Abstract
Newly synthesized mitochondrial proteins are imported into mitochondria with the aid of protein translocator complexes in the outer and inner mitochondrial membranes. We report the identification of yeast Tam41, a new member of mitochondrial protein translocator systems. Tam41 is a peripheral inner mitochondrial membrane protein facing the matrix. Disruption of the TAM41 gene led to temperature-sensitive growth of yeast cells and resulted in defects in protein import via the TIM23 translocator complex at elevated temperature both in vivo and in vitro. Although Tam41 is not a constituent of the TIM23 complex, depletion of Tam41 led to a decreased molecular size of the TIM23 complex and partial aggregation of Pam18 and -16. Import of Pam16 into mitochondria without Tam41 was retarded, and the imported Pam16 formed aggregates in vitro. These results suggest that Tam41 facilitates mitochondrial protein import by maintaining the functional integrity of the TIM23 protein translocator complex from the matrix side of the inner membrane.
Introduction
Eukaryotic cells are divided into many membrane bounded organelles that have unique protein compositions to perform a variety of specialized functions. Mitochondria are such organelles that consist of four compartments, the outer membrane, intermembrane space (IMS), inner membrane, and matrix. Because most mitochondrial proteins are synthesized in the cytosol, they are imported into mitochondria with the aid of translocator complexes in the outer and inner mitochondrial membranes (Endo et al., 2003; Koehler, 2004; Wiedemann et al., 2004; Mokranjac and Neupert, 2005). More than 30 proteins have been identified as translocator components, indicating that pathways of import and sorting of mitochondrial proteins are much more complex than previously envisaged.
The TIM23 complex in the mitochondrial inner membrane, which mediates protein translocation across the inner membrane and protein release into the inner membrane, consists of several different subunits (Jensen and Dunn, 2002; Rehling et al., 2004). Tim23 and -17 constitute the protein-conducting channel through which precursor proteins, usually with an N-terminal cleavable presequence, cross the hydrophobic barrier of the inner membrane in an unfolded state. Tim50 facilitates protein transfer from the TOM40 complex in the outer membrane to the TIM23 complex, and Tim21 is proposed to promote the coupling of the two translocator complexes. Mitochondrial Hsp70 (mtHsp70) in the matrix functions as an import motor to drive vectorial translocation and unfolding of the substrate precursor proteins in cooperation with its partner proteins, mitochondrial Hsp70–associated motor and chaperone (MMC) proteins. Tim44 provides an anchor for mtHsp70 to bind to the translocating polypeptide that emerges from the outlet of the TIM23 channel. Pam18/Tim14 (and Mdj2p) functions as a J protein for mtHsp70, and Pam16/Tim16 mediates association of Pam18 to Tim44. Pam17 is also proposed to facilitate coupling of Pam18 and -16 with Tim44. Yge1/Mge1 and Zim17/Tim15/Hep1 bind to the nucleotide-free form of mtHsp70 to promote its function.
We report the identification and characterization of the gene product of YGR046w, Tam41 (translocator assembly and maintenance 41), which is a peripheral inner mitochondrial membrane protein facing the matrix. The cells without Tam41 are defective in the import of precursor proteins destined for the matrix and the inner membrane via the TIM23 complex both in vitro and in vivo. Tam41 likely mediates maintenance of the functional integrity of the TIM23 complex from the matrix side of the inner membrane.
Results and discussion
Tam41 is a mitochondrial inner membrane protein
On the basis of our analyses of the mitochondrial localization of yeast proteins that are indicated in the database to have essential but unknown functions (Ishikawa et al., 2004; Naoé et al., 2004; Yamamoto et al., 2005), we found Tam41, the gene product of YGR046w, to be a candidate for possible new mitochondrial translocator proteins. The gene product of YGR046w is reported as an essential mitochondrial protein in yeast (Hazbun et al., 2003; Rehling et al., 2003). However, when we deleted the TAM41 gene in diploid cells and subjected them to tetrad analysis, all of the four spores grew normally on YPD at 23°C. The strain with chromosomal TAM41 deletion exhibited slow growth at an elevated temperature (37°C) as compared with that at 23°C, and the temperature-sensitive growth was more prominent on nonfermentable (SCLac) media than on fermentable (SCD) media (Fig. S1, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200603087/DC1).
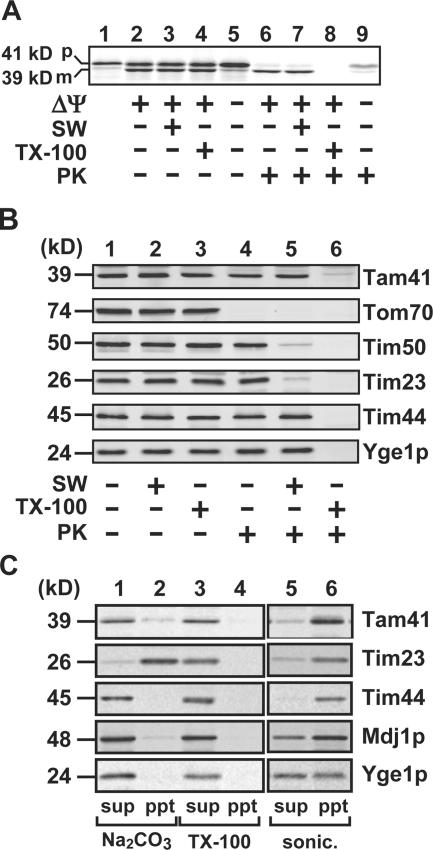
Tam41 comprises 385 amino acid residues with a calculated molecular weight of 44,199 and is predicted to possess a mitochondrial targeting signal at the N terminus. We thus analyzed the in vitro import of Tam41 into isolated yeast mitochondria (Fig. 1 A). When we translated Tam41 with reticulocyte lysate in vitro, a radiolabeled 41-kD protein was synthesized. Upon incubation with isolated yeast mitochondria, it was converted to a 39-kD form in a ΔΨ (membrane potential across the inner membrane)–dependent manner. The 39-kD form was resistant to proteinase K (PK) in mitochondria and in mitoplasts with ruptured outer membrane by osmotic swelling but was digested in mitochondria solubilized with Triton X-100, indicating that the 39-kD form is Tam41 imported into the matrix. We also confirmed that the 41-kD Tam41 precursor is converted to the 39-kD mature form in vivo and that the N-terminal 34 residues of the Tam41 precursor are sufficient to direct nonmitochondrial protein to mitochondria in vitro (Fig. S1, C and D). A search of the database revealed that Tam41 has homologues in a wide range of eukaryotic organisms from yeast to human (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200603087/DC1).
Figure 1.
Tam41 is a mitochondrial inner membrane protein. (A) In vitro import of the radiolabeled precursor of Tam41 into isolated yeast mitochondria (D273-10B) at 30°C for 20 min with or without ΔΨ. The mitochondria were then subjected to osmotic swelling (SW) or treatment with 0.5% Triton X-100 (TX-100) and further treated with or without 100 μg/ml PK for 30 min on ice. The mitochondria or mitoplasts were reisolated by centrifugation, and proteins were analyzed by SDS-PAGE and radioimaging. p, precursor form; m, mature form. (B) Mitochondria and mitoplasts generated by osmotic swelling were treated with 200 μg/ml PK for 30 min on ice in the presence or absence of 0.1% Triton X-100. Proteins were detected by immunoblotting. (C) Mitochondria were treated with sonication, 0.1 M Na2CO3, or 1% Triton X-100 for 30 min on ice. Pellets (ppt) and supernatants (sup) were then separated by centrifugation (100,000 g for 30 min). Proteins were detected by immunoblotting.
We then analyzed the properties of endogenous Tam41 in mitochondrial association by immunoblotting with anti-Tam41 antibodies (Fig. 1 B). Tam41 was inaccessible to protease added to intact mitochondria (−SW) or to mitoplasts (+SW) but was accessible to protease added to mitochondria solubilized with Triton X-100. This behavior resembled that of Tim44, an inner membrane protein exposing a domain to the matrix, but is different from those of Tom70, a surface-exposed outer membrane protein, or Tim23, an inner membrane protein with the IMS domain (Fig. 1 B). Tam41, like the peripheral membrane proteins Mdj1p and Tim44, was extracted by alkaline (Na2CO3) treatment of mitochondria but was not released to the supernatant by sonication followed by ultracentrifugation (Fig. 1 C). These results indicate that Tam41 is a peripheral membrane protein of the inner membrane facing the matrix.
Tam41 facilitates protein import via the TIM23 complex
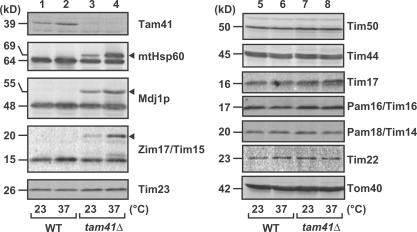
To assess the function of Tam41, we analyzed the effects of Tam41 depletion on mitochondrial protein import in vivo and in vitro. Total lysates were prepared from wild-type (WT) or tam41Δ cells grown in YPD at 23 or 37°C and subjected to immunoblotting for various mitochondrial proteins (Fig. 2). The amounts of the components of the TOM40 complex (Tom40), the TIM23 complex (Tim23, -17, -50, and -44), and the TIM22 complex (Tim22), as well as MMC proteins (Pam18 and -16), were not affected by depletion of Tam41 at 23 or 37°C. However, we could observe accumulation of uncleaved precursor forms of mtHsp60, Mdj1p, and Zim17/Tim15 for tam41Δ cells, and the accumulation was more pronounced at 37 than at 23°C. These results suggest the role of Tam41 in mitochondrial protein import in vivo.
Figure 2.
Tam41 facilitates import of presequence-containing proteins in vivo. Yeast strains W303-1A (WT) and tam41Δ were grown to logarithmic phase in YPD at 23°C and subsequently shifted to 37°C or left at 23°C for 2 h. Cell extracts were then prepared, and proteins were analyzed by SDS-PAGE and immunoblotting. Arrowheads indicate accumulated precursor forms.
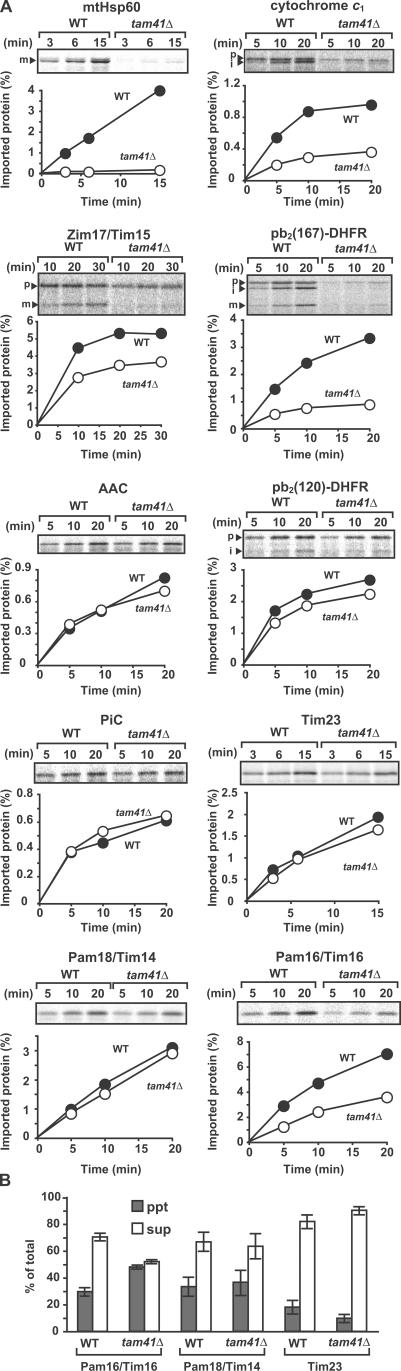
Next, we tested the ability of mitochondria isolated from tam41Δ cells to import various radiolabeled precursor proteins in vitro. Mitochondria isolated from tam41Δ cells did not exhibit a decrease in ΔΨ, which is essential for protein import via the TIM23 and -22 complexes (unpublished data). The steady-state levels of translocator and MMC proteins were similar between WT and tam41Δ mitochondria (Fig. S1 E). We first analyzed the import of matrix-targeting precursor proteins with an N-terminal presequence, which depends on both the TIM23 complex and the import motor machinery comprising mtHsp70 and MMC proteins. The import rates of the precursors to mtHsp60 and Zim17/Tim15 into tam41Δ mitochondria decreased as compared with those into WT mitochondria (Fig. 3). On the other hand, import of presequence-less polytopic inner membrane proteins, ADP/ATP carrier, phosphate carrier, and Tim23, via the TIM22 complex was not retarded in the absence of Tam41 (Fig. 3). These results suggest that Tam41 is involved in the protein import pathway via the TIM23 complex.
Figure 3.
Tam41 facilitates import via the TIM23 complex in vitro. (A) Mitochondria were isolated from yeast strains W303-1A (WT) and tam41Δ after cultivation in YPD at 23°C. Indicated radiolabeled proteins were incubated with the mitochondria at 25°C, which had been preincubated at 37°C for 15 min, and the imported, PK-protected proteins were analyzed by SDS-PAGE and radioimaging. The amounts of radiolabeled proteins added to each reaction are set to 100%. p, precursor form; i, processing-intermediate form; m, mature form. (B) Radiolabeled Pam16, Pam18, and Tim23 were imported into WT and tam41Δ mitochondria at 23°C for 30 min, as in A. The mitochondria treated with PK were solubilized with 1% digitonin and subjected to centrifugation (100,000 g for 30 min). Proteins were analyzed by SDS-PAGE and radioimaging. The sum of pellet (ppt) and supernatant (sup) was set to 100% for each protein, and standard errors were calculated from three independent experiments.
We then tested whether Tam41 facing the matrix plays a role, like Zim17/Tim15 and other MMC proteins (Yamamoto et al., 2005), in the import motor function of mtHsp70. Presequence-containing precursors that possess additional stop-transfer sorting signals are inserted into the inner membrane by the TIM23 complex and ΔΨ but do not require the motor function of mtHsp70. For example, import of pb2(120)–dihydrofolate reductase (DHFR) and pb2(167)-DHFR, fusion proteins between the first 120 and 167 residues, respectively, of the cytochrome b 2 precursor and mouse DHFR, and the precursor to cytochrome c 1 do not require the ATP-dependent motor function of mtHsp70. Indeed, import of pb2(120)-DHFR into the IMS was not affected by depletion of Tam41 (Fig. 3). On the other hand, import of the cytochrome c 1 precursor and pb2(167)-DHFR was strongly retarded by the Tam41 depletion (Fig. 3). These results suggest that the observed defects of the import via the TIM23 complex in the absence of Tam41 cannot be simply ascribed to the defects in the motor functions of mtHsp70 and MMC proteins.
Tam41 is required for integrity of the TIM23 complex
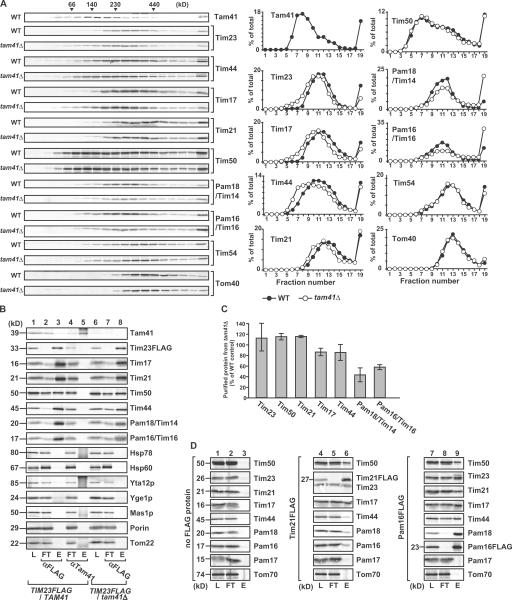
How does the depletion of Tam41 in the matrix affect the protein import via the TIM23 complex? We compared the TIM23 complex structure by glycerol density gradient centrifugation between mitochondria with and without Tam41 (Fig. 4 A). Interestingly, although Tim23 and -17, the channel subunits of the TIM23 complex, were recovered in the fractions corresponding to ∼250 kD in the presence of Tam41, they shifted to the ∼200 kD fractions in the absence of Tam41. Tim44 associating with Tim23 and -17 also shifted to fractions of smaller molecular mass in the absence of Tam41, although the molecular mass of its peak was slightly smaller than that for Tim23 and -17 (∼200 kD), probably reflecting the dynamic association of Tim44 with -23 and -17. On the other hand, Tim50, the peripheral subunits of the TIM23 complex, exhibited a peak at 100–150 kD and a shoulder at ∼250 kD, and they were not affected by the Tam41 depletion. Tim54 in the TIM22 complex and Tom40 in the TOM40 complex were not affected by depletion of Tam41.
Figure 4.
Tam41 depletion affects the TIM23 assembly structure. (A) Mitochondria isolated from yeast strains W303-1A (WT) and tam41Δ, after cultivation in YPD at 23°C, were solubilized with 1% digitonin and analyzed by glycerol density gradient centrifugation (20–40%). After centrifugation, fractions were collected from the top and analyzed by SDS-PAGE and immunoblotting. (B) WT and tam41Δ mitochondria with FLAG-tagged Tim23 were solubilized in 1% digitonin buffer and subjected to immunoprecipitation with the immobilized anti-FLAG and anti-Tam41 antibodies (L, 10% of the loaded proteins). Bound proteins were separated from the supernatant (FT, 10%) by centrifugation and eluted with 100 mM glycine-HCl, pH 2.5 (E, 100%). (C) Quantification of the results of lanes 3 and 8 in B. The amount purified from WT mitochondria was set to 100% for each protein. Standard errors were calculated from three independent experiments. (D) WT mitochondria and those with FLAG-tagged Tim21 or Pam16 were solubilized with 1% digitonin and subjected to immunoprecipitation with the immobilized anti-FLAG antibody as in B except for elution with boiled SDS-PAGE sample buffer.
Although the above results suggest the possibility that Tam41 constitutes the TIM23 core complex with Tim23 and -17, Tam41 was recovered in the fractions corresponding to ∼140 kD after glycerol density gradient centrifugation. When we analyzed the subunit interactions involving Tam41 in the solubilized TIM23 complex containing the FLAG-tagged version of Tim23 by coimmunoprecipitation with the anti-FLAG antibody or anti-Tam41 antibodies, we failed to observe interactions between Tam41 and Tim23, although Tim17, Tim21, Tim44, Tim50, Pam18, and Pam16 were coimmunoprecipitated with Tim23 but not with Tam41 (Fig. 4 B). Therefore, Tam41 is not stably associated with the TIM23 complex, ruling out a trivial possibility that the shifts of Tim23, -17, and -44 toward fractions of lower molecular weight are due to the lack of Tam41 in the TIM23 complex upon depletion of Tam41.
After glycerol density gradient centrifugation, Pam18 and -16, peripheral MMC subunits of the TIM23 complex, were recovered in the fractions corresponding to ∼250 kD in WT mitochondria. Interestingly, however, the amounts of Pam18 and -16 in the ∼250-kD fractions decreased but instead shifted to the bottom upon depletion of Tam41. This suggests that Pam18 and -16 form aggregates in the absence of Tam41. Indeed, when solubilized WT mitochondria were subjected to immunoprecipitation with the anti-FLAG antibody, the amounts of Pam18 and -16 coimmunoprecipitated with the TIM23 complex via FLAG-tagged Tim23 were significantly reduced (Fig. 4 C). These results suggest that Tim23, -17, and -44 shifted to the smaller molecular weight fractions partly because Pam18 and -16 were not properly assembled into the TIM23 complex in the absence of Tam41. When we analyzed the aggregate formation of imported Pam18 and -16 by centrifugation of the mitochondria solubilized with digitonin, the amount of imported Pam16 in the pellet was larger for tam41Δ mitochondria than for WT mitochondria (Fig. 3 B), indicating that Pam16 forms aggregates inside the mitochondria in the absence of Tam41. Therefore, Tam41 is apparently involved in the proper assembly of Pam16 into the TIM23 complex.
Recently, it was proposed that Tim21 and the Pam18–Pam16–Tim44 set are mutually exclusive as constituents of the TIM23 complex; i.e., the TIM23 complex is present in two different forms, one with Tim21 but not with Pam18–Pam16–Tim44, and the other with Pam18–Pam16–Tim44 but not with Tim21 (Chacinska et al., 2005). However, this model is not consistent with our observation that Tim21 at ∼350 kD also shifted to ∼300 kD upon depletion of Tam41, which leads to dissociation of Pam18 and -16 from the TIM23 complex (Fig. 4 A). We thus examined whether Tim21 is in association with Pam18 and -16 by immunoprecipitation of solubilized mitochondria containing the FLAG-tagged Tim21 or Pam16 with anti-FLAG antibodies, and we indeed found that Tim21 was coimmunoprecipitated with Pam16FLAG (Fig. 4 D, lane 9) and Tim44, Pam18, and Pam16 with Tim21FLAG (Fig. 4 D, lane 6).
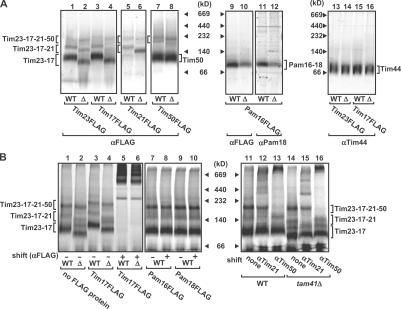
Finally, we analyzed the assembly states of the TIM23 complex in details in the presence or absence of Tam41 by blue-native PAGE (BN-PAGE) separation of digitonin-solubilized mitochondria. Tim23 and -17 were found mainly in a 100-kD core complex (Fig. 5 A, Tim23–17), but additionally in larger TIM23 complexes containing Tim21 (Fig. 5 A, Tim23–17–21) or Tim50 and -21 (Fig. 5 A, Tim23–17–21–50). Constituents of the different forms of the TIM23 complex were identified by immunoblotting to detect the FLAG-tagged Tim23, -17, -21, and -50. Imported radiolabeled Tim23 was found in similar forms of the TIM23 complexes, and antibodies against Tim17FLAG, Tim21, and Tim50 caused shifts of all or a part of those TIM23 complexes (Fig. 5 B). All of the TIM23 complexes became smaller and the amount of the core Tim23–Tim17 complex decreased in mitochondria lacking Tam41 (Fig. 5 A, lane 2), whereas the major fractions of Tim50 and -44 were not affected by Tam41 depletion (Fig. 5 A, lanes 7, 8, and 13–16). Pam16 and -18 were found in the Pam16–Pam18 complex, whose amount was also reduced in the mitochondria without Tam41 (Fig. 5 A, lanes 9–12). These results suggest that depletion of Tam41 leads to partial dissociation of the TIM23 complex as well as destabilization of the Pam16–Pam18 complex.
Figure 5.
BN-PAGE of the TIM23 complex. (A) Mitochondria containing FLAG-tagged Tim23, Tim17, Tim21, Tim50, or Pam16 were isolated from W303-1A (WT) and tam41Δ (Δ) strains, solubilized with 1% digitonin, and subjected BN-PAGE analyses with immunoblotting with the antibodies against the FLAG tag (αFLAG), Pam18 (αPam18), or Tim44 (αTim44). (B) Radiolabeled Tim23 was incubated with mitochondria with or without FLAG-tagged Tim23, Tim17, Tim21, Pam18, Pam16, or Tim50 isolated from the WT or tam41Δ strains for 1 h at 25°C. After stopping the import reaction with valinomycin, the mitochondria were solubilized with 1% digitonin, incubated with the indicated antibodies for 1 h at 4°C (shift), and subjected to BN-PAGE analyses followed by radioimaging.
In conclusion, we identified Tam41 in yeast mitochondria, whose depletion affects protein transport via the TIM23 complex to the matrix and to the inner membrane. Although most TIM23 pathway–associating proteins that reside in the matrix are members of MMC proteins and cooperate with the motor protein mtHsp70, Tam41 facing the matrix does not promote the motor function of mtHsp70 directly but is instead involved in the maintenance of the integrity of the TIM23 complex. In particular, Tam41 affects integration of Pam18 and -16 into the TIM23 complex, and the stability of the TIM23 complex consisting of Tim23, -17, -21, and -50. Elucidation of the detailed mechanisms for Tam41 to maintain the functional integrity of the TIM23 complex from the matrix side of the inner membrane will enhance our understanding of the dynamic nature of the TIM23 complex responsible for its function. During the process of revision of the manuscript, Gallas et al. (2006) reported online a study on the same protein with a similar conclusion.
Materials and methods
Strains and growth conditions
Yeast strains used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200603087/DC1). Cells were grown in YPGal (1% yeast extract, 2% polypeptone, and 2% galactose), YPD (1% yeast extract, 2% polypeptone, and 2% glucose), SD (0.67% yeast nitrogen base without amino acids and 2% glucose), SCD (0.67% yeast nitrogen base without amino acids, 0.5% casamino acid, and 2% glucose), or SCLac (0.67% yeast nitrogen base without amino acids, 0.5% casamino acid, and 2% lactate) media with appropriate supplements.
Plasmids
TAM41 was cloned from the yeast genomic DNA by PCR using primers 5′-GCCGGCTCGAGTGCACTCATAATGCTACTCG-3′ and 5′-CCGCCGGATCCACTGGAACGTATGATCCCCC-3′. The amplified DNA fragment was digested with BamHI and XhoI and introduced into the BamHI and the XhoI sites of pRS316 to produce pRS316-Tam41. The plasmid used for in vitro translation of Tam41 was constructed as follows. A DNA fragment corresponding to the TAM41 gene was amplified from pRS316-Tam41 by PCR using primers 5′-CCGGATCCATAATTTGAATTAATAGGAGCTGCTTT-3′ and 5′-GCGCGTCGACGATACACTAGCTTCTCCTCATCGAT-3′. The amplified DNA fragment was digested with BamHI and SalI and introduced into the BamHI and the SalI sites of pGEM-4Z (Promega) to produce pGEM-Tam41.
Preparation of anti-Tam41 antibodies
A DNA fragment corresponding to full-length ORF of TAM41 was amplified from the yeast genomic DNA by PCR using primers 5′-CCGCCGGATCCATGTTACGAGTTTCTGAAAA-3′ and 5′-GCCGGCTCGAGTGCTTCTCCTCATCGATTTTA-3′. The amplified DNA fragment was digested with BamHI and XhoI and introduced into the BamHI and the XhoI sites of pET-21a (Novagen) to produce pET-21a-Tam41. The fusion protein expressed in the Escherichia coli strain BL21(DE3)/pLysS was recovered in the inclusion body fraction. The protein was solubilized in 8 M urea, 20 mM Tris-HCl, pH 7.5, and 150 mM NaCl and was purified by Ni-NTA agarose chromatography (QIAGEN). The purified protein was used to immunize rabbits with complete Freund's adjuvant followed by four booster immunizations with incomplete Freund's adjuvant.
Import assays
Mitochondria were isolated from D273-10B grown in lactate medium at 30°C and W303-1A and tam41Δ strains grown in YPD at 23°C. Radiolabeled precursor proteins were synthesized with rabbit reticulocyte lysate by coupled transcription/translation in the presence of [35S]methionine. Mitochondria isolated from W303-1A and tam41Δ cells were incubated with radiolabeled precursor proteins in import buffer (250 mM sucrose, 10 mM MOPS-KOH, pH 7.2, 80 mM KCl, 2 mM KPi, 2 mM methionine, 5 mM DTT, 5 mM MgCl2, 2 mM ATP, 2 mM NADH, and 1% BSA) at 25°C. The mitochondria were isolated by centrifugation, and proteins were analyzed by SDS-PAGE and radioimaging. Treatment of mitochondria with PK, sodium carbonate, and Triton X-100 and preparation of mitoplasts were performed as described previously (Naoé et al., 2004).
Glycerol density gradient centrifugation
Mitochondria were solubilized at 2 mg/ml in 1% digitonin buffer (1% digitonin, 20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 0.1 mM EDTA, 10% glycerol, and 1 mM PMSF) for 20 min on ice and were centrifuged at 20,000 g for 15 min. The supernatant was layered onto linear glycerol gradient (20–40%) in 20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 50 mM 6-aminohexanoic acid, 0.1 mM EDTA, 0.1% digitonin, and complete protease inhibitor cocktail (Roche) and centrifuged at 166,000 g for 15 h.
Coimmunoprecipitation
Mitochondria were solubilized at 2 mg/ml in 1% digitonin buffer. After centrifugation at 20,000 g for 15 min, the solubilized proteins were added to the anti-FLAG antibody bound to agarose (Sigma-Aldrich) or anti-Tam41 antibodies bound to protein A–Sepharose (GE Healthcare). The samples were gently rotated for 2 h at 4°C. The resins were washed with wash buffer (0.2% digitonin, 20 mM Tris-HCl, pH 7.4, 50 mM NaCl, 0.1 mM EDTA, 10% glycerol, and protease inhibitor cocktail [Sigma-Aldrich]) three times before bound proteins were eluted with 100 mM glycine-HCl, pH 2.5, or SDS-PAGE sample buffer. The eluted proteins were analyzed by SDS-PAGE and immunoblotting.
BN-PAGE
Mitochondria containing FLAG-tagged Tim23, Tim17, Tim50, or Pam16 were used for BN-PAGE analyses with or without Tam41. The mitochondria (2 mg of protein per milliliter) were solubilized with 1% digitonin buffer for 20 min at 4°C, cleared by centrifugation at 20,000 g for 15 min, diluted 20-fold in 0.2% (wt/vol) CBB G-250 and 5 mM 6-aminohexanoic acid, and subjected to BN-PAGE (Schägger, 2001).
Online supplemental material
Fig. S1 shows characterization of Tam41. Fig. S2 compares amino acid sequences of Tam41 from various organisms. Table S1 shows the yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200603087/DC1.
Supplementary Material
Acknowledgments
We thank members of the Endo Laboratory for discussions and Thomas Langer for anti-Hsp78 and anti-Yta12p antibodies.
Y. Tamura, K. Yamano, and H. Yamamoto are Research Fellows of the Japan Society for the Promotion of Science. This study was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (15207009 and 14037225) and a grant from the Japan Science and Technology Corporation.
Y. Tamura and Y. Harada contributed equally to this paper.
Abbreviations used in this paper: BN-PAGE, blue-native PAGE; DHFR, dihydrofolate reductase; IMS, intermembrane space; MMC, mitochondrial Hsp70–associated motor and chaperone; PK, proteinase K; WT, wild-type.
References
- Chacinska, A., M. Lind, A.E. Frazier, J. Dudek, C. Meisinger, A. Geissler, A. Sickmann, H.E. Meyer, K.N. Truscott, B. Guiard, et al. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor requirement involves Tim21 and Tim17. Cell. 120:817–829. [DOI] [PubMed] [Google Scholar]
- Endo, T., H. Yamamoto, and M. Esaki. 2003. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J. Cell Sci. 116:3259–3267. [DOI] [PubMed] [Google Scholar]
- Gallas, M.R., M.K. Dienhart, R.A. Stuart, and R.M. Long. 2006. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol. Biol. Cell. 10.1091/mbc.E06-04-0366. [DOI] [PMC free article] [PubMed]
- Hazbun, T.R., L. Malmström, S. Anderson, B.J. Graczyk, B. Fox, M. Riffle, B.A. Sundin, J.D. Aranda, W. Hayes, C.-H. McDonald, et al. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 12:1353–1365. [DOI] [PubMed] [Google Scholar]
- Ishikawa, D., H. Yamamoto, Y. Tamura, K. Moritoh, and T. Endo. 2004. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J. Cell Biol. 166:621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R.E., and C.D. Dunn. 2002. Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim. Biophys. Acta. 1592:25–34. [DOI] [PubMed] [Google Scholar]
- Koehler, C.M. 2004. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20:309–335. [DOI] [PubMed] [Google Scholar]
- Mokranjac, D., and W. Neupert. 2005. Protein import into mitochondria. Biochem. Soc. Trans. 33:1019–1023. [DOI] [PubMed] [Google Scholar]
- Naoé, M., Y. Ohwa, D. Ishikawa, C. Ohshima, S. Nishikawa, H. Yamamoto, and T. Endo. 2004. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J. Biol. Chem. 279:47815–47821. [DOI] [PubMed] [Google Scholar]
- Rehling, P., N. Pfanner, and C. Meisinger. 2003. Insertion of hydrophobic membrane proteins into the inner mitochondrial membrane—a guided tour. J. Mol. Biol. 326:639–657. [DOI] [PubMed] [Google Scholar]
- Rehling, P., K. Brander, and N. Pfanner. 2004. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 5:519–530. [DOI] [PubMed] [Google Scholar]
- Schägger, H. 2001. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 65:231–244. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., A.E. Frazier, and N. Pfanner. 2004. The protein import machinery of mitochondria. J. Biol. Chem. 279:14473–14476. [DOI] [PubMed] [Google Scholar]
- Yamamoto, H., T. Momose, Y. Yatsukawa, C. Ohshima, D. Ishikawa, T. Sato, Y. Tamura, Y. Ohwa, and T. Endo. 2005. Identification of a novel member of yeast mitochondrial Hsp70-associated motor and chaperone proteins that facilitates protein translocation across the inner membrane. FEBS Lett. 579:507–511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.