Abstract
Three muscle-specific microRNAs, miR-206, -1, and -133, are induced during differentiation of C2C12 myoblasts in vitro. Transfection of miR-206 promotes differentiation despite the presence of serum, whereas inhibition of the microRNA by antisense oligonucleotide inhibits cell cycle withdrawal and differentiation, which are normally induced by serum deprivation. Among the many mRNAs that are down-regulated by miR-206, the p180 subunit of DNA polymerase α and three other genes are shown to be direct targets. Down-regulation of the polymerase inhibits DNA synthesis, an important component of the differentiation program. The direct targets are decreased by mRNA cleavage that is dependent on predicted microRNA target sites. Unlike small interfering RNA–directed cleavage, however, the 5′ ends of the cleavage fragments are distributed and not confined to the target sites, suggesting involvement of exonucleases in the degradation process. In addition, inhibitors of myogenic transcription factors, Id1-3 and MyoR, are decreased upon miR-206 introduction, suggesting the presence of additional mechanisms by which microRNAs enforce the differentiation program.
Introduction
MicroRNAs (miRNAs) are a class of small noncoding RNAs that are processed by Dicer from precursors with a characteristic hairpin secondary structure (Ambros et al., 2003). Hundreds of miRNAs have been identified from plants, animals, and viruses (miRBase; http://microrna.sanger.ac.uk/sequences/). miRNAs are implicated in various cellular processes, such as cell fate determination, cell death, and tumorigenesis (for review see Bartel, 2004). Many miRNAs are expressed in a tissue-specific manner (Lagos-Quintana et al., 2002; Babak et al., 2004; Barad et al., 2004; Liu et al., 2004; Sempere et al., 2004; Thomson et al., 2004; Baskerville and Bartel, 2005; Wienholds et al., 2005), suggesting a role of the miRNAs in the specification of the tissue during differentiation.
Among the hundreds of miRNAs, only a small fraction have assigned target mRNAs or an established role. Valid target prediction is a major problem in the study of miRNAs. Although several algorithms for target prediction have been based on sequence similarity between targets and miRNAs (Bentwich, 2005), the small size of the miRNAs and the tolerance for mismatches and bulges in the recognition sequence result in most of these algorithms' predicting too many targets.
The mode of action of miRNAs on their targets is controversial. Classic results from lin-4 miRNAs suggested that the miRNAs bind to their targets with imperfect complementarity and decrease the levels of encoded proteins without decreasing the target mRNA (Olsen and Ambros, 1999; Seggerson et al., 2002). In contrast, target mRNA is cleaved specifically at the recognition site by siRNA (Elbashir et al., 2001b), many plant miRNAs (for reviews see Kidner and Martienssen, 2005; Millar and Waterhouse, 2005), and at least one animal miRNA (Yekta et al., 2004). In all cases where the target mRNA is cleaved, the interaction between the small RNA and the target mRNA is nearly perfect. Therefore, the degree of complementarity has been thought to be a major determinant in dictating whether a miRNA promotes mRNA degradation or inhibits protein synthesis. Although this hypothesis is supported by mutation analyses of miRNAs and their target mRNAs (Doench et al., 2003; Saxena et al., 2003), a recent report demonstrated that a miRNA can regulate the levels of several target mRNAs despite mismatches and bulges between the miRNA and the targets (Lim et al., 2005). This was shown true for even lin-4 and let-7 miRNAs (Bagga et al., 2005), which had been thought to block only the translational step.
Differentiation down a specific lineage is characterized by the activation of tissue-specific transcription factors and modulation of gene expression. To study the role of miRNA in such a process and begin the process of identifying potential targets, we studied muscle differentiation using the C2C12 myoblast (MB) cell line as a model system (Yaffe and Saxel, 1977; Andres and Walsh, 1996). Upon serum depletion, muscle-specific transcription factors such as myogenin are induced and many muscle genes are turned on. Subsequently, cells become elongated and fused to each other to form multinucleate myotubes (MTs). Another critical event during the differentiation process is a decrease in DNA synthesis and cell cycle arrest. We show here that miRNAs miR-206, -1, and -133 on their own change the gene expression profile of C2C12 toward the differentiated state and that miR-206 induces many of the markers of differentiation. The miRNAs regulate many target mRNAs as revealed by microarray screening. Antisense oligonucleotides to these miRNAs inhibit muscle differentiation and entry into cell quiescence. We predicted the putative direct targets of miR-206 by intersecting the mRNAs down-regulated in the microarray data with the computational prediction of targets based on sequence match to the miRNA. As an example of the utility of this approach, we identify four mRNAs, including that of the largest subunit (p180 subunit; Pola1) of DNA polymerase α (DNA pol α) as being directly regulated by miR-206. Two of the targets, including DNA pol α, are cleaved at multiple sites by the miRNA. The effect of miR-206 on DNA pol α, thereby DNA synthesis, is a new example of miRNA function connecting the cell quiescence event with the differentiation process. In addition, inhibitors of myogenic transcription factors are indirectly down-regulated by miR-206, thereby further promoting the differentiation process. Recently, another group reported a critical role of miR-1 and -133 in C2C12 differentiation (Chen et al., 2006). Together, we can conclude that all three miRNAs induced during C2C12 differentiation are very important for myogenesis.
Results
Specific expression of miR-1, -133, and -206 during skeletal myogenesis
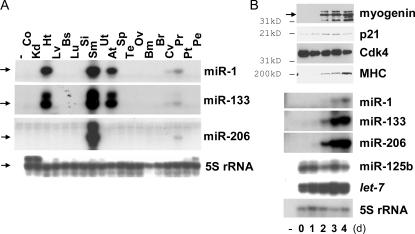
Previous results suggested that miR-1, -133, and -206 are expressed in muscle and heart (Lagos-Quintana et al., 2002; Sempere et al., 2004; Zhao et al., 2005; Chen et al., 2006). Consistent with this, RNase protection assays (Fig. 1 A) showed that miR-1 and -133 are abundant in skeletal muscle and heart and that miR-206 is specifically abundant in skeletal muscle. miR-1 and -206 have an 18/21 match in sequence with each other and complete identity in the first eight nucleotides (miRBase) that constitute the seed sequence for target recognition (Doench and Sharp, 2004).
Figure 1.
The muscle-specific expression of the miRNAs miR-1, -133, and -206. (A) RNase protection assays were performed with RNA from various tissues to detect miR-1, -206, and -133. 5S ribosomal RNA (rRNA) was used as a control. The different panels have different levels of exposure. The protected bands are indicated by arrows on the left. −, no RNA control; Co, colon; Kd, kidney; Ht, heart; Lv, liver; Bs, breast; Lu, lung; Si, small intestine; Sm, skeletal muscle; Ut, uterus; At, atrium; Sp, spleen; Te, testis; Ov, ovary; Bm, bone marrow; Br, brain; Cv, cervix; Pr, prostate tissue; Pt, prostate tumor; Pe, prostate epithelium. (B) During skeletal myogenesis of the C2C12 MB cell line, the miRNAs (bottom six panels) and several marker proteins for muscle differentiation (top four panels) were measured by RNase protection assays or immunoblots, respectively. The numbers at the bottom indicate the days after changing from GM to DM. In the immunoblot panels, molecular mass markers are indicated on the left and the myogenin band is specified by an arrow. The panels for miR-1, -133, and -206 are at comparable levels of exposure.
C2C12 mouse MBs can be induced to differentiate into MTs by serum depletion, as indicated here by the induction of myogenin, cell cycle inhibitor p21, and myosin heavy chain (MHC) with a constant level of Cdk4 as a loading control (Fig. 1 B). miR-1, -133, and -206 were not expressed in undifferentiated C2C12 but were induced during muscle differentiation (Fig. 1 B; Chen et al., 2006), whereas two other miRNAs (let-7 and miR-125b) were expressed constantly throughout the differentiation process. Using this in vitro differentiation of C2C12, we studied whether the miRNAs play an active role during skeletal myogenesis.
Role of the muscle-specific miRNAs during skeletal myogenesis
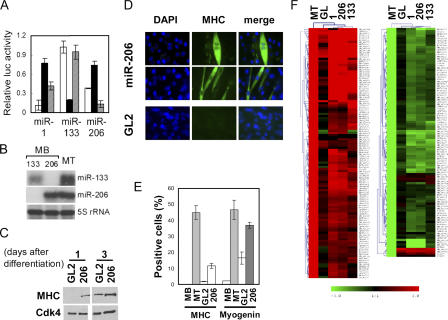
To investigate the function of these miRNAs in myogenesis, we used double-stranded RNA duplexes with miRNA sequence that mimic miRNA function (Fig. 2 A; Hutvagner and Zamore, 2002). Transfection of these duplexes into C2C12 reduced the luciferase expression when the cognate target site is placed at the 3′UTR of luciferase gene, validating the experimental system. The cross-reactivity between miR-1 and -206 can be explained by their similar sequences. To ensure that the transfected miRNAs are not being assayed at supraphysiological levels, we measured the levels of miR-133 and -206 after transfection into MBs and compared the levels of the same miRNA after induction of differentiation into MTs. The levels of the transfected miRNAs were comparable to the levels of the naturally induced miRNAs (Fig. 2 B).
Figure 2.
The muscle-specific miRNAs induce the differentiation of MBs by regulating many genes. (A) Synthetic oligonucleotides complementary to miR-1, -133, and -206 were inserted downstream of Renilla luciferase ORF in pRL-CMV(MCS) to generate pRL1, pRL133, and pRL206, respectively. Luciferase assays were performed with pRL1 (white bar), pRL133 (black bar), and pRL206 (hatched bar) with the cotransfected RNA duplexes mimicking the miRNAs miR-1, -133, and -206, respectively (x axis). Sequential transfection of each RNA duplex (or miR-125b duplex as a control) and reporter plasmids in GM (serum+) was followed by luciferase assays 20 h after transfection. The y axis indicates relative Renilla luciferase activity, which is normalized as described in Materials and methods. (B) RNase protection assays were performed to measure the level of miR-133 (top) and -206 (middle), in MB transfected with the siRNA duplexes mimicking miR-133 or -206. 5S rRNA was used as a loading control (bottom). RNA from differentiated MTs is shown for comparison. (C) Immunoblots at the indicated days after the serum depletion of C2C12. C2C12 was transfected four times at 24-h intervals with miR-206 or GL2 negative control duplex before serum depletion. (D) C2C12 was transfected with miR-206 and GL2 as described in C, split at 5 × 105 cells per well in a 6-well plate, and maintained in GM (serum+) for 3 d before immunostaining for MHC. The density of the cells can also be gauged in Fig. S1 B (available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1). (E) Quantification of MHC and myogenin expression in experiments as in D. MB, C2C12 MB in GM (serum+); MT, MT in DM (serum−); GL2 or 206, C2C12 in GM transfected with GL2 or miR-206 duplexes, respectively. Each value is a mean of triplicate. (F) Hierarchical cluster and heat map to show changes in mRNA relative to MBs in GM. Red and green represent increase and decrease of expression, respectively. Each row represents a single gene in the microarray. MT indicates C2C12 differentiated by serum deprivation, and other rows show C2C12 in GM transfected with the indicated duplexes. The 109 or 92 genes most up- or down-regulated after muscle differentiation are shown.
We began our studies with miR-206 because of the skeletal muscle–specific expression of miR-206 (Fig. 1 A) and because of its similarity to miR-1. miR-206 transfection advanced MHC expression after changing to differentiation medium (DM; Fig. 2 C), whereas miR-133 did not (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1) under the same experimental condition.
Upon transfection in the continued presence of serum, miR-206 markedly up-regulated the percentage of cells expressing MHC and the muscle-specific transcription factor myogenin, relative to cells at comparable density transfected with GL2 control (Fig. 2, D and E; and Fig. S1, B and C). In addition, 28% of MHC-positive cells are multinucleated after miR-206 treatment, whereas no multinucleated cells were detected in GL2 control (Fig. 2 D and not depicted). Therefore, physiological levels of miR-206 induce skeletal myogenesis from C2C12 even without serum depletion.
To measure the extent of differentiation and to extend the studies to the other muscle-specific miRNAs, microarray experiments were used to measure changes in global gene expression profile after muscle differentiation by serum deprivation (DM). The microarray profile after differentiation was compared with that obtained after miRNA transfection in the presence of serum (growth medium [GM]). We focused our analysis on the ∼100 genes that were induced or repressed the most after C2C12 differentiation to MTs by serum depletion (Fig. 2 F). Genes up- or down-regulated after muscle differentiation were changed similarly by transfection of the miRNAs (in GM), with miR-206 and -1 having similar effects and miR-133 affecting a slightly different subset of genes. Given the importance of the first eight nucleotides of miRNA in target interaction (Doench and Sharp, 2004), the identical seed sequences between miR-1 and -206 explain why the two miRNAs had almost identical changes in gene expression. These data suggest that the individual miRNAs change the repertoire of expressed genes in a direction mimicking that seen during muscle differentiation. Further analysis of the microarray data will be addressed in the next section.
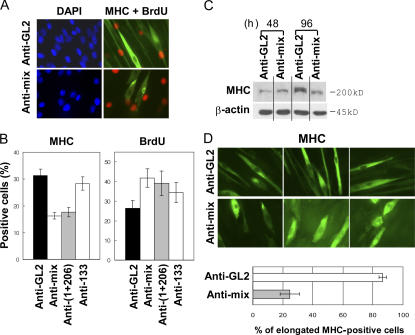
To determine whether the miRNAs were essential for differentiation, C2C12 cells were treated with 2′-O-methyl antisense oligonucleotides against the miRNAs (Hutvagner et al., 2004; Meister et al., 2004) and then induced to differentiate by serum depletion. Inhibition of the miRNAs reduced the number of MHC-positive cells (Fig. 3, A and B) and decreased MHC levels in an immunoblot (Fig. 3 C), with little effect when only miR-133 was inhibited. BrdU immunostaining shows that a significant fraction of the cells remain in active DNA synthesis when the miRNAs are inhibited (42% for the anti-miR mix relative to 26% for the anti-GL2 control; P = 0.0071; Fig. 3, A and B). Inhibition of the miRNAs prevented the elongation of MTs seen during differentiation (Fig. 3 D). The cells remained relatively short and thick even after expression of MHC: 86% of MHC-positive cells in the anti-GL2 control sample were elongated, as opposed to only 25% in the anti-miR mix sample. Although inhibition of miR-1 and -206 (without inhibition of miR-133) was sufficient to decrease MHC expression and derepress DNA synthesis (Fig. 3 B), it was not sufficient to inhibit cell elongation (not depicted), suggesting that miR-133 might have specific targets relevant to MT elongation. Collectively, the results suggest that the three miRNAs are required for complete differentiation to muscle, with miR-206 (and miR-1) being particularly important for induction of cell quiescence.
Figure 3.
Inhibition of the miRNAs by 2′-O-methyl antisense oligonucleotides inhibits differentiation. (A) C2C12 cells transfected with antisense oligonucleotide against GL2 or a mixture of antisense oligonucleotides against miR-1, -133, and -206 (anti-mix). Three transfections at 24-h intervals in GM (serum+) were followed by serum depletion (DM). 96 h after serum depletion, MHC expression (green) and BrdU incorporation (red) were detected by immunostaining. Blue indicates nuclei stained by DAPI. (B) Quantification of percentage of BrdU- and MHC-positive cells in samples prepared as described in panel A. Anti-(1+206) indicates a mixture of 2′-O-methyl antisense oligonucleotides against miR-1 and -206. Each value is a mean of triplicate. (C) Immunoblots at 48 and 96 h after serum depletion (DM). The transfection and induction of differentiation were performed as described in A. (D) Cell morphology visualized by MHC immunostaining as described in A (top). Results are quantitated at the bottom. Cells that were elongated were twofold longer than proliferating C2C12. Each value is a mean of triplicate experiments.
Muscle-specific miRNAs regulate many genes as direct targets, including DNA pol α
Unlike plant miRNAs, animal miRNAs are believed to repress protein synthesis without changing mRNA levels. Yet, the microarrays (Fig. 2 F) revealed a large number of mRNA changes after miRNA introduction. A significant number of up-regulated genes included muscle-specific genes such as myosin light chain (phosphorylatable, fast skeletal muscle), myosin light polypeptide 1, troponin T1 (skeletal, slow), troponin I (skeletal slow 1), myomesin 2, and titin (Schiaffino and Reggiani, 1996; Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1). Because miRNAs are expected to be repressive, the up-regulation of genes is most likely due to indirect effects after the primary differentiation-inducing stimuli of the miRNAs.
In contrast, the list of down-regulated mRNAs (Fig. 2 F and Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1) might include direct targets of the miRNAs if, as in plants, the miRNAs promote target RNA cleavage. To identify such direct targets, we intersected the list of genes down-regulated in the microarrays with the list of predicted genes with miRNA target sites (generated using the miRanda program [Enright et al., 2003]). We focused our analysis on miR-206 because of its similarity with miR-1 and its pronounced effect on muscle differentiation and cell proliferation (Figs. 2 and 3). On this intersection list, we were pleased to find the largest subunit of DNA pol α (Pola1; available from GenBank/EMBL/DDBJ under accession no. NM_008892.1), the replicative polymerase expected to be very important for cell proliferation.
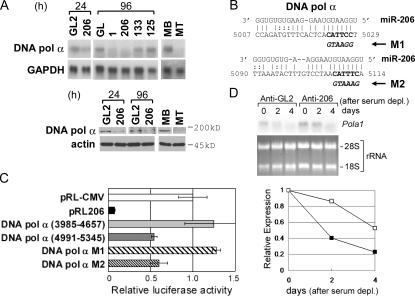
miR-206 alone is sufficient to decrease Pola1 at the mRNA and protein levels as early as 24 h after transfection (Fig. 4 A). During differentiation of MBs to MTs, Pola1 mRNA and protein were also decreased (Fig. 4 A). The Pola1 promoter requires the activity of E2F, a transcription factor that is repressed by pRb during myogenesis (Kalma et al., 2001; Kitzmann and Fernandez, 2001), so we wondered how much of the down-regulation of Pola1 mRNA during differentiation was dependent on miR-206. Antisense to miR-206 significantly delayed the down-regulation of Pola1 mRNA (Fig. 4 D). This result is in agreement with the increase of BrdU-positive cells and decrease of MHC-positive cells when miR-206 was inhibited by antisense oligonucleotide (Fig. 3). The antisense to miR-206 does not, however, completely block differentiation (Fig. 3 B). Therefore, residual inhibition of Cdk2 kinase, hypophosphorylation of Rb, and repression of E2F probably account for the eventual repression of Pola1 in the presence of antisense to miR-206, albeit with delayed kinetics. Together, these results suggest that miR-206 is important for the early down-regulation of Pola1 during differentiation.
Figure 4.
miR-206 directly down-regulates DNA pol α during differentiation. (A) Northern hybridization (top) and immunoblots (bottom) for the largest subunit of DNA pol α (Pola1) at 24 and 96 h after transfection of the indicated duplexes into C2C12 in GM (serum+). MB and MT are shown for comparison. (B) Two predicted target sites of miR-206 in the 3′UTR of Pola1. The nucleotide coordinate of Pola1 is based on the mouse Refseq (available from GenBank/EMBL/DDBJ under accession no. NM_008892.1). M1 and M2, mutated (bold italic) from the seed matches (bold), are indicated. (C) Luciferase assays were performed to measure the effect of miR-206 transfection, as described in Fig. 2 A. pRL206 indicates insertion of a perfectly complementary target of miR-206 downstream of the luciferase gene in pRL-CMV. DNA pol α (3985–4657) and (4991–5345) indicates insertion of the indicated regions of Pola1 mRNA (available from GenBank/EMBL/DDBJ under accession no. NM_008892.1), respectively. M1 and M2 indicate 4991–5345 with point mutations in putative target sequences as described in B. (D) Northern blot of Pola1 (top) and its quantification (bottom) after transfection of antisense oligonucleotide against miR-206 (open squares) or GL2 (closed squares) during C2C12 differentiation as described in Fig. 3 A. The ethidium bromide staining of two rRNA bands is shown as loading control. Each band from Northern hybridization was quantified with ImageQuant 5.2 software and normalized to the total RNA amount from the intensity of 28S and 18S rRNA. The normalized value at day 0 was set at 1.
Pola1 has two potential target sites of miR-206 (Fig. 4 B; Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1; and see Fig. 7 A) in the 4991–5345 segment of the 3′UTR. Fusion of a luciferase reporter to this segment rendered luciferase repressible by miR-206 (Fig. 4 C). The level of repression by cotransfected miRNAs is comparable to the two- to threefold repression by miRNAs in all published experiments where a reporter gene is fused to naturally occurring target 3′UTRs (Krek et al., 2005; Lim et al., 2005; Yu et al., 2005). As a negative control, no repression was seen upon insertion of another part of the 3′UTR of Pola1 without any predicted target sites for miR-206 (DNA pol α 3985–4657). Point mutations in the target sites in pol α 4991–5345 revealed that the one encompassing nucleotides 5007–5029 (M1; Fig. 4, B and C; Fig. S5; and see Fig. 7 A) is necessary for the repression by miR-206, whereas the second site at 5090–5114 (M2) is not required. Therefore, miR-206 directly down-regulates Pola1 mRNA, a down-regulation that probably contributes to the prompt suppression of cell proliferation seen during differentiation. Pola1 is similarly down-regulated by miR-1 but not as much by miR-133 (Fig. 4 A and Fig. S2). However, miR-206 is likely to be more important than miR-1 for this function during C2C12 differentiation because it is present at a much higher level than miR-1 (Fig. 1 B).
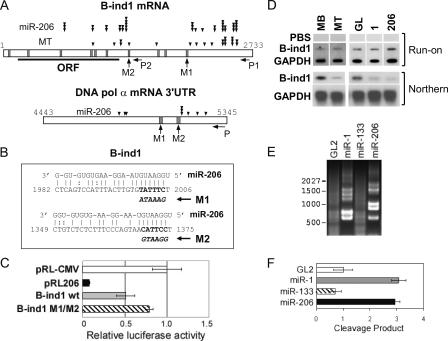
Figure 7.
The down-regulation of direct targets by miR-206 is posttranscriptional, dependent on target sites, and occurs through mRNA cleavage. (A) The schematic of B-ind1 mRNA (top) and 3′UTR of Pola1 mRNA (bottom). Vertical bars show potential binding sites of miR-206, predicted by miRanda program (Δg is less than −14 kcal/mol). M1 and M2 indicate point mutations at indicated sites (used in C and Fig. 4 C and described in B and Fig. 4 B). P, P1, and P2 indicate locations of each set of primers for the RACE-PCR (used in E and F and Figs. S3–S5, available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1). Cleavage sites for each treatment are indicated as described in the figure (see also Figs. S3–S5). The number of arrowheads corresponds to the frequency of cloning from RACE-PCR product. Although MB had very few cleavage products (E), we cloned a few to show that distribution of background cleavage sites is different from that seen in the presence of miRNAs (Fig. S4). (B) Among the several predicted target sites (A) of miR-206 in B-ind1 mRNA (available from GenBank/EMBL/DDBJ under accession no.NM_021345), two sites are shown as described in Fig. 4 B. (C) Luciferase assays were performed as described in Fig. 4 C, with pRL-CMV derivatives containing B-ind1 3′UTR without mutation (B-ind1 wild type [wt]) or with point mutations at both M1 and M2 (B-ind1 M1/M2; described in B). (D) Nuclear run-on assays to measure the transcription of B-ind1 and GAPDH upon differentiation (MB vs. MT) or after transfection of the indicated duplexes in GM (serum+). PBS, pBluescript vector negative control. (E) Agarose gel electrophoresis of RLM-RACE products from primer set P1 (A and Fig. S5 A), showing sites of cleavage of B-ind1 mRNA from C2C12 transfected with indicated duplexes in GM (serum+). Molecular size markers (in nucleotides) are indicated. (F) Quantitation of the amount of the RLM-RACE products from E as described in Materials and methods. The x axis indicates the relative intensity. Each value is a mean of triplicate measurements.
The introduction of miR-206 results in a reduction of DNA synthesis and, eventually, cell cycle arrest
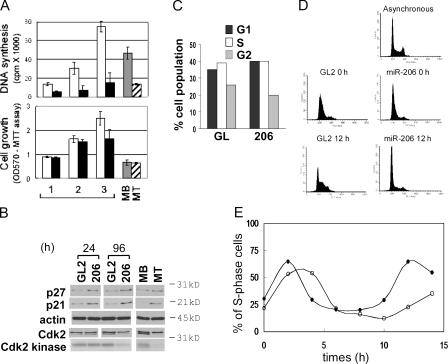
We measured whether down-regulation of DNA synthesis is an early event after miR-206 introduction. In good agreement with the Pola1 decrease, DNA synthesis was significantly inhibited by 24 h after transfection of miR-206, eventually leading to a decrease in cell proliferation that became evident at 72 h (Fig. 5 A). Because cell cycle arrest is a critical step during muscle differentiation, we wondered whether the DNA synthesis inhibition induced by miR-206 leads to the cell cycle arrest. Upon serum depletion of MBs to form MTs, the Cdk inhibitors p21 and p27 are induced and Cdk2 kinase activity is decreased accompanied by a reduction in the amount of the faster moving form of the Cdk2 that is phosphorylated on its activating site, Threonine160 (Fig. 5 B; Guo et al., 1995). miR-206 transfection induces p21 and p27 as early as 24 h after transfection. The FACS profile for DNA content (Fig. 5 C) and the activity of Cdk2 kinase (Fig. 5 B), however, demonstrate that G1 accumulation is not evident at 24 h after transfection, even though DNA synthesis was already reduced significantly (Fig. 5 A). At 96 h after transfection, the miR-206 transfected cells eventually accumulate in G1 with repressed Cdk2 activity (Fig. 5 B), consistent with a G1 arrest (not depicted). Therefore, miR-206–mediated inhibition of DNA synthesis precedes the cell cycle arrest.
Figure 5.
Inhibition of DNA synthesis by the down-regulation of DNA pol α promotes withdrawal from the cell cycle in the continued presence of serum. (A) DNA synthesis (top) and cell growth (bottom) were measured after the transfection of GL2 (white bar) or miR-206 (black bar) duplexes into C2C12 MB in GM (serum+). The same assays were performed at 4 d after serum depletion (MT; hatched bar) with a similar number of cells before serum depletion (MB; gray bar) for comparison. Each value is a mean of four values from two independent transfections. The x axis shows days after transfection, and the y axis shows 3H-thymidine incorporation or absorbance at 570 nm from the MTT assay. (B) Immunoblots as described in Fig. 4 A. Cdk2 kinase activity shows an autoradiogram of phosphorylated histone H1 (see Materials and methods). (C) Propidium iodide staining for DNA content and FACS to determine the number of cells transfected with GL2 or miR-206. The percentage of cells in G1, S, and G2 at 24 h are calculated and plotted. (D) For synchronization of cell cycle, cells were treated with 2 mM of thymidine for 8 h, released for 8 h, and treated with 2 μg/ml of aphidicolin for 8 h. During synchronization, miR-206 (bottom two right panels) or GL2 control duplex (left) was transfected at the onset of thymidine treatment and a second time at 4 h after release from thymidine. Propidium iodide–stained FACS profiles of arrested cells (0 h; middle) and cells at 12 h after release from aphidicolin (bottom) are shown, with that of asynchronous MBs (top) for comparison. (E) The percentage of cells in S phase (y axis) is quantified as described in C before (0 h) or at the indicated time points (x axis) after release from aphidicolin. Closed circles indicate GL2 transfected cells, and open circles indicate miR-206 transfected cells.
Next, we tested whether cells synchronized in the cell cycle responded to miR-206 with an arrest in G1. C2C12 cells were synchronized at the G1/S boundary by thymidine aphidicolin block (Fig. 5 D, middle). During the synchronization, we introduced miR-206 or GL2. Upon removal of aphidicolin, GL2-treated cells were released from G1 through the subsequent stages of the first cell cycle (2–8 h) and proceeded through the second cell cycle (8 h and later; Fig. 5 E). miR-206–treated cells were released into S phase with delayed kinetics (2–8 h), and the delay was even more marked in the second cell cycle (Fig. 5, D [bottom] and E [>8 h]). The time of second G1 (Fig. 5 E, 10–12 h) corresponds to 34–36 h after transfection. Because the kinetics of target repression after miR-206 transfection is likely to be slow, it is entirely expected that the cell cycle effect is more marked as the cell proceeds through successive cell cycles.
Other targets of the miRNAs
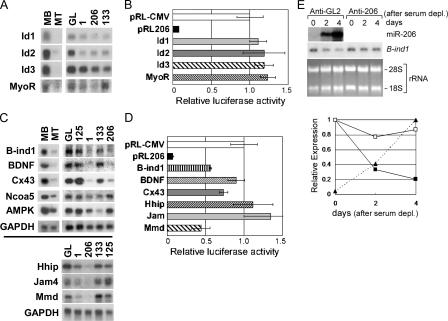
Besides Pola1, we tried to find additional targets of miR-206 by two approaches. First, we reasoned that inhibitors of differentiation could be putative targets of the miRNAs. MyoR (musculin) and Id 1–3 antagonize the action of the bHLH (basic helix-loop-helix) myogenic transcription factors like MyoD, whereas Hedgehog-interacting protein (Hhip) inhibits a factor called Hedgehog that promotes differentiation (Chuang and McMahon, 1999; Puri and Sartorelli, 2000; Li et al., 2004). All these mRNAs were down-regulated after transfection of miR-206 (Fig. 6, A and C; and Table S2). However, Id1-3 and MyoR are unlikely to be regulated directly by miR-206, as indicated by the failure of miR-206 to repress luciferase reporter fused to these genes in transient transfection assays (Fig. 6 B) and the absence of any predicted target site. Hhip has a predicted target site (Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1), but is also not a direct target of miR-206 in the luciferase fusion assay (Fig. 6 D). Indirect repression of these inhibitors of differentiation upon introduction of miR-206 into C2C12 cells indicates how the chain of events initiated by direct down-regulation of key targets by a miRNA can have a profound effect on the differentiation program.
Figure 6.
Examination of down-regulated genes from microarray to determine whether they are direct targets of miR-206. (A and C) Northern blots for MyoD inhibitors or down-regulated genes containing putative miRNA targets, as described in Fig. 4 A. (B and D) The luciferase assays were performed with pRL-CMV derivatives containing the mRNA sequences of the genes in A and C, respectively, as described in Fig. 2 A. The difference from RL-CMV control is significant for B-ind1 (P = 0.0059), Cx43 (P = 0.06), and Mmd (P = 0.0006). (E) Northern blot and quantification of B-ind1 mRNA after transfection of antisense oligonucleotide during C2C12 differentiation, as described in Fig. 4 D. The level of miR-206 during differentiation (of anti-GL2 transfected cells) was measured by RNase protection assays (top) and quantitated (bottom, filled triangle with dotted line).
The second approach to finding direct targets was to focus on genes that were decreased in the microarrays after miRNA introduction and contained putative target sites in the 3′ UTR. These include a Rac1 interacting protein named butyrate-induced transcript 1 (B-ind1), brain-derived neurotrophic factor, the gap-junction protein connexin43 (Cx43), nuclear receptor coactivator 5, AMP activated protein kinase, Hhip, junction adhesion molecule 4, and an adiponectin-related protein expressed in microglia called monocyte-to-macrophage differentiation-associated protein (Mmd). All of these mRNAs were decreased by normal differentiation and by transfection of the miRNA (Fig. 6 C). Although all of them have predicted target sites (Table S3), fusion to luciferase reporter reveals that only B-ind1, Cx43, and Mmd are direct targets of miR-206 like DNA pol α (Fig. 6 D).
Of the four direct targets, Pola1 has already been shown to be important for cell quiescence. B-ind1 might influence differentiation through its interaction with the G protein Rac-1. Down-regulation of B-ind1 mRNA was inversely related to the up-regulation of miR-206 during differentiation and was abrogated when miR-206 was inhibited by anti–miR-206 (Fig. 6 E). We do not have any evidence, however, that B-ind1 down-regulation is critical for differentiation. The known functions of Cx43 and Mmd do not suggest that their repression is critical for differentiation.
Mechanism for down-regulation of the target mRNAs by the miRNAs
Having confirmed a critical role of miR-206 in the down-regulation of at least two targets, Pola1 and B-ind1 mRNA (Figs. 4 and 6), we next turned our attention to the mechanism of the down-regulation. miRNAs, unlike siRNAs, are not perfectly matched to their targets and have been reported to repress their targets at the protein synthesis level (Olsen and Ambros, 1999; Seggerson et al., 2002; Dugas and Bartel, 2004). Plant miRNAs (for reviews see Kidner and Martienssen, 2005; Millar and Waterhouse, 2005) and at least one animal miRNA (Yekta et al., 2004) with perfect match to the target mRNA induce cleavage of the target. Short RNAs have also been shown to alter the chromatin state and thus repress the transcription of some genes (for review see Lippman and Martienssen, 2004). As was the case for Pola1, the addition of B-ind1 mRNA sequence downstream from a luciferase reporter confers suppression by miR-206, suggesting that B-ind1 is a direct target of miR-206 (Fig. 7 C). Point mutations indicate that at least two predicted target sites contribute to the down-regulation (Fig. 7, A, B, and C, M1 and M2; and Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1).
Nuclear run-on experiments were performed to determine whether there is any transcriptional repression of these direct target genes induced during differentiation or after introduction of miRNAs miR-1 or -206. Transcriptional activity of Pola1 was too low to be measured by nuclear run-on (unpublished data), but that of B-ind1 was measured and found to not decrease after C2C12 differentiation or after the introduction of miR-1 and -206 (Fig. 7 D). The Northern blots that accompanied this experiment show that the steady-state mRNA level of B-ind1 is repressed by at least fivefold (Fig. 7 D), suggesting that the miRNAs repress B-ind1 at the posttranscriptional level.
To test if the posttranscriptional reduction is due to the cleavage of mRNA by the miRNAs, we performed modified rapid amplification of cDNA ends (RACE)–PCR (Llave et al., 2002) to map the 5′ ends of potential cleavage fragments of B-ind1 and Pola1. Differentiated MTs (not depicted) and C2C12 MBs transfected with miR-206 and -1 (Fig. 7, E and F) generated more cleavage products from the B-ind1 transcript compared with the MBs or cells transfected with either GL2 control or miR-133. Similar results were obtained from Pola1 (unpublished data), supporting the hypothesis that the mRNAs are cleaved in the presence of the miRNAs. The sizes of RACE-PCR products derived from the B-ind1 (Fig. 7 E) or Pola1 (not depicted) after transfection of miR-206 are distinct from those obtained after transfection of miR-1 (Fig. 7 E and Fig. S3). As miR-1 and -206 recognize similar target sites and similarly decreased B-ind1 (Fig. 7 D), the different cleavage sites between the two miRNAs suggest that the miRNAs do not select the cleavage sites. To support this idea, sequencing of the RACE-PCR products revealed that the 5′ ends of cleavage fragments mapped at multiple sites in the B-ind1 mRNA (Fig. 7 A and Fig. S3) and were not confined to putative target sites of miR-206. B-ind1 mRNA was cleaved similarly during differentiation (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1). DNA pol α mRNA was cleaved by transfection of miR-206, and these cleavage sites also did not match with putative target sites (Fig. 7 A and Fig. S5). In contrast, siRNAs cleave the target mRNA at the center of RNA duplex by RISC (RNA-induced silencing complex; Elbashir et al., 2001b; Yekta et al., 2004). A control experiment with miR-206 siRNA against a perfectly matching site in luciferase 3′UTR showed that cleavage in this case was exactly at the siRNA target site (unpublished data). Thus, although the down-regulation of B-ind1 and Pola1 mRNA depends on miR-206 target sites with multiple mismatches and bulges, and although the target mRNA is cleaved, the 5′ ends of the cleavage fragments are distributed and do not match the target sites.
Discussion
We show that miRNAs not only promote differentiation of MBs in vitro but are required for differentiation. Individual miRNAs like miR-206 produce large changes in gene expression at the mRNA level that mimic changes seen upon differentiation, and the down-regulation of a direct target of miR-206, Pola1, might contribute to the cell cycle quiescence upon differentiation. In addition, several inhibitors of myogenic transcription factors are repressed after miR-206 transfection, leading to further amplification of the prodifferentiation function. At least two of the direct targets are cleaved at the posttranscriptional level, but the 5′ ends of the cleavage fragments suggest the involvement of exonucleases, a finding that has implications for the proposed involvement of processing bodies (P-bodies) in miRNA function. Finally, we describe an efficient strategy for identifying direct targets of miRNAs by intersecting mRNAs down-regulated by miR-206 in microarray screens with computationally predicted targets and then checking the intersection list by transient transfection and luciferase fusion assays.
Transfection of miR-1 in a nonmuscle cell has been reported to alter the gene expression profile toward that of muscle (Lim et al., 2005), but there was no indication of the magnitude of the change or the importance of the miRNAs in the physiology of muscle differentiation. Our results suggest that muscle-specific miRNAs promote skeletal myogenesis through direct down-regulation of several target genes and are required for normal differentiation. The microarray result indicates that the gene expression changes effected by miR-206 or -1 are closer to that of normal differentiation compared with that effected by miR-133. Because miR-206 is specifically expressed in skeletal muscle, whereas miR-1 is present in both skeletal and cardiac muscle, it would be interesting to compare miR-206 and -1 in greater detail to identify overlapping or unique roles in myogenesis.
A block to DNA synthesis through the direct down-regulation of DNA pol α precedes the cell cycle withdrawal and is the first demonstration of miRNAs directly affecting DNA replication. This result is supported by the recent observation that overexpression of miR-1 decreased the pool of proliferating ventricular cardiomyocytes in transgenic mice (Zhao et al., 2005). It is not clear yet whether down-regulation of the three other targets identified, B-ind1, Cx43, and Mmd, contributes in any way to the muscle differentiation program. To fully understand the role of miR-206 in skeletal myogenesis, we expect to uncover more targets of the miRNA in the future. Combinatorial down-regulation of multiple targets may be necessary to fully mimic the effect of miR-206 transfection into MBs.
miR-135b, -338 (Wienholds et al., 2005), and -181 (Naguibneva et al., 2006) are also muscle specific, suggesting that there may be additional miRNA involved in the regulation of myogenesis. miR-181 (Naguibneva et al., 2006), -1, and -133 (Chen et al., 2006) have already been shown to play a role in muscle differentiation by repressing their cognate targets, and our results add miR-206 to the list. We expect that extensive cooperation between several miRNAs and several transcription factors is necessary to effect the complete differentiation program.
The mode of action of miRNAs is still being debated. The multiple mismatches with target sites are believed to prevent cleavage of the target by a siRNA-like mechanism. Our results indicate, however, that at least for four endogenous targets, the repression by a miRNA is accompanied by a decrease in mRNA levels. For one of these direct targets (B-ind1), we rule out any transcriptional component, and for two of them (B-ind1 and Pola1), we show that the repression is accompanied by generation of mRNA cleavage fragments with multiple 5′ ends that do not map to the essential target sites matching the miRNA sequence. The exact 5′ ends vary and do not overlap when B-ind1 is down-regulated by the transfection of miR-206 or -1 or serum deprivation. This variability of the 5′ ends of the cleavage fragments suggests that they may be created by exonuclease activity after an initial endonucleolytic cleavage. RNA P-bodies have recently been proposed as sites where miRNAs sequester their target mRNAs from the protein-synthesis machinery (Liu et al., 2005; Sen and Blau, 2005). P-bodies contain exonucleases, so that the degradation of target mRNAs observed in this paper could be the end stage of the sequestration of targets in P-bodies. By serving as a common platform for both processes, P-bodies might explain how miRNAs can in some cases repress protein levels but not mRNAs and in other cases promote the degradation of mRNAs. An important point is that changes in mRNA profile during differentiation, thought to be mainly the province of transcription factors, can also be dictated by miRNA-mediated posttranscriptional cleavage of mRNAs.
The approach described here of intersecting mRNAs discovered to be down-regulated in microarray screens after miRNA transfection with mRNAs that have computationally predicted matches to miRNAs provides a high yield of potential targets. Fusion of candidate 3′UTRs to luciferase can then be used to quickly narrow down the list of targets to those that are directly down-regulated by miRNAs. Combining this strategy with in vitro differentiation systems is likely to yield many miRNA targets critical for differentiation down specific lineages. We are aware, of course, that this strategy might overlook targets that are exclusively down-regulated at the protein level, such as HoxA11 by miR-181 (Naguibneva et al., 2006) and histone deacetylase by miR-1 (Chen et al., 2006). If, however, down-regulation of protein synthesis by miRNAs involves sequestration of the target mRNAs to P-bodies, we suspect that many direct targets of miRNAs will eventually be down-regulated at the mRNA level and thus be revealed in the microarray screens. Because different computational algorithms predict different sets of targets for a given miRNA, a future extension of this work will be to intersect the microarray data with the output from other target prediction programs.
The ability of miRNAs to affect many mRNAs is similar to the ability of transcription factors to regulate many promoters simultaneously. Thus, just like transcription factors, we predict that miRNAs will induce complex changes in rate-limiting steps of cell metabolism and thus have a profound effect on the differentiation program. An additional point of interest is the interaction between these miRNAs and transcription factors known to be involved in muscle differentiation. For example, MyoD has already been suggested to be involved in the induction of miR-1 in cardiac myogenesis (Zhao et al., 2005). We suggest here that one of the muscle-specific miRNAs, miR-206, indirectly down-regulates Id1-3 and MyoR, inhibitors of myogenic transcription factors like MyoD. Such a positive feedback loop between the myogenic transcription factors and the muscle-specific miRNAs will be expected to push the equilibrium toward differentiation and could be very significant for holding muscle cells in a permanently differentiated state.
Materials and methods
Cell culture
C2C12 (American Type Culture Collection; Yaffe and Saxel, 1977) mouse skeletal MBs were maintained at subconfluent densities in DME supplemented with 20% FCS (GM). Myogenic differentiation (Andres and Walsh, 1996) into MT was induced by changing subconfluent cells to DME containing 2% heat-inactivated horse serum (DM).
Antibodies
Antibodies to p21 (C-19), p27 (C-19), Cdk2 (M2), Cdk4 (C-22), Pola1 (G-16), and myogenin (mouse mAb F5D) were purchased from Santa Cruz Biotechnology, Inc. Mouse mAb against MHC and anti–β-actin antibody were obtained from Sigma-Aldrich.
RNA oligonucleotides and transfection
Double-stranded RNA oligonucleotides containing on one strand the sequences of miR-1 (5′-uggaauguaaagaaguaugua-3′), -133 (5′-uugguccccuucaaccagcugu-3′), -206 (5′-uggaauguaaggaagugugugg-3′), and -125b (5′-ucccugagacccuaacuuguga-3′), as well as GL2 (Elbashir et al., 2001a) were synthesized by Invitrogen. Complementary sequence of each oligonucleotide was designed to produce a two-nucleotide overhang at both the 3′ ends of the duplex.
2′-O-methyl antisense oligonucleotides against miR-1, -133, and -206 and GL2 were synthesized by Dharmacon RNA Technologies. Transfection into C2C12 was performed with Lipofectamine 2000 reagent (Invitrogen), combined with 267 nM of each siRNA duplex or with 133 nM of 2′-O-methyl antisense oligonucleotide.
Immunostaining
Immunostaining was performed as described previously (Andres and Walsh, 1996). Unless otherwise specified, all manipulations were at room temperature. Cells on sterile glass coverslip were fixed with 2% formaldehyde in PBS for 15 min and were permeabilized with 0.2% Triton X-100 and 1% normal goat serum (NGS) in ice-cold PBS for 5 min. After blocking with 1% NGS in PBS two times for 15 min, incubation with primary antibody (1:400 in 1% NGS) for 1 h was followed by the FITC-conjugated anti–mouse IgG (dilution 1:500; DakoCytomation) for 1 h. Control experiment with H-3 (anti-hexahistidine) as a primary antibody ensured no cross-reactivity of the secondary antibodies. In case of sequential anti-BrdU probing, the above steps are repeated, but treatment with 1.5 N HCl for 30 min was included before the incubation with Alexa Fluor 594–conjugated anti-BrdU (Invitrogen) antibody (1:100 in 1% NGS) for 1 h. After washes, nuclei were counterstained with DAPI (H-1200; Vector Laboratories) for 1 min before mounting. Images were visualized using a microscope (Microphot-SA; Nikon) with 60× magnitude, captured using a camera (UFX-DX; Nikon), and processed using SPOT (version 3.5.4 for MacOS; Diagnostic Instruments) software.
Cdk2 kinase assays
50 μg of protein was immunoprecipitated with anti-Cdk2 antibody overnight and then pulled down on protein G–Sepharose beads for 2 h. The beads were washed three times with lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% NP-40, 5 mM EDTA, 50 mM NaF, 1 mM sodium vanadate, and protease inhibitors) and twice with kinase buffer (50 mM Hepes-NaOH, pH 7.4, and 25 mM MgCl2). The beads were then incubated with 25 μl kinase reaction mixture (50 mM Hepes-NaOH, pH 7.4, 25 mM MgCl2, 0.5 mM DTT, 50 μM ATP, 5 μCi γ-[32P]ATP, and 2 μg histone H1). The reactions were incubated at 30°C for 30 min and stopped by the addition of 12.5 μl of 3× SDS sample buffer.
Cell proliferation assay and measurement of DNA synthesis
Cell growth was measured with CellTiter 96 nonradioactive cell proliferation assay kit (Promega). DNA synthesis was measured as described previously (Busino et al., 2003) with minor modifications. Cells were treated with 1 μCi of methyl-[3H]thymidine in 0.25 ml of medium in a 24-well dish. Incubation at 37°C for 1 h was followed by washing with PBS two times. Radioactively labeled cells were treated with ice-cold stop solution containing 10% (vol/vol) TCA and 0.2 M sodium pyrophosphate for 20 min, washed in 95% ethanol two times, and solubilized in 1% (vol/vol) SDS and 10 mM NaOH. Solubilized samples were transferred onto paper (3MM; Whatman) and dried under an infrared lamp. The radioactivity was measured with a liquid scintillation counter.
RNA isolation and RNase protection assay
Total RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer's instructions. Total RNAs from human atrium, breast, brain, colon, cervix, heart, kidney, lung, ovary, prostate tumor, prostate, spleen, small intestine, skeletal muscle, testis, and uterus were purchased from Ambion, and total RNA from human bone marrow was purchased from CLONTECH Laboratories, Inc.
RNase protection assay was performed with miRNA detection kit (Ambion) with minor modifications. 20 μl of reaction contained the indicated amount of RNA, 0.6 μg of yeast tRNA, and the probe RNA, which was in vitro transcribed, labeled with α-[32P]UTP using miRNA probe construction kit (Ambion), and purified from polyacrylamide gel. Digestion products were resolved by electrophoresis in an 18% polyacrylamide gel with 7 M urea.
Microarray
C2C12 cells (MB) were induced to MT or were transfected six times at 24-h intervals with GL2 or miR-1, -133, or -206, respectively. Total RNAs from each sample were isolated on day 7 by using Trizol reagent. Subsequent steps for the hybridization to Affymetrix GeneChip Mouse Genome 430 2.0 Array (containing ∼45,000 transcripts) were done according to standard Affymetrix protocols. The array data were analyzed with the GeneChip Operating Software. The varied genes were identified by comparing the two samples in fold change and/or “present” and “absent” calls. The results of the microarray screen can be found on Gene Expression Open Source System (https://genes.med.virginia.edu/public_data/index.cgi) and are freely available to the public.
Nuclear run-on assay
For nuclear run-on assay, 0.5 μg of each DNA fragment was slot blotted onto a positively charged Nylon membrane (Nytran; Schleicher & Schuell). Nucleus isolation and run-on transcription reaction were performed as described previously (Greenberg and Bender, 2002) with modifications. After the transcription reaction, Trizol reagent was added, and RNA probe was prepared according to the manufacturer's instructions. Membranes were probed with 32P-labeled run-on RNA for 16 h at 68°C in hybridization buffer containing 0.25 M Na2HPO4, pH 7.2, 1 mM EDTA, and 7% (wt/vol) SDS. Membranes were washed twice in 2× SSC at room temperature for 15 min each, followed by a wash in 0.2× SSC and 1% (wt/vol) SDS at 65°C for 20 min.
RACE-PCR (RNA ligase-mediated [RLM]–PCR)
To map the cleavage sites of B-ind1 and Pola1 mRNA, RLM-PCR was performed with GeneRacer kit (Invitrogen) with modifications (Llave et al., 2002). Total RNAs from C2C12 cells transfected with GL2 or with miR-206 were ligated with RNA adaptor without any pretreatment (Yekta et al., 2004). B-ind1– or Pola1-specific primers for the cDNA synthesis, the first-round PCR, and the second-round PCR are indicated in Figs. S3, S4, and S5. The PCR products were separated on 1.1% agarose gel. Each lane on the agarose gel was quantified with ImageQuant 5.2 software and normalized to the B-ind1 mRNA level in the input mRNA. Titration of input mRNA ensured that the amount of RLM-RACE PCR product in a given sample is proportional to the input mRNA. The PCR products were subcloned into PCR4-TOPO vector and sequenced.
Luciferase reporter assays
For ease of subsequent subcloning, pRL-CMV(MCS) was modified from the original vector pRL-CMV (Promega) by inserting a synthetic linker with various restriction sites into the XbaI restriction site downstream of the ORF of Renilla (Renilla reniformis) luciferase gene.
At 20 h after transfection of the miRNAs and the luciferase plasmids into C2C12 cells, luciferase assays were performed with Dual-luciferase reporter assay system (Promega) per the manufacturer's instructions. Luminescent signal was quantified by luminometer (Monolight 3020; BD Biosciences). Each value from Renilla luciferase construct (Rr) was first normalized to the firefly (Photinus pyralis) luciferase assay value (Pp) from the cotransfected pGL3-control vector (Promega). Each Rr/Pp value was again normalized to the Rr/Pp value from miR-125b as a control. Each value is a mean of three transfections.
The luciferase constructs containing various putative target genes include B-ind1 (1248–2334 of accession no. NM_021345.1, available from GenBank/EMBL/DDBJ), brain-derived neurotrophic factor (700–1756 of accession no. NM_007540.3), Cx43 (82–2700 of accession no. BC006894), Hhip (800–2982 of accession no. NM_020259.3), junction adhesion molecule 4 (91–1491 of accession no. NM_028078.1), and Mmd (301–2510 of accession no. NM_026178.2). The indicated regions were PCR amplified and inserted into pRL-CMV.
Online supplemental material
Fig. S1 shows an examination of muscle markers and cell density after transfection of the miRNAs. Fig. S2 shows that miR-1, but not -133, directly down-regulates DNA pol α through the M1 site. Figs. S3, S4, and S5 show cleavage sites on B-ind1 or Pola1 mRNA. Tables S1 and S2 show the 30 most up- and down-regulated genes, respectively, in C2C12 transfected with miR-206 duplex. Table S3 shows that the potential target sequences for miR-206, predicted by miRanda, exist in several putative target genes. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200603008/DC1.
Supplementary Material
Acknowledgments
This work was supported by grant RO1 CA89406 (to A. Dutta) from the National Institutes of Health.
H.K. Kim and Y.S. Lee contributed equally to this paper.
Abbreviations used in this paper: B-ind1, butyrate-induced transcript 1; Cx43, connexin43; DM, differentiation medium; GM, growth medium; MB, myoblast; MHC, myosin heavy chain; miRNA, microRNA; Mmd, monocyte-to-macrophage differentiation-associated protein; MT, myotube; P-body, processing body; RACE, rapid amplification of cDNA ends; RLM, RNA ligase-mediated; rRNA, ribosomal RNA.
References
- Ambros, V., B. Bartel, D.P. Bartel, C.B. Burge, J.C. Carrington, X. Chen, G. Dreyfuss, S.R. Eddy, S. Griffiths-Jones, M. Marshall, et al. 2003. A uniform system for microRNA annotation. RNA. 9:277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres, V., and K. Walsh. 1996. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J. Cell Biol. 132:657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babak, T., W. Zhang, Q. Morris, B.J. Blencowe, and T.R. Hughes. 2004. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 10:1813–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga, S., J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus, and A.E. Pasquinelli. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 122:553–563. [DOI] [PubMed] [Google Scholar]
- Barad, O., E. Meiri, A. Avniel, R. Aharonov, A. Barzilai, I. Bentwich, U. Einav, S. Gilad, P. Hurban, Y. Karov, et al. 2004. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 14:2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297. [DOI] [PubMed] [Google Scholar]
- Baskerville, S., and D.P. Bartel. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 11:241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich, I. 2005. Prediction and validation of microRNAs and their targets. FEBS Lett. 579:5904–5910. [DOI] [PubMed] [Google Scholar]
- Busino, L., M. Donzelli, M. Chiesa, D. Guardavaccaro, D. Ganoth, N.V. Dorrello, A. Hershko, M. Pagano, and G.F. Draetta. 2003. Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature. 426:87–91. [DOI] [PubMed] [Google Scholar]
- Chen, J.F., E.M. Mandel, J.M. Thomson, Q. Wu, T.E. Callis, S.M. Hammond, F.L. Conlon, and D.Z. Wang. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, P.T., and A.P. McMahon. 1999. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 397:617–621. [DOI] [PubMed] [Google Scholar]
- Doench, J.G., and P.A. Sharp. 2004. Specificity of microRNA target selection in translational repression. Genes Dev. 18:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench, J.G., C.P. Petersen, and P.A. Sharp. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas, D.V., and B. Bartel. 2004. MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 7:512–520. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. a. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., W. Lendeckel, and T. Tuschl. 2001. b. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright, A.J., B. John, U. Gaul, T. Tuschl, C. Sander, and D.S. Marks. 2003. MicroRNA targets in Drosophila. Genome Biol. 5:R1.1–R1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, M.E., and T.P. Bender. 2002. Identification of newly transcribed RNA. In Short Protocols in Molecular Biology, vol. 1. F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, editors. John Wiley & Sons, Inc., Indianapolis, IN. 4-25–4-29.
- Guo, K., J. Wang, V. Andres, R.C. Smith, and K. Walsh. 1995. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol. Cell. Biol. 15:3823–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., and P.D. Zamore. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 297:2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., M.J. Simard, C.C. Mello, and P.D. Zamore. 2004. Sequence-specific inhibition of small RNA function. PLoS Biol. 2:E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalma, Y., L. Marash, Y. Lamed, and D. Ginsberg. 2001. Expression analysis using DNA microarrays demonstrates that E2F-1 up-regulates expression of DNA replication genes including replication protein A2. Oncogene. 20:1379–1387. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and R.A. Martienssen. 2005. The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 8:38–44. [DOI] [PubMed] [Google Scholar]
- Kitzmann, M., and A. Fernandez. 2001. Crosstalk between cell cycle regulators and the myogenic factor MyoD in skeletal myoblasts. Cell. Mol. Life Sci. 58:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, A., D. Grun, M.N. Poy, R. Wolf, L. Rosenberg, E.J. Epstein, P. MacMenamin, I. da Piedade, K.C. Gunsalus, M. Stoffel, and N. Rajewsky. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735–739. [DOI] [PubMed] [Google Scholar]
- Li, X., C.S. Blagden, H. Bildsoe, M.A. Bonnin, D. Duprez, and S.M. Hughes. 2004. Hedgehog can drive terminal differentiation of amniote slow skeletal muscle. BMC Dev. Biol. 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L.P., N.C. Lau, P. Garrett-Engele, A. Grimson, J.M. Schelter, J. Castle, D.P. Bartel, P.S. Linsley, and J.M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., and R. Martienssen. 2004. The role of RNA interference in heterochromatic silencing. Nature. 431:364–370. [DOI] [PubMed] [Google Scholar]
- Liu, C.G., G.A. Calin, B. Meloon, N. Gamliel, C. Sevignani, M. Ferracin, C.D. Dumitru, M. Shimizu, S. Zupo, M. Dono, et al. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA. 101:9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., M.A. Valencia-Sanchez, G.J. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Z. Xie, K.D. Kasschau, and J.C. Carrington. 2002. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 297:2053–2056. [DOI] [PubMed] [Google Scholar]
- Meister, G., M. Landthaler, Y. Dorsett, and T. Tuschl. 2004. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 10:544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., and P.M. Waterhouse. 2005. Plant and animal microRNAs: similarities and differences. Funct. Integr. Genomics. 5:129–135. [DOI] [PubMed] [Google Scholar]
- Naguibneva, I., M. Ameyar-Zazoua, A. Polesskaya, S. Ait-Si-Ali, R. Groisman, M. Souidi, S. Cuvellier, and A. Harel-Bellan. 2006. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 8:278–284. [DOI] [PubMed] [Google Scholar]
- Olsen, P.H., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671–680. [DOI] [PubMed] [Google Scholar]
- Puri, P.L., and V. Sartorelli. 2000. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J. Cell. Physiol. 185:155–173. [DOI] [PubMed] [Google Scholar]
- Saxena, S., Z.O. Jonsson, and A. Dutta. 2003. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 278:44312–44319. [DOI] [PubMed] [Google Scholar]
- Schiaffino, S., and C. Reggiani. 1996. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76:371–423. [DOI] [PubMed] [Google Scholar]
- Seggerson, K., L. Tang, and E.G. Moss. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243:215–225. [DOI] [PubMed] [Google Scholar]
- Sempere, L.F., S. Freemantle, I. Pitha-Rowe, E. Moss, E. Dmitrovsky, and V. Ambros. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, G.L., and H.M. Blau. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633–636. [DOI] [PubMed] [Google Scholar]
- Thomson, J.M., J. Parker, C.M. Perou, and S.M. Hammond. 2004. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 1:47–53. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., W.P. Kloosterman, E. Miska, E. Alvarez-Saavedra, E. Berezikov, E. de Bruijn, H.R. Horvitz, S. Kauppinen, and R.H. Plasterk. 2005. MicroRNA expression in zebrafish embryonic development. Science. 309:310–311. [DOI] [PubMed] [Google Scholar]
- Yaffe, D., and O. Saxel. 1977. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 270:725–727. [DOI] [PubMed] [Google Scholar]
- Yekta, S., I.H. Shih, and D.P. Bartel. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596. [DOI] [PubMed] [Google Scholar]
- Yu, Z., T. Raabe, and N.B. Hecht. 2005. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol. Reprod. 73:427–433. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., E. Samal, and D. Srivastava. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 436:214–220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.