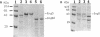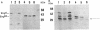Abstract
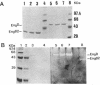
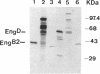
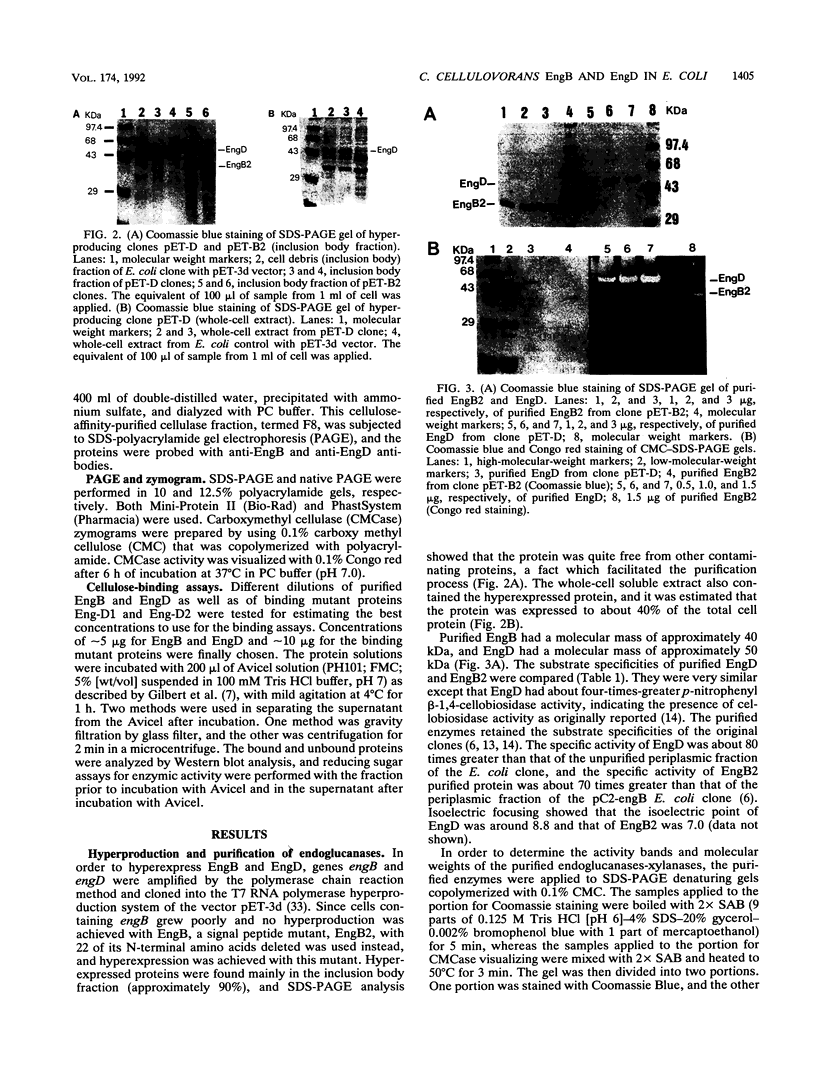
By the use of a T7 expression system, endoglucanases-xylanases EngB and EngD from Clostridium cellulovorans were hyperexpressed and purified from Escherichia coli. The two enzymes demonstrated both endoglucanase and xylanase activities. The substrate specificities of both endoglucanases were similar except that EngD had four-times-greater p-nitrophenyl beta-1,4-cellobiosidase activity. The two proteins were very homologous (80%) up to the Pro-Thr-Thr region which divided the protein into -NH2- and -COOH-terminals. The -COOH- region of EngB has high homology to the endoglucanases and a xylanase from Clostridium thermocellum and to an endoglucanase from Clostridium cellulolyticum and did not show strong binding to cellulose (Avicel). However, the -COOH- region of EngD, which had homology to the cellulose-binding domains of Cellulomonas fimi exo- and endoglucanases and to Pseudomonas fluorescens endoglucanase, demonstrated binding ability to cellulose even when the domain was fused to the N-terminal domain of EngB. By probing the Avicel-purified cellulase complex (F8) with anti-EngB and anti-EngD antibodies, both EngB and EngD were shown to be present on the cellulase complex of C. cellulovorans. Many proteins homologous to EngB and EngD were also present on the complex.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- Cavedon K., Leschine S. B., Canale-Parola E. Cellulase system of a free-living, mesophilic clostridium (strain C7). J Bacteriol. 1990 Aug;172(8):4222–4230. doi: 10.1128/jb.172.8.4222-4230.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavedon K., Leschine S. B., Canale-Parola E. Characterization of the extracellular cellulase from a mesophilic clostridium (strain C7). J Bacteriol. 1990 Aug;172(8):4231–4237. doi: 10.1128/jb.172.8.4231-4237.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B. Y., Doi R. H. Overproduction, purification, and characterization of Bacillus subtilis RNA polymerase sigma A factor. J Bacteriol. 1990 Jun;172(6):3257–3263. doi: 10.1128/jb.172.6.3257-3263.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira L. M., Durrant A. J., Hall J., Hazlewood G. P., Gilbert H. J. Spatial separation of protein domains is not necessary for catalytic activity or substrate binding in a xylanase. Biochem J. 1990 Jul 1;269(1):261–264. doi: 10.1042/bj2690261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foong F., Hamamoto T., Shoseyov O., Doi R. H. Nucleotide sequence and characteristics of endoglucanase gene engB from Clostridium cellulovorans. J Gen Microbiol. 1991 Jul;137(7):1729–1736. doi: 10.1099/00221287-137-7-1729. [DOI] [PubMed] [Google Scholar]
- Gilbert H. J., Hall J., Hazlewood G. P., Ferreira L. M. The N-terminal region of an endoglucanase from Pseudomonas fluorescens subspecies cellulosa constitutes a cellulose-binding domain that is distinct from the catalytic centre. Mol Microbiol. 1990 May;4(5):759–767. doi: 10.1111/j.1365-2958.1990.tb00646.x. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Henrissat B., Kilburn D. G., Miller R. C., Jr, Warren R. A. Domains in microbial beta-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev. 1991 Jun;55(2):303–315. doi: 10.1128/mr.55.2.303-315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes N. R., Kilburn D. G., Miller R. C., Jr, Warren R. A. Structural and functional analysis of a bacterial cellulase by proteolysis. J Biol Chem. 1989 Oct 25;264(30):17802–17808. [PubMed] [Google Scholar]
- Gilkes N. R., Warren R. A., Miller R. C., Jr, Kilburn D. G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988 Jul 25;263(21):10401–10407. [PubMed] [Google Scholar]
- Grépinet O., Chebrou M. C., Béguin P. Nucleotide sequence and deletion analysis of the xylanase gene (xynZ) of Clostridium thermocellum. J Bacteriol. 1988 Oct;170(10):4582–4588. doi: 10.1128/jb.170.10.4582-4588.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Barker P. J., Gilbert H. J. Conserved reiterated domains in Clostridium thermocellum endoglucanases are not essential for catalytic activity. Gene. 1988 Sep 15;69(1):29–38. doi: 10.1016/0378-1119(88)90375-7. [DOI] [PubMed] [Google Scholar]
- Hazlewood G. P., Davidson K., Clarke J. H., Durrant A. J., Hall J., Gilbert H. J. Endoglucanase E, produced at high level in Escherichia coli as a lacZ' fusion protein, is part of the Clostridium thermocellum cellulosome. Enzyme Microb Technol. 1990 Sep;12(9):656–662. doi: 10.1016/0141-0229(90)90004-a. [DOI] [PubMed] [Google Scholar]
- Jauris S., Rücknagel K. P., Schwarz W. H., Kratzsch P., Bronnenmeier K., Staudenbauer W. L. Sequence analysis of the Clostridium stercorarium celZ gene encoding a thermoactive cellulase (Avicelase I): identification of catalytic and cellulose-binding domains. Mol Gen Genet. 1990 Sep;223(2):258–267. doi: 10.1007/BF00265062. [DOI] [PubMed] [Google Scholar]
- Kellett L. E., Poole D. M., Ferreira L. M., Durrant A. J., Hazlewood G. P., Gilbert H. J. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem J. 1990 Dec 1;272(2):369–376. doi: 10.1042/bj2720369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer F., Coughlan M. P., Mori Y., Ljungdahl L. G. Macromolecular Organization of the Cellulolytic Enzyme Complex of Clostridium thermocellum as Revealed by Electron Microscopy. Appl Environ Microbiol. 1987 Dec;53(12):2785–2792. doi: 10.1128/aem.53.12.2785-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin M., Forsberg C. W. Isolation and characterization of endoglucanases 1 and 2 from Bacteroides succinogenes S85. J Bacteriol. 1988 Jul;170(7):2914–2922. doi: 10.1128/jb.170.7.2914-2922.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischak H., Hofer F., Messner R., Weissinger E., Hayn M., Tomme P., Esterbauer H., Küchler E., Claeyssens M., Kubicek C. P. Monoclonal antibodies against different domains of cellobiohydrolase I and II from Trichoderma reesei. Biochim Biophys Acta. 1989 Jan 27;990(1):1–7. doi: 10.1016/s0304-4165(89)80003-0. [DOI] [PubMed] [Google Scholar]
- Morag E., Bayer E. A., Lamed R. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J Bacteriol. 1990 Oct;172(10):6098–6105. doi: 10.1128/jb.172.10.6098-6105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morag E., Halevy I., Bayer E. A., Lamed R. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J Bacteriol. 1991 Jul;173(13):4155–4162. doi: 10.1128/jb.173.13.4155-4162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill G., Goh S. H., Warren R. A., Kilburn D. G., Miller R. C., Jr Structure of the gene encoding the exoglucanase of Cellulomonas fimi. Gene. 1986;44(2-3):325–330. doi: 10.1016/0378-1119(86)90197-6. [DOI] [PubMed] [Google Scholar]
- Shoseyov O., Doi R. H. Essential 170-kDa subunit for degradation of crystalline cellulose by Clostridium cellulovorans cellulase. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2192–2195. doi: 10.1073/pnas.87.6.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov O., Hamamoto T., Foong F., Doi R. H. Cloning of Clostridium cellulovorans endo-1,4-beta-glucanase genes. Biochem Biophys Res Commun. 1990 Jun 15;169(2):667–672. doi: 10.1016/0006-291x(90)90382-w. [DOI] [PubMed] [Google Scholar]
- Sleat R., Mah R. A., Robinson R. Isolation and Characterization of an Anaerobic, Cellulolytic Bacterium, Clostridium cellulovorans sp. nov. Appl Environ Microbiol. 1984 Jul;48(1):88–93. doi: 10.1128/aem.48.1.88-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Ståhlberg J., Johansson G., Pettersson G. A binding-site-deficient, catalytically active, core protein of endoglucanase III from the culture filtrate of Trichoderma reesei. Eur J Biochem. 1988 Apr 5;173(1):179–183. doi: 10.1111/j.1432-1033.1988.tb13982.x. [DOI] [PubMed] [Google Scholar]
- Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988 Jan 4;170(3):575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]