Abstract
In many animals, the bipolar spindle of the first zygotic division is established after the contribution of centrioles by the sperm at fertilization. To avoid the formation of a multipolar spindle in the zygote, centrosomes are eliminated during oogenesis in most organisms, although the mechanism of this selective elimination is poorly understood. We show that cki-2, a Caenorhabditis elegans cyclin-dependent kinase (Cdk) inhibitor, is required for their appropriate elimination during oogenesis. In the absence of cki-2, embryos have supernumerary centrosomes and form multipolar spindles that result in severe aneuploidy after anaphase of the first division. Moreover, we demonstrate that this defect can be suppressed by reducing cyclin E or Cdk2 levels. This implies that the proper regulation of a cyclin E–Cdk complex by cki-2 is required for the elimination of the centrosome that occurs before or during oogenesis to ensure the assembly of a bipolar spindle in the C. elegans zygote.
Introduction
Experiments performed by Boveri (1900) over a century ago revealed the essential requirement for accurate centrosome inheritance and its role in regulating genome integrity in the developing embryo. In many metazoans, the establishment of the bipolar spindle during the first zygotic cell division is dependent on the paternal contribution of a microtubule organizing center. After fertilization, this organelle will recruit pericentriolar material present within the oocyte cytoplasm to assemble the two functional centrosomes that will define the first mitotic spindle. In addition to this essential role of the centrosome in organizing the spindle, in Caenorhabditis elegans, this structure is also required to specify the anterior/posterior axis after sperm entry in a microtubule-dependent and -independent manner (O'Connell et al., 2000; Wallenfang and Seydoux, 2000; Cowan and Hyman, 2004a). Therefore, the appropriate regulation of centrosome number is pivotal because aberrations in these controls result in asymmetrical chromosome segregation and/or severe polarity defects.
Although centrosomes are associated with most nuclei in C. elegans, including those in the germ line, they are absent in oocytes, whereas they are clearly detectable and required for fertility in the sperm (Kemp et al., 2004). The loss of the centrosome from the oocyte is common to many species, but the mechanism responsible for this elimination is currently unknown. During our characterization of a C. elegans Cdk inhibitor (CKI; cki-2) we noticed that compromise of cki-2 function caused embryos to arrest at the one-cell stage with a multipolar spindle. We show that this defect is due to a role of cki-2 in centrosome elimination, and our data provide pioneering evidence on how centrosomes are appropriately eliminated from the developing oocyte.
Results and discussion
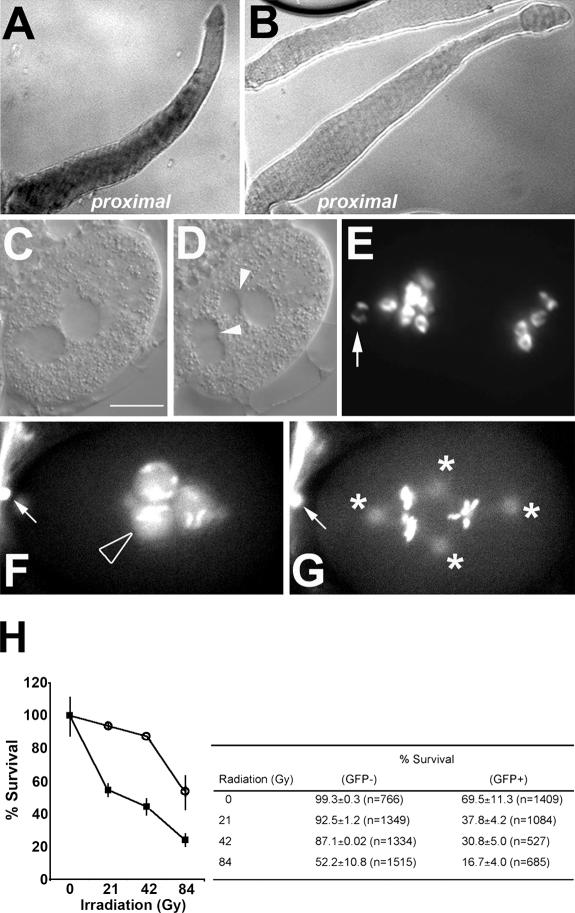
Recently, large-scale screens using RNAi-based strategies have provided a framework for understanding many maternally controlled embryonic processes (Sonnichsen et al., 2005). However, not all genes respond equally to RNAi. Our initial use of RNAi analysis to understand the role of a C. elegans CKI called cki-2 was not informative because of the variable penetrance and frequency of the RNAi-related phenotypes. Furthermore, no loss-of-function cki-2 alleles are currently available. We therefore turned to an alternative reverse genetic approach called cosuppression, which is an RNAi-related posttranscriptional gene-silencing mechanism that is conserved among many phyla (Ketting and Plasterk, 2000). In wild-type animals, cki-2 mRNA is normally present in the hermaphrodite germ line but is excluded from the distal mitotic zone (Fig. 1 A). To test whether cki-2 could be compromised through the cosuppression pathway, we expressed the 3′ portion of the cki-2 gene (Dernburg et al., 2000), which could not encode a functional protein and shared a very low degree of sequence conservation with cki-1, a second C. elegans CKI (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200512160/DC1). The cosuppression transgenic array included a GFP marker facilitating our detection of animals that possessed the transgene. We obtained several transgenic lines in different genetic backgrounds, all of which indicated that reduction of cki-2 consistently resulted in reproducible embryonic lethality wherein ∼60% of the GFP transgene-bearing embryos (GFP+) failed to complete embryogenesis (Table I). The abundance of cki-2 mRNA was reduced substantially throughout the gonad in these GFP+ animals (Fig. 1 B), whereas the observed embryonic lethality could be reversed by genetically disrupting this silencing mechanism using mutants in the downstream components of the cosuppression pathway (mut-7 and rde-2), indicating that the observed lethality was specifically due to the reduction of cki-2 through cosuppression (Table I). We therefore refer to these GFP+ animals as cki-2 cosuppressed (cki-2cs). Although ∼40% of the cki-2cs embryos survive embryogenesis and continue larval development without visible abnormalities, we found that these animals are irradiation sensitive (Fig. 1 H). This indicates that despite their wild-type appearance, the DNA damage response in cki-2cs animals is nonetheless compromised. Therefore, reduction of cki-2 function results in cell cycle–related abnormalities that reflect the various thresholds of cki-2 activity required to appropriately execute these cellular processes. Among the embryonically arrested embryos, we noticed that 7% of the embryos (n = 558) arrested at the one-cell stage with multiple micronuclei (9.1%; n = 66), consistent with abnormal chromosome segregation and/or cytokinesis (Fig. 1, C–E). Examination of the affected zygotes by differential interference contrast indicated that early events (contractions of the anterior membrane or ruffling and pseudocleavage) before the pronuclear meeting were not significantly different from wild type (unpublished data). Shortly after nuclear envelope breakdown, however, the two pronuclei reformed and several de novo micronuclei became apparent. Cleavage furrows appeared occasionally but would regress, and ∼50% (n = 18) of the micronuclei-containing embryos did not form a cleavage furrow. The remaining 50% were defective in cleavage plane orientation, although both classes did undergo multiple rounds of karyokinesis (Fig. 1, C–E).To better understand the basis of the “one-cell” arrest phenotype, we imaged cki-2cs embryos that harbored GFP-histone and GFP–β-tubulin transgenes. In some embryos, we observed a second maternal pronucleus (4.5%; n = 66), a meiotic defect that arises because of abnormal polar body exclusion (Fig. 1 F). We also noted that chromosomes failed to align correctly after nuclear envelope breakdown, whereas the spindle microtubules appeared to be organized around multiple foci, typical of extra microtubule organizing centers or centrosome-like structures (Fig. 1 G and Video 1).
Figure 1.
cki-2cs causes multiple phenotypes typical of a negative cell cycle regulator. (A and B) in situ RNA hybridization using an antisense cki-2 probe on wild-type (A) or cki-2cs (B) gonads extruded from adult hermaphrodites. (C and D) Sequential differential interference contrast images of a cki-2cs one-cell embryo showing normal pronuclear meeting (C) and nuclear divisions without appropriate cytokinesis giving rise to supernumerary nuclei (D, arrowheads) with variable DNA content based on staining with DAPI (E). (F and G) A sequential GFP fluorescence image of cki-2cs one-cell–arrested embryo that expresses [H2B∷GFP; β-tubulin∷GFP]. The open arrowhead indicates an extra maternal pronucleus, asterisks mark centrosomes, and the arrows indicate polar bodies. (H) Irradiation sensitivity of cki-2cs (GFP+; closed square) or wild-type sibling (GFP−) animals (open circle). The values are presented as the percentage of embryos that hatched from a total population of embryos laid from irradiated or not parents that were examined at each point. At point zero in each experiment, the survival percentage was normalized to 100%. The error bars represent the standard deviation of two independent experiments (P < 0.05; 95% confidence). Bar, 10 μm.
Table I.
cki-2 cosuppression causes embryonic lethality
| Embryonic lethality
|
||
|---|---|---|
| Genotype | GFP+ | GFP− |
| % | % | |
| N2 | NA | 0.29 (n = 1384) |
| N2; cki-2 (RNAi) | NA | 5.5 (n = 710) |
| N2; [fem-1∷GFP] (0/4) | 0 (n = 244)a | ND |
| N2; [fem-1∷cki-2C] (3/3) | ||
| line #1 | 26.9 (n = 466)a | 0.7 (n = 280) |
| line #2 | 23.3 (n = 103) | ND |
| line #3 | 8.1 (n = 186) | ND |
| rrf-3; [fem-1∷cki-2C] (2/2) | ||
| line #1 | 55.3 (n = 159)a | 27.6 (n = 116) |
| line #2 | 42.2 (n = 436) | 25.1 (n = 231) |
| TH27 (pie-1∷γ-tub∷GFP); [fem-1∷cki-2C] (5/5) | ||
| line #1 | 29.1 (n = 1257)a | 1.7 (n = 232) |
| line #2 | 21.5 (n = 395) | ND |
| line #3 | 19.2 (n = 198) | ND |
| rde-2; [fem-1∷cki-2C] (0/2) | ||
| line #1 | 5.7 (n = 357) | 7.5 (n = 374) |
| line #2 | 11.6 (n = 404) | 17.7 (n = 561) |
| mut-7; [fem-1∷cki-2C] (0/3) | ||
| line #1 | 17.9 (n = 313) | 20.2 (n = 325) |
| line #2 | 11.4 (n = 245) | 12.1 (n = 440) |
| line #3 | 12.7 (n = 181) | 9.4 (n = 276) |
A C. elegans strain that harbors an extrachromosomal array containing the [fem-1∷cki-2C] cosuppression transgene segregates animals that possess the array (GFP+) or not (GFP−), as indicated by the presence of the dominant elt-2∷GFP cotransformation marker. Embryonic lethality was presented as the percentage of unhatched embryos from total progeny obtained from GFP+ or GFP− young adult animals. The frequency of the embryonic lethality phenotype in the various trangenic lines obtained is shown in parentheses. The embryonic lethality from GFP− animals was determined from only one transgenic line of each tested genotype.
The transmission frequency (%) of the transgenic array in these strains was scored as the number of GFP+ progeny from the total number of progeny, and the transmission rate of the cki-2cs strain used throughout the study was ∼50%.
To confirm that this unique multipolar spindle phenotype was due to the reduction of cki-2 and not due to cosuppression-related phenomena or nonspecific effects on cki-1, we used an RNAi-sensitive strain (Simmer et al., 2002) to reduce either cki-1 or -2 levels to reproduce the cki-2cs–associated multipolar spindle phenotype. We did detect one-cell embryos with supernumerary centrosomes after cki-2(RNAi) in rrf-3 (Table II and see Fig. 3, E and F), although the penetrance of the defect was considerably lower than that observed in cki-2cs animals. On the other hand, despite causing a high frequency of embryonic arrest in the rrf-3 background, cki-1(RNAi) never caused a one-cell arrest or a multipolar spindle phenotype (Table II). Therefore, we conclude that the supernumerary centrosomes and the resulting multipolar spindle defect observed in cki-2cs embryos were not due to effects on cki-1 function or due to cosuppression per se but, rather, to a loss or reduction of cki-2 function.
Table II.
Supernumerary centrosomes are present in the one-cell embryo of cki-2cs animals
| Genotype | Embryonic lethality | Supernumerary centrosomea |
|---|---|---|
| % | % | |
| rrf-3 | 23.0 ± 1.2 (n = 374) | 0 (n = 76) |
| rrf-3; cki-1 (RNAi) | 94.7 (n = 570) | 0 (n = 40) |
| rrf-3; cki-2 (RNAi) | 27.5 ± 3.7 (n = 734) | 4.5 (n = 111) |
| N2; [fem-1∷cki-2C]b | 26.9 (n = 466) | 13.5 (n = 133) |
| TH27; [fem-1∷cki-2C]b | 29.1 (n = 1257) | 6.7 (n = 60) |
One-cell embryos obtained from the cki-2 cosuppression transgene-bearing animals (GFP+) were examined to score the frequency (%) of the supernumerary centrosomes. For RNAi of cki-1 or -2, each dsRNA was injected into rrf-3 hermaphrodites as described (see Materials and methods), and the frequency (%) of both the embryonic lethality and the supernumerary centrosomes was scored. The embryonic lethality was presented as the percentage of unhatched embryos from total progeny obtained from the RNAi-treated mothers.
Embryos were stained with anti–SPD-2 or γ-tubulin∷GFP, and the results are presented as the percentage of the total number of one-cell stage embryos examined. All one-cell embryos examined were at or before the first cell division.
The frequency of the supernumerary centrosome defect was determined in the most penetrant cosuppressed lines (line #1 of N2; [fem-1∷cki-2C] and TH27; [fem-1∷cki-2C]) for comparison.
Figure 3.
cki-2(RNAi) causes defects in the elimination of the maternal centrosome. (A–C) Early wild-type one-cell embryo (A; prepronuclear migration stage) or cki-2cs embryos that express GFP–γ-tubulin to visualize centrosomes (B and C). (D–F) Early one-cell embryos (prepronuclear migration stage) from rrf-3 (D) or rrf-3; cki-2(RNAi) (E and F) adult hermaphrodites stained with anti–SPD-2 antibody. The arrows indicate polar bodies stained with DAPI (anterior). Asterisks mark centrosomes (maternal [m] and paternal [p]). The white rectangular box in A shows the paternal centrosome that could not be observed in the same focal plane. The rectangular boxed regions in B and D–F were magnified to show greater detail.
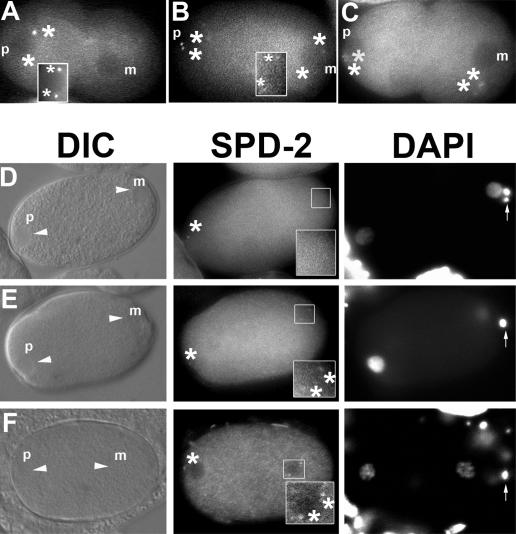
To address whether cki-2 affected the centrosome cycle during spermatogenesis or, alternatively, during oogenesis, we examined centrosome numbers in early pronuclear stage embryos using an antibody against SPD-2, a coiled-coil protein that associates with the centrosome (Kemp et al., 2004). We noticed that unlike wild-type embryos, strong SPD-2 expression was visible at distinct foci in both the paternal and maternal pronuclei (pronuclear meeting stage; Fig. 2, A and B). To ascertain whether the presence of the extra centrosomes was indeed due to their contribution from the maternal pronucleus, as opposed to defects associated with failed cytokinesis (Skop et al., 2004), we imaged embryos from meiosis to pronuclear meeting using GFP–γ-tubulin, revealing that GFP–γ-tubulin was associated with the maternal pronucleus in prepronuclear migration stage embryos obtained from cki-2cs animals (6.7%; n = 60; Fig. 3, B and C), whereas we never observed GFP–γ-tubulin associated with the maternal pronucleus in wild-type embryos (n = 80; Fig. 3 A).
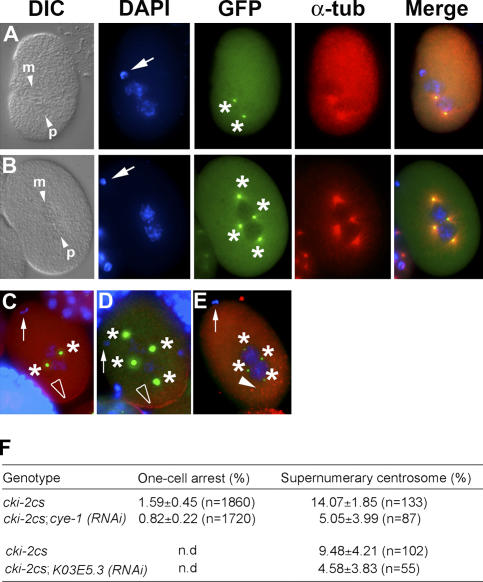
Figure 2.
Supernumerary centrosomes observed in cki-2cs embryos are contributed by the maternal pronucleus in a cyclin E/Cdk2-dependent manner. (A and B) Late pronuclear stage wild-type (A) or cki-2cs (B) one-cell embryo stained with DAPI (blue), anti–SPD-2 (green), and anti–α-tubulin (red). The small arrowheads indicate the pronuclei at different stages. Arrows indicate polar bodies, and asterisks indicate centrosomes. p and m, paternal and maternal pronuclei, respectively. (C and D) PAR-2∷GFP (red) in the posterior cortex (open arrowheads) of a wild-type (C) or a cki-2cs (D) one-cell embryo. (E) Anti–P-granule staining (red spots; closed arrowhead) of a cki-2cs one-cell embryo. The arrows indicate polar bodies (anterior), and the asterisks mark centrosomes. (F) Frequency (%) of cki-2cs–associated one-cell arrest and the persistence of maternal centrosome after cye-1(RNAi) or K03E5.3(RNAi). Standard deviation of at least three independent experiments is shown, and asterisks represent significant differences compared with cki-2cs controls (P < 0.05; 95% confidence). The one-cell arrest phenotype was presented as the percentage of unhatched one-cell embryos from the total number of progeny (embryos and larvae). The embryos from injected or uninjected (control) animals were labeled with DAPI and anti–SPD-2 antibody 24 h after dsRNA microinjection, and the resulting one-cell embryos were examined for supernumerary centrosomes. The results are presented as the percentage of the total number of embryos examined at the one-cell stage. All one-cell embryos examined were at or before the first cell division. The variation observed in the penetrance of the centrosome defect is due to the progressive silencing of the cosuppression transgene over time.
Collectively, these results indicate that the supernumerary centrosomes were already associated with the maternal pronucleus at the time of fertilization in cki-2cs embryos, possibly because they were not appropriately eliminated in the maternal germ line as a result of a reduction in cki-2 function. However, because we could not show definitive live images of an embryonic cell division beginning in the prepronuclear stage to the first mitotic division, we cannot formally rule out the possibility that the supernumerary centrosomes may arise from a cytokinesis failure after the first mitotic division.
Therefore, to test whether centrosome elimination is defective in cki-2cs oocytes, we stained the gonads of affected (GFP+) and unaffected (GFP−) animals with an anti–SAS-4 antibody to determine whether centrioles were abnormally present in the oocytes of cki-2cs animals. SAS-4 is associated with all centrioles in C. elegans and is required for their duplication (Leidel and Gonczy, 2003). In wild-type animals, SAS-4 is associated with all germ cell nuclei, although SAS-4 staining foci were noticeably absent from oocytes (Fig. 4 A). The absence of the SAS-4/centriole staining in oocytes is consistent with previous observations that the centrosomes are eliminated from the germ cell nuclei at or around the stage of oocyte commitment (Albertson and Thomson, 1993).
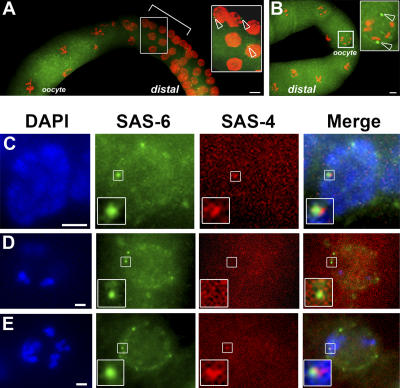
Figure 4.
Centrioles are not appropriately eliminated during oogenesis in cki-2cs animals. (A and B) Extruded gonads from wild-type (A) or cki-2cs (B) adult hermaphrodites stained with DAPI (red) and anti–SAS-4 (green). The bracket in A delineates the region that corresponds to oocyte commitment, where ∼50% of the germ cell nuclei stain positively for SAS-4. The region within the rectangular box is shown in detail, and the open arrowheads indicate SAS-4 foci (centrioles) in this inset and throughout. The inset in B shows a magnified oocyte (from the white frame) with two SAS-4 staining foci. (C–E) A wild-type meiotic germ cell (C), a wild-type oocyte (D), or an oocyte from a cki-2cs adult hermaphrodite (E). All were stained with DAPI (blue), Cy3-conjugated anti–SAS-6 (green), or Cy5-conjugated anti–SAS-4 (red). The region within the rectangular box is shown at higher magnification. Bars: (A and B) 10 μm; (C–E) 2.5 μm.
Anti–SAS-4 staining of the oocytes from the cki-2cs hermaphrodite animals revealed that SAS-4 staining structures were present next to the oocyte nuclei at a frequency consistent with the penetrance of the extra centrosome defect caused by the cki-2cs transgene (8.9%; n = 79), whereas no obvious SAS-4 foci were ever observed in oocytes in wild-type animals (Fig. 4 B and not depicted). Although this is the strongest evidence that cki-2 is required for appropriate centriole elimination during oogenesis, we wanted to further confirm that the anti–SAS-4 staining recognized bona fide centrioles and not simply SAS-4 aggregates in the oocyte. We therefore stained the oocytes of wild-type and cki-2cs animals using anti–SAS-4 and anti–SAS-6, both of which recognize the centriole ( Dammermann et al., 2004; Leidel and Gonczy, 2005). Both antibodies recognized the centrioles of embryos, where they colocalize with γ-tubulin (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200512160/DC1). After double staining, we compared the number of overlapping signals between wild-type and cki-2cs germ lines (Fig. 4, C–E). Consistent with our previous observation (Fig. 4 B), we noted that significantly more SAS-6 staining oocytes showed overlapping positive signals with anti–SAS-4 in the cki-2cs animals (14/55 SAS-6–positive oocytes) compared with wild-type (1/29 SAS-6–positive oocytes; this single overlapping SAS-4 signal may be due to juxtaposition of the signals during the deconvolution process; Fig. 4, D and E). Therefore, our staining with two independent centriole-specific antibodies suggests that the observed foci are indeed centrioles, which are not appropriately eliminated in the cki-2cs oocytes.
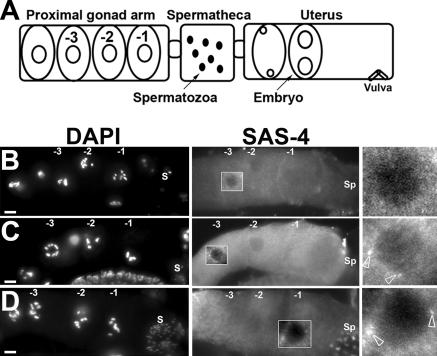
In C. elegans, oogenesis occurs in an assembly line–like fashion (Fig. 5 A; Schedl, 1997). We observed that the SAS-4 staining structures persisted into the late stages of oogenesis in cki-2cs hermaphrodites (Fig. 5, B–D). These data are consistent with cki-2 playing a critical role in the timely elimination of the maternal centrioles during oogenesis, and when its activity is reduced below a critical threshold, the centrioles persist and eventually will give rise to the supernumerary centrosomes. Although our results strongly argue that cki-2 is involved in the elimination of maternal centrioles, ultrastructural studies would provide more definitive evidence of centriolar perdurance. Intriguingly, although the maternally contributed centrosomes are the likely cause of the abnormal division observed in the one-cell–arrested cki-2cs embryos, we have been unable to show that these supernumerary centrosomes can nucleate microtubules and/or duplicate beyond the first division. We also noticed that the polarity of the affected embryos seems consistently normal based on GFP–PAR-2 (100%; n = 17; Fig. 2, C and D) or P-granule staining (Fig. 2 E; Cowan and Hyman, 2004b). Our observation that anterior/posterior polarity does not seem to be affected in cki-2cs zygotes suggests that although the maternally contributed centrosomes appear competent to organize a mitotic spindle, they are seemingly not equivalent to the paternal centrosome in providing the polarity cue in the zygote. The basis of this difference between the centrosome pairs is currently unknown, as no difference in centrosomal morphology or molecular composition has been identified between the centrosomes of paternal and maternal origin.
Figure 5.
Centrioles persist into the later stages of oogenesis in cki-2cs animals. (A) Diagram of late-stage oogenesis in the proximal gonad arm. The number indicates the position of the oocyte undergoing meiotic maturation. Oocytes in diakinesis of meiotic prophase I before maturation (−3, −2); the oocyte adjacent to the spermatheca is designated as −1. (B–D) A proximal gonad arm from a wild-type animal (B) or cki-2cs animals (C and D) stained with anti–SAS-4 antibody. S, Spermatozoa and/or Spermatids; Sp, Spermatheca. Open arrowheads indicate SAS-4 foci detected in the oocyte nuclei (C and D). The white rectangular boxed region was magnified to provide greater detail. Bars, 10 μm.
Our observations, although obtained with fixed embryos, suggest that a functional difference may distinguish the maternal and the paternal centrosome in establishing the anterior/posterior polarity at fertilization. However, we have been unsuccessful in imaging the maternally contributed centrosomes into and beyond the first division while simultaneously monitoring the establishment of the PAR-2 domain. Therefore, we cannot formally rule out the possibility that the polarity is established early by the sperm and that the extra centrosomes we observe in the multinucleate embryos are paternal in origin that have duplicated and appear later due to cytokinesis defects (Fig. 2, A–E).
Because meiotic defects were also observed in cki-2cs embryos, we determined whether the abnormal presence of centrosomal components on the meiotic spindle might disrupt the normal mechanism of the acentriolar meiotic division. We found that the morphology of the meiotic spindle in early cki-2cs zygotes is disorganized (Fig. S2 C, available at http://www.jcb.org/cgi/content/full/jcb.200512160/DC1), whereas SPD-2 was detectable as a diffuse haze surrounding the spindle (Fig. S2, A and B). We also found that ZYG-1, a protein that is also required for centrosomal duplication (O'Connell et al., 2001), was similarly present on the meiotic spindle in cki- 2cs zygotes (unpublished data), suggesting that the atypical presence of these ectopic centrosomal materials may be responsible for the meiotic spindle abnormalities and the consequent meiotic defects observed in cki-2cs embryos.
The loss of cki-2 could result in misregulated levels of Cdk activity within the oocyte, causing a centrosomal anlage to persist and eventually form the tetrapolar spindle that results in a one-cell arrest. To test this scenario, we compromised G1/S Cdk activity by performing cye-1(RNAi), which is the only E-type cyclin in C. elegans (Fay and Han, 2000). Loss of cyclin E has no effect on the first cell division in C. elegans (Fay and Han, 2000). However, after cye-1(RNAi) in cki-2cs animals, the characteristic one-cell arrest phenotype was suppressed substantially, which was also reflected in the nearly twofold reduction in the frequency of the multipolar spindle defect (Fig. 2 F). A similar degree of suppression was also observed after K03E5.3(RNAi), where K03E5.3 is the predicted C. elegans Cdk2 homologue (Liu and Kipreos, 2000; Fig. 2 F). Control animals injected with double-stranded (dsRNA) corresponding to cyclin D showed no such effect (unpublished data).
That this effect of cyclin E occurs independently of Cdk activity (Matsumoto and Maller, 2004) seems unlikely based on the current accepted mechanism of CKI function and our observation that K03E5.3(RNAi) suppressed the frequency of the persistence of the maternal centrosomes to levels comparable to cye-1(RNAi). Our data are thus consistent with the loss of cki-2 resulting in misregulated cyclin E/Cdk2 activity in the germ line that consequently allows centrioles to perdure into the developing oocyte.
That both ZYG-1 and SPD-2 persist during oogenesis and are present on the meiotic spindle in cki-2cs embryos suggests that their levels may be regulated by cyclin E/Cdk activity, in a manner similar to Mps1 (Fisk and Winey, 2001). The loss of cki-2 therefore reveals a previously undescribed function of cyclin E–Cdk complexes in centrosome stabilization in the C. elegans germ line. Through the timely regulation of this activity, the maternal centrosomes are eliminated as the germ cell acquires its oocyte fate.
This novel function of Cdks and CKIs in centrosome inheritance would probably not have been uncovered through conventional gene targeting in mouse models. Unlike most animals, the sperm does not contribute the centrioles in the mouse; instead, they arise de novo in the fertilized zygote (Schatten, 1994). Why, then, do most metazoans selectively eliminate the centrosomes within the maternal germline? The answer may come from species that can develop parthenogenetically, where the oocyte is thought to harbor a centriolar anlage (Delattre and Gonczy, 2004). This would be selected against in species that undergo a biparental mode of development based on sperm-specific centriolar contribution. The elimination of the maternal centrosomes, either through CKI-mediated or related mechanisms, would block the ability of the oocyte to develop parthenogenetically and strongly favor the union of sperm and egg to trigger the onset of cell division in the zygote. Because the mode of centrosome inheritance in C. elegans shares considerable parallels with that of many animals, identification of the Cdk targets in this model may provide invaluable insight pertinent to the mode of centrosome inheritance shared by most metazoans, including humans.
Materials and methods
Nematode strains
The following C. elegans strains were used: N2 Bristol was used as the wild type throughout. MR258 (N2; rrEx258 [fem-1∷cki-2C; elt-2∷GFP]), MR306 (N2; rrEx306 [fem-1∷GFP; elt-2∷GFP]), MR294 (rde-2; rrEx294 [fem-1∷cki-2C; elt-2∷GFP]), MR303 (mut-7; rrEx303 [fem-1∷cki-2C; elt-2∷GFP]), NL917 (mut-7 [pk204]), WM29 (rde-2 [ne221]), MR446 (unc-119; ruIs32 [unc-119(+); pie-1∷GFP∷H2B]; ojIs1 [unc-119(+); pie-1∷GFP∷ TBB-2]; rrEx258 [fem-1∷cki-2C; elt-2∷GFP]), XA3501 (unc-119; ruIs32 [unc119(+); pie-1∷GFP∷H2B]; ojIs1 [unc-119(+); pie-1∷GFP∷TBB-2]), TH27 (unc-119; ddIs6 [unc-119(+); pie-1∷GFP∷ TBG-1]), MR628 (itIS153 [rol-6(+); pie-1∷PAR-2∷GFP]; rrEx258 [fem-1∷cki-2C; elt-2∷GFP]), MR824 (unc119; ddIs6 [unc-119(+); pie-1∷GFP∷TBG-1]; rrEx824 [fem-1∷cki-2C; elt-2∷GFP]), NL2099 (rrf-3(pk1426)), and KK866 (itIS153 [rol-6(+); pie-1∷PAR-2∷GFP]). All C. elegans strains were cultured using standard techniques and maintained at 20°C unless stated otherwise (Brenner, 1974).
Constructs
For cki-2 cosuppression, 3 kb of genomic sequence upstream of the fem-1 translational start site was PCR amplified from N2 genomic DNA followed by SphI–Pst1 digestion and insertion into pPD49.26 to yield pMR220. The cki-2C fragment (amino acids 116–259; lacking a translational start site; Fig. S1) was prepared by PCR and then inserted into pMR220 at the BamHI–XmaI sites to create pMR221. The fem-1 promoter fragment was inserted into pPD95.77 at SphI–PstI sites to yield pMR266. For RNAi of cki-2, a cki-2 template for dsRNA synthesis was generated by subcloning the cki-2 cDNA into the PstI–KpnI sites of pBluescript II to generate pMR215. cye-1 dsRNA was prepared as described previously (Fay and Han, 2000). cki-1 dsRNA was prepared as described previously (Hong et al., 1998). K03E5.3 dsRNA template was amplified from a clone of the bacterial feeding RNAi library (I-1D09) using PCR and inserted into the SacI–SacII sites of pBluescript II to generate pMR330.
cki-2 cosuppression and RNAi
pMR220 and pMR221 were coinjected (50 μg/ml) with 100 μg/ml elt-2∷GFP as a coinjection marker into N2 hermaphrodites as described previously (Mello et al., 1991). F1 progeny expressing elt-2∷GFP were singled, and their progeny (F2) were scored for transmission of the extrachromosomal array. Embryonic lethality was scored from each transgenic line. dsRNA was obtained by in vitro transcription reactions, annealing, and injection as described previously (Fire et al., 1998). Injected animals were transferred to new plates every 24 h, and the F1 progeny was examined for visible abnormalities that affected development or cell division.
Antibodies and immunological methods
The following primary antibodies were used: anti–α-tubulin (Sigma-Aldrich), polyclonal anti–rabbit SPD-2 (a gift from K. O'Connell, National Institutes of Health, Bethesda, MD), rabbit polyclonal anti–SAS-4 (a gift from P. Gonczy, Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland), Cy3-conjugated anti–SAS-6 and Cy5-conjugated anti–SAS-4 (a gift from K. Oegema, University of California, San Diego, La Jolla, CA), and rabbit polyclonal anti–P-granule (a gift from S. Strome, Indiana University, Bloomington, IN). Secondary antibodies were anti–rabbit or anti–mouse Texas red or FITC-conjugated secondary antibodies or anti–rabbit Alexa Fluor 594 secondary antibody (all obtained from Invitrogen). DAPI (Sigma-Aldrich) was used to counterstain slides to reveal DNA. Embryos or hermaphrodite gonads were fixed and stained as described elsewhere (Couteau et al., 2004). Indirect immunofluorescence microscopy was performed using a 60× oil-immersion objective lens in a compound microscope (DMR; Leica) equipped with a digital camera (C4742-95; Hamamatsu), imaging an ∼0.5-μm-thick optical section. Image analysis, computational deconvolution, and pseudocoloring were performed using Openlab 4.0.2 software (Improvision). All images using Cy3-conjugated anti–SAS-4 and Cy5–conjugated anti–SAS-6 were acquired (using a 60× oil-immersion objective lens) and deconvolved using an image restoration system (DeltaVision; Applied Precision). Data were collected as a series of 35 optical sections in increments of 0.25 μm under standard parameters using the SoftWoRx 3.0 program (Applied Precision). Images were processed using Photoshop 8.0 (Adobe). All microscopic works were performed at 20°C.
In situ hybridization
Digoxigenin-labeled antisense and sense probes were generated using T7 and T3 kits with digoxigenin-11-UTP (Roche). In situ hybridization was performed on the gonads dissected from wild-type or cki-2cs (GFP+) adult hermaphrodites as described previously (Feng et al., 1999).
Online supplemental material
Fig. S1 shows protein sequence alignment of CKI-2 with -1. Fig. S2 depicts centrosomal material persisting on the meiotic spindle in cki-2cs one-cell embryos. Fig. S3 shows an embryonic cell labeled with GFP–γ-tubulin, anti–SAS-6, and anti–SAS-4. Video 1 shows a cki-2cs one-cell embryo labeled with GFP histones and GFP–β-tubulin. Video 2 shows a wild-type one-cell embryo (pronuclear migration stage) labeled with GFP–γ-tubulin. Video 3 shows a cki-2cs one-cell embryo (pronuclear migration stage) labeled with GFP–γ-tubulin. Video 4 shows a cki-2cs one-cell embryo (prepronuclear migration stage) labeled with GFP–γ-tubulin. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200512160/DC1.
Supplementary Material
Acknowledgments
We are very grateful to Kevin O'Connell for discussion and support; to Pierre Gonczy, Tony Hyman, Karen Oegema, David Fay, and Susan Strome for reagents and comments; to Tony Gartner for suggesting cosuppression; to Jackie Vogel, Monique Zetka, and Jean-Claude Labbe for comments on the manuscript; to Shaolin Li for technical assistance; and to the Caenorhabditis Genetics Center for their continued support.
This work was funded by the Natural Sciences and Engineering Research Council and a research award from the Canadian Cancer Society. D.Y. Kim is a Fonds de Recherche en Santé du Québec scholar, and R. Roy is a Canadian Institutes of Health Research new investigator.
Abbreviations used in this paper: CKI, Cdk inhibitor; dsRNA, double-stranded RNA.
References
- Albertson, D.G., and J.N. Thomson. 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1:15–26. [DOI] [PubMed] [Google Scholar]
- Boveri, T. 1900. Zellen-Studien: Ueber die Natur der Centrosomen. Fisher, Jena, Germany. 220 pp.
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau, F., K. Nabeshima, A. Villeneuve, and M. Zetka. 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14:585–592. [DOI] [PubMed] [Google Scholar]
- Cowan, C.R., and A.A. Hyman. 2004. a. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 431:92–96. [DOI] [PubMed] [Google Scholar]
- Cowan, C.R., and A.A. Hyman. 2004. b. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 20:427–453. [DOI] [PubMed] [Google Scholar]
- Dammermann, A., T. Muller-Reichert, L. Pelletier, B. Habermann, A. Desai, and K. Oegema. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 7:815–829. [DOI] [PubMed] [Google Scholar]
- Delattre, M., and P. Gonczy. 2004. The arithmetic of centrosome biogenesis. J. Cell Sci. 117:1619–1630. [DOI] [PubMed] [Google Scholar]
- Dernburg, A.F., J. Zalevsky, M.P. Colaiacovo, and A.M. Villeneuve. 2000. Transgene-mediated co-suppression in the C. elegans germ line. Genes Dev. 14:1578–1583. [PMC free article] [PubMed] [Google Scholar]
- Fay, D.S., and M. Han. 2000. Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell-cycle control and vulval development. Development. 127:4049–4060. [DOI] [PubMed] [Google Scholar]
- Feng, H., W. Zhong, G. Punkosdy, S. Gu, L. Zhou, E.K. Seabolt, and E.T. Kipreos. 1999. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1:486–492. [DOI] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M.K. Montgomery, S.A. Kostas, S.E. Driver, and C.C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811. [DOI] [PubMed] [Google Scholar]
- Fisk, H.A., and M. Winey. 2001. The mouse Mps1p-like kinase regulates centrosome duplication. Cell. 106:95–104. [DOI] [PubMed] [Google Scholar]
- Hong, Y., R. Roy, and V. Ambros. 1998. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 125:3585–3597. [DOI] [PubMed] [Google Scholar]
- Kemp, C.A., K.R. Kopish, P. Zipperlen, J. Ahringer, and K.F. O'Connell. 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell. 6:511–523. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., and R.H. Plasterk. 2000. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 404:296–298. [DOI] [PubMed] [Google Scholar]
- Leidel, S., and P. Gonczy. 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 4:431–439. [DOI] [PubMed] [Google Scholar]
- Leidel, S., and P. Gonczy. 2005. Centrosome duplication and nematodes: recent insights from an old relationship. Dev. Cell. 9:317–325. [DOI] [PubMed] [Google Scholar]
- Liu, J., and E.T. Kipreos. 2000. Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): differential conservation of CAKs in yeast and metazoa. Mol. Biol. Evol. 17:1061–1074. [DOI] [PubMed] [Google Scholar]
- Matsumoto, Y., and J.L. Maller. 2004. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 306:885–888. [DOI] [PubMed] [Google Scholar]
- Mello, C.C., J.M. Kramer, D. Stinchcomb, and V. Ambros. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, K.F., K.N. Maxwell, and J.G. White. 2000. The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev. Biol. 222:55–70. [DOI] [PubMed] [Google Scholar]
- O'Connell, K.F., C. Caron, K.R. Kopish, D.D. Hurd, K.J. Kemphues, Y. Li, and J.G. White. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 105:547–558. [DOI] [PubMed] [Google Scholar]
- Schatten, G. 1994. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev. Biol. 165:299–335. [DOI] [PubMed] [Google Scholar]
- Schedl, T. 1997. Developmental genetics of the germ line. In C. elegans II. D.L. Riddle, T. Blumenthal, B.J. Meyer, and J.R. Priess, editors. Cold Spring Harbor Laboratory Press, New York. 241–269. [PubMed]
- Simmer, F., M. Tijsterman, S. Parrish, S.P. Koushika, M.L. Nonet, A. Fire, J. Ahringer, and R.H. Plasterk. 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12:1317–1319. [DOI] [PubMed] [Google Scholar]
- Skop, A.R., H. Liu, J. Yates III, B.J. Meyer, and R. Heald. 2004. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 305:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen, B., L.B. Koski, A. Walsh, P. Marschall, B. Neumann, M. Brehm, A.M. Alleaume, J. Artelt, P. Bettencourt, E. Cassin, et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 434:462–469. [DOI] [PubMed] [Google Scholar]
- Wallenfang, M.R., and G. Seydoux. 2000. Polarization of the anterior-posterior axis of C. elegans is a microtubule-directed process. Nature. 408:89–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.