Abstract
The regulation of microtubule dynamics is attributed to microtubule-associated proteins that bind to the microtubule outer surface, but little is known about cellular components that may associate with the internal side of microtubules. We used cryoelectron tomography to investigate in a quantitative manner the three dimensional structure of microtubules in intact mammalian cells. We show that the lumen of microtubules in this native state is filled with discrete, globular particles with a diameter of 7 nm and spacings between 8 and 20 nm in neuronal cells. Cross-sectional views of microtubules confirm the presence of luminal material in vitreous sections of brain tissue. Most of the luminal particles had connections to the microtubule wall, as revealed in tomograms. A higher accumulation of particles was seen near the retracting plus ends of microtubules. The luminal particles were abundant in neurons, but were also observed in other cells, such as astrocytes and stem cells.
Introduction
The systematic ultrastructural analysis of intact cellular components in their native, hydrated state has only recently become possible because of major improvements of noninvasive EM methods for specimen preservation and imaging—cryoelectron tomography (cryo-ET) and cryoelectron microscopy of vitreous sections (CEMOVIS). In CEMOVIS, vitrified biological material is sectioned and imaged at a low temperature (for review see Al-Amoudi et al., 2004). Cryo-ET can directly image thin regions of cells that adhere or grow on EM grids (Medalia et al., 2002), and it is becoming the method of choice for providing 3D information about intact intracellular structures at molecular resolution (Baumeister, 2005). Recently, these emerging techniques have provided valuable insights into cellular architecture, as well as into complex macromolecular assemblies (for reviews see Al-Amoudi et al., 2004; Lučić et al., 2005). Microtubules have been studied by ET in sections from freeze-substituted and plastic-embedded material (O'Toole et al., 2003), and recently, tomograms of frozen-hydrated sea urchin sperm flagella revealed details of their microtubules and associated proteins (Nicastro et al., 2005; Sui and Downing, 2006).
Microtubule-associated proteins (MAPs) maintain the stability of microtubules and regulate their dynamics, whereas microtubule-based motors mediate the transport of cargo along microtubule tracks. Although all MAPs and motors studied so far bind to the outside of the microtubule wall, small molecules such as taxol can associate with the luminal side of the microtubule (Nogales et al., 1999). It has also been proposed that a short repeat motif of the MAP tau can be localized on the inner surface of the microtubule lattice (Kar et al., 2003). Intriguingly, the presence of electron-dense material was observed within the microtubule lumen in plastic-embedded and heavy metal–stained preparations of insect epithelia and spermatids (Bassot and Martoja, 1966; Afzelius, 1988) and blood platelets (Behnke, 1967; Xu and Afzelius, 1988). Such luminal material appears to be especially prominent in neuronal cells (Peters et al., 1968; Rodriguez Echandia et al., 1968; Burton, 1984). However, none of these studies has revealed details about the form and distribution of this material along microtubules or the nature of its association with the microtubule wall.
We used cryo-ET and CEMOVIS to examine the microtubules of neuronal and other cells in a state of optimal structural preservation. We demonstrate that these microtubules contain within their lumens discrete particles with connections to the microtubule wall, and we analyze the occurrence, size, and distribution of these particles.
Results and discussion
To study microtubules in their native state, we acquired cryo-ETs of thin processes of cultured cells, as well as images of vitrified sections, both depicting the morphology of the cytoplasm as it is believed to occur in living cells (Al-Amoudi et al., 2004; Baumeister, 2005). For cryo-ET we cultivated hippocampal neurons and other mammalian cells directly on the carbon support of gold EM grids. These cells have processes or peripheral areas that are thinner than 0.5 μm, and thus, are amenable to being studied in detail by ET without the need for sectioning. The morphology of neurons cultured on these EM grids was similar to that of neurons grown on classical glass coverslips, as assessed by light microscopy (Fig. 1, A and B). The use of “finder” EM grids facilitated correlating light microscopy images of living neurons with EM images and tomograms that were acquired after preservation by rapid freezing in liquid ethane to localize the imaged region of the cell (Fig. 1, C–G). Among a variety of membrane organelles and complexes, the cytoskeletal elements are visible on electron micrographs (Fig. 1, F and G) and could be analyzed in detail in tomograms (Fig. 2, A–C, and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200606074/DC1). Close examination of microtubules from >20 tilt series revealed electron-dense material within the lumens that comprised discrete, closely spaced globular particles (Fig. 2, A–C). These particles were reliably detected only when analyzing the tomograms and not in projection images (compare Fig. 1 [F and G] to Fig. 2 A and Video 1). The typical distances between particles were 8, 12, 16, and, occasionally, 20 nm (Fig. 2, C and D). This distribution, with an apparent multiple of 4 nm (Fig. 2 D) corresponding to the size of a tubulin monomer, suggests that the luminal particles could systematically interact with tubulin along the microtubule wall. It also raises the possibility that the particles might associate with both α and β tubulin or bind to a site located between the α and β subunits, possibly along stretches of the same protofilament. Occasionally, stretches devoid of luminal particles were also observed over distances of several tens of nanometers.
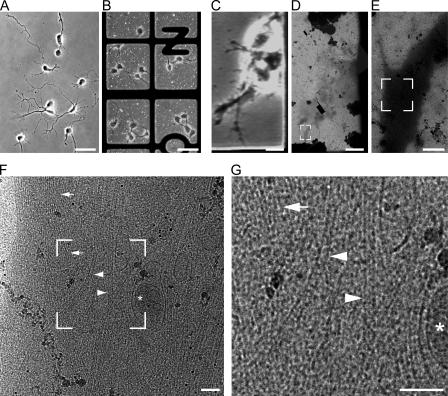
Figure 1.
Cultivation and imaging of neurons on EM grids. (A and B) Phase-contrast light microscopy images of 2-d-old rat embryonic hippocampal neurons grown on a glass coverslip (A) and on the carbon support of an EM grid (B). (C) Phase-contrast light microscopy image of a neuronal process. (D) A montage of low-magnification cryo-EM images showing the same region as in C, but after plunge-freezing the grid in liquid ethane. (E–G) Cryo-EM images showing subsequent enlargements in the area along the major process. The boxes in D–F indicate the area magnified in the subsequent image. (G) A projection image recorded at 0° tilt, at the magnification typically used for tilt series (43,000 on the CCD camera). Microtubules (arrowheads), actin bundles (arrows), and membrane organelles (asterisk) are indicated. Bars: (A and B) 50 μm; (C and D) 10 μm; (E) 1 μm; (F and G) 0.1 μm.
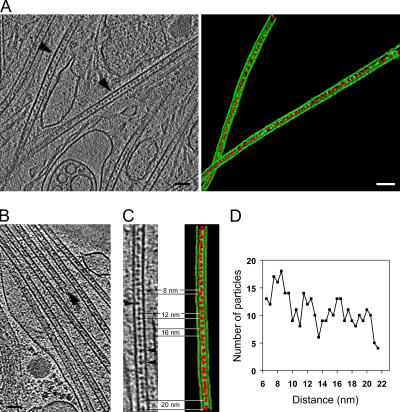
Figure 2.
Cryo-ET of intact neuronal processes reveals luminal particles inside microtubules. (A) A 6-nm-thick slice through a tomogram of a neuronal process. Particulate densities are clearly detectable within the lumen of microtubules (left). The microtubules indicated with arrowheads were surface-rendered in green, with the luminal particles highlighted in red (right). (B) A cross-section through a reconstruction from another neuronal process, showing a bundle of microtubules. The arrow indicates a microtubule end. (C) Magnified view (left) and surface-rendered representation (right) of one microtubule from the tomogram in B. Different distances between neighboring particles (red) can be measured within a short stretch of a single microtubule, typically ranging between 8 and 20 nm. The arrowheads indicate luminal particles with apparent connections to the microtubule wall. (D) Plot of the distribution of the distances between neighboring luminal particles in microtubules, showing peaks at 8, 12, 16, and possibly 20 nm.
Some tomograms revealed microtubules with widely flared ends (Fig. 3 A; compare with the nondepolymerizing end of the microtubule in Fig. 2 B), suggesting that they were captured during shrinking, which can take place as a result of the naturally occurring cycles of neurite extension and retraction (Dotti et al., 1988). In such shrinking microtubules, the luminal particles showed ∼1.5-fold higher packing densities compared with microtubules in growing neurites (Fig. 3 A and Fig. 2, A and B). The luminal particles at the depolymerizing ends reached the minimum measured interparticle distance of ∼8 nm. This packing may explain the “beaded fiber” appearance of some classical transmission EM preparations (Peters et al., 1968). However, the particulate character of the structures becomes obvious in cryotomograms (Fig. 3 A).
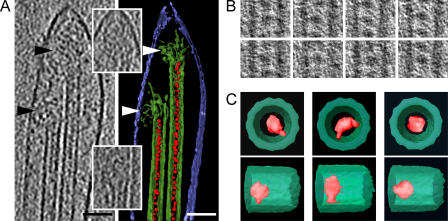
Figure 3.
Enrichment of luminal particles at depolymerizing microtubule ends and modes of binding to the microtubule wall. (A) A slice through a tomogram, corrected for the CTF (left), and a surface-rendered representation (right), showing the end of a shrinking neuronal process with depolymerizing microtubules. The depolymerizing state of the microtubules is indicated by the flared ends of the protofilaments, which are shown in different tomographic sections in the insets (the left and right microtubule flares are displayed in the bottom and top insets, respectively). The color coding is as in Fig. 2, with the plasma membrane depicted in blue. The arrowheads indicate the flared protofilaments. Bar, 50 nm. (B) Examples of luminal particles used for averaging. Most of the particles have apparent connections to the microtubule wall. (C) Surface-rendered averaged volumes in top views (top row) and side views (bottom row). Two types of interactions with the tubulin wall could be detected; single contact (left) and double contact (middle); some particles had no detectable contact (right). All individual panels in B and C are 35 nm wide.
In side views, some of the luminal particles show distinct connections with the microtubule wall (Fig. 2 C and Fig. 3 B). The visual inspection was strongly reinforced by particle-averaging procedures (Beck et al., 2004; Förster et al., 2005). We analyzed 518 luminal particles using 3D averaging (Fig. 3 B). All analyzed particles had a very similar overall size of 7 nm; however, the alignment and averaging of such small particles in noisy tomograms failed to uncover fine structural details. Nevertheless, it revealed three subtypes of particles, based on their association with the microtubule wall (Fig. 3 C). The particles belonging to the first group (n = 193) appeared rodlike and were elongated normal to the microtubule axis. The bulk of the averaged density was located centrally in the microtubule lumen, whereas an elongated protrusion pointed toward and contacted the microtubule wall. The particles assigned to the second group (n = 169) appeared crescent shaped, with two arms protruding radially from a central mass at an angle of ∼140° relative to each other, and with their ends attached to the inner aspect of the microtubule. Particles with no evident connection to the microtubule wall (n = 156) were classified in a third group. The lack of a visible contact does not necessarily indicate that the particles bear no attachment points. One limitation of ET is the restricted range of viewing angles (Baumeister, 2005), resulting in incomplete sampling of data. Connecting stalks could incline parallel to the tomographic Z axis so as to fall into the “missing wedge” of data (Lučić et al., 2005), and thus, appear less distinctly in the tomograms. In support of such an explanation, there were ∼30% of “unconnected” particles within microtubules, and the missing wedge within the tomographic data comprises a third of the complete volume.
Because microtubules in cells grown on flat supports for cryo-ET are principally oriented perpendicularly to the imaging direction, we could obtain relatively complete representations of longitudinal views. On the other hand, top views of microtubules can be particularly well demonstrated by sectioned material. Therefore, we imaged vitreous sections of neuronal cells in organotypic cultures derived from hippocampus (Fig. 4, A and B). In top views of microtubules (Fig. 4, A and B), the luminal material appeared as discrete densities, some of which displayed connections to the tubulin wall. In 50-nm-thick sections, we expect an average of 3–4 luminal particles (Table I) superimposed in a projected image along the microtubule axis. Occasionally, we also observed transversely sectioned microtubules that were devoid of distinct luminal material (Fig. 4 A), presumably corresponding to the empty stretches we detected by cryo-ET. In longitudinally sectioned microtubules, the internal material was also clearly observed as discrete particles (Fig. 4 A), which is consistent with the cryo-ET observations. The luminal densities were also detected with similar frequencies within the microtubules in cell bodies on vitreous sections (Fig. 4 B), indicating that they are a general microtubule constituent in neuronal cells, and not restricted to the neuronal processes. In contrast to neural tissue, distinct luminal material was not commonly seen in transverse sections of microtubules from cultured rat hepatoma (HTC) cells (Fig. 4 C).
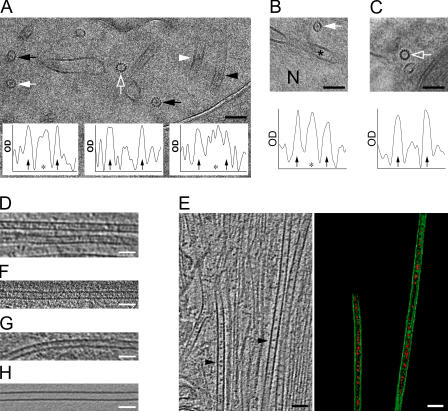
Figure 4.
Luminal particles in different cell types revealed by CEMOVIS and cryo-ET. (A) Cryo-EM projection image of a vitrified section (CEMOVIS) of neuronal tissue, showing an area within a process containing microtubules with luminal material, seen in both microtubule top views (black arrows) and side views (black arrowhead). Some microtubules have little detectable luminal material (empty arrow). The insets show density profiles across the microtubules indicated with a white arrow (left), an empty white arrow (middle), and white arrowhead (right). The arrows in the plots indicate the position of the microtubule protofilaments; asterisks denote the central density (if detected), generated by the luminal material. (B) CEMOVIS of neuronal tissue, showing a region within the cell body, as indicated by the proximity of the nucleus (N), where microtubules containing luminal material are also observed (white arrow). Asterisk, nuclear envelope. The plot shows the density profile through the microtubule indicated with the arrow. (C) Microtubules from rat HTC cells. The plot shows a density profile through the transversely sectioned microtubule (indicated with an empty white arrow). The density of material inside the microtubule lumen is similar to that of the surrounding cytoplasm. The images in A–C were CTF corrected. (D) Tomographic slice, showing microtubules in an astroglial cell in dissociated hippocampal culture. (E) Tomographic slice, showing a region in a process of a P19 cell grown on an EM grid (left). The two microtubules indicated with arrowheads were surface rendered (right). A 3D view of this tomogram is shown in Video 2. (F and G) Tomographic slices of a microtubule from a PtK2 cell (F) or HeLa cell (G) cultured on EM grids, lacking regular dense particles in the lumen. (H) A tomographic slice of a microtubule nucleated in vitro, devoid of any luminal material. Video 2 is available at http://www.jcb.org/cgi/content/full/jcb.200606074/DC1. Bars, 50 nm.
Table I.
Occurrence and distribution of the luminal particles
| MT length | Number of particles | Particles per micrometer | Mean distance between particles | |
|---|---|---|---|---|
| μm | nm | |||
| Minor neurites, proximal part | 6.704 | 458 | 68.3 | 14.6 |
| Minor neurites, distal part | 6.401 | 489 | 76.3 | 13.1 |
| Axons, proximal part | 5.163 | 326 | 63.3 | 15.8 |
| Axons, distal part | 5.756 | 439 | 76.3 | 13.1 |
| Depolymerizing MTs, at plus ends (≤ 1 μm) | 1.922 | 212 | 111.0 | 9.0 |
| Depolymerizing MTs, middle part (>1 μm) | 2.596 | 203 | 78.1 | 12.8 |
| Stem cells (P19) | 4.655 | 241 | 51.8 | 19.3 |
The luminal particles were quantified in the microtubules of axons and minor neurites in the regions close to the cell body (proximal parts) or close to the neurite tips (distal parts). In the case of processes of P19 pluripotent precursor cells, no distinction was made. Regions of depolymerizing microtubules (MTs) that were closer than 1 μm to the flared end were counted as “plus ends,” and regions further than 1 μm from the flared end were counted as the “middle part.”
The particulate luminal material was abundant in the microtubules of all examined tomograms from neuronal processes. It was also observed in microtubules from astroglial cells, which are present as a minor constituent in our dissociated hippocampal cultures (Fig. 4 D). We further examined whether it could also be detected by cryo-ET within the microtubules of stem cells using the pluripotent P19 embryonal carcinoma cell line (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200606074/DC1; McBurney et al., 1982). The microtubules of P19 cells contained discrete internal particles (Fig. 4 E; Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200606074/DC1; and Table I) that were very similar in size to the ones observed in neuronal cells (Fig. 2, A–C). However, inside P19 microtubules, the typical stretches of closely spaced particles were more frequently separated by longer particle-free intervals of up to 50 nm.
To determine whether the luminal particles are a common constituent of the microtubule lumen in other cell types, we applied similar data collection and tomographic analyses to microtubules in epithelial cell lines, such as PtK2 (Fig. 4 F) and HeLa (Fig. 4 G), as well as to microtubules nucleated in vitro from purified pig brain tubulin (Fig. 4 H). There was no evidence of any electron-dense material in the lumen of in vitro–nucleated microtubules (Fig. 4 H). Consistent with the observations made using CEMOVIS (Fig. 4 C), the lumen of microtubules in tomograms of epithelial cell lines was mostly free from distinct, particulate densities (Fig. 4, F and G); however, occasional traces of electron-dense material could be detected inside microtubules. These densities were irregularly distributed, varied in size, and displayed no obvious symmetry. Based on the known constraints on the signal and resolution, as well as the relatively high level of noise in images and tomograms of intact cells (Baumeister, 2005), we can presently neither confirm nor conclusively rule out the presence of luminal material in such microtubules. In contrast, in neuronal, astroglial, and stem cells we can systematically observe and analyze luminal particles with a consistent size and distribution, irrespective of the noise levels in the tomograms.
Measurements both on original tomograms and on the volumes reconstructed by cross-correlation and averaging revealed that the luminal particles had a roughly globular shape with a diameter of 6–7 nm (Fig. 3, B and C). A particle of such dimensions would have a molecular mass of at least 200 kD. As the mean distance between particles was 14 nm (Table I), we estimated that ∼6–7% of the total volume available within the neuronal microtubule lumen was occupied by the observed luminal material. At depolymerizing microtubule plus ends with mean distances of 8–9 nm (Table I), the occupied volume increased to ∼10%. The particles were also enriched by 12–21% in the proximal segments of both minor neurites and nascent axons compared with the distal regions of the same processes (Table I). The luminal particles inside the microtubules of P19 cells were also abundant, but the average distance between them (19.3 nm) was somewhat larger than in neurons.
We have demonstrated that cryo-ET and CEMOVIS are reliable methods for analyzing the macromolecular architecture of microtubules, as they occur in the cytoplasm of living cells. Currently, it is an open question as to whether the luminal particles we describe represent novel types of MAPs that would be involved in modulating microtubule stability. Another possibility is suggested by the fact that the sole acetylated residue of tubulin (α-tubulin Lys40) has been predicted to reside on the internal surface of microtubules (Nogales et al., 1999). It would therefore be intriguing to investigate whether the luminal particles we observed may represent the hitherto elusive tubulin acetyltransferase or a tubulin deacetylase (Westermann and Weber, 2003). The alternative hypothesis that the luminal material (protein or mRNA) may be transported inside the microtubules has already been put forward (Rodriguez Echandia et al., 1968; Burton, 1984). Our estimates of the size, shape, and molecular mass of the particles do not support a classical motor–cargo complex; however, novel mechanisms of transport, or even the use of the lumen as a storage space, cannot be excluded. Our work provides a basis for future studies to characterize the biological role and the structure of the luminal particles within microtubules.
Materials and methods
Cell culture on EM grids and cryopreparation
For primary neuronal cultures, the hippocampi of 17.5-d-old rat embryos were dissected, trypsinized, and dissociated by trituration (de Hoop et al., 1998). 5 × 104 cells were plated in 35-mm tissue culture dishes containing either EM grids, which were processed as described in the following paragraph, or poly-l-lysine–coated coverslips. The cultures were first imaged at ∼30°C in HBSS containing 7 mM Hepes, pH 7.25, with an inverted light microscope (Axiovert 135 TV; Carl Zeiss MicroImaging, Inc.), with a 32× air objective (Achrostigmat, NA 0.40; Carl Zeiss MicroImaging, Inc.), using a high-performance charged-couple device (CCD) camera (model 4912; Cohu) and Scion Image 4.0.2 software (Scion Corporation). Detailed maps of the cultivated cells were recorded from every grid before preparing them for cryo-EM by rapid freezing.
P19 cells were cultivated on EM grids using a synthetic medium and serum replacement (Knockout D-MEM and Knockout SR, respectively; both from Invitrogen) to minimize spontaneous differentiation.
HeLa and PtK2 cells were grown on EM grids in MEM containing 10% fetal bovine serum.
Rat HTC cells were grown at 37°C, 5% CO2, in 50-ml tissue culture flasks (Falcon) containing D-MEM supplemented with 10% fetal calf serum, 2% l-glutamine, 50 μg penicillin, and 50 U streptomycin per ml.
400-μm-thick transverse hippocampal slices were prepared from 6–7-d-old rats and maintained for 10–15 d in culture as previously described (Stoppini et al., 1991). They were high-pressure frozen after a 5-min immersion in medium supplemented with 20% dextran (40 kD) and 5% sucrose, as previously described (Zuber et al., 2005). This treatment did not affect the viability of the slices.
Cryopreparation and imaging
For cell cultivation and cryopreparation we used finder gold EM grids of 200 mesh, covered with a carbon support containing widely spread small holes (either self-made or obtained from Jena [Quantifoil-R5/20]). The grids were sterilized (UV light for 15 min), coated in a 1 mg/ml solution of poly-l-lysine, washed in water, and incubated in MEM containing 10% horse serum, which was substituted with N2 medium before plating the neurons (de Hoop et al., 1998). During preparation, the grids were kept at a temperature of 31–36.5°C. The grids containing cells were mounted in a plunger equipped with a custom-made humidifying device (Cyrklaff et al., 1990). After adding fiducial markers (3 μl protein A-gold; Sigma-Aldrich; in N2 medium) the excess liquid was removed from the grids by blotting with filter paper (Whatman Nr 4) from the underside for 30–40 s. The grids were rapidly frozen in liquid ethane slush, cooled in liquid nitrogen to a temperature of −180°C, mounted in a 70° tilt cryospecimen holder (model 626; Gatan, Inc.), and examined in a cryoelectron microscope (CM 300; FEI) equipped with field emission gun and a postcolumn GIF 2002 energy filter (Gatan, Inc.), and slow-scan CCD camera (Gatan, Inc.) with 2048 × 2048 pixels. Low electron–dose series (4,000–5,000 electrons/nm2) of typically 60–70 images were recorded using the Digital Micrograph package (Gatan, Inc.) in tilt ranges of ±60 to ±70°, with 2° tilt intervals, at nominal magnifications of 43,000 (0.82 nm/pixel) or 52,000 (0.68 nm/pixel), and with objective lens defocus of 6–10 μm. The areas previously imaged in the light microscope were relocated in the electron microscope using the symbols on the finder grids.
Tomographic reconstruction and image processing
The images in tilt series were aligned using fiducial markers and merged in 3D reconstructions by weighted back-projection using the EM program package (Hegerl, 1996). We used this package, as well as the TOM package (Nickell et al., 2005), for postprocessing the volumes.
Particle averaging
We extracted 518 luminal particles from tomograms of microtubules that were differently oriented with respect to the tilt axis (only side views; no microtubule top views were available) using the TOM tools in MatLab (The MathWorks). For the first round of alignment, an in silico–created cylindrical density model was used. The resulting mean volume was used as a reference for further iterative missing wedge–weighted correlation averaging (Förster et al., 2005). For the quantitative analysis, the alignment was focused on the luminal densities using a mask excluding the neighboring luminal densities during the cross correlation. The positions of the luminal densities were then quantified based on the shifts determined by the converged single-particle alignment, providing a list of distances between neighboring particles. Based on previous work indicating that the overwhelming majority of cellular microtubules are composed of 13 protofilaments (Tilney et al., 1973), we imposed 13-fold symmetry for alignment and averaging of the microtubule wall. The luminal densities, on the other hand, were averaged without imposing any symmetry. The aligned single particles were sorted into three general groups, based on cross-correlation values, supported by their visual appearance. Averaged volumes within each group were separately refined by further iterative refinements (Förster et al., 2005). For visualization, these averages were then merged together with the 3D density map of the microtubule wall.
Visualization
We used the AMIRA visualization package (Mercury Computer Systems) for surface rendering the microtubules and the luminal densities in the original reconstructions, as well as for displaying the results of particle averaging. The volumes for color displays were selected by adjusting the threshold, and then by removing the noise-dominated parts using automated procedures in AMIRA. The final threshold was set so as to match the 4-nm thickness of microtubule walls.
CEMOVIS
Sample vitrification, cryosectioning, and imaging were carried out as previously described in detail (Al-Amoudi et al., 2004; Zuber et al., 2005; Bouchet-Marquis et al., 2006). In brief, vitreous samples obtained by high-pressure freezing were mounted in an FCS cryochamber of a microtome (Ultracut UCT; Leica). 50-nm-thick cryosections (nominal thickness; because of compression the final thickness increased to ∼75 nm) were obtained using a 45° cryodiamond knife (Diatome) with a clearance angle of 6°. The sections were observed on a cryotransmission electron microscope (CM100; FEI) at 80 or 100 kV under minimal beam exposure conditions (<1,000 electrons/nm2/micrograph). The images of vitreous sections were recorded on film (SO-163 film; Kodak) at various magnifications, and the negatives were scanned on a PRO film scanner (Expression 1680; Epson) with 1600 dpi resolution. Fourier transform calculations of the images were low-pass filtered with a mask, the radius of which corresponded to the first zero of the contrast transfer function (CTF), 1/3 nm−1, and back projected to real space. Density profiles were determined on rectangular selections of inverted images using the Plot Profile function of ImageJ (National Institutes of Health).
Online supplemental material
Fig. S1 depicts an example of P19 cells in culture. Videos 1 and 2 provide 3D representations of the tomograms shown in Fig. 2 A and Fig. 4 E, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200606074/DC1.
Supplementary Material
Acknowledgments
We thank Liane Meyn for expert help with neuronal cultures, Jacomine Krijnse-Locker and Iosune Ibiricu for preparations of control microtubules, Dominique Muller's laboratory for providing organotypic slices, and Ilona Grunwald and Rüdiger Klein for involvement in early attempts to image neuronal cultures by cryo-EM.
B.K. Garvalov, M. Kudryashev, M. Gruska, W. Baumeister, M. Beck, A. Leis, F. Bradke, F. Frischknecht, and M. Cyrklaff produced and analyzed the cryo-ET data; C. Bouchet-Marquis, B. Zuber, and J. Dubochet produced and analyzed the CEMOVIS data.
This work was supported by the Deutsche Forschungsgemeinschaft (grants SFB 391 to F. Bradke, and SFB 544 to F. Frischknecht), a Human Frontier Science Program Career Development Award (to F. Bradke), the BioFuture Program of the German Bundesministerium für Bildung und Forschung (to F. Frischknecht and M. Kudryashev), and the European Commission's 3D-EM Network of Excellence (M. Gruska, A. Leis, W. Baumeister, C. Bouchet-Marquis, B. Zuber, and J. Dubochet).
B. Zuber and C. Bouchet-Marquis contributed equally to this paper.
Abbreviations used in this paper: CCD, charged-couple device; CEMOVIS, cryoelectron microscopy of vitreous sections; cryo-ET, cryoelectron tomography; CTF, contrast transfer function; MAP, microtubule-associated protein.
References
- Afzelius, B.A. 1988. Microtubules in the spermatids of stick insects. J. Ultrastruct. Mol. Struct. Res. 98:94–102. [DOI] [PubMed] [Google Scholar]
- Al-Amoudi, A., J.J. Chang, A. Leforestier, A. McDowall, L.M. Salamin, L.P. Norlen, K. Richter, N.S. Blanc, D. Studer, and J. Dubochet. 2004. Cryo-electron microscopy of vitreous sections. EMBO J. 23:3583–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassot, J.M., and R. Martoja. 1966. Données histologiques et ultrastructurales sur les microtubles cytoplasmiques du canal éjaculateur des insectes orthoptéres. Z. Zellforsch. Mikrosk. Anat. 74:145–181. [PubMed] [Google Scholar]
- Baumeister, W. 2005. From proteomic inventory to architecture. FEBS Lett. 579:933–937. [DOI] [PubMed] [Google Scholar]
- Beck, M., F. Förster, M. Ecke, J.M. Plitzko, F. Melchior, G. Gerisch, W. Baumeister, and O. Medalia. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 306:1387–1390. [DOI] [PubMed] [Google Scholar]
- Behnke, O. 1967. Incomplete microtubules observed in mammalian blood platelets during microtubule polymerization. J. Cell Biol. 34:697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchet-Marquis, C., J. Dubochet, and S. Fakan. 2006. Cryoelectron microscopy of vitrified sections: a new challenge for the analysis of functional nuclear architecture. Histochem. Cell Biol. 125:43–51. [DOI] [PubMed] [Google Scholar]
- Burton, P.R. 1984. Luminal material in microtubules of frog olfactory axons: structure and distribution. J. Cell Biol. 99:520–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyrklaff, M., M. Adrian, and J. Dubochet. 1990. Evaporation during preparation of unsupported thin vitrified aqueous layers for cryo-electron microscopy. J. Electron Microsc. Tech. 16:351–355. [DOI] [PubMed] [Google Scholar]
- de Hoop, M.J., L. Meyn, and C.G. Dotti. 1998. Culturing hippocampal neurons and astrocytes from fetal rodent brain. In Cell Biology: A Laboratory Handbook. Vol. 1. J.E. Celis, editor. Academic Press, San Diego. 154–163.
- Dotti, C.G., C.A. Sullivan, and G.A. Banker. 1988. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8:1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster, F., O. Medalia, N. Zauberman, W. Baumeister, and D. Fass. 2005. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. USA. 102:4729–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegerl, R. 1996. The EM program package: a platform for image processing in biological electron microscopy. J. Struct. Biol. 116:30–34. [DOI] [PubMed] [Google Scholar]
- Kar, S., J. Fan, M.J. Smith, M. Goedert, and L.A. Amos. 2003. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J. 22:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lučić, V., F. Förster, and W. Baumeister. 2005. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74:833–865. [DOI] [PubMed] [Google Scholar]
- McBurney, M.W., E.M. Jones-Villeneuve, M.K. Edwards, and P.J. Anderson. 1982. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 299:165–167. [DOI] [PubMed] [Google Scholar]
- Medalia, O., I. Weber, A.S. Frangakis, D. Nicastro, G. Gerisch, and W. Baumeister. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science. 298:1209–1213. [DOI] [PubMed] [Google Scholar]
- Nicastro, D., J.R. McIntosh, and W. Baumeister. 2005. 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl. Acad. Sci. USA. 102:15889–15894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell, S., F. Förster, A. Linaroudis, W.D. Net, F. Beck, R. Hegerl, W. Baumeister, and J.M. Plitzko. 2005. TOM software toolbox: acquisition and analysis for electron tomography. J. Struct. Biol. 149:227–234. [DOI] [PubMed] [Google Scholar]
- Nogales, E., M. Whittaker, R.A. Milligan, and K.H. Downing. 1999. High-resolution model of the microtubule. Cell. 96:79–88. [DOI] [PubMed] [Google Scholar]
- O'Toole, E.T., K.L. McDonald, J. Mantler, J.R. McIntosh, A.A. Hyman, and T. Muller-Reichert. 2003. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans. J. Cell Biol. 163:451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, A., C.C. Proskauer, and I.R. Kaiserman-Abramof. 1968. The small pyramidal neuron of the rat cerebral cortex. The axon hillock and initial segment. J. Cell Biol. 39:604–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Echandia, E.L., R.S. Piezzi, and E.M. Rodriguez. 1968. Dense-core microtubules in neurons and gliocytes of the toad Bufo arenarum Hensel. Am. J. Anat. 122:157–166. [DOI] [PubMed] [Google Scholar]
- Stoppini, L., P.A. Buchs, and D. Muller. 1991. A simple method for organotypic cultures of nervous tissue. J. Neurosci. Methods. 37:173–182. [DOI] [PubMed] [Google Scholar]
- Sui, H., and K.H. Downing. 2006. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature. 442:475–478. [DOI] [PubMed] [Google Scholar]
- Tilney, L.G., J. Bryan, D.J. Bush, K. Fujiwara, M.S. Mooseker, D.B. Murphy, and D.H. Snyder. 1973. Microtubules: evidence for 13 protofilaments. J. Cell Biol. 59:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, S., and K. Weber. 2003. Post-translational modifications regulate microtubule function. Nat. Rev. Mol. Cell Biol. 4:938–947. [DOI] [PubMed] [Google Scholar]
- Xu, Z., and B.A. Afzelius. 1988. The substructure of marginal bundles in human blood platelets. J. Ultrastruct. Mol. Struct. Res. 99:244–253. [DOI] [PubMed] [Google Scholar]
- Zuber, B., I. Nikonenko, P. Klauser, D. Muller, and J. Dubochet. 2005. The mammalian central nervous synaptic cleft contains a high density of periodically organized complexes. Proc. Natl. Acad. Sci. USA. 102:19192–19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.