Abstract
A pulse of the steroid hormone ecdysone triggers the destruction of larval salivary glands during Drosophila metamorphosis through a transcriptional cascade that converges on reaper (rpr) and head involution defective (hid) induction, resulting in caspase activation and cell death. We identify the CREB binding protein (CBP) transcriptional cofactor as essential for salivary gland cell death. We show that CBP acts 1 d before the onset of metamorphosis in apparent response to a mid-third instar ecdysone pulse, when CBP is necessary and sufficient for down-regulation of the Drosophila inhibitor of apoptosis 1 (DIAP1). It is only after DIAP1 levels are reduced that salivary glands become competent to die through rpr/hid-mediated cell death. Before this time, high levels of DIAP1 block salivary gland cell death, even in the presence of ectopic rpr expression. This study shows that naturally occurring changes in inhibitor of apoptosis levels can be critical for regulating cell death during development. It also provides a molecular mechanism for the acquisition of competence in steroid signaling pathways.
Introduction
Competence has long been recognized as a critical mechanism for restricting the developmental potential of cells to specific fates (Waddington, 1940). Competence is progressively regulated such that cells achieve a series of competent states, each of which sets up the subsequent developmental response during the life cycle. In this manner, widespread signals are refined to direct spatially and temporally restricted biological responses. Although often invoked, relatively little is known about how competence is achieved at a molecular level.
We are studying the programmed cell death of larval tissues during Drosophila metamorphosis as a model system for understanding the molecular basis of competence and its role in determining the appropriate temporal progression of steroid-triggered biological responses during development. Programmed cell death plays a central role in animal development, eliminating unwanted tissues, controlling cell numbers, and sculpting complex structures. The decision to live or die is determined by each cell based on a critical balance between evolutionarily conserved death activators and death inhibitors (Martin, 2002; Danial and Korsmeyer, 2004). Several signals can affect this balance, regulating the patterns of programmed cell death in a precise temporal and spatial manner. In frogs, thyroid hormone signals the destruction of the tadpole tail and remodeling of the intestine as the animal progresses from a juvenile to adult form (Shi et al., 2001). Similarly, steroid hormones regulate mammalian apoptotic pathways, including the glucocorticoid-induced apoptosis of immature thymocytes and mature T cells (Winoto and Littman, 2002).
A dramatic manifestation of steroid-triggered cell death can be seen in the fruit fly Drosophila, where the steroid hormone ecdysone directs the massive and rapid destruction of larval tissues during metamorphosis. Although relatively little is known about the mechanisms of hormone-regulated programmed cell death in vertebrates, considerable insights into this pathway have been gained in Drosophila. A high titer pulse of ecdysone at the end of larval development acts through the ecdysone receptor (EcR)/Ultraspiracle nuclear receptor heterodimer to signal puparium formation and the destruction of several larval tissues, including the midgut (Baehrecke, 2003; Yin and Thummel, 2005). A second ecdysone pulse, ∼10 h after puparium formation (APF), triggers adult head eversion, marking the prepupal–pupal transition and signaling the rapid destruction of the larval salivary glands (Robertson, 1936; Jiang et al., 1997). Destruction of the larval midguts and salivary glands is accompanied by classic hallmarks of apoptosis, including acridine orange staining, TUNEL staining, and caspase activation (Jiang et al., 1997). These larval tissues, however, undergo a distinct form of programmed cell death referred to as autophagy, characterized by the formation of intracellular autophagic vesicles (Lee and Baehrecke, 2001; Baehrecke, 2005).
Three related death activator genes play a central role in the control of Drosophila programmed cell death: reaper (rpr), head involution defective (hid), and grim (White et al., 1994; Grether et al., 1995; Chen et al., 1996). Elimination of these genes completely blocks programmed cell death, whereas ectopic expression of any is sufficient to trigger a cell death response. The Rpr, Hid, and Grim proteins interact with the cell death inhibitor Drosophila inhibitor of apoptosis 1 (DIAP1), disrupting its interaction with caspases and targeting DIAP1 for degradation, allowing caspase activation and cell death (for review see Martin, 2002). A similar regulatory pathway is used in the destruction of larval salivary glands during metamorphosis, where the prepupal pulse of ecdysone triggers a transcriptional cascade that culminates in rpr and hid induction, overcoming the inhibitory effects of DIAP1 and initiating tissue destruction (Jiang et al., 1997, 2000; Lee et al., 2002; Yin and Thummel, 2004; Cao et al., 2007).
Here, we show that during most of larval development, DIAP1 cannot be overcome by death activator expression in larval salivary glands. This is due to high levels of DIAP1 in this tissue at early stages, substantially higher than are present at the onset of metamorphosis. This switch in DIAP1 levels occurs in the mid-third instar and depends on EcR and the CREB binding protein (CBP), defining a new transcriptional hierarchy that regulates salivary gland cell death. CBP is both necessary and sufficient to down-regulate DIAP1, indicating a central role for this factor in mediating the switch in DIAP1 levels. The resulting threshold level of DIAP1 is sufficient to hold back cell death, but low enough to be sensitive to subsequent ecdysone-induced rpr and hid expression in prepupae. Larval salivary gland cell death is thus a two-step temporally regulated response that involves sequential effects on DIAP1 levels. The first step, mediated by CBP, reduces DIAP1 levels, providing the competence to die. The second step, mediated by rpr and hid, eliminates residual DIAP1 and triggers tissue destruction. This work provides a new context for understanding how steroid hormones regulate programmed cell death during development. It also provides a molecular basis for understanding how competence is achieved to allow the appropriate temporal progression of hormone-regulated biological responses during development.
Results
CBP is required for larval salivary gland cell death
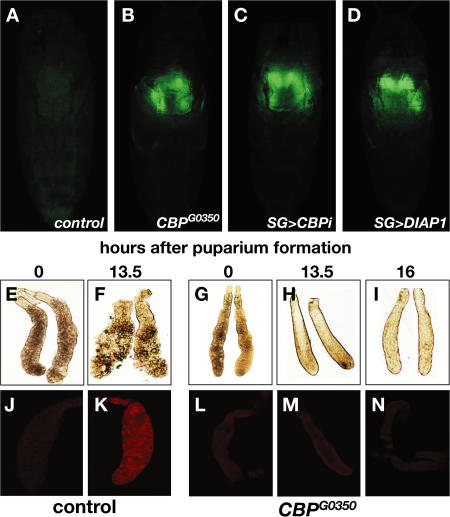
To further our understanding of the regulation of steroid-triggered cell death, we screened for mutations that block the destruction of the larval salivary glands in living animals using a salivary gland–specific GFP reporter (Ward et al., 2003). Starting with a collection of P-element–induced lethal mutations (Peter et al., 2002), we identified several complementation groups that resulted in persistent larval salivary glands (unpublished data). One complementation group of three alleles mapped to an ∼400-bp region near the 5′ end of the gene that encodes the Drosophila homologue of CBP/Nejire (Nej), adjacent to a previously uncharacterized exon of CBP (CG15321, linked to CBP by RT-PCR; unpublished data). All three mutations (CBPG0350, CBPG0112, and CBPG0470) fail to complement a known CBP mutation, nejQ7. The nej3 CBP-null allele, like the three P-element–induced mutations, leads to developmental arrest soon after the prepupal–pupal transition, with persistent larval salivary glands (Fig. 1 B). This block in cell death can be phenocopied by salivary gland–specific inactivation of CBP using RNAi, demonstrating that the effects of CBP on cell death are tissue autonomous (Fig. 1 C).
Figure 1.
Mutations in CBP block ecdysone-triggered programmed cell death of larval salivary glands. (A–D) Living animals assayed 24 h APF with a GFP reporter expressed in larval salivary glands. (A) Salivary glands are no longer detectable in control pupae (w; UAS-GFP; SG-GAL4) but persist in CBP mutants (CBPG0350; UAS-GFP/+; SG-GAL4/+) (B) or salivary glands in which CBP is inactivated by RNAi (w; UAS-CBP-RNAi/ UAS-GFP; SG-GAL4/+) (C) or in which diap1 is overexpressed (w; UAS-diap1/UAS-GFP; SG-GAL4/+) (D). (E and F) Larval salivary glands dissected at 0 or 13.5 h APF from controls or 0-, 13.5-, or 16-h CBPG0350 animals (G–I). (J and K) Control and CBPG0350 mutant (L–N) salivary glands stained for the cleaved active form of caspase-3.
CBP mutant pupae are virtually indistinguishable from their wild-type counterparts at 12 h APF. The absence of defects in morphogenesis of adult structures and the lack of developmental delay between puparium formation and the prepupal–pupal transition suggest that the global ecdysone-regulated control of metamorphosis is normal in CBP mutant animals. Similarly, CBP mutant salivary glands synthesize glue proteins at the appropriate time during third-instar larval (L3) development and secrete their contents in response to ecdysone at puparium formation, as normal (unpublished data). In contrast to wild-type glands, however, which show clear signs of cell death at ∼13.5 h APF (Fig. 1 F), CBP mutant salivary glands survive for at least 72 h, with 70% of CBPG0350 mutant pupae displaying persistent glands (n = 60). The mutant glands fail to display morphological signs of tissue breakdown, showing tight cellular association at 13.5 and 16 h APF (Fig. 1, H and I). Moreover, although wild-type glands show staining for active caspase-3 at 13.5 h APF (Fig. 1 K), CBP mutant salivary glands do not display this response, consistent with their block in programmed cell death (Fig. 1, L–N). This phenotype is specific to the CBP locus, as it can be rescued by the expression of a wild-type UAS-CBP transgene in combination with a SGS3-GAL4 driver, which is induced in mid-L3 salivary glands and expressed through puparium formation (Andres et al., 1993). Of 51 mutant pupae examined from a control cross of CBPG0350/FM7i, Act-GFP; [SGS3-GAL4], [UAS-GFP] and FM7i, Act-GFP flies, 47% had persistent salivary glands. In contrast, of 12 mutants examined from the rescue cross of CBPG0350/FM7i, Act-GFP; [SGS3-GAL4], [UAS-GFP] and FM7i, Act-GFP; [UAS-CBP] flies, 17% had persistent salivary glands, a threefold reduction from the control.
Death activators are expressed normally in CBP mutant salivary glands
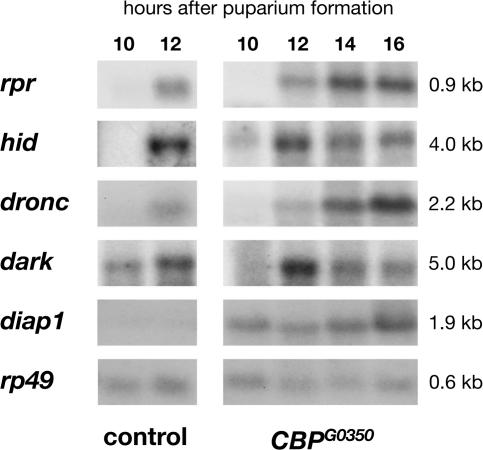
CBP is a key transcriptional coactivator, bridging transcription factors with the basal transcriptional machinery and directing histone acetylation. CBP can physically interact with a large number of transcription factors, including nuclear receptors, and has been implicated in regulating cell death (Giordano and Avantaggiati, 1999). Given its general role in transcriptional control, we predicted that CBP mutations would act at the top of the ecdysone-triggered death cascade, disrupting induction of the key death activators rpr and hid at the prepupal–pupal transition. Surprisingly, however, Northern blot analysis of RNA isolated from control and CBP mutant salivary glands revealed that rpr and hid are induced normally in mutant glands, as are the dronc apical caspase gene and the Apaf-1 adaptor encoded by dark (Fig. 2). Expression of any one of these four death regulators is sufficient to trigger cell death, an observation that appears inconsistent with the healthy morphology of CBP mutant salivary glands (Grether et al., 1995; White et al., 1996; Dorstyn et al., 1999; White, 2000). This apparent paradox, however, can be explained by up-regulation of diap1, the only known death inhibitor that acts parallel to or downstream from these death activators (Fig. 2; Wang et al., 1999; Goyal et al., 2000; Lisi et al., 2000; Yin and Thummel, 2004). In addition, salivary gland–specific overexpression of diap1 phenocopies the persistent salivary gland phenotype of CBP mutants, indicating that selective up-regulation of diap1 is sufficient to block salivary gland cell death (Fig. 1 D).
Figure 2.
CBP regulates diap1 mRNA levels in larval salivary glands. Northern blots from control (left) and CBPG0350 mutant (right) larval salivary glands staged at 10, 12, 14, or 16 h APF, probed for key cell death regulators. Death activators rpr and hid, the caspase gene dronc, and the Apaf-1 gene dark are all induced normally in mutant salivary glands, whereas diap1 mRNA is selectively up-regulated. rp49 is a loading control. The kinetics of rpr and hid induction in control and mutant salivary glands were confirmed by Northern blot analysis of four independent RNA samples.
CBP down-regulates DIAP1 in mid-third instar salivary glands
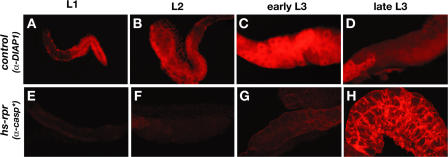
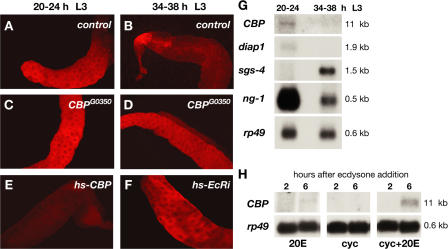
The observation that diap1 levels are up-regulated in CBP mutant salivary glands (Fig. 2) suggested that diap1 levels may be high early in development and that CBP may down-regulate diap1 at a specific time, achieving the final level seen at the onset of metamorphosis. We thus shifted our attention to examining DIAP1 protein levels and looking at earlier stages of wild-type development. Consistent with this model, DIAP1 is abundant in the salivary glands of control first (L1), second (L2), and early third instar larvae (see Fig. 4, A–C), but drops to low levels by the end of L3 (see Fig. 4 D). Staining of salivary glands from staged L3 with DIAP1 antibodies revealed that this switch in DIAP1 expression levels occurs between 20–24 and 34–38 h after the L2–L3 molt (Fig. 3, A and B) and fails to occur in CBP mutant salivary glands (Fig. 3, C and D). Moreover, ectopic expression of CBP in early L3 is sufficient to direct premature DIAP1 down-regulation, defining CBP as a critical regulator of DIAP1 levels and suggesting that stage-specific expression of CBP in mid-L3 directs this switch in diap1 expression (Fig. 3 E).
Figure 4.
Down-regulation of diap1 confers competence for rpr-mediated cell death. Salivary glands dissected from control (w) (A–D) first (L1), second (L2), early third and late third (L3) instar larvae or animals at the same stages after heat-induced rpr overexpression (w; hs-rpr) (E–H) were stained for DIAP1 protein (A–D) or cleaved active caspase-3 (E–H). DIAP1 levels drop during L3 development. Cell death in response to rpr overexpression only occurs when DIAP1 levels are low in late L3.
Figure 3.
CBP is necessary and sufficient for diap1 down-regulation in response to ecdysone during the mid-third instar transition. (A–F) DIAP1 antibody stains of larval salivary glands dissected from animals that span the mid-L3 transition (larva staged relative to the L2–L3 molt). (A and B) DIAP1 levels drop between 20–24 and 34–38 h in control glands (w) but not in CBPG0350 mutant salivary glands (C and D). (E) Ectopic expression of CBP (w; hs-CBP) in 12-h L3 animals is sufficient to down-regulate DIAP1 expression at 20–24 h, whereas removal of EcR by RNAi blocks DIAP1 down-regulation (F). (G) Northern blots of RNA isolated from wild-type salivary glands from before (20–24 h) or after (34–38 h) the mid-L3 transition, probed to detect CBP, diap1, ng-1, and Sgs-4 mRNA, with rp49 as a control. (H) Organs dissected from L3 were cultured in the presence of 20-hydroxyecdysone (20E), cycloheximide (cyc), or 20E and cycloheximide for 2 or 6 h, and total RNA was extracted for Northern blot hybridization to detect CBP and rp49 mRNA.
DIAP1 down-regulation occurs at the mid-third instar transition
A critical developmental transition occurs about 1 d after the L2–L3 molt, in preparation for puparium formation 1 d later, defining what has been called the mid-third instar transition (Andres and Cherbas, 1992; Andres et al., 1993; Cherbas et al., 2003). This transition is manifested by a global switch in gene expression as well as behavioral and metabolic changes that culminate in larval wandering and puparium formation. The mid-L3 transition occurs in synchrony with a low titer pulse of ecdysone, and at least some aspects are dependent on EcR (Cherbas et al., 2003). Northern blot analysis of RNA isolated from staged larval salivary glands revealed that both CBP and diap1 are expressed at 20–24 h after the L2–L3 molt and down-regulated by 34–38 h, in synchrony with the switch from ng to Sgs glue gene expression, a hallmark of the mid-third instar transition (Fig. 3 G; Andres et al., 1993; Furia et al., 1993; Mougneau et al., 1993). This change in CBP levels, combined with our genetic studies, indicates that although CBP is required for DIAP1 down-regulation in mid-L3, it is not needed to maintain diap1 expression in salivary glands. The observation that diap1 transcript levels and protein levels drop at the same time is consistent with the short half-life of DIAP1 protein (Yoo et al., 2002). A more detailed understanding, however, of the precise timing and mechanism of diap1 transcriptional repression remains to be addressed.
The down-regulation of diap1 fails to occur when EcR function is disrupted by RNAi (Fig. 3 F) or upon expression of a dominant-negative form of EcR (Cherbas et al., 2003; unpublished data). In addition, low levels of CBP mRNA can be induced by ecdysone in cultured larval organs, and this response is enhanced upon addition of the protein synthesis inhibitor cycloheximide, defining it as a primary response to the hormone (Fig. 3 H). Collectively, these observations suggest that ecdysone-induced expression of CBP in larval salivary glands leads to diap1 down-regulation as part of the mid-L3 transition.
High levels of DIAP1 during early larval stages effectively block programmed cell death
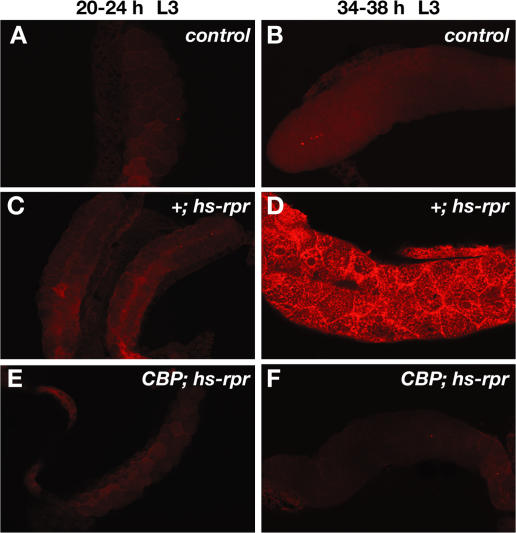
The high level of DIAP1 in early larval salivary glands raises the interesting possibility that this tissue may be resistant to cell death, even in the presence of ectopic death activator expression. Consistent with this hypothesis, control L1, L2, and early L3 salivary glands are resistant to ectopic rpr overexpression, surviving normally and showing no staining for active caspase-3 (Fig. 4, E–G; Fig. 5 C; and not depicted). In contrast, salivary glands from animals after the mid-L3 transition are susceptible to rpr-induced cell death (Fig. 4 H and Fig. 5 D). This sensitization for rpr-mediated cell death fails to occur in CBP mutant salivary glands (Fig. 5, E and F). Thus, although wild-type salivary glands do not display caspase activation immediately after the mid-L3 transition (Fig. 5 B), 77% of salivary glands are positive for this assay at the same stage in development in the presence of ectopic rpr expression (heat-treated hs-rpr animals; n = 30; Fig. 5 D). This susceptibility to rpr-mediated cell death drops to 8% in CBP mutant salivary glands (heat-treated CBPG0350; hs-rpr animals; n = 26; Fig. 5 F). These observations demonstrate that CBP expression at the mid-L3 transition is a prerequisite for destruction of this tissue by death activators and suggest that this sensitization is conferred through specific down-regulation of DIAP1.
Figure 5.
Competence for rpr-mediated cell death occurs after the mid-third instar transition. Salivary glands were dissected from heat-treated third instar larvae (L3) at either 20–24 h after the L2–L3 molt (A, C, and E) or 34–38 h after the molt (B, D, and F) and stained with antibodies directed against cleaved active caspase-3. Heat-treated control (w) L3 (A and B) and hs-rpr transformants at 20–24 h after the molt (C) display no staining for active caspase-3, whereas salivary glands at 34–38 h after the molt are susceptible to hs-rpr–mediated cell death (D). This switch in competence to respond to rpr, however, fails to occur in CBP mutant glands (CBPG0350; hs-rpr) (E and F).
Discussion
Prior studies have shown that the destruction of larval tissues is triggered by a pulse of the steroid hormone ecdysone via a transcriptional cascade that results in rpr and hid death activator expression during the early stages of Drosophila metamorphosis. In this paper, we identify a new regulator in this pathway, CBP, and show that this transcriptional cofactor acts at an earlier stage in development, contributing to an unexpected level of ecdysone-triggered cell death control, providing competence for death through down-regulation of DIAP1. In the following section, we present a model to explain the roles of CBP and DIAP1 in larval tissue destruction and discuss how this study provides a new basis for understanding the molecular mechanisms of competence and steroid-triggered programmed cell death.
A two-step model for steroid-triggered programmed cell death
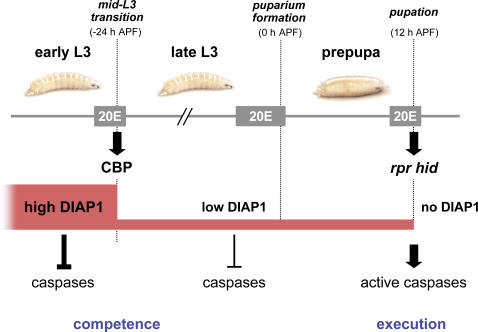
Before this work, reverse genetic studies had implicated rpr and hid induction in late prepupae as the central control point for larval tissue cell death (Baehrecke, 2003; Yin and Thummel, 2005). This study shows that diap1 is also a critical target for transcriptional control by ecdysone and that the switch in DIAP1 levels occurs in mid-L3 and is dependent on EcR and CBP function. We propose that high levels of DIAP1 effectively block larval tissue cell death before the mid-L3 transition, preventing environmental factors from disrupting the essential role of these tissues in larval feeding and growth (Fig. 6). Cells in this protected state need to acquire competence to become susceptible to death activators. Only after commitment to metamorphosis, during the mid-L3 transition, do the larval salivary glands become dispensable and acquire this competence to die through down-regulation of DIAP1. This reduced level of DIAP1 is at a critical threshold where it is sufficient to effectively hold back cell death but low enough to be sensitive to subsequent ecdysone-induced rpr and hid expression. The steroid-triggered destruction of larval salivary glands thus appears to be a two-step transcriptional response, in which both steps of this temporally regulated pathway are triggered by the hormone ecdysone. In the first step, the mid-L3 ecdysone pulse induces CBP, directing DIAP1 down-regulation in larval salivary glands, providing competence for rpr/hid-mediated cell death (Fig. 6). It is likely that CBP indirectly regulates diap1 transcription in mid-L3 salivary glands because CBP is known to function as a coactivator rather than a repressor (Giordano and Avantaggiati, 1999). We propose that CBP exerts its effects on diap1 through one or more ecdysone-inducible transcription factors that act in a mid-L3 salivary gland regulatory cascade, as part of a larger organism-wide genetic program that defines the end of larval life and prepares the animal for entry into metamorphosis. The second steroid-triggered step in salivary gland cell death occurs in early pupae, 36 h later, in response to the prepupal ecdysone pulse. At this stage, hormone-mediated transcriptional induction of rpr and hid targets the basal DIAP1 protein for degradation, allowing caspase activation and cell death (Fig. 6).
Figure 6.
A model for the temporal regulation of salivary gland cell death by sequential pulses of ecdysone. Ecdysone (20E)–induced CBP expression at the mid-L3 transition directs down-regulation of DIAP1 in larval salivary glands to a critical threshold level, establishing competence for cell death. Later, ecdysone-induced rpr and hid expression at pupation (12 h APF) eliminates the remaining DIAP1, allowing caspase activation and salivary gland destruction.
Changes in DIAP1 levels are critical for cell death
Most studies of Drosophila cell death regulation involve the constant presence of DIAP1 to block caspase activation until this effect is overcome by death activators that direct DIAP1 degradation and trigger cell death (Wang et al., 1999; Goyal et al., 2000; Lisi et al., 2000; Martin, 2002). An exception to this is the down-regulation of diap1 during stages 7 and 8 of oogenesis, a time when egg chambers undergo degeneration (Foley and Cooley, 1998). This timing correlates with the ecdysone-dependent up-regulation of two transcription factors that also act in ecdysone hierarchies at the onset of metamorphosis, E74 and E75, raising the interesting possibility that ecdysone signaling may also direct diap1 down-regulation during ovarian development (Buszczak et al., 1999). More recently, the Hippo pathway has been shown to regulate DIAP1 levels, at least in part through effects on diap1 transcription, although there is disagreement on this level of control (for review see Edgar, 2006). We have not found evidence that this pathway contributes to DIAP1 down-regulation at the mid-L3 transition (unpublished data). Moreover, the Hippo pathway is not known to regulate changes in DIAP1 levels during development but rather is required to maintain an appropriate threshold level of DIAP1 to allow cell death. In contrast, our data show that DIAP1 levels can change during development and that this change is critical for a naturally occurring cell death response.
A molecular mechanism for the acquisition of competence
We show here that activation of the death machinery, as represented by rpr, hid, dark, and dronc induction, is not sufficient to kill. Rather, salivary gland cells must first acquire the competence to die through down-regulation of DIAP1. Despite its central role in development, molecular mechanisms for the acquisition of competence have been defined in only a few general areas, all at the transcriptional level. One fundamental way to achieve competence is through expression of the appropriate receptor or cofactors that directly regulate receptor activity (Rosenfeld et al., 2006). In the absence of the receptor or its cofactor, cells are incapable of responding to the signal. Competence can also be regulated through dynamic expression of histone H1 subtypes or by phosphorylation of histone H1 (Steinbach et al., 1997; Lee and Archer, 1998). These changes allow transcription by relieving the repressive function of higher order chromatin. Stage-specific expression of the βFTZ-F1 competence factor has been shown to direct appropriate genetic and biological responses to ecdysone during the early stages of Drosophila metamorphosis and the female mosquito vitellogenic response (Woodard et al., 1994; Broadus et al., 1999; Lee et al., 2002; Zhu et al., 2003). Most recently, down-regulation of the Fork head (Fkh) transcription factor at puparium formation has been shown to be critical for the subsequent transcriptional induction of rpr and hid in doomed salivary glands (Cao et al., 2007). The work described here further extends our understanding of the molecular basis of competence, acting at an earlier stage than βFTZ-F1 and Fkh and through a different mechanism, down-regulation of a death inhibitor.
Implications for the regulation of programmed cell death
Our studies provide an explanation for earlier observations that indicate that ectopic death activator expression is not always sufficient to trigger programmed cell death. Embryonic cells, for example, show an insensitivity to expression of the death activator rpr during the later stages of embryogenesis (White et al., 1996). In addition, widespread heat-induced rpr overexpression in larvae has no effect on viability before the mid-L3 transition (unpublished data). Thus, certain cells are actively protected against ectopic cell death, even if those same cells are fated to die later during development. Protection of specific cells against death by up-regulation of survival factors may have evolved as a general strategy to preserve postmitotic, terminally differentiated cells during development, protecting them from cell death until they are no longer essential to the organism. A similar protection against cell death has been seen in vertebrate postmitotic neurons, which appear to acquire competence to death activators only after removal of target-derived growth factors (Deshmukh and Johnson, 1998). Failure to regulate this process may contribute to the etiology of human diseases. For example, escape from programmed cell death is a hallmark of cancer, and one mechanism through which tumor cells appear to acquire resistance to cell death is through overexpression of IAPs (Liston et al., 2003). IAPs were discovered in viruses where they are used to prevent the death of host cells, facilitating viral propagation. We propose that regulating survival factor levels like IAPs is a universal mechanism to protect irreplaceable cells from inappropriate activation of programmed cell death during development.
Materials and methods
Stocks and developmental staging
Further information on the CBP alleles described in this paper can be found on FlyBase using the Bloomington stock numbers: CBPG0350 (11978), CBPG0112 (11821), and CBPG0470 (12265; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200703206/DC1). The nej3 allele was provided by S. Smolik (Oregon Health & Science University, Portland, OR). Embryos were collected in 6-h intervals on molasses/agar plates supplemented with yeast paste, transferred to fresh egg caps, and allowed to age as necessary. For analysis of the mid-third instar transition, larvae were staged relative to the second-to-third instar molt. For analysis of the prepupal–pupal transition, animals were staged relative to the white prepupal stage (0 h APF). For heat shock–driven expression of transgenes, staged animals were transferred to a 1.5-ml microcentrifuge tube plugged with cotton, subjected to a 30-min heat treatment in a 37.5°C water bath, and allowed to recover at 25°C for at least 6 h before dissection and fixation.
Immunohistochemistry
Larval salivary glands were dissected from the appropriate stage and fixed with 4% formaldehyde, 1× PBST (PBS and 0.1% Tween-20), and 3 vol of heptane for 30 min at room temperature. Samples were washed four times in 1× PBST and blocked in 1× PBST and 4% normal goat serum (NGS) for 2 h at room temperature. The samples were stained with antibodies directed against either DIAP1 (a gift from B. Hay, California Institute of Technology, Pasadena, CA) at 1:500 dilution or against the cleaved/active form of caspase-3 (Cell Signaling Technologies) at 1:1,000 in 1× PBST and 4% NGS overnight at 4°C. Samples were washed four times in 1× PBST and stained with either Cy3 donkey anti–mouse secondary antibody (Jackson ImmunoResearch Laboratories) for DIAP1 or Cy3 donkey anti–rabbit secondary antibody (Jackson ImmunoResearch Laboratories) for active caspase-3, at 1:400 in 1× PBST and 4% NGS for 2 h at room temperature. Samples were mounted in Vectashield (Vector Laboratories).
Microscopy and image capture
Images in Figs. 1 (A–D) were obtained using a 2.5×/0.075 Plan-neofluar objective (Carl Zeiss MicroImaging, Inc.) on a microscope (Axiophot; Carl Zeiss MicroImaging, Inc.) and captured with a digital charge-coupled device camera (SensiCamQE; Cooke Corp.) and Slidebook 3.0 software (Intelligent Imaging Innovations, Inc.). Images in Fig. 1 (E–I) were obtained using a 10×/0.30 Plan-neofluar objective (Carl Zeiss MicroImaging, Inc.) on a microscope (Axioskop2; Carl Zeiss MicroImaging, Inc.) and captured with a charge-coupled device camera (CoolSNAP-Pro; Media Cybernetics) and Image-Pro Plus software (Media Cybernetics). The remaining images were captured as a z series with a confocal laser-scanning microscope (MRC1024; Bio-Rad Laboratories) using 10×/0.30 Plan-neofluar (Fig. 1, J–N) or 20×/0.50 Plan-neofluar (Fig. 3, A–F; Fig. 4; and Fig. 5) objectives. All images were acquired at room temperature. Each experiment was performed using identical laser intensity and acquisition parameters to allow direct comparison of control and mutant salivary glands. All figures were processed in parallel with Photoshop CS (Adobe) and assembled in Illustrator CS (Adobe).
Organ culture
Wild-type Canton-S late third-instar larvae, 18 h before puparium formation, were dissected and cultured in Schneider's Drosophila medium (Invitrogen) in the presence of 5 μM 20-hydroxyecdysone (Sigma-Aldrich), 85 μM cycloheximide (Sigma-Aldrich), or both compounds, for 2 or 6 h. 20-hydroxyecdysone is the physiologically active form of ecdysone. Total RNA was extracted from whole animals and analyzed by Northern blot analysis, as described previously (Andres et al., 1993).
Northern blot hybridizations
Larval salivary glands were dissected from animals staged at 20–24 or 34–38 h from the second-to-third instar larval molt or relative to puparium formation in 2-h intervals. Equal amounts of total RNA, isolated using Tripure (Roche), were fractionated on 1% formaldehyde gels and transferred to nylon membranes for Northern blot hybridization. Probes were prepared as described previously (Andres et al., 1993).
CBP RNAi
A pair of oligonucleotides, 5′-CTCTGTCAACGTCGGTGGC-3′ and 5′-CTGTTGCTGCTGTCCT-3′, was used to amplify a 942-bp fragment spanning the coding region for the conserved KIX domain (nt 2778–3720; available from GenBank/EMBL/DDBJ under accession no. NM_079903) from adult genomic DNA with EcoRI and XbaI sites at the ends. Amplification with a second pair of oligonucleotides, 5′-CTCTGTCAACGTCGGTGGC-3′ and 5′-GCCTGCTGCCGTCTGTTGGC-3′, generated the same fragment with KpnI and EcoRV sites at the ends. Each fragment was sequentially inserted into the appropriate restriction sites of the vector pUAST, generating a construct with an inverted repeat of the 942-bp fragments in a tail-to-tail orientation under the control of the Gal4-dependent UAS promoter. The P-element carrying the CBP inverted repeats was introduced into the w1118 germline using standard protocols. Multiple independent lines of UAS-CBP-RNAi were isolated and tested. Expression from these transgenes using a larval salivary gland–specific GAL4 (w; UAS-CBP-RNAi/ UAS-GFP; SG-GAL4) showed a low penetrance of persistent larval salivary glands (27%; n = 33), probably a result of inefficient RNAi processing of CBP (persistent larval salivary glands at 24 h APF is never seen in wild-type animals). The w1118 allele was used in all white mutant stocks. SG-GAL4 was provided by A. Andres (University of Nevada, Las Vegas, Las Vegas, NV).
Functional assay for death competence
For broad assessment of death competence, animals carrying a heat-shock inducible rpr transgene (w; hs-rpr; provided by H. Steller, The Rockefeller University, New York, NY; White et al., 1996) were staged from egg lay and subjected to a single 30-min heat treatment during first, second, early third, or late third larval instars to induce rpr expression and allowed to recover for 6 h before dissecting and staining salivary glands with antibodies to detect either DIAP1 or the cleaved active form of caspase-3. Salivary glands dissected from control larvae (w1118) did not exhibit signs of necrosis or apoptosis. For a more precise assessment of the change in death competence during the third larval instar, control and mutant animals (w or w; hs-rpr or CBPG0350; hs-rpr) were staged from the second-to-third instar larval molt in 4-h intervals, subjected to a single 30-min heat treatment, and allowed to recover for 6 h before dissecting salivary glands and staining.
Disruption of EcR function by RNAi
Transgenic expression of heat-inducible double-stranded RNA common to all EcR mRNA isoforms (hs-EcR-RNAi-11) was used to disrupt EcR function, as described previously (Lam and Thummel, 2000). Animals expressing EcR-RNAi that were allowed to continue in their development displayed a fully penetrant block in puparium formation, demonstrating efficient inactivation of EcR (Lam and Thummel, 2000; unpublished data).
Online supplemental material
Fig. S1 shows the genomic location of the P-element–induced CBP mutations described in the text. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200703206/DC1.
Supplementary Material
Acknowledgments
We thank the Bloomington Stock Center for P-element–induced lethal mutations and other stocks, S. Smolik for nej3 and hs-CBP, B. Hay for antibodies against DIAP1, H. Steller for hs-rpr, and A. Andres for SG-GAL4.
This work was supported by the National Institutes of Health (GM073670).
V.P. Yin's present address is Department of Cell Biology, Duke University Medical Center, Durham, NC 27710.
A. Bashirullah's present address is School of Pharmacy, Division of Pharmaceutical Sciences, University of Wisconsin–Madison, Madison, WI 53705.
Abbreviations used in this paper: APF, after puparium formation; CBP, CREB binding protein; DIAP1, Drosophila inhibitor of apoptosis 1; EcR, ecdysone receptor; hid, head involution defective; nej, nejire; rpr, reaper.
References
- Andres, A.J., and P. Cherbas. 1992. Tissue-specific ecdysone responses: regulation of the Drosophila genes Eip28/29 and Eip40 during larval development. Development. 116:865–876. [DOI] [PubMed] [Google Scholar]
- Andres, A.J., J.C. Fletcher, F.D. Karim, and C.S. Thummel. 1993. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev. Biol. 160:388–404. [DOI] [PubMed] [Google Scholar]
- Baehrecke, E.H. 2003. Autophagic programmed cell death in Drosophila. Cell Death Differ. 10:940–945. [DOI] [PubMed] [Google Scholar]
- Baehrecke, E.H. 2005. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 6:505–510. [DOI] [PubMed] [Google Scholar]
- Broadus, J., J.R. McCabe, B. Endrizzi, C.S. Thummel, and C.T. Woodard. 1999. The Drosophila βFTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol. Cell. 3:143–149. [DOI] [PubMed] [Google Scholar]
- Buszczak, M., M.R. Freeman, J.R. Carlson, M. Bender, L. Cooley, and W.A. Segraves. 1999. Ecdysone response genes govern egg chamber development during mid-oogenesis in Drosophila. Development. 126:4581–4589. [DOI] [PubMed] [Google Scholar]
- Cao, C., Y. Liu, and M. Lehmann. 2007. Fork head controls the timing and tissue selectivity of steroid-induced developmental cell death. J. Cell Biol. 176:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P., W. Nordstrom, B. Gish, and J.M. Abrams. 1996. grim, a novel cell death gene in Drosophila. Genes Dev. 10:1773–1782. [DOI] [PubMed] [Google Scholar]
- Cherbas, L., X. Hu, I. Zhimulev, E. Belyaeva, and P. Cherbas. 2003. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 130:271–284. [DOI] [PubMed] [Google Scholar]
- Danial, N.N., and S.J. Korsmeyer. 2004. Cell death: critical control points. Cell. 116:205–219. [DOI] [PubMed] [Google Scholar]
- Deshmukh, M., and E.M. Johnson Jr. 1998. Evidence of a novel event during neuronal death: development of competence-to-die in response to cytoplasmic cytochrome c. Neuron. 21:695–705. [DOI] [PubMed] [Google Scholar]
- Dorstyn, L., P.A. Colussi, L.M. Quinn, H. Richardson, and S. Kumar. 1999. DRONC, an ecdysone-inducible Drosophila caspase. Proc. Natl. Acad. Sci. USA. 96:4307–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, B.A. 2006. From cell structure to transcription: Hippo forges a new path. Cell. 124:267–273. [DOI] [PubMed] [Google Scholar]
- Foley, K., and L. Cooley. 1998. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 125:1075–1082. [DOI] [PubMed] [Google Scholar]
- Furia, M., P.P. D'Avino, S. Crispi, D. Artiaco, and L.C. Polito. 1993. Dense cluster of genes is located at the ecdysone-regulated 3C puff of Drosophila melanogaster. J. Mol. Biol. 231:531–538. [DOI] [PubMed] [Google Scholar]
- Giordano, A., and M.L. Avantaggiati. 1999. p300 and CBP: partners for life and death. J. Cell. Physiol. 181:218–230. [DOI] [PubMed] [Google Scholar]
- Goyal, L., K. McCall, J. Agapite, E. Hartwieg, and H. Steller. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19:589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether, M.E., J.M. Abrams, J. Agapite, K. White, and H. Steller. 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9:1694–1708. [DOI] [PubMed] [Google Scholar]
- Jiang, C., E.H. Baehrecke, and C.S. Thummel. 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development. 124:4673–4683. [DOI] [PubMed] [Google Scholar]
- Jiang, C., A.F. Lamblin, H. Steller, and C.S. Thummel. 2000. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell. 5:445–455. [DOI] [PubMed] [Google Scholar]
- Lam, G., and C.S. Thummel. 2000. Inducible expression of double-stranded RNA directs specific genetic interference in Drosophila. Curr. Biol. 10:957–963. [DOI] [PubMed] [Google Scholar]
- Lee, C.Y., and E.H. Baehrecke. 2001. Steroid regulation of autophagic programmed cell death during development. Development. 128:1443–1455. [DOI] [PubMed] [Google Scholar]
- Lee, C.Y., C.R. Simon, C.T. Woodard, and E.H. Baehrecke. 2002. Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev. Biol. 252:138–148. [DOI] [PubMed] [Google Scholar]
- Lee, H.L., and T.K. Archer. 1998. Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J. 17:1454–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisi, S., I. Mazzon, and K. White. 2000. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 154:669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston, P., W.G. Fong, and R.G. Korneluk. 2003. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 22:8568–8580. [DOI] [PubMed] [Google Scholar]
- Martin, S.J. 2002. Destabilizing influences in apoptosis: sowing the seeds of IAP destruction. Cell. 109:793–796. [DOI] [PubMed] [Google Scholar]
- Mougneau, E., D. von Seggern, T. Fowler, J. Rosenblatt, T. Jongens, B. Rogers, D. Gietzen, and S.K. Beckendorf. 1993. A transcriptional switch between the Pig-1 and Sgs-4 genes of Drosophila melanogaster. Mol. Cell. Biol. 13:184–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, A., P. Schottler, M. Werner, N. Beinert, G. Dowe, P. Burkert, F. Mourkioti, L. Dentzer, Y. He, P. Deak, et al. 2002. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3:34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, C.W. 1936. The metamorphosis of Drosophila melanogaster, including an accurately timed account of the principal morphological changes. J. Morphol. 59:351–399. [Google Scholar]
- Rosenfeld, M.G., V.V. Lunyak, and C.K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405–1428. [DOI] [PubMed] [Google Scholar]
- Shi, Y.B., L. Fu, S.C. Hsia, A. Tomita, and D. Buchholz. 2001. Thyroid hormone regulation of apoptotic tissue remodeling during anuran metamorphosis. Cell Res. 11:245–252. [DOI] [PubMed] [Google Scholar]
- Steinbach, O.C., A.P. Wolffe, and R.A. Rupp. 1997. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 389:395–399. [DOI] [PubMed] [Google Scholar]
- Waddington, C.H. 1940. Organisers & Genes. Cambridge University Press, Cambridge, UK. 160 pp.
- Wang, S.L., C.J. Hawkins, S.J. Yoo, H.A. Muller, and B.A. Hay. 1999. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 98:453–463. [DOI] [PubMed] [Google Scholar]
- Ward, R.E., P. Reid, A. Bashirullah, P.P. D'Avino, and C.S. Thummel. 2003. GFP in living animals reveals dynamic developmental responses to ecdysone during Drosophila metamorphosis. Dev. Biol. 256:389–402. [DOI] [PubMed] [Google Scholar]
- White, K. 2000. Cell death: Drosophila Apaf-1—no longer in the (d)Ark. Curr. Biol. 10:R167–R169. [DOI] [PubMed] [Google Scholar]
- White, K., M.E. Grether, J.M. Abrams, L. Young, K. Farrell, and H. Steller. 1994. Genetic control of programmed cell death in Drosophila. Science. 264:677–683. [DOI] [PubMed] [Google Scholar]
- White, K., E. Tahaoglu, and H. Steller. 1996. Cell killing by the Drosophila gene reaper. Science. 271:805–807. [DOI] [PubMed] [Google Scholar]
- Winoto, A., and D.R. Littman. 2002. Nuclear hormone receptors in T lymphocytes. Cell. 109(Suppl.):S57–S66. [DOI] [PubMed] [Google Scholar]
- Woodard, C.T., E.H. Baehrecke, and C.S. Thummel. 1994. A molecular mechanism for the stage specificity of the Drosophila prepupal genetic response to ecdysone. Cell. 79:607–615. [DOI] [PubMed] [Google Scholar]
- Yin, V.P., and C.S. Thummel. 2004. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. Proc. Natl. Acad. Sci. USA. 101:8022–8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, V.P., and C.S. Thummel. 2005. Mechanisms of steroid-triggered programmed cell death in Drosophila. Semin. Cell Dev. Biol. 16:237–243. [DOI] [PubMed] [Google Scholar]
- Yoo, S.J., J.R. Huh, I. Muro, H. Yu, L. Wang, S.L. Wang, R.M. Feldman, R.J. Clem, H.A. Muller, and B.A. Hay. 2002. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 4:416–424. [DOI] [PubMed] [Google Scholar]
- Zhu, J., L. Chen, and A.S. Raikhel. 2003. Posttranscriptional control of the competence factor βFTZ-F1 by juvenile hormone in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA. 100:13338–13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.