Abstract
Phosphoinositide 3-kinase (PI3K)γ and Dictyostelium PI3K are activated via G protein–coupled receptors through binding to the Gβγ subunit and Ras. However, the mechanistic role(s) of Gβγ and Ras in PI3K activation remains elusive. Furthermore, the dynamics and function of PI3K activation in the absence of extracellular stimuli have not been fully investigated. We report that gβ null cells display PI3K and Ras activation, as well as the reciprocal localization of PI3K and PTEN, which lead to local accumulation of PI(3,4,5)P3. Simultaneous imaging analysis reveals that in the absence of extracellular stimuli, autonomous PI3K and Ras activation occur, concurrently, at the same sites where F-actin projection emerges. The loss of PI3K binding to Ras–guanosine triphosphate abolishes this PI3K activation, whereas prevention of PI3K activity suppresses autonomous Ras activation, suggesting that PI3K and Ras form a positive feedback circuit. This circuit is associated with both random cell migration and cytokinesis and may have initially evolved to control stochastic changes in the cytoskeleton.
Introduction
Phosphoinositide 3-kinase (PI3K) is conserved across eukaryotic organisms and regulates many facets of pathways involving cellular growth, survival, metabolism, vehicle trafficking, and chemotaxis. PI3Ks are classified into Class I, II, and III based on their structures and substrate preferences. Class I PI3K is primarily responsible for the production of PI(3,4,5)P3 via phosphorylation of PI(4,5)P2 in response to extracellular stimulation (Vanhaesebroeck et al., 2005; Engelman et al., 2006; Sasaki and Firtel, 2006). All mammalian Class I PI3Ks contain a Ras binding domain (RBD) and can be activated by interacting with GTP-Ras (Pacold et al., 2000).
The Class IB PI3K, PI3Kγ, is expressed most highly in neutrophils and activated by binding to Gβγ subunits and Ras upon G protein–coupled receptor activation (Stephens et al., 1994, 1997; Stoyanov et al., 1995; Suire et al., 2005). Overexpression of Gβγ subunits in HEK293 cells leads to PI3Kγ activation via its interaction with the p101 catalytic subunit (Krugmann et al., 2002; Brock et al., 2003). Recently, p101 knockout mice and RBD-mutated PI3Kγ knock-in mice have been generated. In the neutrophils from these mutant mice, chemoattractant-induced PI3Kγ activation is significantly decreased (Suire et al., 2006). These in vitro and in vivo studies demonstrate a linkage between chemoattractant stimulation and PI3Kγ activation. Interestingly, lipid PI3K assays demonstrated that cells have a basal level of PI3K activity in the absence of chemoattractants or serum (Huang et al., 2003; Suire et al., 2005). It is unclear whether this basal activity of PI3K is actively controlled or merely a passive property of this enzyme.
Dictyostelium Class I PI3Ks are considered to be the functional counterpart of PI3Kγ on the basis of biochemical and structural characteristics (Janetopoulos et al., 2005; Sasaki and Firtel, 2006). As implicated in PI3Kγ regulation in mammalian cells, Dictyostelium cells lacking Gβ cannot activate PI3K in response to chemoattractant or GTPγS stimulation (Meili et al., 1999; Huang et al., 2003). Dictyostelium cells, however, manage to divide and undergo random motility in the absence of functional heterotrimeric G proteins (Wu et al., 1995). It is known that these complex cell shape changes involve the polymerization of F-actin, but the upstream signals regulating these pathways have yet to be determined.
Cells have the intrinsic ability to produce pseudopodia and move in the absence of chemoattractants or nutrients (Wessels et al., 1988; Condeelis and Segall, 2003). This random cell migration allows cells to explore their environment and is associated with metastasis of tumor cells. Although much progress has been made in elucidating the molecular mechanisms of chemoattractant-mediated migration (chemotaxis), random cell migration has barely been investigated. In this study, we genetically separate the GPCR heterotrimeric G protein–dependent cellular events that regulate chemotaxis from the basal regulatory signaling loops that control random cell movement and cytokinesis. We demonstrate that both PI3K and Ras activation occur actively, at the same sites of new pseudopod formation in the absence of extracellular stimuli and without heterotrimeric G protein input. PI3K and Ras activation also occur at the poles of dividing cells in wild-type strains and in cells lacking functional heterotrimeric G proteins, implying that cell shape changes during cytokinesis are controlled by a similar mechanism. We suggest that this “unprompted” Ras activity and PI(3,4,5)P3 accumulation, which are independent of external stimuli, constitute a core regulatory pathway involved in a variety of physiological responses and provide the basis for many ligand- or substrate-mediated processes, such as chemotaxis and phagocytosis.
Results and discussion
Gβ-independent PI(3,4,5)P3 accumulation without extracellular stimuli
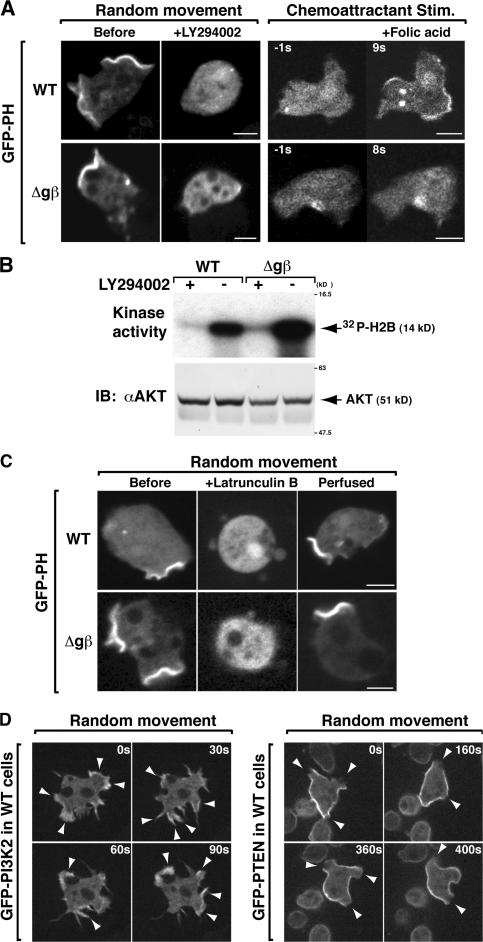
In this study, we used vegetative cells rather than starved, developmentally competent cells, which produce and respond to cAMP and are normally used to investigate chemoattractant-mediated cell movement in Dictyostelium, as Gβγ signaling is required for cells to become developmentally competent. We expressed the GFP-tagged PH domain of CRAC (PHcrac), a reporter for PI(3,4)P2 and PI(3,4,5)P3, in the KAx-3 wild-type strain and a null strain of Gβ, the sole Gβ subunit in Dictyostelium (Wu et al., 1995; Comer et al., 2005). Consistent with previous studies (Parent 1998; Meili et al., 1999), gβ null (gβ−) cells show no detectable folic acid–mediated translocation of PHcrac to the cell cortex (Fig. 1 A). However, we noticed that, in the buffer containing only Na/KPO4, the PH domains spontaneously accumulated at both pseudopodial extensions and to macropinosomes (the structures responsible for fluid-phase uptake) of randomly moving KAx-3 cells (Lee and Knecht, 2002). The PHcrac accumulation in randomly moving cells is a consequence of PI3K activation because its membrane localization was abolished ∼1 min after the addition of LY294002, a PI3K inhibitor (Fig. 1 A). Importantly, gβ− cells also exhibited spontaneous PHcrac accumulation, which was sensitive to the PI3K inhibitor. This finding indicates that PI3K is spontaneously activated without heterotrimeric G protein signaling.
Figure 1.
G protein–independent PI3K activation in the absence of extracellular stimuli. (A) The vegetative state cells were plated in Na/KPO4 buffer. Fluorescent images of GFP-PHcrac in wild-type and gβ− cells before or after the addition of 50 μM LY294002 (left). The cells were exposed to a uniform concentration of chemoattractant (50 μM folic acid; right). The folic-induced PH translocation is weak compared with the spontaneous PH accumulation at the plasma membrane. (B) Spontaneous activation of Akt/PKB is shown. (C) Fluorescent images of GFP-PHcrac in wild-type and gβ− cells before or after the addition of 5 μM LatB and after the removal of LatB. The LatB-treated cells increased GFP intensity due to the loss of membrane PH, and the cell became round and shrank. (D) Fluorescent images of GFP-tagged PI3K (left) and PTEN (right) in wild-type cells. The arrows indicate PI3K accumulation sites (left) and PTEN dissociation sites (right). Bars, 5 μm.
To validate whether gβ− cells activate PI3K signaling, we measured the kinase activity of Akt, a downstream effector of PI3K. Fig. 1 B illustrates that gβ− cells have a robust Akt activity in the absence of a chemoattractant or nutrients. The treatment of cells with LY294002 completely suppresses this spontaneous Akt activity in both strains to background levels (akt − cells; unpublished data). Notably, basal Akt activity in gβ− cells is higher than that in the wild-type cells, suggesting a possibility that Gβ attenuates on the basal PI3K/Akt signaling in the absence of stimuli. Although the importance of the Gβ subunit in receptor-mediated PI3K activation has been demonstrated (Krugmann et al., 2002; Brock et al., 2003), these data reveal that PI3K and its downstream signaling are activated without a chemoattractant or nutrients and Gβ, whereas Gβ is essential for ligand-induced PI3K activation.
As spontaneous PI(3,4,5)P3 accumulation appears to occur strictly at sites of F-actin protrusion, including both the sites of macropinosomes and pseudopodial extensions, we examined whether spontaneous PI3K activation requires F-actin synthesis. (In many instances, we could not resolve whether protrusions began as macropinosomes or pseudopodia, but consider them pseudopodia if they ultimately protruded from the cell and gave rise to a net movement.) After treatment with Latrunculin B (LatB), an inhibitor of F-actin polymerization, spontaneous PI(3,4,5)P3 accumulation (as visualized through PHcrac recruitment) was lost and cells rounded up. After washout of LatB, cells regained the spontaneous PI(3,4,5)P3 accumulation at the sites of new pseudopodial projections (Fig. 1 C). This finding suggests that Gβ-independent, spontaneous PI3K activation requires and occurs at sites of F-actin polymerization. Furthermore, we found that GFP-PI3K2 labeled the sites of new F-actin projections in both wild-type and gβ− cells (Fig. 1 D and Video 1, which is available at http://www.jcb.org/cgi/content/full/jcb.200611138/DC1), whereas PTEN-GFP detached from the membrane in pseudopodial extensions in both wild-type cells and gβ− cells (unpublished data). The gβ− cells did not exhibit PI3K translocalization in response to a chemoattractant, whereas wild-type cells did (unpublished data). Thus, although PI3K and PTEN localization is regulated by extracellular stimuli in the chemotaxing cells, the reciprocal localization of PI3K and PTEN occurs during random cell movement, even in the absence of chemoattractant, nutrients, and heterotrimeric G proteins.
Gβ-independent, PI3K-dependent Ras activation in the absence of extracellular stimuli
As Ras is required for PI3K activation in response to extracellular stimuli in neutrophils and Dictyostelium, we monitored the localization of activated Ras during random movement using a GFP fusion of the human Raf1 RBD (GFP-RBD; Sasaki et al., 2004). As we demonstrated for PI(3,4,5)P3 accumulation, spontaneous localization of GFP-RBD occurs at the sites of F-actin projections in wild-type, gβ−, and pten − cells. Surprisingly, the Ras activation was abrogated with LY294002 treatment and recovered after removal of the drug (Fig. 2 A and Video 2, which is available at http://www.jcb.org/cgi/content/full/jcb.200611138/DC1). Biochemical assays confirmed a basal Ras activity in both wild-type and gβ− vegetative cells in the absence of exogenous stimulation or nutrients (Fig. 2 B). The basal Ras and PI3K activation do not require cellular attachment to the substratum, as cells in suspension display the LY294002- sensitive spontaneous Ras and Akt activations (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200611138/DC1). The basal Ras activation is elevated in pten − cells (Fig. 2 C), supporting our notion of the requirement of PI3K. Furthermore, like PI3K activation, this unprompted Ras activation requires F-actin (Fig. 2 D and Video 3), although it is possible that a very low level of Ras and PI3K activity exists in these drug-treated cells. These drug-treated wild-type cells can activate Ras in response to chemoattractant stimulation (Fig. 2 E; Sasaki et al., 2004).
Figure 2.
G protein–independent, PI3K-dependent Ras activation without extracellular stimuli. (A) Fluorescent images of GFP-RBD in wild-type and gβ− cells before or after the addition of 50 μM LY294002 and after the removal of LY294002. (B and C) Spontaneous activation of Ras in indicated cells is shown. Cells were treated or not treated with 50 μM LY294002 (LY) or DMSO for 5 min. (D) Fluorescent images of GFP-RBD in wild-type and gβ− cells before or after the addition of 5 μM LatB and after the removal of LatB. (E) Translocation of GFP-RBD in a wild-type cell treated with 50 μM LY294002 for 10 min or 5 μM LatB for 20 min before folic acid stimulation. (F) Ras activation level in response to folic acid. (G) Translocation of GFP-RBD in wild-type and gβ null cells in response to folic acid. Bars, 5 μm.
Next, we examined whether Gβ was required for chemoattractant-induced Ras activation. Fig. 2 F illustrates that chemoattractant-induced Ras activation occurs in wild-type cells but not in gβ− cells. There was no detectable translocation of GFP-RBD in response to chemoattractant stimulation in cells lacking the Gβ subunit (Fig. 2 G). These results uncover two different pathways that lead to detectable levels of activated Ras: a receptor-mediated, Gβ-dependent pathway and a Gβ-independent, PI3K-dependent pathway.
It is worth noting that we previously observed spontaneous PI3K and Ras activation in developed PTEN-deficient Dictyostelium cells (Sasaki et al., 2004). The findings here differ because the developed pten − cells likely increase the level of the chemoattractant (cAMP) secretion (Iijima and Devreotes, 2002), which would evoke a GPCR/heterotrimeric G protein–mediated autocrine Ras activation (Comer et al., 2005).
Spontaneous activation of Ras and PI3K occurs simultaneously at the same sites
To compare the sites and timing of Ras activation to those of PI3K in randomly moving cells, we simultaneously imaged both GFP-RBD and RFP-PH by using a dual-wavelength beam splitter with high time resolution (∼200 msec). The RBD and PH domain appeared concurrently (within the limits of our resolution) at the same sites where pseudopodia start to form (Fig. 3 A, arrow, and Video 4, which is available at http://www.jcb.org/cgi/content/full/jcb.200611138/DC1). The kinetics of the disappearance of the RBD and PHcrac probes were also closely correlated (Fig. 3 B). Interestingly, we observed this synchronized localization of RBD and PHcrac in the pten − cells. RBD and PHcrac also appear at micropinosome cups (clathrin-dependent small invaginating pits) with the same timing; however, PHcrac is retained considerably longer at these sites than RBD, suggesting differential regulation of Ras at later stages of micropinocytosis (Fig. S1 B). These data demonstrate that the timing of PI3K and Ras activation occurs in parallel and at the same sites during random movement.
Figure 3.
Tight correlation of the sites and timing of autonomous Ras and PI3K activation. (A and B) Fluorescent images of GFP-RBD and RFP-PHcrac in a wild-type cell. Simultaneous imaging of spontaneous RBD and PH domain accumulation (A) and disappearance (B) are shown. (C) Translocation of GFP-RBD and PHcrac in developed wild-type cells by cAMP is depicted. (D) Translocation of GFP-RBD and PHcrac in vegetative (left) and developed pten null cells by folic acid and cAMP is depicted.
In experiments in which cells were stimulated with a chemoattractant, the RBD translocated ∼0.6 s faster than PHcrac to the plasma membrane (Fig. 3 C, cAMP, and Fig. S2 C, folic acid, which is available at http://www.jcb.org/cgi/content/full/jcb.200611138/DC1). Thus, Ras activation is not synchronized with PI3K activity in the initial phases (∼2 s) of chemoattractant signaling. Furthermore, in pten − cells stimulated with either folic acid or cAMP, PHcrac was retained at the plasma membrane for >1 min, whereas RBD returned to the cytosol in ∼20 s (Fig. 3 D). These results suggest that a chemoattractant/Gβ-dependent pathway induces the signaling response that overrides or disrupts the intrinsic feedback activation of Ras/PI3K. In response to a chemoattractant gradient, cells activate a localized response at the site on the plasma membrane closest to the chemoattractant source while inhibiting the spontaneous activation of Ras/PI3K at the lateral sides of cells, thereby repressing random cell movement.
PI3K and Ras form a G protein–independent circuit and regulate random cell movement
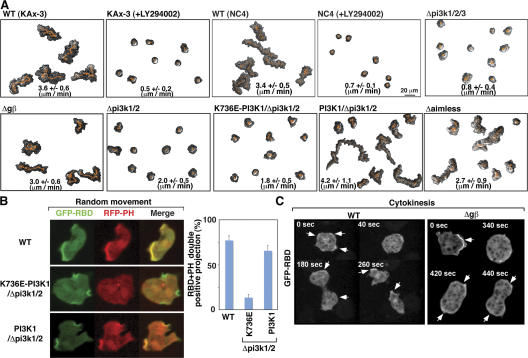
We further investigated whether Gβ-independent PI3K activation is involved in random motility. Fig. 4 A illustrates that random movement of two strains of wild-type cells (KAx-3 and NC-4) is rapidly blocked by 50 μM LY294002 (Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200611138/DC1; similar results are observed with 30 μM LY294002, unpublished data). As NC-4 cells do not have macropinosomes, the cellular movement is driven by LY294002-sensitive pseudopodial extensions. This LY294002-sensitive random cell movement does not require heterotrimeric G proteins because the gβ− cells exhibit random cell movement (Fig. 4 A). To clarify whether the random movement inhibition by LY294002 is due to the inhibition of PI3K signaling, we used pi3k1/2/3 − cells (a strain lacking the three major PI3Ks responsible for PIP3 production in Dictyostelium [Takeda et al., 2007]). Strikingly, pi3k1/2/3 − cells exhibit defects in random cell motility (Fig. 4 A and Video 6). LY294002-treated cells and pi3k1/2/3 − cells still produce small pseudopodial projections, suggesting that a low level of PI3K-independent F-actin synthesis pathway is active. On the other hand, wild-type cells expressing membrane-bound PI3K2 (Myr-PI3K2; Funamoto et al., 2002), thus bypassing F-actin–induced PI3K translocalization, periodically exhibit sudden robust protrusions that are associated with rapid cell movement (Video 7). This finding suggests that F-actin–mediated PI3K translocation plays an important role in amplifying F-actin projection during random movement. Overall, these data demonstrate that PI3K regulates random cell movement and the extent of F-actin projections in the absence of extracellular stimuli.
Figure 4.
Spontaneous PI3K activation requires Ras binding and regulates random cell movement. (A) Time-lapse recording of random movement of the indicated strains or wild-type cells treated with LY294002. The vegetative state cells and the nonaxenic NC4 cells grown on bacteria were placed in Na/KPO4 buffer. (B) Fluorescent images of GFP-RBD and RFP-PHcrac in the indicated strains. We note that the spontaneous RBD accumulation in the pi3k mutant cells was completely blocked by LY294002. (C) Time sequence of GFP-RBD in wild-type and gβ− cells during cytokinesis.
In randomly moving wild-type cells, RFP-PHcrac and GFP-RBD colocalize on F-actin projection sites (Fig. 4 B and Video 8). The pi3k1/2 − strain is a PI3K hypomorphic cell line and displays reduced random cell movement and abolished PH domain localization at the plasma membrane. The overexpression of wild-type PI3K1, but not PI3K1K736E, which harbors a mutation in its RBD that abrogates binding to Ras-GTP (Funamoto et al., 2002), in pi3k1/2 − cells complements this random cell movement defect and spontaneous PI(3,4,5)P3 accumulation at sites where Ras is activated (Fig. 4, A and B). Furthermore, an isogenic strain lacking Aimless, one of the RasGEFs responsible for chemoattractant-induced Ras activation (Insall et al., 1996; Sasaki et al., 2004), displays reduced random movement (Fig. 4 A). Collectively, these results show that Gβ-independent activation of PI3K requires interaction with Ras-GTP, and that Ras activation is required for basal cell motility. This suggests that cells use a Gβ-independent Ras/PI3K feedback amplification pathway to form pseudopodia.
As Dd-target of rapamycin (Dd-TOR) may be another LY294002-sensitive kinase, we examined the role of TOR in random cell movement. We found that inhibition of TOR complex (TORC)1 by 1 μM rapamycin and disruption of TORC2 using pianissimo − cells, which lack the ortholog of mammalian Rictor/mAVO3 (Lee et al., 2005), did not result in random movement defects (unpublished data and Video 9, which is available at http://www.jcb.org/cgi/content/full/jcb.200611138/DC1). Akt is under the regulation of TORC2. We find that akt −cells, which have a defect in macropinocytosis, display normal RFP-PHcrac and GFP-RBD colocalization on F-actin projections linked to cellular movement, consistent with TORC2 not playing a major role in random cell movement (Rupper et al., 2001; Video 10).
The Ras/PI3K circuit regulates cell morphology during cytokinesis
We previously demonstrated that the regulation of PI(3,4,5)P3 plays a central role in cell shape changes during cytokinesis (Janetopoulos et al., 2005). We show here that Ras activation as examined by RBD cortical localization, like PI3K activation, was uniformly suppressed at the onset of cytokinesis as cells rounded up. As cells progressed through mitosis, the Ras activity reporter gradually localized to the polar ruffles during spindle assembly, cell elongation, and cytokinesis (Fig. 4 C). When the daughter cells separate from one another, cortically localized RBD and PI(3,4,5)P3 activity increase dramatically, resulting in high levels of random pseudopod extension. The gβ− cells exhibit activation of Ras indistinguishable from that of wild-type cells during cytokinesis, suggesting that the Gβ-independent Ras/PI3K circuit plays a fundamental role in cytokinesis.
Conclusions
Our studies reveal a critical role of a Gβ-independent Ras/PI3K/PTEN/F-actin feedback loop in regulating random cell movement, a basic cellular function, and showed a linkage to cytokinesis. We dissect two distinct pathways that regulate Ras and PI3K activation and PTEN localization: (1) cells use Gβγ-dependent signaling to evoke Ras/PI3K activation and PTEN relocalization in response to chemoattractant stimulation; and (2) cells exploit Gβ-independent machinery to induce stochastic Ras and PI3K activation and PTEN dissociation, for example, during random cell movement and cytokinesis. We assume that the regulators for Ras/PI3K/PTEN and F-actin polymerization/disassembly can influence the initiation and decay of the circuit (Fig. S2). As the process is stochastic, we hypothesize that an increase in the level of any of the responses over a threshold level may be sufficient to trigger the feedback loops and pseudopod formation, whereas components such as GAPs and phosphatases regulate the threshold and level/time of activation.
The pathways controlling random movement parallel those of the amplification step of chemotaxis that is controlled through a regulatory loop containing Ras, PI3K, PTEN, and F-actin (Sasaki and Firtel, 2006). We suggest that, in chemotaxis, the directed activation of the pathway by the chemoattractant/G proteins biases the localized activation of the intrinsic Ras/PI3K circuit and locally restricts the positive feedback loop that is the basis for random cell movement (Fig. S2). The output from the sensing mechanism activates Ras and generates PI3K binding sites and simultaneously results in the loss of PTEN binding sites. This reciprocal regulation, along with the activation of Ras, leads to the local production of PI(3,4,5)P3 and pseudopod extension.
RasG may be one of Ras isoforms that plays a role in the Ras/PI3K feedback circuit because the GFP-RBD recognizes RasG-GTP and random movement of rasG − cells is decreased (Tuxworth et al., 1997; Sasaki et al., 2004). There are likely other Ras isoforms that are integrated into the PI3K feedback loop, as rasG null cells show modest defects. The challenge for future investigation will be to elucidate the molecular mechanisms by which Ras, RasGEF, and RasGAP regulate basic cell motility in a Gβ-independent fashion in the absence of extracellular stimuli.
Materials and methods
Materials
We obtained folic acid, LatB, and LY294002 from Sigma-Aldrich, and monoclonal anti-Ras (Ab-3) antibody from Oncogene Research Products. We used 50 μM LY294002 for the experiments because this concentration has become the standard and at this concentration Akt/protein kinase B (PKB) activity and GFP-PH translocation are blocked, but PKBR1 (which is not PI3K dependent but is a TORC2-dependent, Akt/PKB-related kinase in Dictyostelium) is not blocked. We observed that 30 μM LY294002 similarly suppressed random movement of wild-type cells. GST-RBD, GFP-RBD, GFP-PHcrac, N-PI3K-GFP, and PTEN-GFP were described previously (Funamoto et al., 2002; Iijima et al., 2002; Sasaki et al., 2004). RFP-PHcrac was cloned into a hygromycin-resistant CV5 vector (pYu34).
We created an aimless null strain in the KAx-3 background using a targeting vector similar to that used to generate aimless null in a KAx2 background (Insall et al., 1996). The gβ, pten, and pi3k1/2 null strains were described previously (Wu et al., 1995; Funamoto et al., 2001; Iijima and Devreotes, 2002).
Cell culture
All cell lines except for NC-4 were cultured axenically in HL5 medium at 22°C. NC-4 cells were grown on bacterial lawns. Transformants were maintained in 40 μg/ml G418, 50 μg/ml hygromycin, or both as required.
Biochemical assays
PKB activation and Ras activation were measured as described previously (Meili et al., 1999; Sasaki et al., 2004). Cells were treated or not treated with 50 μM LY294002 for 5 min. Endogenous Akt was immunoprecipitated and subjected to an in vitro kinase assay using H2B as a substrate. To monitor the Akt and Ras activation in vegetative cells, cells were harvested at 2–4 × 106 cells/ml, washed twice with 12 mM sodium/potassium phosphate buffer, pH 6.1, buffer A, and then resuspended in buffer A at 107 cells/ml. After 30 min of starvation, cells were subjected to the assay. For comparing Ras activation of wild-type cells to that of the mutant cells, washed cells were placed on the plate for 1 h and stimulated with 50 μM folic acid for the indicated duration, and then lysed on the plate.
Assays for random movement and cytokinesis
In a random movement assay, vegetative cells growing on plates were harvested and seeded onto a chambered coverglass in starvation buffer. Cells were rinsed three times with an excess amount of buffer A at 10 min after seeding, and then sat for 1 h. Images were collected on a microscope (model TE300; Nikon) with DIC and 40x/0.60 objectives. Initial images were captured using Metamorph software and analyzed with the DIAS program (Wessels et al., 1988). Speed refers to the speed of the cell's centroid movement along the total path. The cell movement during the 1 min between measured frames was measured to calculate speed so that genuine movement was measured rather than cytoplasmic rearrangement. Parallel experiments were performed with cells in HL5 axenic growth medium or cells starved for 2 h. No differences in the results were observed under these three conditions.
Cytokinesis was measured as described previously (Janetopoulos et al., 2005). Confocal images were obtained by using a CSU10 scanner unit (Yokogawa) on a Leica inverted DMIRE2 microscope with a 63x/1.4 objective using an ORCA-ER camera (Hamamatsu) or a Dual-View OI-11-EM–equipped EM-CCD camera (Hamamatsu) for simultaneous imaging. Imaging was described previously (Sasaki et al., 2004; Janetopoulos et al., 2005).
Online supplemental material
Fig. S1 A shows spontaneous Ras and PI3K activation in low density suspended cells, and Fig. S1 B shows the differences of RBD and PH accumulation kinetics in micropinosome formation from those in pseudopodial formation. Fig. S1 C shows translocation of GFP-RBD and PHcrac in vegetative wild-type cells by folic acid. Fig. S2 illustrates a model for the Ras/PI3K circuit during random movement and chemotaxis. Video 1 shows GFP-N-PI3K1 localization in gβ null cells without extracellular stimuli. In Video 2, GFP-RBD localization in a gβ null cell corresponds to Fig. 2 A in the text. In Video 3, GFP-RBD localization in a gβ null cell corresponds to Fig. 2 D in the text. In Video 4, simultaneous imaging of GFP-RBD and RFP-PHcrac in the wild-type vegetative cells corresponds to Fig. 3 A in the text. In Video 5, random movement analysis of nonaxenic NC4 cells corresponds to Fig. 4 A in the text. In Video 6, random movement analysis of cells corresponds to Fig. 4 A in the text. Video 7 shows random movement analysis of myristoylated PI3K2-overexpressing wild-type cells. In Video 8, GFP-RBD and RFP-PHcrac in the wild-type cells correspond to Fig. 4 B in the text. Video 9 shows random movement analysis of pianissimo null cells. Video 10 shows GFP-RBD and RFP-PHcrac in the akt null cells. The online supplemental material can be found at http://www.jcb.org/cgi/content/full/jcb.200611138/DC1.
Supplementary Material
Acknowledgments
We gratefully acknowledge the members of Firtel laboratory for their stimulating discussions and helpful suggestions; Yung Duong for excellent technical assistance; and Jennifer Roth, Michelle Mendoza, and Andrew Wilkins for critical reading of this manuscript.
A.T. Sasaki was supported, in part, by a Japanese Society for the Promotion of Science Research Fellowship for Research Abroad. This work was funded by research grants from the USPHS to P.N. Devreotes (GM28007, GM34933, and GM071920) and R.A. Firtel (GM037830, GM06847, and GM24279).
Abbreviations used in this paper: LatB, Latrunculin B; PHcrac, PH domain of CRAC; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; RBD, Ras binding domain; TOR, target of rapamycin; TORC, TOR complex.
References
- Brock, C., M. Schaefer, H. Reusch, C. Czupalla, M. Michalke, K. Spicher, G. Schultz, and B. Nurnberg. 2003. Roles of Gβγ in membrane recruitment and activation of p110γ/p101 phosphoinositide 3-kinase γ. J. Cell Biol. 160:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer, F.I., C.K. Lippincott, J.J. Masbad, and C.A. Parent. 2005. The PI3K-mediated activation of CRAC independently regulates adenylyl cyclase activation and chemotaxis. Curr. Biol. 15:134–139. [DOI] [PubMed] [Google Scholar]
- Condeelis, J., and J.E. Segall. 2003. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer. 3:921–930. [DOI] [PubMed] [Google Scholar]
- Engelman, J.A., J. Luo, and L.C. Cantley. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7:606–619. [DOI] [PubMed] [Google Scholar]
- Funamoto, S., R. Meili, S. Lee, L. Parry, and R. Firtel. 2002. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 109:611–623. [DOI] [PubMed] [Google Scholar]
- Funamoto, S., K. Milan, R. Meili, and R. Firtel. 2001. Role of phosphatidylinositol 3′ kinase and a downstream pleckstrin homology domain–containing protein in controlling chemotaxis in Dictyostelium. J. Cell Biol. 153:795–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., M. Iijima, C. Parent, S. Funamoto, R. Firtel, and P. Devreotes. 2003. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P3 to the leading edge of chemotaxing cells. Mol. Biol. Cell. 14:1913–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima, M., and P. Devreotes. 2002. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 109:599–610. [DOI] [PubMed] [Google Scholar]
- Insall, R.H., J. Borleis, and P.N. Devreotes. 1996. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. Curr. Biol. 6:719–729. [DOI] [PubMed] [Google Scholar]
- Janetopoulos, C., J. Borleis, F. Vazquez, M. Iijima, and P. Devreotes. 2005. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell. 8:467–477. [DOI] [PubMed] [Google Scholar]
- Krugmann, S., M.A. Cooper, D.H. Williams, P.T. Hawkins, and L.R. Stephens. 2002. Mechanism of the regulation of type IB phosphoinositide 3OH-kinase by G-protein betagamma subunits. Biochem. J. 362:725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E., and D.A. Knecht. 2002. Visualization of actin dynamics during macropinocytosis and exocytosis. Traffic. 3:186–192. [DOI] [PubMed] [Google Scholar]
- Lee, S., F.I. Comer, A. Sasaki, I.X. McLeod, Y. Duong, K. Okumura, J.R. Yates, C.A. Parent, and R.A. Firtel. 2005. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol. Biol. Cell. 16:4572–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili, R., C. Ellsworth, S. Lee, T. Reddy, H. Ma, and R. Firtel. 1999. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 18:2092–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold, M., S. Suire, O. Perisic, S. Lara-Gonzalez, C. Davis, E. Walker, P. Hawkins, L. Stephens, J. Eccleston, and R. Williams. 2000. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 103:931–943. [DOI] [PubMed] [Google Scholar]
- Parent, C.A., B.J. Blacklock, W.M. Froehlich, D.B. Murphy, and P.N. Devreotes. 1998. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 95:81–91. [DOI] [PubMed] [Google Scholar]
- Rupper, A.C., J.M. Rodriguez-Paris, B.D. Grove, and J.A. Cardelli. 2001. p110-related PI 3-kinases regulate phagosome-phagosome fusion and phagosomal pH through a PKB/Akt dependent pathway in Dictyostelium. J. Cell Sci. 114:1283–1295. [DOI] [PubMed] [Google Scholar]
- Sasaki, A.T., C. Chun, K. Takeda, and R.A. Firtel. 2004. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J. Cell Biol. 167:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A.T., and R.A. Firtel. 2006. Regulation of chemotaxis by the orchestrated activation of Ras, PI3K, and TOR. Eur. J. Cell Biol. 85:873–895. [DOI] [PubMed] [Google Scholar]
- Stephens, L., A. Smrcka, F.T. Cooke, T.R. Jackson, P.C. Sternweis, and P.T. Hawkins. 1994. A novel phosphoinositide 3 kinase activity in myeloid-derived cells is activated by G protein beta gamma subunits. Cell. 77:83–93. [DOI] [PubMed] [Google Scholar]
- Stephens, L.R., A. Eguinoa, H. Erdjument-Bromage, M. Lui, F. Cooke, J. Coadwell, A.S. Smrcka, M. Thelen, K. Cadwallader, P. Tempst, and P.T. Hawkins. 1997. The G beta gamma sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. Cell. 89:105–114. [DOI] [PubMed] [Google Scholar]
- Stoyanov, B., S. Volinia, T. Hanck, I. Rubio, M. Loubtchenkov, D. Malek, S. Stoyanova, B. Vanhaesebroeck, R. Dhand, B. Nurnberg, et al. 1995. Cloning and characterization of a G protein-activated human phosphoinositide-3 kinase. Science. 269:690–693. [DOI] [PubMed] [Google Scholar]
- Suire, S., J. Coadwell, G.J. Ferguson, K. Davidson, P. Hawkins, and L. Stephens. 2005. p84, a new Gbetagamma-activated regulatory subunit of the type IB phosphoinositide 3-kinase p110gamma. Curr. Biol. 15:566–570. [DOI] [PubMed] [Google Scholar]
- Suire, S., A.M. Condliffe, G.J. Ferguson, C.D. Ellson, H. Guillou, K. Davidson, H. Welch, J. Coadwell, M. Turner, E.R. Chilvers, et al. 2006. Gbetagammas and the Ras binding domain of p110gamma are both important regulators of PI3Kgamma signalling in neutrophils. Nat. Cell Biol. 8:1303–1309. [DOI] [PubMed] [Google Scholar]
- Takeda, K., A. Sasaki, H. Ha, H. Seung, and R.A. Firtel. 2007. Role of PI3 kinases in chemotaxis in Dictyostelium. J. Biol. Chem. 282:11874–11884. [DOI] [PubMed] [Google Scholar]
- Tuxworth, R.I., J.L. Cheetham, L.M. Machesky, G.B. Spiegelmann, G. Weeks, and R.H. Insall. 1997. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J. Cell Biol. 138:605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck, B., K. Ali, A. Bilancio, B. Geering, and L.C. Foukas. 2005. Signalling by PI3K isoforms: insights from gene-targeted mice. Trends Biochem. Sci. 30:194–204. [DOI] [PubMed] [Google Scholar]
- Wessels, D., D.R. Soll, D. Knecht, W.F. Loomis, A. De Lozanne, and J. Spudich. 1988. Cell motility and chemotaxis in Dictyostelium amebae lacking myosin heavy chain. Dev. Biol. 128:164–177. [DOI] [PubMed] [Google Scholar]
- Wu, L., R. Valkema, P.J. Van Haastert, and P.N. Devreotes. 1995. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J. Cell Biol. 129:1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.