longest neurites in the neurite-bearing cells with SEM. The neurite length in the control cells was taken as 100%. The number of cells analyzed in each experiment is as follows: control, n = 106; SHIP2 knockdown, n = 96; PTEN knockdown, n = 75; double knockdown, n = 74. (E) The length of neurites in neurite-bearing cells was divided into six segments, as indicated under the horizontal axis. A histogram plotting the percentage of cell number in each segment is presented as in C.
To further explore the role of SHIP2 and PTEN on the number and the length of neurites, we varied the intervals between the transfection of shRNA vectors and NGF stimulation (Fig. S3, E–H). The decay of SHIP2 and PTEN after the transfection was determined in a preliminary experiment (Fig. S3 D). When pSUPER-SHIP2 and pSUPER-PTEN were transfected on the day of NGF stimulation (KD 0 d), neither the number nor the length of neurites did not increase, whereas the number and the length of neurites increased substantially when the shRNA vectors were transfected 2 or 3 d before NGF stimulation (KD 2 and 3 d; Fig. S3, F–H). Intriguingly, the transfection of the shRNA vectors 1 d before NGF stimulation increased the neurite length without affecting its number (Fig. S3, F–H). These observations indicate that the increase in PIP3 and the following cytoskeletal changes observed immediately after NGF stimulation play a pivotal role in the determination of the number of neurites in the differentiated PC12 cells.
NGF-dependent positive feedback loop from Rac1 to PI3-kinase
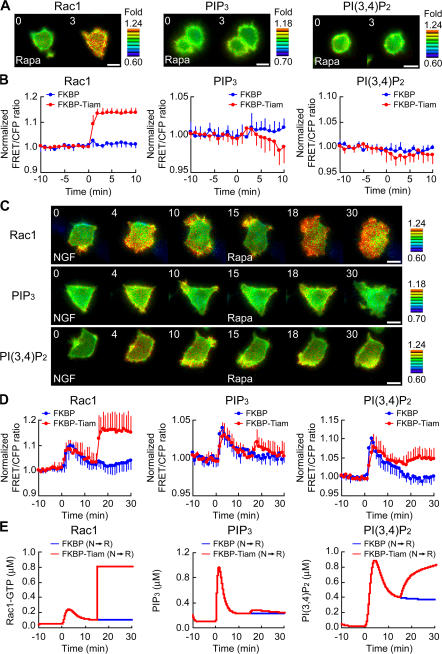
We investigated Rac1-mediated PI3-kinase activation by using the inducible translocation technique to rapidly activate Rac1 (Inoue et al., 2005). Rapamycin triggers the heterodimerization of a Lyn N-terminal sequence-tagged FRB (LDR) with an FKBP-fused protein. FKBP-Tiam1 translocated to the plasma membrane after rapamycin addition and activated Rac1 within 2 min (Fig. 7 A, left). However, we could not detect a rapamycin-triggered production of PIP3 (Fig. 7 A, middle) or PI(3,4)P2 (Fig. 7 A, right). These findings clearly demonstrated that Rac1 activation alone is not sufficient for PI3-kinase activation and argue against a positive feedback loop composed of Rac1 and PI3-kinase. Therefore, we next treated the PC12 cells expressing LDR, FKBP-Tiam1, and the FRET probe (Raichu-Rac1, Pippi-PIP3, or Pippi-PI[3,4]P2) with rapamycin at 15 min after NGF addition. Again, Rac1 was rapidly activated by rapamycin-triggered Tiam1 translocation within 2 min (Fig. 7, C [top] and D [left]). Of note, PIP3 was also increased by the rapamycin-triggered activation of Rac1 within 3 min (Fig. 7, C and D, middle). This provided direct evidence of an NGF-dependent positive regulation from Rac1 to PI3-kinase. Although rapamycin-triggered Rac1 activation was prolonged for >15 min (Fig. 7 C, top), rapamycin- triggered PIP3 production was transient (Fig. 7 C, middle). Meanwhile, PI(3,4)P2 was gradually produced by rapamycin-triggered Rac1 activation (Fig. 7, C [bottom] and D [right]). To explain this observation in our kinetic simulation model, we integrated the positive feedback loop from Rac1 to PI3-kinase and its dependency to NGF by assuming that the production of PIP3 is partly catalyzed by the complex of Rac1 and phosphorylated PI3-kinase. According to the rapid activation of Rac1 at 15 min after NGF addition in silico (Fig. 7 E, left, red line), PIP3 was generated via the positive feedback from Rac1 to PI3-kinase and then degraded by the negative feedback from Rac1 to SHIP2, which gradually produced PI(3,4)P2 (Fig. 7 E, middle and right, red lines). These computational and experimental studies together supported the existence of NGF-dependent positive and negative feedback loops in PC12 cells (Fig. 8 A).
Figure 7.
NGF-dependent positive feedback loop. (A) LDR and FKBP (blue) or FKBP-Tiam1 (red) were transfected with pRaichu-Rac1 (left), pPippi-PIP3 (middle), or pPippi-PI(3,4)P2 (right). After serum starvation, the cells were treated with 50 nM rapamycin. The means of normalized FRET/CFP ratios are shown as described in the legend to Fig. 1 (D and E). The number of experiments is as follows: Rac1 (FKBP, n = 3; FKBP-Tiam1, n = 7), PIP3 (FKBP, n = 5; FKBP-Tiam1, n = 7), PI(3,4)P2 (FKBP, n = 6; FKBP-Tiam1, n = 8). Error bars indicate SD. (B) PC12 cells expressing LDR, FKBP-Tiam1, and the indicated FRET probe were serum starved, stimulated with NGF, and treated with rapamycin at 15 min after NGF addition. Representative images of normalized FRET/CFP ratios of Rac1 (top), PIP3 (middle), and PI(3,4)P2 (bottom) are shown at the indicated time points (in minutes). Bars, 10 μm. (C) PC12 cells expressing LDR, FKBP (blue), or FKBP-Tiam1 (red) and the indicated FRET probe were serum starved, stimulated with NGF, and treated with rapamycin at 15 min after NGF addition. The means of normalized FRET/CFP ratios of Rac1 (left), PIP3 (middle), and PI(3,4)P2 (right) are shown. The number of experiments is as follows: Rac1 (FKBP, n = 10; FKBP-Tiam1, n = 5), PIP3 (FKBP, n = 7; FKBP-Tiam1, n = 8), PI(3,4)P2 (FKBP, n = 8; FKBP-Tiam1, n = 6). Error bars indicate SD. (D) Results of computer simulation for Rac1 (left), PIP3 (middle), and PI(3,4)P2 (right) are shown. Blue or red lines indicate the results of NGF addition (at 0 min) followed by rapamycin treatment (at 15 min) in FKBP- or FKBP-Tiam1–expressing cells, respectively.
Figure 8.
Mechanism of determining neurite-budding sites in PC12 cells. (A) In response to NGF, NGF/TrkA signaling drives a cycling of positive and negative feedback loops to control spatiotemporal PIP3 level and Rac1/Cdc42 activity. All reactions and parameters are listed in Fig. S3 (available at http://www.jcb.org/cgi/content/full/jcb.200609017/DC1). (B and C) PC12 cells expressing Vav2-PAGFP, SHIP2-PAGFP, or PTEN-PAGFP were treated with NGF for 3–30 min. At time 0, PA-GFP proteins in a region of interest in NGF-induced protrusions were photoactivated by UV irradiation (Fig. S5, G and H). (B) Mean of relative fluorescence intensities in the region of interest after photoactivation (PA) was plotted with SD. In C, the bars represent the mean ± SD of time constant (1/e) of Vav2-PAGFP (red; n = 5), SHIP2-PAGFP (blue; n = 8), and PTEN-PAGFP (green; n = 7). (D) A hypothetical model of neuritogenesis in PC12 cells. During initial neuritogenesis, positive and negative feedback loops at cell periphery generate a local activation of PI3-kinase (red) and a long-range activation of SHIP2 (blue), respectively. PTEN activity is uniformly distributed (green). As a result, a sharp peak of PIP3 level and Rac1/Cdc42 activity (purple) is generated and promotes the budding of neurites.
Mechanism of initial budding of neurites in PC12 cells
Finally, we investigated the mechanism of determining neurite-budding sites from cell bodies. During the chemotaxis of Dictyostelium, the combination of a PIP3-dependent positive feedback at the cell front and localization of PTEN at the side and back of the cell has been shown to establish cell polarity (Iijima et al., 2002; Merlot and Firtel, 2003). In PC12 cells, Vav2 and Vav3 were translocated to the plasma membrane in response to NGF (Aoki et al., 2005). Thus, we examined the localization of SHIP2 and PTEN during the neuritogenesis. Upon NGF stimulation, SHIP2 was rapidly recruited to lamellipodia and protrusions, whereas PTEN did not show any translocation (Fig. S5, A–F, available at http://www.jcb.org/cgi/content/full/jcb.200609017/DC1). This observation indicates that the mechanism of initial neurite budding in PC12 cells is different from that of the polarity formation in Dictyostelium. On the other hand, the existence of both positive and negative feedback loops in the induction of neurites raises the possibility that the polarity of PC12 cells may be generated by Turing's reaction-diffusion system (Fig. 8 A); the combination of local positive feedback loop and long-range negative feedback loop generates a spatial pattern autonomously (Meinhardt, 1999; Meinhardt and Gierer, 2000). Therefore, we examined the diffusion rate of a positive regulator, Vav2, and negative regulators, SHIP2 and PTEN, in the NGF-induced protrusions, where these feedbacks were operating. We found that the diffusion of Vav2 was slower than those of SHIP2 and PTEN (Fig. 8, B and C; and Fig. S5, G and H), supporting the model that the polarity of PC12 cells was generated by Turing's reaction-diffusion system (Fig. 8 D).
Discussion
In this study, we have shown that the PI-5-phosphatase SHIP2 constitutes a critical component of a negative feedback loop regulating the PIP3 level in NGF-stimulated PC12 cells and that the PIP3 level in PC12 cells is primarily regulated by SHIP2 and PTEN, a constitutively active PI-3-phosphatase. This proposal is supported by the finding that the double knockdown of SHIP2 and PTEN caused a PIP3 overproduction, a Rac1/Cdc42 overactivation, and an increase in the number and length of neurites in NGF-treated PC12 cells, whereas no marked change was observed in SHIP2 or PTEN single-knockdown cells. Our finding is in line with previous reports showing that the depletion of PTEN by antisense oligonucleotide did not affect the NGF-induced neurite outgrowth (Lachyankar et al., 2000) but that overexpression of PTEN blocks NGF-induced neurite outgrowth of PC12 cells (Musatov et al., 2004). Meanwhile, SHIP2 has been shown to be phosphorylated and form a complex with Shc after NGF treatment (Habib et al., 1998). This finding agrees with our observation that depletion of SHIP2 impaired PI(3,4)P2 generation upon NGF stimulation (Fig. 3 E).
Based on the identification of the SHIP2-mediated negative feedback loop and our previous finding that a positive feedback loop composed of PI3-kinase, Vav2/Vav3, and Rac1/Cdc42 is operated in NGF-treated PC12 cells (Aoki et al., 2005), we presumed that the initial budding sites of neurites from cell bodies may be determined by the Turing's reaction-diffusion system. In this system, long-range lateral inhibition (a negative feedback loop that diffuses rapidly), in conjunction with local self-activation (a positive feedback loop that diffuses slowly), autonomously generates spatial patterns (Meinhardt, 1999; Meinhardt and Gierer, 2000). In accordance with this model, we found that the diffusion of Vav2 is slower than that of SHIP2 in the NGF-induced membrane protrusions. Therefore, NGF stimulation of PC12 cells generates the slowly diffusing positive and the rapidly diffusing negative feedback loops regulating the PIP3 level, which fulfill the conditions of Turing's spatial pattern formation (Fig. 8 D). We propose that this pattern formation in the PIP3 level and the following Rac1/Cdc42 activation contributes to the initial neuritogenesis of PC12 cells.
A positive feedback loop involving PIP3 and Rac1 has also been demonstrated in leukocytes (Wang et al., 2002; Weiner et al., 2002) and Dictyostelium (Merlot and Firtel, 2003); however, because these studies used dominant-negative mutants or inhibitors, direct evidence of the positive feedback loop has not been provided. Here, we directly validated the presence of the positive feedback loop from Rac1 to PI3-kinase using the rapamycin-induced acute activation system. Intriguingly, however, we found that this positive feedback loop only operated in the presence of NGF (Fig. 7 C). Which model can reconcile these findings? Based on a previous report that the p85 regulatory subunit of PI3-kinase binds to GTP-loaded Rac1 through its RhoGAP homology domain (Zheng et al., 1994), we hypothesized that the active PI3-kinase is actually a complex of phosphorylated PI3-kinase and Rac1-GTP. In this way, Rac1-GTP promotes the production of PIP3 by two related mechanisms; Rac1-GTP increases the active complex and competitively inhibits tyrosine phosphatases. In our model integrating this assumption, we could faithfully reproduce the experimental results in silico (Fig. 7, C and D).
To consider the mechanism of the negative feedback from Rac1 to SHIP2, the regulation of OCRL1, another PI-5-phosphatase, may provide some clues. After growth factor stimulation, OCRL1 has been shown to bind to GTP-Rac1 through its RhoGAP domain and to be translocated to the plasma membrane (Faucherre et al., 2005). Although SHIP2 does not bind to GTP-Rac1 directly, SHIP2 binds to several proteins associated with cytoskeletal reorganization, i.e., Cas, Filamin, and Vinexin (Dyson et al., 2001; Prasad et al., 2001; Paternotte et al., 2005). Because Rac1 stimulates actin assembly, it is possible that these SHIP2 binding proteins are relocated to the actin accumulation sites, accompanied by SHIP2 recruitment, where SHIP2 can then dephosphorylate PIP3 at these sites.
In morphogenesis, similar but different mechanisms exist that coordinate multiple signaling pathways, depending on the cellular context. During the chemotaxis of Dictyostelium, the combination of a PIP3-dependent positive feedback loop at the cell front and localization of PTEN at the side and back of the cell has been shown to establish cell polarity and promote the forward movement (Iijima et al., 2002; Merlot and Firtel, 2003). On the other hand, the present study demonstrates the presence of PIP3-dependent positive and negative feedback loops in NGF-stimulated neuritogenesis. Further differences can be pointed out. In neutrophils, the PIP3-dependent positive feedback loop is independent of receptor-mediated signals because the delivery of exogenous PIP3 into these cells induces the formation of cell polarity (Weiner et al., 2002). In contrast, an NGF-TrkA signal was required for the positive feedback loop in PC12 cells (Fig. 7).
During the elongation process, SHIP2 and PTEN down-regulated neurite protrusion (Fig. 6). Considering the negative feedback from Rac1 to SHIP2, the inhibitory role of SHIP2 should be prominent at the neurite tips, which showed the recurrent activation of Rac1 during neurite elongation (Aoki et al., 2004). In fact, the depletion of SHIP2 considerably increased the length of neurites (Fig. 6, D and E). The inhibitory role of the PIP3 phosphatases might be involved in the repeated resetting of the concentration of PIP3 to the basal levels to allow sensing and adapting to an environment by changing the extending direction, although neurite tips of PC12 cells have lost such guidance ability during transformation.
We developed a method to measure the level of PIP3 semiquantitatively in living cells. FRET efficiency obtained by Pippi-PIP3 is dependent on both the amount of endogenous PIP3 and the expression level of the FRET probe. In addition, the detection range of Pippi-PIP3 is affected by the dissociation constant of the PH domain for PIP3, like Ca2+ indicators. In Fig. 4 D, we could not observe the enhancement of NGF-induced PIP3 production in the SHIP2 and PTEN double-knockdown cells, suggesting that the level of PIP3 in these cells exceeded the upper limit of detection of Pippi-PIP3. Although many groups use the translocation of the PH domain to measure the level of phosphoinositides (Lemmon and Ferguson, 2000; Cullen et al., 2001; Halet, 2005), the amount of translocation strongly depends on the expression level of the PH domain and cell morphology. Further, Akt phosphorylation is used as an indicator of PIP3 levels. However, the level of Akt phosphorylation reflects the balance of upstream kinases and phosphatases, and not the level of PIP3. Our new method of PIP3 quantification used in this study basically utilizes the dose dependency of PIP3-induced translocation of GRP and obtains the amount of PIP3 as an asymptotic value. Thereby, this method overcomes the aforementioned problems.
In this study, we used in silico simulation of kinetic reactions as a tool to help generate hypotheses and validate the reliability of experimental results. As shown here, combining in vivo analyses with in silico simulation helps us understand the systemic properties of signaling networks. In fact, many groups have been trying to dissect complex signaling pathways using computational simulation (Wiley et al., 2003; Bhalla, 2004; Kholodenko, 2006). Here, we built a simple kinetic model of NGF–PIP3–Rac1 signaling. Most of the parameters were estimated arbitrarily except the concentration of all proteins. And no spatial information was taken into account in this model, although many reactions in NGF–PIP3–Rac1 signaling occur on the plasma membrane. Nevertheless, this model could reproduce the results of several experiments, i.e., perturbation with inhibitors (K252a and LY294002) and knockdown experiments (SHIP2 and PTEN). Therefore, we emphasize a potential of in silico analysis as a powerful tool in the research of complicated signal transduction pathways.
Materials and methods
FRET probes
The plasmids encoding the FRET probes, Raichu-Rac1/1011x, Raichu-Cdc42/1054x (Itoh et al., 2002), Pippi-PIP3/fllip-pm (Sato et al., 2003; Aoki et al., 2005), and Pippi-PI(3,4)P2 (Yoshizaki et al., 2007) have been described previously.
Plasmids
The RNA targeting constructs were generated using pSUPER.retro.puro vector (OligoEngine). The 19-nucleotide sequences used to target rat SHIP2 and PTEN mRNAs were 5′-GGCCTACATTGAGTTTGAG-3′ and 5′-GTCAGAGGCGCTATGTATA-3′, respectively. Rat GRP cDNA was obtained by RT-PCR and subcloned into the pCXN2-FLAG vector fused with EGFP at the C terminus (Fujioka et al., 2006). pCAGGS-ERed-NES encoded Express red (Invitrogen) fused to nuclear export sequence of HIV-1 rev protein (LQLPPLERLTLD). pIRM21-FLAG-Rac1N17 has been described previously (Aoki et al., 2004). The cDNAs for LDR (Lyn11-targeted FRB), FKBP, and FKBP-Tiam1 (Inoue et al., 2005) were prepared by PCR and subcloned into pERedNLS and pCAGGS-3HA (Aoki et al., 2005). The cDNAs for TrkA, p85, Vav2, SHIP2, PTEN, and Rac1 were subcloned into pCAGGS-EGFP and pCXN2-Flag-EGFP. The cDNA for PA-GFP (Patterson and Lippincott-Schwartz, 2002), which was provided by T. Nagai (Hokkaido University, Hokkaido, Japan), was used to prepare pCAGGS-Flag-Vav2-PAGFP, pCAGGS-Flag-SHIP2-PAGFP, and pCAGGS-Flag-PTEN-PAGFP.
Cells, reagents, and antibodies
PC12 cells were maintained in RPMI (Invitrogen) supplemented with 10% horse serum and 5% fetal bovine serum. The cells were plated on 35-mm glass-based dishes (Asahi Techno Glass), which were coated with polyethyleneimine (Sigma-Aldrich). NGF and K252a were purchased from Calbiochem. LY294002 and puromycin were obtained from Sigma-Aldrich. Rapamycin was purchased from LC Laboratories. Fluorescent microsphere for calibration of EGFP proteins was a gift from S. Okabe (Tokyo Medical and Dental University, Tokyo, Japan; Sugiyama et al., 2005). Anti-GFP rabbit serum was prepared in our laboratory (Ohba et al., 2003). Anti-Trk (I-20), anti-Vav2 (H-200), and anti-SHIP2 (I-20) polyclonal antibodies were purchased from Santa Cruz Biotechnology, Inc. Anti-PTEN, anti-Akt, anti–phospho-Akt (Thr308), and anti–phospho-TrkA (Tyr490) were obtained from Cell Signaling Technology, and anti-tubulin monoclonal antibody was obtained from Calbiochem. Anti-Rac1, anti- Cdc42, and anti-p85 monoclonal antibodies came from BD Biosciences.
RNA interference experiments
PC12 cells were transfected with the desired pSUPER constructs by using Lipofectamine 2000 (Invitrogen). After recovery, the cells were selected by a 2-d incubation with 3 μg/ml puromycin and then used for further analysis. For FRET imaging, the indicated pRaichu plasmids were transfected into the shRNA-expressing cells 1 d after the addition of puromycin as described. After an additional 1-d incubation with puromycin, the cells were starved and used for imaging.
Time-lapse FRET imaging
PC12 cells expressing FRET probes were starved for 6–12 h with phenol red–free DME/F12 medium containing 0.1% BSA and then treated with 50 ng/ml NGF. The medium was covered with mineral oil (Sigma-Aldrich) to preclude evaporation. Cells were imaged with an inverted microscope (IX81; Olympus) at 37°C, equipped with a cooled charge-coupled device camera (Cool SNAP-HQ; Roper Scientific), a laser-based auto-focusing system (IX2-ZDC; Olympus), and an automatically programmable XY stage (MD-XY30100T-Meta; SIGMA KOKI), which allowed us to obtain time-lapse images of several view fields in a single experiment. The filters used for the dual-emission imaging were obtained from Omega Optical: an XF1071 (440AF21) excitation filter, an XF2034 (455DRLP) dichroic mirror, and two emission filters (XF3075 [480AF30] for CFP and XF3079 [535AF26] for FRET). Cells were illuminated with a 75-W Xenon lamp through a 12% ND filter and viewed through a 60× oil-immersion objective lens (PlanApo 60×/1.4). The exposure times for 4 × 4 binning were 400 ms for CFP and FRET images and 100 ms for differential interference contrast images. After background subtraction, FRET/CFP ratio images were created with MetaMorph software (Universal Imaging Corp.), and the images were used to represent FRET efficiency. FRET/CFP ratio images were shown after normalization as follows. First, in each sample, we determined the mean ratio over the whole cell before NGF addition and used that ratio as the reference value. Then, the raw FRET/CFP ratio of each pixel was divided by the reference value, and this normalized value was used to generate a normalized ratio image.
Neurite extension assay
PC12 cells were transfected with the indicated pSUPER constructs and selected with 3 μg/ml puromycin for 2 d. Then, neurite outgrowth was stimulated with 50 ng/ml NGF and allowed to proceed for 60 h in DME/F12 medium containing 0.1% BSA and 3 μg/ml puromycin. Quantification of neurite outgrowth was performed as described previously (Nakamura et al., 2002).
Photoactivation experiments
PC12 cells on polyethyleneimine-coated 35-mm glass-based dishes were transfected with PA-GFP fusion plasmids. After a 36-h incubation, the cells were serum starved for 6 h and stimulated with 50 ng/ml NGF. The cells were imaged with a confocal microscope (FV1000; Olympus) equipped with an argon laser, a 405-nm laser diode, and a 60× oil-immersion objective lens (UPlanSApo). For photoactivation, a region of interest (3-μm diameter) in an NGF-induced protrusion was illuminated with 405-nm laser light. Images were obtained every 0.065 s by illuminating with the argon laser. The filters used were a DM405/488 dichroic mirror and an emission filter (BA505IF; Olympus).
Numerical simulation of biochemical reactions and block diagram
All reactions were represented by linear molecule–molecule interactions and enzymatic reactions. The biochemical reactions and the rate constants used in this study are shown in Fig. S3 and Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200609017/DC1), respectively. GENESIS simulator (version 2.2) with a Kinetikit interface (version 9.0) was used for solving the ordinary differential equations with a time step of 100 ms as described previously (Sasagawa et al., 2005; Fujioka et al., 2006).
Online supplemental material
Fig. S1 shows the NGF-induced phosphorylation of TrkA in various conditions and the effect of K252a or LY294002 treatment on Rac1/Cdc42 activity and the levels of PIP3 and PI(3,4)P2 in the absence of NGF. Fig. S2 shows the schematic representation of all reactions in the model of the NGF–PIP3–Rac1 signaling pathway and the quantification of endogenous TrkA, p85, Vav2, SHIP2, PTEN, and Rac1. Fig. S3 shows the NGF-induced PI(3,4)P2 production in control, SHIP2- or PTEN-depleted PC12 cells, and the effect of depletion of SHIP2 and PTEN on neurite outgrowth of PC12 cells. Fig. S4 depicts the method of semiquantitative measurement of intracellular PIP3 levels. Fig. S5 demonstrates the subcellular localization of SHIP2 and PTEN in PC12 cells stimulated with NGF and the different diffusion rate among Vav2, SHIP2, and PTEN. Table S1 describes the parameters of the molecule–molecule interactions in the computational model. Table S2 describes the parameters of the enzymatic reactions. Table S3 describes the parameters of transitions. Table S4 describes the initial concentration of NGF and PI(4,5)P2 in the computational model. The supplemental text gives details of the quantification of endogenous TrkA, p85, PTEN, SHIP2, Vav2, and Rac1 and the semiquantitative measurement of intracellular PIP3 level. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200609017/DC1.
Supplementary Material
Acknowledgments
We would like to thank S. Okabe for the provision of fluorescent microspheres; N. Yoshida, N. Fujimoto, A. Nishiyama, K. Fukuhara, and Y. Kasakawa for technical assistance; and members of the Matsuda laboratory for their technical advice and helpful input.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the New Energy and Industrial Technology Development Organization, Mitsubishi Foundation, and the Japan Health Science Foundation. K. Aoki was supported by research fellowships from the Japan Society for the Promotion of Science for Young Scientists.
Abbreviations used in this paper: FRET, fluorescence resonance energy transfer; GAP, GTPase-activating protein; GRP, general receptor of phosphoinositides; PH, pleckstrin homology; PI(3,4)P2, phosphatidylinositol (3,4) bisphosphate; PI3-kinase, phosphatidylinositol 3-kinase; PIP3, phosphatidylinositol (3,4,5) trisphosphate; PTEN, phosphate and tensin homologue; SHIP2, Src homology 2 domain–containing inositol phosphatase 2; shRNA, short hairpin RNA.
References
- Aoki, K., T. Nakamura, and M. Matsuda. 2004. Spatio-temporal regulation of Rac1 and Cdc42 activity during nerve growth factor-induced neurite outgrowth in PC12 cells. J. Biol. Chem. 279:713–719. [DOI] [PubMed] [Google Scholar]
- Aoki, K., T. Nakamura, K. Fujikawa, and M. Matsuda. 2005. Local phosphatidylinositol 3,4,5-trisphosphate accumulation recruits Vav2 and Vav3 to activate Rac1/Cdc42 and initiate neurite outgrowth in nerve growth factor-stimulated PC12 cells. Mol. Biol. Cell. 16:2207–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla, U.S. 2004. Models of cell signaling pathways. Curr. Opin. Genet. Dev. 14:375–381. [DOI] [PubMed] [Google Scholar]
- Bourne, H.R., and O. Weiner. 2002. A chemical compass. Nature. 419:21. [DOI] [PubMed] [Google Scholar]
- Cantley, L.C. 2002. The phosphoinositide 3-kinase pathway. Science. 296:1655–1657. [DOI] [PubMed] [Google Scholar]
- Cullen, P.J., G.E. Cozier, G. Banting, and H. Mellor. 2001. Modular phosphoinositide-binding domains—their role in signalling and membrane trafficking. Curr. Biol. 11:R882–R893. [DOI] [PubMed] [Google Scholar]
- da Silva, J.S., and C.G. Dotti. 2002. Breaking the neuronal sphere: regulation of the actin cytoskeleton in neuritogenesis. Nat. Rev. Neurosci. 3:694–704. [DOI] [PubMed] [Google Scholar]
- Dowler, S., R.A. Currie, D.G. Campbell, M. Deak, G. Kular, C.P. Downes, and D.R. Alessi. 2000. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson, J.M., C.J. O'Malley, J. Becanovic, A.D. Munday, M.C. Berndt, I.D. Coghill, H.H. Nandurkar, L.M. Ooms, and C.A. Mitchell. 2001. The SH2-containing inositol polyphosphate 5-phosphatase, SHIP-2, binds filamin and regulates submembraneous actin. J. Cell Biol. 155:1065–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucherre, A., P. Desbois, F. Nagano, V. Satre, J. Lunardi, G. Gacon, and O. Dorseuil. 2005. Lowe syndrome protein Ocrl1 is translocated to membrane ruffles upon Rac GTPase activation: a new perspective on Lowe syndrome pathophysiology. Hum. Mol. Genet. 14:1441–1448. [DOI] [PubMed] [Google Scholar]
- Fujioka, A., K. Terai, R.E. Itoh, K. Aoki, T. Nakamura, S. Kuroda, E. Nishida, and M. Matsuda. 2006. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J. Biol. Chem. 281:8917–8926. [DOI] [PubMed] [Google Scholar]
- Govek, E.E., S.E. Newey, and L. Van Aelst. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19:1–49. [DOI] [PubMed] [Google Scholar]
- Habib, T., J.A. Hejna, R.E. Moses, and S.J. Decker. 1998. Growth factors and insulin stimulate tyrosine phosphorylation of the 51C/SHIP2 protein. J. Biol. Chem. 273:18605–18609. [DOI] [PubMed] [Google Scholar]
- Halet, G. 2005. Imaging phosphoinositide dynamics using GFP-tagged protein domains. Biol. Cell. 97:501–518. [DOI] [PubMed] [Google Scholar]
- Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science. 279:509–514. [DOI] [PubMed] [Google Scholar]
- Iijima, M., Y.E. Huang, and P. Devreotes. 2002. Temporal and spatial regulation of chemotaxis. Dev. Cell. 3:469–478. [DOI] [PubMed] [Google Scholar]
- Inoue, T., W.D. Heo, J.S. Grimley, T.J. Wandless, and T. Meyer. 2005. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, R.E., K. Kurokawa, Y. Ohba, H. Yoshizaki, N. Mochizuki, and M. Matsuda. 2002. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22:6582–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko, B.N. 2006. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 7:165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K., S. Hattori, Y. Kabuyama, Y. Shizawa, J. Takayanagi, S. Nakamura, S. Toki, Y. Matsuda, K. Onodera, and Y. Fukui. 1994. Neurite outgrowth of PC12 cells is suppressed by wortmannin, a specific inhibitor of phosphatidylinositol 3-kinase. J. Biol. Chem. 269:18961–18967. [PubMed] [Google Scholar]
- Kita, Y., K.D. Kimura, M. Kobayashi, S. Ihara, K. Kaibuchi, S. Kuroda, M. Ui, H. Iba, H. Konishi, U. Kikkawa, et al. 1998. Microinjection of activated phosphatidylinositol-3 kinase induces process outgrowth in rat PC12 cells through the Rac-JNK signal transduction pathway. J. Cell Sci. 111:907–915. [DOI] [PubMed] [Google Scholar]
- Kobayashi, M., S. Nagata, Y. Kita, N. Nakatsu, S. Ihara, K. Kaibuchi, S. Kuroda, M. Ui, H. Iba, H. Konishi, et al. 1997. Expression of a constitutively active phosphatidylinositol 3-kinase induces process formation in rat PC12 cells. Use of Cre/loxP recombination system. J. Biol. Chem. 272:16089–16092. [DOI] [PubMed] [Google Scholar]
- Lachyankar, M.B., N. Sultana, C.M. Schonhoff, P. Mitra, W. Poluha, S. Lambert, P.J. Quesenberry, N.S. Litofsky, L.D. Recht, R. Nabi, et al. 2000. A role for nuclear PTEN in neuronal differentiation. J. Neurosci. 20:1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, M.A., and K.M. Ferguson. 2000. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350:1–18. [PMC free article] [PubMed] [Google Scholar]
- Luo, L. 2000. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1:173–180. [DOI] [PubMed] [Google Scholar]
- Meinhardt, H. 1999. Orientation of chemotactic cells and growth cones: models and mechanisms. J. Cell Sci. 112:2867–2874. [DOI] [PubMed] [Google Scholar]
- Meinhardt, H., and A. Gierer. 2000. Pattern formation by local self-activation and lateral inhibition. Bioessays. 22:753–760. [DOI] [PubMed] [Google Scholar]
- Merlot, S., and R.A. Firtel. 2003. Leading the way: directional sensing through phosphatidylinositol 3-kinase and other signaling pathways. J. Cell Sci. 116:3471–3478. [DOI] [PubMed] [Google Scholar]
- Musatov, S., J. Roberts, A.I. Brooks, J. Pena, S. Betchen, D.W. Pfaff, and M.G. Kaplitt. 2004. Inhibition of neuronal phenotype by PTEN in PC12 cells. Proc. Natl. Acad. Sci. USA. 101:3627–3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T., M. Komiya, K. Sone, E. Hirose, N. Gotoh, H. Morii, Y. Ohta, and N. Mori. 2002. Grit, a GTPase-activating protein for the Rho family, regulates neurite extension through association with the TrkA receptor and N-Shc and CrkL/Crk adapter molecules. Mol. Cell. Biol. 22:8721–8734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T., K. Aoki, and M. Matsuda. 2005. FRET imaging in nerve growth cones reveals a high level of RhoA activity within the peripheral domain. Brain Res. Mol. Brain Res. 139:277–287. [DOI] [PubMed] [Google Scholar]
- Ohba, Y., K. Kurokawa, and M. Matsuda. 2003. Mechanism of the spatio-temporal regulation of Ras and Rap1. EMBO J. 22:859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J., Y.L. Kao, S. Joshi, S. Jeetendran, D. Dipette, and U.S. Singh. 2005. Activation of Rac1 by phosphatidylinositol 3-kinase in vivo: role in activation of mitogen-activated protein kinase (MAPK) pathways and retinoic acid-induced neuronal differentiation of SH-SY5Y cells. J. Neurochem. 93:571–583. [DOI] [PubMed] [Google Scholar]
- Paternotte, N., J. Zhang, I. Vandenbroere, K. Backers, D. Blero, N. Kioka, J.M. Vanderwinden, I. Pirson, and C. Erneux. 2005. SHIP2 interaction with the cytoskeletal protein Vinexin. FEBS J. 272:6052–6066. [DOI] [PubMed] [Google Scholar]
- Patterson, G.H., and J. Lippincott-Schwartz. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877. [DOI] [PubMed] [Google Scholar]
- Prasad, N., R.S. Topping, and S.J. Decker. 2001. SH2-containing inositol 5′-phosphatase SHIP2 associates with the p130Cas adapter protein and regulates cellular adhesion and spreading. Mol. Cell. Biol. 21:1416–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa, S., Y. Ozaki, K. Fujita, and S. Kuroda. 2005. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat. Cell Biol. 7:365–373. [DOI] [PubMed] [Google Scholar]
- Sato, M., Y. Ueda, T. Takagi, and Y. Umezawa. 2003. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nat. Cell Biol. 5:1016–1022. [DOI] [PubMed] [Google Scholar]
- Shi, S.H., L.Y. Jan, and Y.N. Jan. 2003. Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell. 112:63–75. [DOI] [PubMed] [Google Scholar]
- Sugiyama, Y., I. Kawabata, K. Sobue, and S. Okabe. 2005. Determination of absolute protein numbers in single synapses by a GFP-based calibration technique. Nat. Methods. 2:677–684. [DOI] [PubMed] [Google Scholar]
- Van Aelst, L., and C. D'Souza-Schorey. 1997. Rho GTPases and signaling networks. Genes Dev. 11:2295–2322. [DOI] [PubMed] [Google Scholar]
- Wang, F., P. Herzmark, O.D. Weiner, S. Srinivasan, G. Servant, and H.R. Bourne. 2002. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Biol. 4:513–518. [DOI] [PubMed] [Google Scholar]
- Weiner, O.D., P.O. Neilsen, G.D. Prestwich, M.W. Kirschner, L.C. Cantley, and H.R. Bourne. 2002. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 4:509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley, H.S., S.Y. Shvartsman, and D.A. Lauffenburger. 2003. Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol. 13:43–50. [DOI] [PubMed] [Google Scholar]
- Yoshizaki, H., N. Mochizuki, Y. Gotoh, and M. Matsuda. 2007. Akt-PDK1 complex mediates epidermal growth factor-induced membrane protrusion through Ral activation. Mol. Biol. Cell. 18:119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., S. Bagrodia, and R.A. Cerione. 1994. Activation of phosphoinositide 3-kinase activity by Cdc42Hs binding to p85. J. Biol. Chem. 269:18727–18730. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.