Abstract
Ciliated epithelial cells have the unique ability to generate hundreds of centrioles during differentiation. We used centrosomal proteins as molecular markers in cultured mouse tracheal epithelial cells to understand this process. Most centrosomal proteins were up-regulated early in ciliogenesis, initially appearing in cytoplasmic foci and then incorporated into centrioles. Three candidate proteins were further characterized. The centrosomal component SAS-6 localized to basal bodies and the proximal region of the ciliary axoneme, and depletion of SAS-6 prevented centriole assembly. The intraflagellar transport component polaris localized to nascent centrioles before incorporation into cilia, and depletion of polaris blocked axoneme formation. The centriolar satellite component PCM-1 colocalized with centrosomal components in cytoplasmic granules surrounding nascent centrioles. Interfering with PCM-1 reduced the amount of centrosomal proteins at basal bodies but did not prevent centriole assembly. This system will help determine the mechanism of centriole formation in mammalian cells and how the limitation on centriole duplication is overcome in ciliated epithelial cells.
Introduction
The mammalian centrosome contains two barrel-shaped centrioles made of nine microtubule triplets, surrounded by a proteinaceous pericentriolar matrix. In 1898, Henneguy and Lenhossék independently observed that the centrioles of the centrosome and basal bodies that anchor ciliary and flagellar axonemes are identical structures (Chapman et al., 2000); we will use centriole to refer to the free structure and basal body to refer to the structure at the base of cilia. In addition to flagellated sperm cells, many other animal cells generate cilia (Olsen, 2005). The majority of cells produce a single immotile cilium, the primary cilium, that transduces mechanical and chemical signals from the extracellular environment (Praetorius and Spring, 2005). Sensory cilia on specialized retinal, olfactory, and auditory cells are also essential for communicating sensory stimuli to the nervous system. Multiple motile cilia are made by ciliated protists, flagellated sperm of lower plants, and certain animal epithelial cell types. In mammals, multiciliated epithelium is found in the airways, the oviduct, and the ventricular system of the brain.
Each of hundreds of basal bodies in multiciliated epithelial cells anchors a motile cilium; the concerted beating of cilia propels substances over the epithelial surface. Receptor proteins have been found in the ciliary membrane of motile cilia as well, suggesting that both types of cilia might function in signaling (Christensen et al., 2003; Teilmann et al., 2005). Intraflagellar transport (IFT), which involves the bidirectional trafficking of molecules along the axonemal microtubules, is common to all types of cilia and is required for axoneme formation and ciliary signal transduction (Scholey and Anderson, 2006). In mammals, a hypomorphic mutation in polaris (also known as IFT88/Tg737), a core component of the IFT machinery, results in shorter or absent primary cilia in kidney epithelial cells and leads to polycystic kidney disease (Pazour et al., 2000). Polaris mutation also results in sparser, shorter motile cilia in ventricular epithelial cells (Taulman et al., 2001). However, because of the embryonic lethality of the polaris-null mutation in mouse (Murcia et al., 2000), the function of polaris and IFT in general has not been fully characterized in ciliated epithelial cells.
In contrast to cycling cells, multiciliated cells have the ability to assemble hundreds of centrioles. EM shows that these centrioles arise through two parallel pathways initiated in the vicinity of the cell's existing centrosome (Dirksen, 1991; Hagiwara et al., 2004). In the centriolar pathway, multiple new centrioles form around an existing mother centriole, similar to the process in cycling cells, with the exception that only a single centriole is generated there. In the acentriolar pathway, by which the majority of centrioles in multiciliated cells are generated, new centrioles form around the deuterosome, a non–microtubule-based structure. In both cases, protein-rich fibrous granules are found surrounding the elongating centrioles. Centrioles assemble in the cytoplasm and then move to the apical cell surface, where they align at the plasma membrane and begin forming the ciliary axoneme.
Centriole formation in ciliating cells differs from centrosome duplication in normal cycling cells in four key ways: (1) more than two daughter centrioles are generated in the presence of the existing centrosome, (2) a mother centriole simultaneously nucleates more than one daughter centriole, (3) noncentriolar structures (deuterosomes) nucleate multiple centrioles, and (4) centrioles are generated in nondividing cells. Despite these differences, ciliogenesis and centrosome duplication produce seemingly identical structures, raising the possibility of a common regulatory mechanism. A potential common regulator is SAS-6, a conserved centrosomal protein that is required for the initial steps of centriole formation in Caenorhabditis elegans (Dammermann et al., 2004; Leidel et al., 2005; Pelletier et al., 2006). In human cells, HsSAS-6 depletion by RNAi blocks centriole assembly, and overexpression leads to the formation of excess centrosomal foci (Leidel et al., 2005).
The creation of hundreds of centrioles is likely to require a dramatic increase in the expression and transport of constituent proteins. In cycling cells, some centrosomal proteins rely on dynein-mediated transport for localization. Ninein, centrin, pericentrin, and other centrosomal proteins are found in centriolar satellites (Dammermann and Merdes, 2002), dynein-containing protein complexes that traffic toward microtubule minus ends (Kubo et al., 1999; Kubo and Tsukita, 2003). Two centriolar satellite proteins, PCM-1 and BBS4, are thought to tether cargo proteins to dynein, and in their absence, assembly of cargo proteins at the centrosome is decreased (Dammermann and Merdes, 2002; Kim et al., 2004). PCM-1 also localizes to the abundant fibrous granules found in the ciliating cell cytoplasm (Kubo et al., 1999) and, therefore, might be particularly important for ciliogenesis.
Comparative genomic (Avidor-Reiss et al., 2004; Li et al., 2004), proteomic (Keller et al., 2005; Pazour et al., 2005), and gene expression (Ross et al., 2007) studies have identified conserved ciliary components and potential regulators; however, mechanistic understanding of centriole formation in ciliated epithelial cells remains limited. The forkhead family transcription factor, Foxj1, is uniquely expressed in cells with flagella or motile cilia (Tichelaar et al., 1999). Ciliated epithelial cells in FOXJ1 −/− mice make centrioles but lack motile multicilia due to failure to anchor centrioles at the plasma membrane (Gomperts et al., 2004). Several proteins important for centrosome structure and function, including γ-tubulin, centrin, and pericentrin, localize to the basal body region of ciliated epithelial cells (Muresan et al., 1993; Levy et al., 1996; Jurczyk et al., 2004), but their role in ciliogenesis has not been tested. Many genes required for basal body and axoneme formation have been characterized in the biflagellate alga Chlamydomonas (Taillon et al., 1992; Dutcher and Trabuco, 1998; Preble et al., 2001) and in multiciliated cells, such as Paramecium and Tetrahymena (Garreau de Loubresse et al., 2001; Stemm-Wolf et al., 2005). Despite many conserved elements, ciliated epithelial cells possess features that cannot be modeled by lower organisms. For example, Chlamydomonas has only two basal bodies, and in multiciliated protists, basal bodies assemble on the cell cortex rather than in the cytoplasm.
Here, we report the development of a model system using in vitro differentiated, multiciliated epithelial cells to study the pathway of centriole assembly during ciliogenesis. We define the localization of centrosomal proteins during the process and examine the role of the IFT component polaris, the mouse orthologue of the centrosomal protein SAS-6, and PCM-1–containing fibrous granules in centriole formation.
Results
In vitro system for studying centriole formation
We adapted the mouse tracheal epithelial cell (MTEC) culture system developed by You et al. (2002) to study centriole formation during ciliogenesis. The culture is started by seeding freshly isolated tracheal cells onto a porous filter suspended in medium. Cells proliferate into a confluent, polarized epithelium while submerged in medium. Although ciliated cells are present in the isolated tracheal cell population at the time of plating, most are unable to attach to the filter; therefore, the ciliated cells that appear later in the culture are due to in vitro differentiation (You et al., 2002).
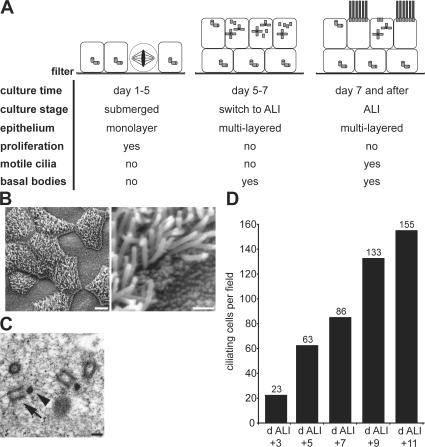
Ciliogenesis is initiated by altering the medium and creating an air–liquid interface (ALI) by supplying medium only from below the filter. We defined three phases of the culture based on landmark events observed by light and electron microscopy (Fig. 1 A). No ciliogenesis occurs during the pre-ALI phase comprising the first 5 d of culture. Ciliogenesis begins in the second phase, a period of 2–3 d after ALI creation when centriole formation begins, but cilia are not yet detected at the surface. The third phase consists of a period of active ciliogenesis leading to a maximally ciliated epithelium at ∼14 d after ALI creation. The timing of experiments in this paper is reported in days relative to ALI creation (noted as “day ALI ± n of culture”).
Figure 1.
In vitro MTEC culture system for centriole formation. (A) Schematic description of ciliogenesis and MTEC culture progression. The three key phases of the culture are depicted in cartoon form. Ciliogenesis begins during the second phase with the appearance of centrioles at ∼2 d after the switch to ALI culture. (B) Scanning EM of in vitro–cultured MTECs at day ALI + 14 of culture showing ciliated and nonciliated cells. Bars: (left) 5 μm; (right) 1 μm. (C) TEM of in vitro–cultured MTECs at day ALI + 2 of culture showing nascent centrioles (arrow) and deuterosomes (arrowhead). Bar, 200 nm. (D) MTEC cultures were fixed at 48-h intervals starting at day ALI + 3 of culture, and the number of ciliating cells per field was counted based on γ-tubulin signal (see the supplemental text, available at http://www.jcb.org/cgi/content/full/jcb.200703064/DC1, for details).
The mature culture contains many fully ciliated cells (Fig. 1 B) with occasional cells at earlier steps of ciliogenesis. In day ALI + 14 cultures, typically 40–60% of cells are ciliated, consistent with a previous report (Toskala et al., 2005). Transmission EM (TEM) of day ALI + 2 cultures revealed deuterosomes and fibrous granules in the apical cytoplasm of ciliating cells (Fig. 1 C), similar to structures seen in vivo (Sorokin, 1968). These results indicate that in vitro ciliogenesis proceeds through the same steps as in vivo. Finally, cultured MTECs acquire cilia over the course of several days (Fig. 1 D), similar to the timing of ciliogenesis during airway development and tracheal epithelium reformation in vivo after damage (Rawlins et al., 2007).
Pathway of centriole formation during ciliogenesis
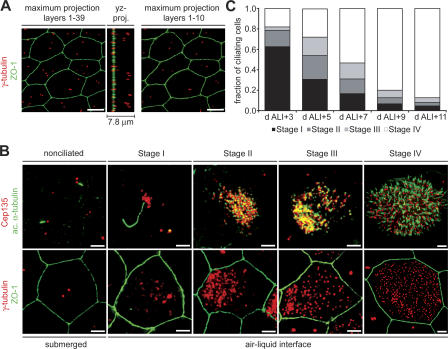
Centriole formation during in vitro ciliogenesis was characterized by immunofluorescence localization of the marker proteins γ-tubulin and Cep135 (centrosomes and centrioles), acetylated α-tubulin (centrioles, cytoplasmic, and axonemal microtubules), and ZO-1 (epithelial cell boundaries). Before ciliogenesis, MTECs formed a confluent, polarized epithelium that appeared to consist of multiple cell layers (Fig. 2 A), resembling the pseudostratified tracheal epithelium in vivo. Because of the multilayered nature of the in vitro cultures, a maximum projection of deconvolved image planes through the entire epithelium showed a variable number of γ-tubulin–labeling centrosomes within a cell boundary (Fig. 2 A, image layers 1–39). However, a yz projection showed that the centrosomes are actually found at different depths, as expected for multiple cell layers. Most subsequent depictions include only the apical portion of the image stack (Fig. 2 A, image layers 1–10). Although cell boundaries are not always shown, all images are of fully confluent epithelia, except where noted.
Figure 2.
Centriole and axoneme formation in in vitro–cultured MTECs. (A) Nonciliated MTECs at ALI creation were labeled with γ-tubulin (red) and ZO-1 (green) antibodies. The yz projection of the image stack shows that the γ-tubulin–labeled centrosomes originate from different depths in the epithelium. The maximum projection of layers 1–10 shows that no more than two centrosomes per cell are present at the apical surface. Bars, 5 μm. (B) MTECs were labeled with either γ-tubulin (red) and ZO-1 (green) or Cep135 (red) and acetylated α-tubulin (green) antibodies as indicated. The numbered stages of culture correspond to those described in the text. Bars, 2 μm. (C) MTEC cultures were fixed at 48-h intervals starting at day ALI + 3 of culture, and the fraction of ciliating cells in each stage of ciliogenesis was determined based on γ-tubulin signal (n = 300 ciliating cells per interval).
We identified four stages (stages I–IV) of centriole formation in MTECs during ciliogenesis (Fig. 2 B). At the time of ALI creation, the culture consisted of nonciliated cells that were no longer proliferating. Most cells had two separated centrosomes; however, each had only a single centrin-labeling centriole, consistent with G1 cells in which the centriole pair had separated (not depicted). Most cells had a primary cilium extending from one of the two centrioles (Fig. 2 B, nonciliated). The first detectable sign of centriole formation was at stage I, when foci of centrosomal proteins appeared near the centrosome in the apical cytoplasm (Fig. 2 B, stage I). The foci formed at approximately day ALI + 2 and preceded the formation of centrioles, based on the absence of acetylated α-tubulin labeling at the foci. The appearance of cytoplasmic foci coincided with an increase in the amount of centrosomal proteins at the existing centrosome (Fig. 2 B, stage I). Primary cilia in these cells were approximately fivefold longer than in nonciliated cells (nonciliated = 0.98 ± 0.09 μm, stage I = 5.39 ± 0.04 μm; see the supplemental text, available at http://www.jcb.org/cgi/content/full/jcb.200703064/DC1). Ciliating cells also had more cytoplasmic microtubules during stage I (Fig. S1 A), and these microtubules were more resistant to depolymerization than those of neighboring nonciliating cells (Fig. S1 B).
During stage II, centrosomal proteins began to localize to a single dense cluster per cell (Fig. 2 B, stage II). Acetylated α-tubulin labeling indicated that centrioles were present in these clusters. In most ciliating cells, this nascent centriole cluster was closer to one side of the cell, as judged by cell boundary labeling, but the orientation of this eccentric localization in neighboring ciliating cells appeared to be random. Primary cilia were no longer present on ciliating cells beginning in stage II, although they remained on adjacent nonciliating cells (unpublished data). In stage III, centrioles dispersed from the cytoplasmic cluster toward the plasma membrane (Fig. 2 B, stage III). Axoneme formation began during stage IV, shortly after the centrioles reached the plasma membrane but before all centrioles were distributed evenly at the surface. Basal bodies in mature ciliated cells were evenly distributed at the apical membrane, and each anchored a cilium (Fig. 2 B, stage IV) of 2.89 ± 0.09 μm mean length. Cells in the stages defined above appeared sequentially during the culture period (Fig. 2 C), suggesting that these morphological states represent stages in the pathway of ciliogenesis.
Centrosomal proteins localize to centrioles and are up-regulated in ciliating cells
The above results suggest that the accumulations of material in ciliating cells previously observed by EM likely represent the accumulation of centrosomal material before assembly into centrioles. In addition to γ-tubulin and Cep135, we examined the localization of many centrosomal proteins during ciliogenesis (Fig. S1 C, centriolin). All tested proteins localized to centrosomes in nonciliated cells and to cytoplasmic foci and centrioles during ciliogenesis (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200703064/DC1).
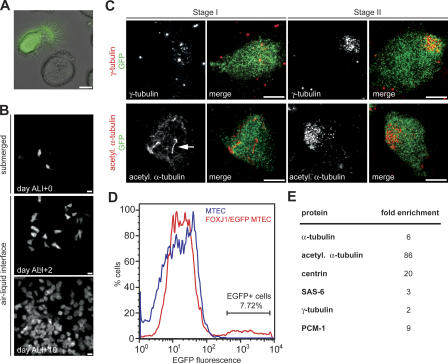
Ciliogenesis is accompanied by an increase in centrin expression (Laoukili et al., 2000), and we tested whether other centrosomal proteins are also up-regulated. The MTEC culture contains both ciliated and nonciliated cells, and to analyze ciliated cells specifically, we cultured tracheal epithelial cells from a transgenic FOXJ1/EGFP mouse strain that expresses EGFP under the control of the ciliated cell–specific Foxj1 promoter (Ostrowski et al., 2003; Fig. 3 A). We found that in mature cultures of these cells, all ciliated cells were EGFP+, and all EGFP+ cells were ciliated (unpublished data). FOXJ1/EGFP expression began at about day ALI + 2 of culture (Fig. 3 B), coinciding with the first signs of centriole formation (Fig. 3 C, stages I and II). Thus, FOXJ1/EGFP is a useful marker for both early ciliating and mature ciliated cells.
Figure 3.
Centrosomal proteins are more abundant in ciliating cells. (A) Phase image overlayed with EGFP signal (green) from live tracheal epithelial cells isolated from FOXJ1/EGFP transgenic mice. Only multiciliated cells express cytoplasmic EGFP. Bar, 5 μm. (B) In vitro–cultured FOXJ1/EGFP MTECs were labeled with GFP antibody at days ALI + 0, ALI + 2, and ALI + 10 of culture. Images show confluent epithelia; the few EGFP+ cells at day ALI + 0 are original ciliated cells from the trachea that occasionally persist in the MTEC culture and are not the product of in vitro ciliogenesis. Bars, 20 μm. (C) FOXJ1/EGFP MTECs were labeled at day ALI + 4 of culture with γ-tubulin or acetylated α-tubulin (red) and GFP (green) antibodies as indicated. Stage I and II ciliating cells were always EGFP+, indicating that FOXJ1/EGFP is a useful early marker of ciliogenesis. Arrow points to the enlarged primary cilium. Bars, 5 μm. (D) MTECs and FOXJ1/EGFP MTECs at day ALI + 4 of culture were analyzed by flow cytometry based on EGFP fluorescence. EGFP+ and EGFP− cells were obtained by sorting FOXJ1/EGFP MTECs. The culture at this stage contained relatively few EGFP+ cells, with the majority in the early stages of ciliogenesis. (E) Protein levels for centrosomal proteins in EGFP+ and EGFP− MTECs obtained by FACS were determined by Western blotting, normalized to cell number. Fold enrichment is the ratio of EGFP+/EGFP− protein levels (mean of at least two experiments).
To assay centrosomal protein abundance in ciliating cells, EGFP+ cells from a ciliating FOXJ1/EGFP culture were obtained by FACS at day ALI + 4 of culture (Fig. 3 D). Relative protein levels were compared by Western blotting for the centrosomal proteins indicated in Fig. 3 E from cell lysate prepared from equal numbers of cells. The examined proteins were 2- to 86-fold more abundant in ciliating (EGFP+) than nonciliated (EGFP−) cell types (Fig. 3 E). In sum, these results indicate that many centrosomal proteins are up-regulated during ciliogenesis, appear in cytoplasmic foci at the site of centriole assembly, and are recruited to the centrioles.
Interfering with ciliogenesis
To determine the role of individual proteins in ciliogenesis, we used the in vitro culture system to interfere with their function. For this purpose, we developed a means of efficiently introducing RNAi constructs into MTECs using lentiviral infection (see Materials and methods). We chose to focus on three proteins, representing different functional classes: the IFT component polaris; the centrosome component SAS-6; and PCM-1, a component of fibrous granules.
Depletion of the IFT protein polaris disrupts axoneme and basal body formation
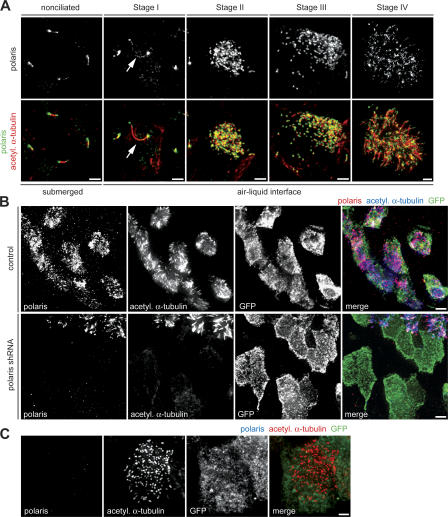
The localization of the IFT component polaris in MTECs was determined by immunofluorescence (Fig. 4 A). Polaris localized along both primary cilia and motile cilia (Fig. 4 A, nonciliated, stages I and IV) in a punctate pattern with enrichment at the base and tip of the axoneme, consistent with previous work (Taulman et al., 2001). During ciliogenesis, polaris was present on the enlarged primary cilium in stage I but was not in pericentrosomal cytoplasmic foci, like most other centriolar components (Fig. 4 A, stage I). Polaris colocalized with nascent centrioles during stages II and III and then with axonemes in stage IV (Fig. 4 A, stages II–IV). Polaris was fivefold more abundant in ciliating cells (FOXJ1/EGFP+) than in nonciliated cells (FOXJ1/EGFP−) at day ALI + 4 and 14-fold more abundant at day ALI + 10 (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200703064/DC1).
Figure 4.
Polaris is required for ciliogenesis. (A) MTECs were labeled with polaris (green) and acetylated α-tubulin (red) antibodies. Arrows indicate polaris signal in the enlarged primary cilium at stage I. Bars, 2 μm. (B) FOXJ1/EGFP MTECs were infected with lentivirus with (polaris shRNA) or without (control) polaris shRNA at day ALI − 2 of culture. Infected cells were observed at day ALI + 10 of culture by labeling with polaris (red), GFP (green), and acetylated α-tubulin (blue) antibodies. Bars, 5 μm. (C) Polaris-depleted FOXJ1/EGFP MTEC (from Fig. 4 B) labeled with polaris (blue), GFP (green), and acetylated α-tubulin (red) antibodies make no axonemes, but contain some basal bodies. Bar, 2 μm.
A lentivirally expressed short hairpin RNA (shRNA) construct targeting polaris was used to address its role in motile multicilia formation. The lentivirus effectively depleted polaris from NIH/3T3 cells (Fig. S2 B) and disrupted the formation of primary cilia in NIH/3T3 cells and MTECs before ciliogenesis (unpublished data). To examine polaris function during ciliogenesis, MTECs derived from FOXJ1/EGFP mice were infected on day ALI − 2 and were assayed on day ALI + 10 of culture. Depletion by RNAi was demonstrated by decrease in polaris labeling in cells, with most having no detectable polaris signal (Fig. S2 C). Control and shRNA-treated cultures had similar numbers of FOXJ1/EGFP+ cells (unpublished data), indicating that polaris depletion did not affect the adoption of the ciliated cell fate. Complete polaris depletion blocked axoneme formation in FOXJ1/EGFP+ cells (156/156 cells; Fig. 4 B). FOXJ1/EGFP+ cells with a partial depletion of polaris had normal (60/127), short and sparse (54/127), or absent (13/127) axonemes. At the same culture stage, virtually all FOXJ1/EGFP+ cells in control infected cultures had fully formed axonemes (182/189), indicating that ciliogenesis was nearly complete. In contrast to the effect on ciliary axoneme assembly, basal bodies were still present in FOXJ1/EGFP cells without detectable polaris signal (Fig. 4 C), although they were fewer in number (128 per cell; n = 10) when compared with control MTECs (325 per cell; n = 10) and less evenly distributed on the cell surface (Fig. 4 C).
The centrosomal protein, SAS-6, is required for centriole formation
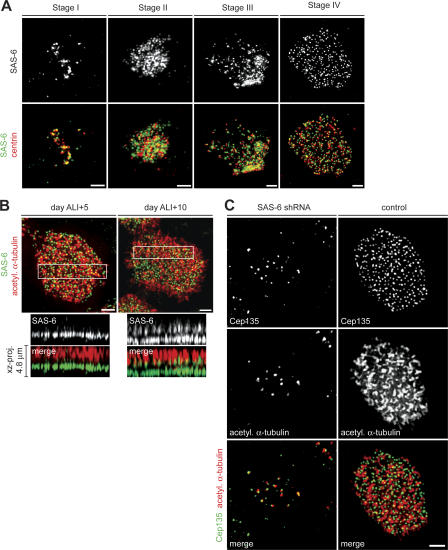
Extensive similarities between basal bodies and centrioles suggest that proteins involved in centrosome duplication may also be required for centriole assembly during ciliogenesis. We chose to focus on SAS-6, as it is a highly conserved centriolar protein that appears to specifically regulate centriole formation (Dammermann et al., 2004; Leidel et al., 2005; Pelletier et al., 2006), and our initial assessment showed that it localizes to basal bodies (Table S1) and is up-regulated during ciliogenesis (Fig. 3 E). Immunofluorescence and lentiviral expression of a SAS-6–GFP construct in MTECs showed that SAS-6 distribution was similar to that of other centrosomal proteins throughout ciliogenesis (Fig. 5 A and Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200703064/DC1). SAS-6 localized to pericentrosomal cytoplasmic foci during stage I and to nascent centrioles from stage II and on (Fig. 5 A and Fig. S3 A). SAS-6 did not colocalize precisely with either centrin or γ-tubulin on centrioles, suggesting that these proteins reside in different structural domains. Surprisingly, in fully ciliated cells from mature MTEC cultures, SAS-6 localized both to basal bodies and to the proximal region of axonemes (Fig. 5 B); this localization was confirmed with a second antibody to SAS-6 (not depicted). An xz projection through a mature ciliated cell from day ALI + 10 of culture revealed two distinct regions of SAS-6 labeling (Fig. 5 B, right, xz projection), with overlap between the top domain and the acetylated α-tubulin signal marking the axonemal microtubules. In a newly formed ciliated cell from day ALI + 5, the xz projection showed only the characteristic basal body localization, a single SAS-6–labeling region distinct from the axoneme (Fig. 5 B, left, xz projection). This axonemal localization in mature cells was unique to SAS-6 among the analyzed basal body components and suggested that it might be involved in both basal body and axoneme formation.
Figure 5.
SAS-6 is required for centriole formation. (A) MTECs were labeled with SAS-6 (green) and centrin (red) antibodies. Bars, 2 μm. (B) MTECs from day ALI + 5 (left) and ALI + 10 (right) were labeled with SAS-6 (green) and acetylated α-tubulin (red) antibodies. The xz projection of the boxed area shows that in younger ciliated cells (left), SAS-6 does not colocalize with axonemes, whereas in more mature cells (right), SAS-6 localizes to basal bodies and axonemes. Note that the acetylated α-tubulin signal in centrioles is fainter than in axonemal microtubules and is not apparent in images exposed for the axonemal signal. Bars, 2 μm. (C) MTECs were infected with lentivirus with (SAS-6 shRNA) or without (control) SAS-6 shRNA at day ALI − 2 of culture. Infected cells were observed at day ALI + 10 of culture by labeling with Cep135 (green) and acetylated α-tubulin (red) antibodies. Bar, 2 μm.
A lentivirally expressed shRNA targeting SAS-6 was used to determine its role during ciliogenesis. The lentivirus effectively depleted SAS-6 from NIH/3T3 cells (Fig. S3 B), and consistent with published results (Leidel et al., 2005), SAS-6 depletion blocked centriole formation during mitotic cycles in NIH/3T3 cells (Fig. S3 C). To examine SAS-6 function during ciliogenesis, MTECs were infected on day ALI − 2 and were assayed on day ALI + 10 of culture. RNAi resulted in a substantial, but not complete, depletion of SAS-6 in most infected cells, as judged by decrease in the intensity of SAS-6 antibody signal (Fig. S3 D). Cep135 antibody labeling showed that most of the ciliated cells (147/150) from the control population had a full complement of mature basal bodies (Fig. 5 C). In contrast, most of the SAS-6 shRNA–depleted ciliated cells (112/150) had only a small number of Cep135-labeling dots at the cell surface (Fig. 5 C). These dots also contained acetylated α-tubulin, confirming that they were centrioles and not foci of centrosomal material (Fig. 5 C). Control cells contained a mean of 317 centrioles/cell (n = 10), whereas SAS-6 shRNA–treated cells had a mean of 33 centrioles/cell (n = 10). Interestingly, ciliary axonemes were also absent in depleted cells (Fig. 5 C), raising the possibility that SAS-6 is also involved in axoneme formation, although it is also possible that the few basal bodies that form under SAS-6 depletion are abnormal and are not capable of initiating axoneme formation.
Depletion of the centriolar satellite protein PCM-1 has no effect on centriole assembly
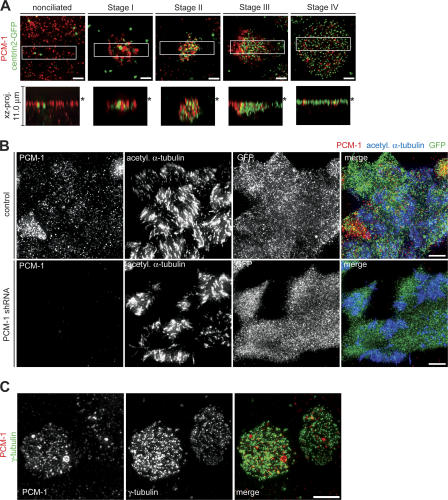
A hallmark of centriole formation in ciliated epithelial cells is the presence of PCM-1–containing fibrous granules in close proximity to nascent centrioles (Kubo et al., 1999). These might be transporting centriolar proteins by analogy with the purported role of centriolar satellites in dividing cells (Dammermann and Merdes, 2002). In nonciliated cells, PCM-1 localized to an apical layer of disperse cytoplasmic granules similar to centriolar satellites (Fig. 6 A, nonciliated). Unlike in cycling cells, these granules were not clustered around the centrioles marked by lentivirally expressed centrin2-GFP. In the transition to stage I, PCM-1 formed several large aggregates around the existing centrioles (Fig. 6 A, stage I). These PCM-1 aggregates appeared at the same time as, and partially colocalized with, the cytoplasmic foci of centrosomal proteins in stage I. In stage II cells, PCM-1 aggregates were smaller and more dispersed, in close association with the nascent centrioles (Fig. 6 A, stage II). These aggregates tracked with the centrioles to the plasma membrane through stage III, while continuing to decrease in size and abundance (Fig. 6 A, stage III). In mature ciliated cells, the little remaining PCM-1 was associated with basal bodies at the apical surface (Fig. 6 A, stage IV). PCM-1 distribution was supported by exact colocalization of the endogenous protein with lentivirally expressed, GFP-tagged PCM-1 (Fig. S4 A, available at http://www.jcb.org/cgi/content/full/jcb.200703064/DC1; nonciliated). PCM-1 also localized to fibrous granules in ciliating MTECs by immuno-EM, confirming previous results for PCM-1 localization (Kubo et al., 1999; Fig. S4 B). Furthermore, similar to cycling cells (Dammermann and Merdes, 2002), PCM-1 also colocalized with cytoplasmic granules of ninein and centrin in both nonciliated cells and during stage I of ciliogenesis (Fig. S4 C [ninein] and not depicted [centrin]). These results are consistent with the observed PCM-1–labeling structures in ciliating cells being analogous to centriolar satellites.
Figure 6.
PCM-1 granules during ciliogenesis. (A) MTECs were infected with lentivirus containing centrin2-GFP at day ALI − 3 of culture and observed at multiple time points by labeling with PCM-1 (red) and GFP (green) antibodies. The xz projections of the boxed areas show the position of the signal relative to the cell surface (asterisk). Exposure of PCM-1 signal was optimized for the ciliating cell in each image, obscuring the signal in neighboring cells with fainter labeling. Bars, 2 μm. (B) FOXJ1/EGFP MTECs were infected with lentivirus with (PCM-1 shRNA) or without (control) PCM-1 shRNA at day ALI − 2 of culture. Infected cells were observed at day ALI + 10 of culture by labeling with PCM-1 (red), GFP (green), and acetylated α-tubulin (blue) antibodies. Bars, 5 μm. (C) MTECs were infected at day ALI − 2 of culture with PCM-1 shRNA–containing lentivirus. Infected cells were observed at day ALI + 10 of culture by labeling with PCM-1 (red) and γ-tubulin (green) antibodies. Centriole formation was not altered in cells depleted of PCM-1 (cell on the right), but accumulation of γ-tubulin at the basal bodies was substantially reduced in stage IV cells with mature basal bodies. Bar, 5 μm.
A lentivirally expressed shRNA construct targeting PCM-1 was used to assess its role in centriole generation. The lentivirus effectively depleted PCM-1 from NIH/3T3 cells (Fig. S4 D), and consistent with published results (Dammermann and Merdes, 2002), PCM-1 depletion led to decreased centrosomal accumulation of ninein and centrin in shRNA-treated NIH/3T3 cells (unpublished data). To determine the function of PCM-1 during ciliogenesis, MTECs derived from FOXJ1/EGFP mice were infected on day ALI − 2 and were assayed on day ALI + 10 of culture. RNAi resulted in a range of depletion as judged by PCM-1 antibody labeling, with most cells having no or very few PCM-1 granules. Similar to NIH/3T3 cells, PCM-1–depleted MTECs had decreased amounts of centrosomal ninein and γ-tubulin (Fig. S4 E [ninein] and not depicted [γ-tubulin]). Remarkably, PCM-1–depleted FOXJ1/EGFP+ cells showed no observable defects in either centriole or axoneme formation (Fig. 6 B). All examined centrosomal proteins were present on mature basal bodies, although often at reduced levels (Fig. 6 C, γ-tubulin), suggesting that PCM-1 depletion did indeed affect protein targeting in fully ciliated cells but that this did not prevent centriole formation.
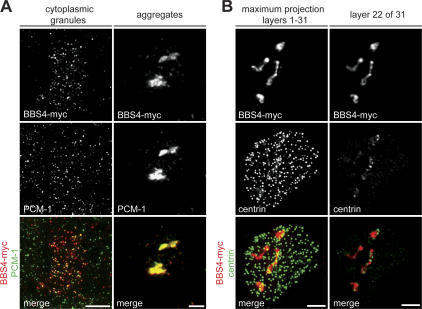
As substantial depletion of PCM-1 had no effect on ciliogenesis, we attempted to interfere with PCM-1 by lentivirus-mediated overexpression of BBS4, which results in the formation of large aggregates trapping PCM-1 and associated cargo proteins in cycling cells (Kim et al., 2004). In some infected cells, BBS4-myc perfectly colocalized with PCM-1 granules (Fig. 7 A, left), but in the majority of cells, infection resulted in aggregates containing BBS4-myc, PCM-1, and some centrin and ninein (Fig. 7 A, right [PCM-1], Fig. 7 B, image layer 22 of 31 [centrin], and not depicted [ninein]). Although the aggregates effectively trapped all visible PCM-1, cells with BBS4-myc–induced aggregates still assembled centrioles (Fig. 7 B). Individual z slices of fully ciliated cells revealed that the induced aggregates still contained centrosomal proteins and occasionally trapped centrioles in the cytoplasm (Fig. 7 B, image layer 22 of 31). The combination of the RNAi results and BBS4-induced aggregation suggests that normal levels of PCM-1 are dispensable for centriole formation and ciliogenesis.
Figure 7.
Overexpression of BBS4 in MTECs disrupts PCM-1 granules but not ciliogenesis. (A) MTECs were infected at day ALI − 3 of culture with lentivirus containing myc-tagged BBS4 cDNA. Infected cells were observed at day ALI + 5 of culture by labeling with myc (red) and PCM-1 (green) antibodies. In some cells, BBS4-myc and PCM-1 colocalized in cytoplasmic granules of normal appearance (left), whereas in most infected cells, BBS4-myc and PCM-1 formed aggregates (right). Bars, 2 μm. (B) BBS4-myc–infected MTECs were labeled with myc (red) and anti-centrin (green) antibodies. The maximum projection of an infected cell with BBS4-myc–containing aggregates shows that basal bodies still form. However, the individual image layer of the aggregate shows that centrosomal material and some centrioles are contained within the aggregates. Bars, 2 μm.
Discussion
We have investigated the process of ciliogenesis in MTECs, as a model for centriole formation. These cells are unique in that they generate hundreds of centrioles during differentiation, whereas dividing cells produce only two centrioles per cell cycle. We adapted an established in vitro culture system for these cells and examined centriole formation with molecular markers. We found that proteins defined as centrosomal in dividing cells also localized to the basal bodies of ciliated cells and that these proteins were up-regulated during ciliogenesis. Based on the localization of these proteins, we identified four stages of centriole assembly, which are consistent with previous EM studies of the process. We used the culture system to investigate the role of three proteins in ciliogenesis—polaris, SAS-6, and PCM-1—and found that polaris and SAS-6 are required for distinct stages in ciliogenesis. Although PCM-1, a component of fibrous granules, colocalized with particles containing centrosomal proteins during ciliogenesis, normal levels of PCM-1 were not required for ciliogenesis. Here, we consider the implications of these findings.
We characterized the pathway of centriole and axoneme formation during ciliogenesis by observing the localization of centrosomal proteins; this process had been previously studied only by EM (Dirksen, 1991; Hagiwara et al., 2004). We have shown that most centrosomal proteins behave as expected for being components of the protein-rich particles observed by EM during ciliogenesis, appearing first as small foci in the apical domain of cells, before the appearance of centrioles. Similar concentrations of centrosomal proteins have been observed during de novo centriole formation in Naegleria, in which a protein complex containing γ-tubulin, pericentrin, and myosin II forms before centrioles appear (Kim et al., 2005). Also, cytoplasmic foci of centrin were seen before the de novo generation of centrioles in HeLa cells in which the original centrioles were destroyed (La Terra et al., 2005). Given that centrosomal protein levels increased greatly during the early phase of ciliogenesis, this suggests that pools of precursor material are amassed in particulate form and deposited at the site of assembly before incorporation into centrioles. It will be important to determine the full extent of gene expression changes that take place specifically in ciliating cells, as it is possible that the transcriptional program is ultimately responsible not just for increased levels of structural components of centrioles, but also of the regulators that allow the assembly of hundreds of centrioles.
We found that centrioles formed in ciliating cells and centrosomes in dividing cells have similar protein constituents, including both centriolar and pericentriolar matrix proteins. This suggests that centrioles, even when acting as basal bodies, are associated with proteins normally thought of as being limited to the cycling cell centrosome. We noted a key difference in centriole maturation during ciliogenesis: proteins that are specific to the mature mother centriole in cycling cells, such as ninein and ɛ-tubulin, localized to new centrioles in ciliating cells with timing similar to that of other components. Thus, the normal maturation cycle by which a centriole acquires these proteins is bypassed in ciliating cells, perhaps because the relative abundance of these proteins in ciliating cells drives their association with centrioles.
Similar to published results from mouse mutants (Pazour et al., 2000; Taulman et al., 2001), we found that the IFT component polaris is required for ciliary axoneme formation in MTECs. In addition, polaris depletion caused a modest decrease in centriole number in MTECs. It is possible that polaris directly affects centriole assembly, as it localizes to nascent centrioles during ciliogenesis, and it has recently been described as a functionally important component of the centrosome (Robert et al., 2007). Another possibility is that polaris is involved in generating a regulatory signal for induction of the ciliogenesis transcriptional program; this could, for example, be transduced by the elongated primary cilium found during stage I of ciliogenesis. Finally, decreased numbers of centrioles could result from degeneration of basal bodies that failed to form axonemes. Future experiments are required to determine whether the polaris depletion phenotype is unique to polaris or is common to disruption of IFT in general. Our results show that polaris, and possibly IFT, is required for ciliogenesis in the multiciliated epithelium. Because several axonemal components have been implicated in human disease, ciliopathies should be examined to investigate the possible phenotypic contribution of motile cilium defects, in addition to the established defects in primary cilium function.
We found that SAS-6 is a direct effector of centriole assembly during ciliogenesis. This is consistent with the role of SAS-6 in centrosome duplication in human cells and in C. elegans embryos (Dammermann et al., 2004; Leidel et al., 2005). In worms, SAS-6 is required for the early steps of procentriole formation, and SAS-6 depletion results in failure to form complete centrioles (Pelletier et al., 2006). Our results show that SAS-6–depleted MTECs are able to assemble only a few centrioles, presumably because of residual SAS-6 protein in depleted cells. Interestingly, SAS-6 was found at the basal bodies in newly formed ciliated cells but localized to both basal bodies and axonemes in more mature ciliated cells. During this period, the ciliary axoneme is thought to become fully functional by acquiring additional length, motility, and perhaps other components associated with ciliary signaling. This redistribution was unique to SAS-6 among basal body proteins, and it is possible that SAS-6, in addition to regulating centriole formation, might also function later in axoneme maturation.
Fibrous granules, defined by EM, have been observed in ciliating epithelial cells in many tissues and organisms. They are known to contain PCM-1 and are likely to be identical to the granules in cycling cells containing PCM-1, dynein, and centrosomal components that traffic on cytoplasmic microtubules and are enriched around the centrosome (Kubo and Tsukita, 2003). However, the formation of centrioles and axonemes proceeded normally in PCM-1–depleted cells. This is consistent with the presence of respiratory cilia in mice deficient for BBS4, another component of PCM-1 granules (Mykytyn et al., 2004). Although we interfered with PCM-1 function in two ways, it is possible that residual PCM-1 activity was sufficient to support the formation of centrioles or that we were not able to detect subtle kinetic differences in ciliogenesis. Conclusive results regarding the role of PCM-1 in ciliogenesis will likely require examination of the process in the absence of the protein in PCM-1–null cells.
Our results from SAS-6 depletion suggest that common mechanisms control centriole assembly during centrosome duplication and ciliogenesis. However, there are some differences in the processes that will ultimately have to be resolved. For example, recent results (Tsou and Stearns, 2006b) suggest that in cycling cells, separase activity is required to disengage centrioles at the end of mitosis to allow duplication in the subsequent cell cycle, thus limiting centriole assembly to two new centrioles per cell cycle. In contrast, in ciliating epithelial cells, many centrioles grow orthogonally to existing centrioles or deuterosomes and then dissociate from these structures as ciliogenesis progresses, without passage through mitosis. It is unknown whether this dissociation is the same as anaphase disengagement of centrioles and whether separase activity is required. If the processes are analogous, then disengagement might control the availability of sites on the organizing structures and thus play a role in centriole number control during ciliogenesis. Ultimately, some mechanism must limit the number of centrioles formed in ciliating cells. Interestingly, we have noticed that although most ciliated cells in the MTEC culture have ∼300 basal bodies, there are occasional cells with a large apical surface area with >1,000 basal bodies, but distributed with a similar density (unpublished data). This suggests that cell size or apical surface area might control centriole number, but how this is communicated to the centriole assembly machinery is unknown.
We have developed an in vitro culture system that undergoes ciliogenesis, can be manipulated by infection with viral vectors, and from which ciliated cells can be sorted on the basis of FOXJ1/EGFP expression. We believe that this is an ideal model system for addressing many of the outstanding questions in centriole and ciliary biology. Much recent attention has been focused on centriole generation, including number control and pathways of assembly (Pelletier et al., 2006; Tsou and Stearns, 2006a), and on centriole function in microtubule organization and cilium formation (Marshall and Nonaka, 2006; Luders and Stearns, 2007). These processes have been studied individually in cycling mammalian cells, often under nonphysiological conditions, whereas in ciliated epithelial cells, they can all be observed as part of the naturally occurring ciliogenesis pathway.
Materials and methods
Animals
MTECs were derived from wild-type C3H × C57Bl/6J F1 hybrid or FOXJ1/EGFP transgenic mice (a gift from L. Ostrowski, University of North Carolina at Chapel Hill, Chapel Hill, NC) generated on C3H × C57Bl/6J F1 hybrid background (Ostrowski et al., 2003). FOXJ1/EGFP mice were bred by mating transgenic heterozygous males to wild-type C3H × C57Bl/6J F1 hybrid females (Taconic). Offspring were genotyped using PCR with EGFP-specific PCR primers. All procedures involving animals were approved by the Institutional Animal Care and Use Committee in accordance with established guidelines for animal care.
Cell culture and media
NIH/3T3 and 293T cells were grown in DME with 10% FBS (Invitrogen). MTEC culture was based on You et al. (2002). Mice were killed at 4–6 mo of age, and trachea were excised, trimmed of excess tissue, opened longitudinally to expose the lumen, and placed in 1.5 mg/ml pronase E in F-12K nutrient mixture (Invitrogen) at 4°C overnight. Tracheal epithelial cells were dislodged by gentle agitation and collected in F-12K with 10% FBS. Cells were treated with 0.5 mg/ml DNase I for 5 min on ice and centrifuged at 4°C for 10 min at 400 g. Cells were resuspended in DME/F-12 (Invitrogen) with 10% FBS and plated in a tissue culture dish for 3 h at 37°C and 5% CO2 to adhere contaminating fibroblasts. Nonadhered cells were resuspended in an appropriate volume of MTEC Plus medium (You et al., 2002) and seeded onto Transwell-Clear (Corning) permeable filter supports at 105 cells/cm2. The ALI was created ∼2 d after cells reached confluence, by feeding MTEC Serum-free or MTEC NuSerum medium (You et al., 2002) only from below the filter. Cells were cultured at 37°C and 5% CO2 and fed fresh medium every 2 d. Beating cilia were observed by phase microscopy 2–3 d after ALI creation. All chemicals were obtained from Sigma-Aldrich unless otherwise indicated. All media were supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin, and 0.25 mg/ml Fungizone (all obtained from Invitrogen).
Lentiviral constructs and lentivirus production
HIV-derived recombinant lentivirus expressing GFP-tagged constructs were made from the lentiviral transfer vector pRRL.sin-18.PPT.PGK.GFP.pre (Follenzi et al., 2000) by inserting the ORF in frame with the GFP cassette at the AgeI site using PCR. Additional tagged cDNA constructs were made by inserting a PCR fragment of the tagged cDNA into the AgeI site of the lentiviral transfer vector pRRL.sin-18.PPT.PGK.IRES.GFP.pre (Follenzi et al., 2000) after removing the IRES and GFP sequences by digestion with NheI and BsrGI, blunting, and religation. Lentivirus encoding the mouse polaris shRNA (targeting nt 2164–2182; available from GenBank/EMBL/DDBJ under accession no. NM_009376) was made using the pSicoR PGK puro (Ventura et al., 2004) lentiviral vector. Lentiviruses encoding the mouse SAS-6 (targeting nt 1273–1291; accession no. NM_028349) and PCM-1 shRNAs (targeting nt 1213–1231; accession no. NM_023662) were made using the pLentiLox3.7 (Rubinson et al., 2003) lentiviral transfer vector that also expresses GFP from a separate CMV promoter to mark infected cells. For infecting FOXJ1/EGFP MTECs, the modified lentiviral vector, pLentiRFP3.7, was generated by replacing the GFP cassette in pLentiLox3.7 with monomeric RFP using the NheI and EcoRI sites. shRNA constructs were verified by sequencing. Lentiviral vectors were propagated in XL2-Blue cells (Stratagene) and isolated from bacteria using the QIAfilter Maxi Plasmid Purification kit (QIAGEN).
Recombinant lentivirus was produced by transient cotransfection of 293T cells with the appropriate transfer and lentiviral helper plasmids (pCMVDR8.74 packaging vector and pMD2.VSVG envelope vector; a gift from P. Kowalski, Stanford University, Stanford, CA; Dull et al., 1998) using the FuGENE6 transfection reagent (Roche Applied Science) or the calcium phosphate coprecipitation method. 18 h after transfection, cells were given fresh medium. The lentiviral supernatant was harvested 48–72 h after transfection and filtered though a 0.45-μm PES filter (Nalgene). Some lentiviral supernatants were concentrated 100- to 500-fold by ultracentrifugation at 20°C for 180 min at 50,000 g. Lentiviruses were titered on NIH/3T3 cells by flow cytometry or immunofluorescence. Titers for preparations used were 107–108 infectious units/ml.
Lentiviral infection
All lentiviral constructs were verified in NIH/3T3 cells before introduction into MTECs. NIH/3T3 cells were seeded onto 24-well tissue culture plates the day before infection. To infect, the medium was removed and replaced by a mix of lentivirus, 5 μg/ml hexadimethrine bromide (Sigma-Aldrich), and medium in 60% of the normal plating volume. Virus was removed 24 h after infection. Cells were assayed at least 48 h after infection.
To infect MTECs, medium was removed and cells were rinsed twice with PBS. Efficient lentiviral transduction of polarized airway epithelial cells only occurs at the basolateral surface (Borok et al., 2001). To allow access to the basolateral surface, epithelial tight junctions were disrupted by treating cells with 12 mM EGTA in 10 mM Hepes, pH 7.4, at 37°C for 20 min. Cells were rinsed twice with PBS. Fresh medium was added to the bottom of the dish, and a mix of lentivirus, 5 μg/ml hexadimethrine bromide, and medium was placed on top of the cells. The plate was sealed with parafilm and centrifuged at 32°C for 80 min at 1,500 g. After centrifugation, the plate was unsealed and placed at 37°C. Centrifugation greatly enhanced transduction efficiency in MTECs and had no adverse effects on cell morphology or viability. Epithelial junctions were completely reformed by 24 h after infection as monitored by ZO-1 antibody signal. Virus was removed 24 h after infection. Cells were assayed at least 48 h after infection; based on cytoplasmic GFP or monomeric RFP expression from the lentivirus, 20–50% of cells at the surface of the epithelium were transduced. Control infections were performed using virus made from transfer vectors without the transgene or short hairpin construct of interest.
Immunofluorescence
For indirect immunofluorescence, MTECs were rinsed twice with PBS and fixed in either methanol at −20°C for 7 min or 4% paraformaldehyde in PBS at room temperature for 10 min, depending on antigen. To preserve both cytoplasmic GFP signal and epitopes sensitive to aldehyde cross-linking, cells were fixed in 0.5% paraformaldehyde in PBS at room temperature for 5 min, followed by methanol at −20°C for 7 min. After fixation, cells were rinsed twice with PBS and filters were excised from plastic supports. Filters were cut in quarters to provide multiple equivalent samples for consistent observation. Cells were incubated two times for 5 min each in 0.2% Triton X-100 in PBS and blocked for 1 h at room temperature in 5% normal goat serum (Invitrogen) and 3% BSA (Sigma-Aldrich) in PBS. Primary antibodies were applied to filters at 37°C for 1 h or 4°C overnight. Alexa dye–conjugated goat secondary antibodies (Invitrogen) were applied to filters at room temperature for 30 min. Cells were incubated two times for 5 min each in 0.2% Triton X-100 in PBS between changes of antibody. Filters were mounted with 12-mm coverslips (1.5; Erie Scientific) using Mowiol mounting medium containing N-propyl gallate (Sigma-Aldrich). Cells were observed using Openlab 4.0.4 (Improvision) controlling a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.). Image stacks were collected with a z-step size of 0.2 μm and were then deconvolved and processed with AutoDeblur 9.3 and AutoVisualize 9.3 (AutoQuant Imaging). For a list of antibodies and appropriate fixation conditions used, see the supplemental text.
EM
For scanning EM of MTEC cultures, filters were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.2 M Hepes buffer, pH 7.4, for 20 min at room temperature followed by 40 min at 4°C. Fixed samples were stained with osmium tetroxide for 1 h, dehydrated with a graded ethanol series, and dried using a critical point drier. Filters were mounted onto stubs and sputter coated with Gold/Palladium. Samples were visualized using a microscope (SEM525; Philips).
For TEM of MTEC cultures, filters were fixed in 3% paraformaldehyde, 0.1% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.2, at room temperature for 1 h. Fixed samples were dehydrated with a graded ethanol series and infiltrated with either EMbed-812 or LR White resin (Electron Microscopy Sciences). For TEM, 80–100-nm sections were mounted onto nickel grids and analyzed with a microscope (TEM1230; JEOL). For immuno-EM using the PCM-1 antibody, 80–100-nm sections were incubated with 0.1 M glycine and blocked with 5% normal goat serum and 3% BSA in PBS for 30 min. Sections were incubated with antibody or PBS for 1 h and then with 5 nm gold–conjugated goat secondary antibody (Ted Pella) for 30 min. Sections were postfixed in 4% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.2, for 5 min, and poststained with 7% uranyl acetate/acetone (1:1) for 15 s followed by a brief incubation on a drop of lead citrate. The sections were mounted onto nickel grids and analyzed with the TEM1230 microscope.
FACS and Western blotting
To prepare a single cell suspension of MTECs and FOXJ1/EGFP MTECs for FACS, filters were incubated with a 1:1 mix of 0.5% Trypsin/EDTA (Invitrogen) and Cell Dissociation Solution (Sigma-Aldrich) at 37°C for 1 h. Cells were gently pipetted up and down every 15 min. Cells were washed in 1× PBS and resuspended in ice-cold PBS + 10% FBS at 107 cells/ml. EGFP+ and EGFP− cells were sorted with a FACStar (Becton Dickinson) sorter using a 488-nm Argon ion laser. For Western blotting on total cell lysates, sorted cells were rinsed in PBS and resuspended directly in SDS sample buffer at 103 cells/μl; 104 cells were loaded per well. Blots were blocked for 1 h in 5% milk in TBS + 0.05% Tween-20 and incubated overnight with primary antibody. Primary antibodies were detected with Alexa 635– conjugated goat secondary antibody (Invitrogen) and scanning with a Typhoon 9210 Variable Mode Imager using a 633-nm HeNe laser and an emission filter (670 BP 30; GE Healthcare). Images were quantitated with ImageQuant 5.2 (GE Healthcare).
Online supplemental material
Fig. S1 shows microtubule and centrosomal protein distribution in ciliating cells. Fig. S2 shows polaris expression during ciliogenesis and polaris depletion by lentiviral RNAi. Fig. S3 shows SAS-6 localization during ciliogenesis and SAS-6 depletion by lentiviral RNAi. Fig. S4 shows PCM-1– containing fibrous granules during ciliogenesis and PCM-1 depletion by lentiviral RNAi. Table S1 shows localization of centrosomal proteins in MTECs. Table S2 lists antibodies used for immunofluorescence and Western blotting. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200703064/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Lawrence Ostrowski for the gift of the FOXJ1/EGFP transgenic mice; Timothy Stowe (Stanford University) for generating the polaris shRNA construct; Dr. Bryan Tsou (Stanford University) and Dr. Alexander Dammermann (University of California, San Diego) for SAS-6 reagents; Dr. Andreas Merdes (Centre National de la Recherche Scientifique/Pierre Fabre) for the PCM-1 reagents; Dr. Nicholas Katsanis (Johns Hopkins University) for the BBS4 cDNA; Dr. Paul Kowalski for the lentiviral vectors; Dr. Jean-Francois Fortin and Dr. Roland Wolkowicz (Stanford University) for advice on lentivirus generation; Dr. Steven Brody (Washington University) for advice on MTEC culture; Dr. Guowei Fang (Stanford University) for advice on image processing; Emily Hatch (Stanford University) and John Perrino at the Cell Sciences Imaging Facility (Stanford University) for assistance with EM; and the Fang, Nolan, Cohen, Nelson, and Stearns laboratories at Stanford University for additional reagents, access to equipment, and helpful advice.
This work was supported by National Institutes of Health grant GM52022 to T. Stearns.
Abbreviations used in this paper: ALI, air–liquid interface; IFT, intraflagellar transport; MTEC, mouse tracheal epithelial cell; shRNA, short hairpin RNA; TEM, transmission EM.
References
- Avidor-Reiss, T., A.M. Maer, E. Koundakjian, A. Polyanovsky, T. Keil, S. Subramaniam, and C.S. Zuker. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 117:527–539. [DOI] [PubMed] [Google Scholar]
- Borok, Z., J.E. Harboe-Schmidt, S.L. Brody, Y. You, B. Zhou, X. Li, P.M. Cannon, K.J. Kim, E.D. Crandall, and N. Kasahara. 2001. Vesicular stomatitis virus G-pseudotyped lentivirus vectors mediate efficient apical transduction of polarized quiescent primary alveolar epithelial cells. J. Virol. 75:11747–11754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, M.J., M.F. Dolan, and L. Margulis. 2000. Centrioles and kinetosomes: form, function, and evolution. Q. Rev. Biol. 75:409–429. [DOI] [PubMed] [Google Scholar]
- Christensen, S.T., C.F. Guerra, A. Awan, D.N. Wheatley, and P. Satir. 2003. Insulin receptor-like proteins in Tetrahymena thermophila ciliary membranes. Curr. Biol. 13:R50–R52. [DOI] [PubMed] [Google Scholar]
- Dammermann, A., and A. Merdes. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann, A., T. Muller-Reichert, L. Pelletier, B. Habermann, A. Desai, and K. Oegema. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 7:815–829. [DOI] [PubMed] [Google Scholar]
- Dirksen, E.R. 1991. Centriole and basal body formation during ciliogenesis revisited. Biol. Cell. 72:31–38. [DOI] [PubMed] [Google Scholar]
- Dull, T., R. Zufferey, M. Kelly, R.J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher, S.K., and E.C. Trabuco. 1998. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell. 9:1293–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follenzi, A., L.E. Ailles, S. Bakovic, M. Geuna, and L. Naldini. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217–222. [DOI] [PubMed] [Google Scholar]
- Garreau de Loubresse, N., F. Ruiz, J. Beisson, and C. Klotz. 2001. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts, B.N., X. Gong-Cooper, and B.P. Hackett. 2004. Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. J. Cell Sci. 117:1329–1337. [DOI] [PubMed] [Google Scholar]
- Hagiwara, H., N. Ohwada, and K. Takata. 2004. Cell biology of normal and abnormal ciliogenesis in the ciliated epithelium. Int. Rev. Cytol. 234:101–141. [DOI] [PubMed] [Google Scholar]
- Jurczyk, A., A. Gromley, S. Redick, J. San Agustin, G. Witman, G.J. Pazour, D.J. Peters, and S. Doxsey. 2004. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J. Cell Biol. 166:637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, L.C., E.P. Romijn, I. Zamora, J.R. Yates III, and W.F. Marshall. 2005. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15:1090–1098. [DOI] [PubMed] [Google Scholar]
- Kim, H.K., J.G. Kang, S. Yumura, C.J. Walsh, J.W. Cho, and J. Lee. 2005. De novo formation of basal bodies in Naegleria gruberi: regulation by phosphorylation. J. Cell Biol. 169:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.C., J.L. Badano, S. Sibold, M.A. Esmail, J. Hill, B.E. Hoskins, C.C. Leitch, K. Venner, S.J. Ansley, A.J. Ross, et al. 2004. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 36:462–470. [DOI] [PubMed] [Google Scholar]
- Kubo, A., and S. Tsukita. 2003. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 116:919–928. [DOI] [PubMed] [Google Scholar]
- Kubo, A., H. Sasaki, A. Yuba-Kubo, S. Tsukita, and N. Shiina. 1999. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147:969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra, S., C.N. English, P. Hergert, B.F. McEwen, G. Sluder, and A. Khodjakov. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 168:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili, J., E. Perret, S. Middendorp, O. Houcine, C. Guennou, F. Marano, M. Bornens, and F. Tournier. 2000. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 113:1355–1364. [DOI] [PubMed] [Google Scholar]
- Leidel, S., M. Delattre, L. Cerutti, K. Baumer, and P. Gonczy. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125. [DOI] [PubMed] [Google Scholar]
- Levy, Y.Y., E.Y. Lai, S.P. Remillard, M.B. Heintzelman, and C. Fulton. 1996. Centrin is a conserved protein that forms diverse associations with centrioles and MTOCs in Naegleria and other organisms. Cell Motil. Cytoskeleton. 33:298–323. [DOI] [PubMed] [Google Scholar]
- Li, J.B., J.M. Gerdes, C.J. Haycraft, Y. Fan, T.M. Teslovich, H. May-Simera, H. Li, O.E. Blacque, L. Li, C.C. Leitch, et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 117:541–552. [DOI] [PubMed] [Google Scholar]
- Luders, J., and T. Stearns. 2007. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 8:161–167. [DOI] [PubMed] [Google Scholar]
- Marshall, W.F., and S. Nonaka. 2006. Cilia: tuning in to the cell's antenna. Curr. Biol. 16:R604–R614. [DOI] [PubMed] [Google Scholar]
- Murcia, N.S., W.G. Richards, B.K. Yoder, M.L. Mucenski, J.R. Dunlap, and R.P. Woychik. 2000. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 127:2347–2355. [DOI] [PubMed] [Google Scholar]
- Muresan, V., H.C. Joshi, and J.C. Besharse. 1993. Gamma-tubulin in differentiated cell types: localization in the vicinity of basal bodies in retinal photoreceptors and ciliated epithelia. J. Cell Sci. 104:1229–1237. [DOI] [PubMed] [Google Scholar]
- Mykytyn, K., R.F. Mullins, M. Andrews, A.P. Chiang, R.E. Swiderski, B. Yang, T. Braun, T. Casavant, E.M. Stone, and V.C. Sheffield. 2004. Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc. Natl. Acad. Sci. USA. 101:8664–8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, B. 2005. Nearly all cells in vertebrates and many cells in invertebrates contain primary cilia. Matrix Biol. 24:449–450. [DOI] [PubMed] [Google Scholar]
- Ostrowski, L.E., J.R. Hutchins, K. Zakel, and W.K. O'Neal. 2003. Targeting expression of a transgene to the airway surface epithelium using a ciliated cell-specific promoter. Mol. Ther. 8:637–645. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., B.L. Dickert, Y. Vucica, E.S. Seeley, J.L. Rosenbaum, G.B. Witman, and D.G. Cole. 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151:709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., N. Agrin, J. Leszyk, and G.B. Witman. 2005. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, L., E. O'Toole, A. Schwager, A.A. Hyman, and T. Muller-Reichert. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. [DOI] [PubMed] [Google Scholar]
- Praetorius, H.A., and K.R. Spring. 2005. A physiological view of the primary cilium. Annu. Rev. Physiol. 67:515–529. [DOI] [PubMed] [Google Scholar]
- Preble, A.M., T.H. Giddings Jr., and S.K. Dutcher. 2001. Extragenic bypass suppressors of mutations in the essential gene BLD2 promote assembly of basal bodies with abnormal microtubules in Chlamydomonas reinhardtii. Genetics. 157:163–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins, E.L., L.E. Ostrowski, S.H. Randell, and B.L. Hogan. 2007. Lung development and repair: contribution of the ciliated lineage. Proc. Natl. Acad. Sci. USA. 104:410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, A., G. Margall-Ducos, J.E. Guidotti, O. Bregerie, C. Celati, C. Brechot, and C. Desdouets. 2007. The intraflagellar transport component IFT88/polaris is a centrosomal protein regulating G1-S transition in non-ciliated cells. J. Cell Sci. 120:628–637. [DOI] [PubMed] [Google Scholar]
- Ross, A.J., L.A. Dailey, L.E. Brighton, and R.B. Devlin. 2007. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. DOI:10.1165/rcmb.2006-0466OC. [DOI] [PubMed]
- Rubinson, D.A., C.P. Dillon, A.V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, D.L. Rooney, M.M. Ihrig, M.T. McManus, F.B. Gertler, et al. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401–406. [DOI] [PubMed] [Google Scholar]
- Scholey, J.M., and K.V. Anderson. 2006. Intraflagellar transport and cilium-based signaling. Cell. 125:439–442. [DOI] [PubMed] [Google Scholar]
- Sorokin, S.P. 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3:207–230. [DOI] [PubMed] [Google Scholar]
- Stemm-Wolf, A.J., G. Morgan, T.H. Giddings Jr., E.A. White, R. Marchione, H.B. McDonald, and M. Winey. 2005. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 16:3606–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon, B.E., S.A. Adler, J.P. Suhan, and J.W. Jarvik. 1992. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119:1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulman, P.D., C.J. Haycraft, D.F. Balkovetz, and B.K. Yoder. 2001. Polaris, a protein involved in left-right axis patterning, localizes to basal bodies and cilia. Mol. Biol. Cell. 12:589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teilmann, S.C., A.G. Byskov, P.A. Pedersen, D.N. Wheatley, G.J. Pazour, and S.T. Christensen. 2005. Localization of transient receptor potential ion channels in primary and motile cilia of the female murine reproductive organs. Mol. Reprod. Dev. 71:444–452. [DOI] [PubMed] [Google Scholar]
- Tichelaar, J.W., S.E. Wert, R.H. Costa, S. Kimura, and J.A. Whitsett. 1999. HNF-3/forkhead homologue-4 (HFH-4) is expressed in ciliated epithelial cells in the developing mouse lung. J. Histochem. Cytochem. 47:823–832. [DOI] [PubMed] [Google Scholar]
- Toskala, E., S.M. Smiley-Jewell, V.J. Wong, D. King, and C.G. Plopper. 2005. Temporal and spatial distribution of ciliogenesis in the tracheobronchial airways of mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 289:L454–L459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou, M.F., and T. Stearns. 2006. a. Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 18:74–78. [DOI] [PubMed] [Google Scholar]
- Tsou, M.F., and T. Stearns. 2006. b. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 442:947–951. [DOI] [PubMed] [Google Scholar]
- Ventura, A., A. Meissner, C.P. Dillon, M. McManus, P.A. Sharp, L. Van Parijs, R. Jaenisch, and T. Jacks. 2004. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. USA. 101:10380–10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Y., E.J. Richer, T. Huang, and S.L. Brody. 2002. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell. Mol. Physiol. 283:L1315–L1321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.