Abstract
As a latent transcription factor, nuclear factor κB (NF-κB) translocates from the cytoplasm into the nucleus upon stimulation and mediates the expression of genes that are important in immunity, inflammation, and development. However, little is known about how it is regulated inside the nucleus. By a two-hybrid approach, we identify a prefoldin-like protein, ubiquitously expressed transcript (UXT), that is expressed predominantly and interacts specifically with NF-κB inside the nucleus. RNA interference knockdown of UXT leads to impaired NF-κB activity and dramatically attenuates the expression of NF-κB–dependent genes. This interference also sensitizes cells to apoptosis by tumor necrosis factor-α. Furthermore, UXT forms a dynamic complex with NF-κB and is recruited to the NF-κB enhanceosome upon stimulation. Interestingly, the UXT protein level correlates with constitutive NF-κB activity in human prostate cancer cell lines. The presence of NF-κB within the nucleus of stimulated or constitutively active cells is considerably diminished with decreased endogenous UXT levels. Our results reveal that UXT is an integral component of the NF-κB enhanceosome and is essential for its nuclear function, which uncovers a new mechanism of NF-κB regulation.
Introduction
Nuclear factor κB (NF-κB) is a widely expressed primary transcription factor composed of a heterodimeric complex (p65–p50). A myriad of unrelated exogenous and endogenous stimuli are capable of inducing NF-κB activity. In turn, NF-κB regulates the expression of an equally diverse array of cellular genes that are important in immunity, inflammation, and development (Ghosh et al., 1998; Li and Verma, 2002). Aberrance of its function has been linked to such pathological processes as cancer and abnormal development (for review see Rayet and Gelinas, 1999). Determining molecular mechanisms that regulate the activation of NF-κB is crucial to understand how multiple intracellular signaling pathways converge to activate a single transcription factor.
NF-κB is normally localized in the cytoplasm as an inactive complex through physically associating with its inhibitory molecule IκBα. Extensive studies have been performed to address how various stimuli trigger its translocation from the cytoplasm into the nucleus (Hayden and Ghosh, 2004). Seminal works from several laboratories have determined a sequence of biochemical events that result in the ubiquitin-dependent degradation of IκB proteins (Chen et al., 1996; Hatakeyama et al., 1999; Spencer et al., 1999). Consequently, this releases NF-κB to move into the nucleus and switch on the expression of target genes (Ghosh and Karin, 2002; Li and Verma, 2002). NF-κB belongs to the Rel homology domain (RHD) family of transcription factors that exploit similar strategies to achieve initial activation. Recently, an alternative pathway was identified to regulate another member (p100) of this family (Senftleben et al., 2001).
Compared with the fruitful know-how about the molecular events of NF-κB activation in cytoplasm, much less is understood concerning its active regulation and functional interaction with other proteins inside the nucleus. Recent progress has shed light on the importance of nuclear events in shaping the strength and duration of the NF-κB transcriptional response, which is achieved partly by posttranslational modification of the NF-κB transcription factor complex or the histones that surround various NF-κB target genes (Chen and Greene, 2004). For example, I κ-B kinase α (IKKα) was demonstrated to accelerate both the turnover of NF-κB and its removal from proinflammatory gene promoters (Lawrence et al., 2005). This kinase could also phosphorylate histone H3 and was critical for NF-κB–responsive gene expression (Yamamoto et al., 2003). Additionally, the acetyltransferase activity of p300/CREB-binding protein (CBP) was required for the activation of NF-κB–dependent transcriptions. p300/CBP proteins were also found to directly associate with NF-κB, forming a bridge to the basal transcriptional machinery (Gerritsen et al., 1997; Chan and La Thangue, 2001). Although a couple of cofactors were recently shown to reside in the NF-κB enhanceosome, much remains to be done to understand their specific functions and regulatory mechanisms. This indicates that NF-κB assembles a much higher order transcription complex than once expected and that there are additional important layers of regulation for the NF-κB transactivation process.
Ubiquitously expressed transcript (UXT) is ∼18 kD and predominantly localizes in the nucleus (Markus et al., 2002). It was demonstrated to be widely expressed in human and mouse tissues, and its expression was markedly elevated in some human tumors (Schroer et al., 1999; Zhao et al., 2005). Computer modeling predicted that UXT was an α-class prefoldin (PFD) family protein (Gstaiger et al., 2003). Most members of this family are small molecular mass proteins (14–23 kD) and are composed of coiled-coil structures. Yeast and human PFDs 1–6 were previously found to assemble into a hexameric complex that functioned as a new type of molecular chaperone (Vainberg et al., 1998; Siegert et al., 2000).
Until now, the functional characterization of UXT was scarce. One recent study indicated that UXT bound to the N terminus of the androgen receptor and regulated androgen receptor–responsive genes that are important in prostate growth suppression and differentiation (Markus et al., 2002; Taneja et al., 2004). Another investigation suggested UXT to be a component of the centrosome (Zhao et al., 2005). However, much remains to be done as to the in vivo function of UXT and its regulatory roles in cellular processes.
During a systematic screening for proteins that interacted with components of the NF-κB enhanceosome, we identified UXT as a novel p65-interacting protein. This interaction is confirmed both in vitro and in vivo. In this study, we show that RNAi knockdown of UXT leads to impaired NF-κB activity and dramatically attenuates the expression of NF-κB– dependent genes. This interference also sensitizes cells to apoptosis by TNF-α. Furthermore, UXT forms a signal- dependent complex with NF-κB and is recruited to the NF-κB enhanceosome upon stimulation. Interestingly, the UXT protein level correlates with constitutive NF-κB activity in human prostate cancer cell lines. The presence of NF-κB within the nucleus of stimulated or constitutively active cells is considerably diminished with decreased endogenous UXT protein levels. Collectively, our investigation reveals that UXT is an integral component of the NF-κB enhanceosome and is essential for its nuclear function, which uncovers a new mechanism of NF-κB regulation.
Results
Identification of UXT as a p65-interacting protein
To identify new components of the NF-κB enhanceosome, we performed a systematic yeast two-hybrid screening in which the cDNA fragment harboring the RHD of p65 (amino acids 1–312) was used as bait. Several positive clones were identified to encode full-length UXT (Fig. 1 A). In addition, previously confirmed p65-interacting proteins (e.g., IκBα and PIAS3) were screened out.
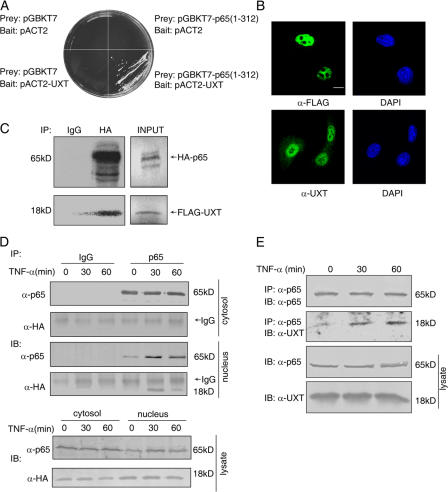
Figure 1.
UXT interacts with p65 in vitro and in vivo. (A) Interaction between p65 and UXT in a yeast two-hybrid assay. (B) Subcellular localization of endogenous and exogenous UXT. 293T cells were transfected with (top) or without (bottom) FLAG-UXT. Immunofluorescentmicroscopy was performed with the indicated primary antibodies. (C) Full-length HA-p65 and FLAG-UXT proteins were labeled with [35S]methionine by in vitro translation. The products were mixed and immunoprecipitated with the indicated antibodies coupled onto protein A/G beads. The immunoprecipitates were resolved by SDS-PAGE and visualized by autoradiography. 10% of the input proteins for pull down are shown at the left. (D) 293T cells were transfected with HA-UXT. 24 h after transfection, cells were treated with 10 ng/ml TNF-α for the indicated times and fractionated to cytoplasmic and nuclear fractions, which were immunoprecipitated and immunoblotted with the indicated antibodies, respectively. (E) 293T cells were treated with 10 ng/ml TNF-α for the indicated times. Whole cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies. Bar, 10 μm.
UXT was previously reported to be expressed almost exclusively inside the nucleus of most cells (Markus et al., 2002). This was confirmed in our investigation for either endogenous or overexpressed UXT (Fig. 1 B). To further substantiate its interaction with p65, an in vitro coimmunoprecipitation assay was applied in which full-length HA-p65 and FLAG-UXT proteins were generated and labeled, respectively, with [35S]methionine by in vitro translation. The products were mixed and immunoprecipitated with either control IgG or anti-HA antibody. As shown in Fig. 1 C, UXT could be coprecipitated by antibody against the HA epitope but not by control IgG, which suggests that UXT indeed interacts directly with full-length p65.
To address the physiological relevance of this interaction in mammalian cells, we expressed HA-UXT in 293T cells and then stimulated cells with or without TNF-α for the indicated times. The fractionated cytoplasmic or nuclear extracts were immunoprecipitated with either anti-p65 antibody or IgG as a control, respectively. There was no detectable UXT that interacted with cytoplasmic p65 in the presence or absence of TNF-α (Fig. 1 D), which was consistent with the unique subcellular location of UXT. In addition, there was only a marginal amount of endogenous p65 in the nucleus devoid of TNF-α treatment. Consequently, no UXT was coimmunoprecipitated from this nuclear extract even though there existed a large amount of UXT. In contrast, there exhibited a strong interaction between nuclear p65 and UXT upon TNF-α stimulation. Furthermore, we tested whether endogenous UXT and p65 could interact in response to TNF-α. As shown in Fig. 1 E, endogenous UXT was coimmunoprecipitated by p65 antibody from cells treated with TNF-α. In contrast, UXT was barely detected in the immunoprecipitates without TNF-α treatment. One possible explanation for this phenomenon is that only after p65 translocation into the nucleus could UXT have access to p65. However, we could not formally rule out the possibility that posttranslational modifications of either protein were prerequisites for this interaction in vivo. Collectively, these results indicate that UXT interacts in vivo with p65 upon TNF-α stimulation.
RHD of p65 mediates its interaction with UXT
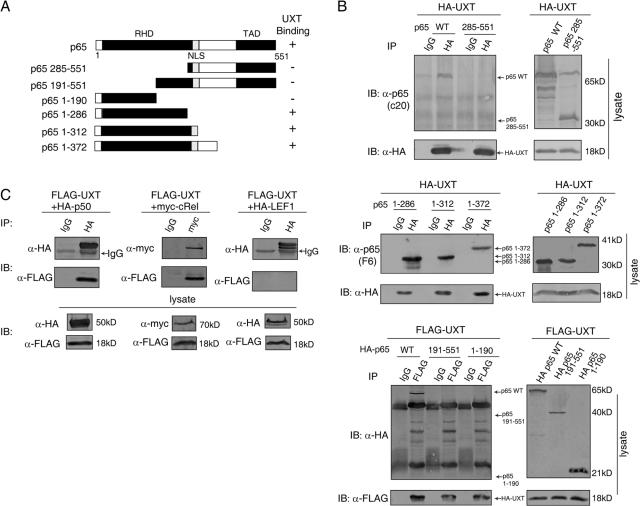
The p65 subunit of NF-κB harbors an N-terminal conserved region (∼300 amino acid residues) known as RHD and a C-terminal transactivation domain. To explore the UXT-binding region within p65, we constructed a series of p65 deletion mutants (Fig. 2 A). It was found that the loss of amino acids 1–285 at the N terminus of p65 resulted in its complete inability to interact with UXT (Fig. 2 B, top). In contrast, p65 fragments spanning amino acids 1–286, 1–312, or 1–372 fully retained their binding capability and interacted with UXT as well as the wild type (Fig. 2 B, middle). Because the RHD of p65 consisted of two Ig-like domains (Chen et al., 1998), we made two additional deletion mutants of p65 (amino acids 1–190 and 191–551), each of which contained only one Ig-like domain. Immunoprecipitation assays revealed that neither of them was able to interact with UXT (Fig. 2 B, bottom). In addition, we generated several point mutations of UXT and did not observe a considerable change on UXT and p65 interaction. We also had attempted in vain to express truncation mutants of UXT in mammalian cells, which prevented us from further dissecting UXT. Collectively, these data suggest that the intact RHD of p65 is both essential and sufficient to mediate interaction with UXT.
Figure 2.
RHD of NF-κB mediates its interaction with UXT. (A) Schematic illustration of p65 and its mutants. RHD, Rel homology domain; NLS, nuclear localization signal; TAD, transactivation domain. (B) Tagged full-length UXT was transfected into 293T cells along with p65 and its deletion mutants as indicated. Whole cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies. (C) 293T cells were transfected with FLAG-UXT together with HA-p50, myc-cRel, and HA–lymphoid enhancer binding factor 1. Cell lysates were immunoprecipitated and immunoblotted with the indicated antibodies.
The RHD structure defined a highly conserved family of transcription factors (Rel family) that are important in immunity and inflammation (Ghosh et al., 1998). This led us to wonder whether this interaction was also applicable to other proteins of this family. To explore this possibility, we transfected UXT into 293T cells along with p50 or c-Rel. Interestingly, UXT was also capable of binding to p50 or c-Rel specifically and strongly (Fig. 2 C). In contrast, lymphoid enhancer binding factor 1, a transcription factor unrelated to the Rel family, did not have any affinity to UXT. This phenomenon suggested that UXT recognized a consensus structure fold and that there was probably a unified theme for its interaction with p65 and other members of the Rel family.
Overexpression of UXT does not markedly affect NF-κB activation
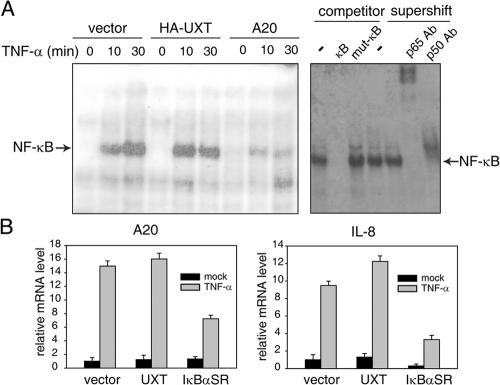
Because UXT interacted specifically inside the nucleus with p65 in a signal-dependent manner, we went on to address whether this interaction was important for regulating NF-κB activity and genes responsive to it. We transfected 293T cells with UXT or other control plasmids, prepared nuclear extracts, and performed electrophoretic mobility shift assay (EMSA) as indicated. Consistently, there was no NF-κB binding to its cognate probe without TNF-α treatment. Notably, the overexpression of UXT alone did not induce any detectable basal NF-κB binding activity. In contrast, robust NF-κB binding activity was induced upon TNF-α stimulation. Expectedly, this activity was severely impaired by A20, a potent inhibitor of NF-κB signaling (Wertz et al., 2004). However, in response to TNF-α, we observed neither inhibitory nor synergically stimulatory effects on NF-κB binding affinity (Fig. 3 A).
Figure 3.
Overexpression of UXT does not markedly affect NF-κB activation induced by TNF-α. (A) 293T cells were transfected with an equal amount of UXT, A20, or control vector. 24 h after transfection, cells were stimulated with 10 ng/ml TNF-α for the indicated times. Equal amounts (10 μg) of nuclear extracts were subjected to EMSA. For competition analysis, 100-fold excess of unlabeled wild-type or mutant κB probes were added to the reaction mixtures. For supershift assays, nuclear extracts were incubated with antibody as indicated. (B) 293T cells were transfected with equal amounts of UXT, IκBαSR, or empty vector. 24 h after transfection, cells were stimulated with 10 ng/ml TNF-α for 30 min or left untreated. Relative mRNA levels of A20 and IL-8 were analyzed by real-time RT-PCR. Data represent means ± SD (error bars) of at least three independent experiments.
Alternatively, we explored whether overexpressing UXT had any measurable effects on inducible genes such as A20 or interleukin-8 (IL-8) that were regulated by NF-κB. On transfection of UXT alone, we did not observe any change on the basal expression of these two genes (unpublished data). Thus, we transfected 293T cells with UXT or IκBα super repressor (SR) and analyzed the amount of A20 or IL-8 mRNA induced by TNF-α, respectively, via quantitative real-time RT-PCR. IκBαSR was a well-established potent inhibitor of NF-κB activation (Diao et al., 2005). Consistently, A20 or IL-8 inductions by TNF-α were severely attenuated in the presence of IκBαSR. Interestingly, we observed marginal synergic inductions of both A20 and IL-8 by TNF-α in the presence of UXT (Fig. 3 B), which suggested that UXT might modulate NF-κB function.
Knockdown of UXT sharply attenuates NF-κB activation by multiple stimuli
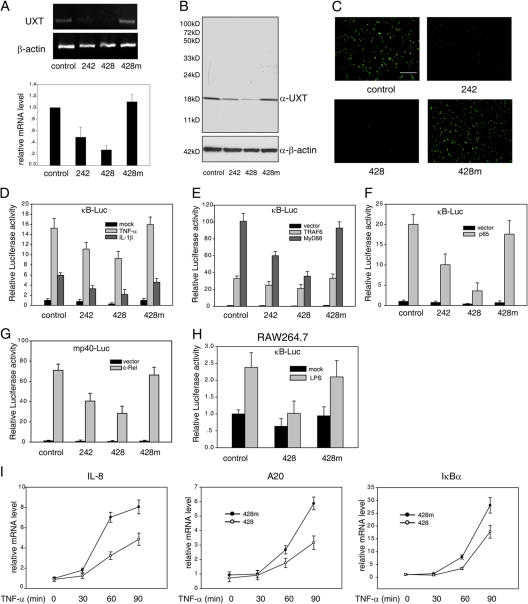
Because we failed via overexpressing UXT to convincingly demonstrate the involvement of UXT in NF-κB regulation, we turned to investigate the effect if endogenous UXT expression was reduced via RNAi. Two UXT siRNAs were fished out, which are designated as 242 and 428 hereafter. We confirmed the effectiveness of them against UXT by monitoring the mRNA level (Fig. 4 A), protein level (Fig. 4 B), and cellular immunofluorescence of UXT (Fig. 4 C). 428 was reproducibly better than 242 and was used more frequently in later experiments. In addition, two negative control siRNAs were used. One was a nonspecific siRNA and was named as control; the other is a mutant form of siRNA 428 (428m) that could partially bind to UXT mRNA but lost the interfering ability.
Figure 4.
Knockdown of UXT attenuates NF-κB activation induced by specific stimuli. (A) 293T cells were transfected with the indicated siRNA. 48 h after transfection, the endogenous expression of UXT mRNA was monitored by RT-PCR (top) or by real-time RT-PCR (bottom). (B) Cell lysates from A were subjected to Western blotting for determining endogenous UXT protein levels after siRNA transfection. (C) FLAG-UXT was cotransfected with the indicated siRNA. 24 h after transfection, UXT protein levels were monitored by immunofluorescence. (D) 293T cells were cotransfected with 3×κB-Luc and siRNA as indicated. 48 h after transfection, cells were stimulated with 10 ng/ml TNF-α or 20 ng/ml IL-1 for 7 h before luciferase assays were performed. (E) The indicated siRNA and 3×κB-Luc were transfected into 293T cells along with MyD88 or TRAF6. 48 h after transfection, cells were assayed as in D. (F) p65 was transfected into 293T cells along with the indicated siRNA and 3×κB-Luc. Cells were treated and assayed as in E. (G) c-Rel was transfected into 293T cells along with the indicated siRNA and mp40-Luc. Cells were treated and assayed as in E. (H) RAW264.7 cells were cotransfected with 3×κB-Luc and the indicated siRNA. 48 h after transfection, cells were stimulated with 500 ng/ml lipopolysaccharide for 7 h before luciferase assays were performed. (I) 293T cells were transfected with the indicated siRNA and stimulated by 10 ng/ml TNF-α for the indicated times. Endogenous mRNA expressions of IL-8, A20, and IκBα were measured by real-time RT-PCR. Data represent means ± SD (error bars) of at least three independent experiments. Bar, 200 μm.
Initially, we used a κB-Luc reporter gene to evaluate the effect of UXT knockdown on NF-κB activation status. Excitedly, luciferase assay revealed that the decrease of endogenous UXT considerably inhibited NF-κB transcriptional activity induced by TNF-α, IL-1β (Fig. 4 D), or lipopolysaccharide (Fig. 4 H). Notably, this phenomenon also held true for basal luciferase expression. Likewise, similar effects were observed in cells with reduced UXT expression when the cells were stimulated by overexpressing MyD88 or TRAF6, which are well known to induce NF-κB activity (Fig. 4 E; Muzio et al., 1997; Deng et al., 2000). In addition, gene activation induced by p65 alone was also markedly attenuated in cells with a decreased expression of endogenous UXT (Fig. 4 F).
Our data have shown that UXT could interact with other Rel family proteins like c-Rel (Fig. 2 C). c-Rel was previously found to regulate the expression of the IL-12 p40 subunit by specifically interacting with its promoter (Gri et al., 1998). Thus, a reporter gene was used that harbored firefly luciferase under the control of the mouse p40 promoter. Consistently, c-Rel activation was severely impaired in cells expressing reduced amounts of UXT. This attenuation was in direct proportion to the intensity of RNAi, as evidenced in oligonucleotides 428 versus 242 (Fig. 4 G).
To make it more physiologically relevant, we also investigated how the induction of NF-κB–dependent genes (IL-8, A20, and IκBα) was influenced by knocking down endogenous UXT expression. For cells transfected with control siRNA or UXT siRNA, quantitative real-time RT-PCR was used to measure endogenous mRNA levels of IL-8, A20, and IκBα induced by TNF-α during a time course. Consistently, these inductions were sharply attenuated when endogenous UXT expression was suppressed (Fig. 4 I). Collectively, our data strongly indicate that UXT is required for inducing genes tightly regulated by NF-κB and that it plays an essential role in NF-κB function.
Knockdown of UXT attenuates NF-κB binding to its cognate promoter
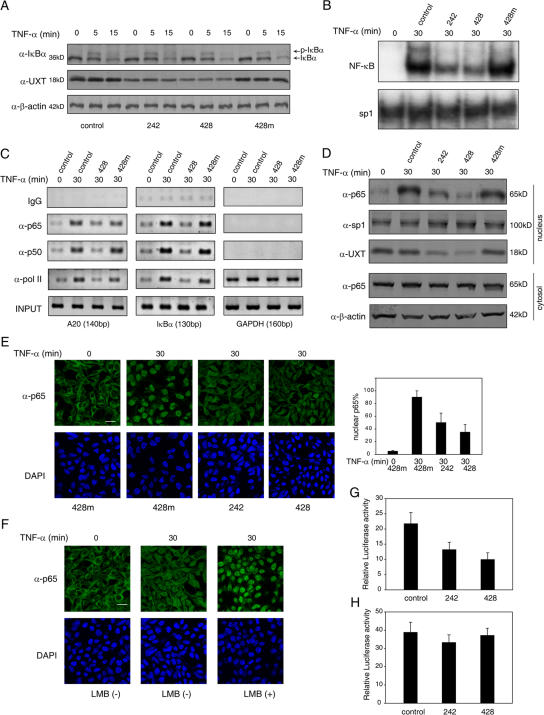
As was stated in the Introduction, NF-κB activation involved a series of molecular events both in the cytoplasm and in the nucleus. To rule out the possibility that UXT might act on processes other than directly on NF-κB itself, we examined the effects of UXT knockdown on the phosphorylation and degradation of IκBα. We did not find any differences in this regard between UXT-deficient and normal cells during TNF-α stimulation (Fig. 5 A). This was further supported by the observation that IKK kinase activity was not affected at all in terms of UXT knockdown (unpublished data). Given that UXT was expressed almost exclusively inside the nucleus and interacted directly with NF-κB, these data strongly indicate that UXT performed its function via targeting NF-κB itself.
Figure 5.
Knockdown of UXT attenuates the activity and amount of nuclear NF-κB. (A) 293T cells were transfected with the indicated siRNAs. After 48 h, cells were induced by 10 ng/ml TNF-α for the indicated times. Western blotting was performed on the cell extracts to check the phosphorylation and degradation of IκBα. (B) 293T cells were treated as in A. EMSA was performed to test endogenous NF-κB or sp1 binding to their cognate probes. (C) 293T cells were treated as in A. The ChIP assays were performed in terms of A20, IκBα, or GAPDH promoters using antibodies and corresponding primers as described. (D) 293T cells were transfected with the indicated siRNAs. After 48 h, cells were induced by 10 ng/ml TNF-α for 30 min. Cytoplasmic and nuclear fractions were prepared and immunoblotted with the indicated antibodies, respectively. (E) Cells were treated as in D and stained with anti-p65 primary antibody and FITC-conjugated secondary antibody. The nucleus was counterstained with DAPI. Quantification was performed to 100–200 cells in the same ranges of microscopy field for the presence of an appreciable nuclear signal of p65. Only those showing typical focused nuclear p65 were counted. The data are presented as percentages of cells with nuclear p65 versus total cells. (F) 293T cells were transfected with siRNA 428. After 48 h, cells were induced by 10 ng/ml TNF-α or 10 ng/ml TNF-α plus 20 ng/ml LMB for the indicated times and stained with anti-p65 primary antibody and FITC-conjugated secondary antibody. The nucleus was counterstained with DAPI. (G) 293T cells were cotransfected with the indicated siRNAs, 3×κB-Luc, and chimeric p65 (1–312)-VP16. Luciferase assays were performed 48 h after transfection. (H) 293T cells were cotransfected with the indicated siRNAs, Gal4-Luc, and chimeric Gal4 BD-p65 (285–551). Luciferase assays were performed 48 h after transfection. Data represent means ± SD (error bars) of at least three independent experiments. Bars, 20 μm.
Via EMSA, we then analyzed endogenous NF-κB DNA binding activity in cells with reduced UXT expression. Expectedly, TNF-α alone induced endogenous NF-κB to bind to its cognate probe strongly and specifically. Considerably, this interaction was markedly diminished in nuclear extracts from UXT-specific knockdown cells. In addition, this reduction was correlated with the effectiveness of the siRNA administered (Fig. 5 B).
To substantiate this finding, we took advantage of the chromatin immunoprecipitation (ChIP) assay to examine whether binding of NF-κB to its endogenous promoter was influenced in vivo when the expression of UXT was diminished. Consistently, deficiency of the endogenous UXT level resulted in considerable decreases in the amount of p65 associated with its endogenous cognate promoters upon TNF-α stimulation. In addition, this was also true for other components of the NF-κB transcriptional enhanceosome such as p50 and RNA polymerase II. Understandably, deficiency of the endogenous UXT level had no effect on the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcriptional complex (Fig. 5 C). Collectively, these finding indicate that UXT plays an important role in the NF-κB enhanceosome.
UXT maintains the presence of NF-κB inside the nucleus
A previous study has predicted UXT as an α-class PFD family protein via computer modeling (Gstaiger et al., 2003). Some members of this family were previously found to assemble into a hexameric complex that functioned as a molecular chaperone in protein folding and stability (Vainberg et al., 1998). Thus, we hypothesized that UXT performed its function by serving as a specific molecular chaperone for NF-κB inside the nucleus.
To explore this hypothesis, we investigated whether the nuclear presence of p65 was influenced by UXT. Therefore, 293T cells were transfected with or without the indicated siRNAs and stimulated with or without TNF-α as shown. Interestingly, the amount of nuclear p65 was markedly diminished in cells that were treated with siRNA against UXT even though these cells were stimulated with TNF-α. Notably, siRNA 428 was better than siRNA 242 in causing the loss of nuclear p65. In contrast, control siRNA displayed no such effect on the nuclear presence of p65. The transcription factor sp1 as a negative control was expressed constitutively inside the nucleus and remained intact in response to UXT knockdown. The cytoplasmic reservoir of p65 did not seem to be affected by siRNA against UXT (Fig. 5 D). This suggested that UXT positively regulated p65 inside the nucleus, and the loss of it affected the sustained action of p65 in the nucleus, which led to the attenuated EMSA results as observed in Fig. 5 B. Alternatively, we performed immunofluorescence analysis to support this speculation. Interestingly, when transfecting cells with siRNA against UXT and stimulating them with TNF-α, there were an apparently decreased percentage of cells that displayed focused nuclear p65. Approximately 90% of control cells displayed p65 inside the nucleus upon stimulation, whereas only 35–50% displayed p65 in the case of the UXT knockdown specimen (Fig. 5 E). Collectively, these data indicate that UXT is a stabilizing factor for the NF-κB transcriptional enhanceosome.
One possible mechanism for the aforementioned phenomenon is that the loss of UXT releases NF-κB from the enhanceosome, which consequently causes NF-κB to be exported out into the cytoplasm. To address this hypothesis, leptomycin B (LMB), a specific inhibitor of nuclear export (Ullman et al., 1997; Ghosh and Karin, 2002), was used. Cells were transfected with siRNA against UXT and stimulated with TNF-α in the presence or absence of LMB. Immunofluorescence analysis indicated that LMB considerably increased the nuclear accumulation of p65 even though the endogenous UXT was knocked down, which suggests that UXT contributed to the sustained presence of p65 inside the nucleus. We have also tested other possibilities, but no obvious correlation could be drawn at present (see Discussion).
Our data formerly indicated that RHD of p65 was essential and sufficient to mediate the interaction between p65 and UXT (Fig. 2 B). We wondered whether this stabilizing effect of UXT was applicable to any proteins containing this domain. To address this speculation, we constructed two fusing proteins, p65 (1–312)-VP16 and Gal4 BD-p65 (285–551): the former one harbors the RHD, whereas the latter one spans all of p65 except RHD. Luciferase assays were performed to monitor the effect of UXT knockdown on activations of indicated reporters induced by these chimeric proteins. Interestingly, the knockdown of UXT inhibited the activity induced by p65 (1–312)-VP16 (Fig. 5 G) but not that induced by Gal4 BD-p65 (285–551) (Fig. 5 H). Collectively, these data indicate that UXT directly modulates the nuclear function of NF-κB.
The siRNA-resistant form of UXT rescues the phenotype of its endogenous knockdown
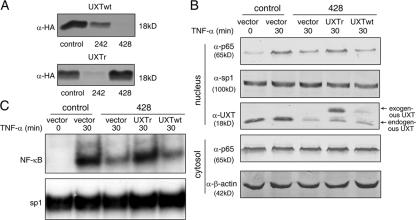
To rule out possible off-target effects of the siRNA against UXT, a siRNA-resistant UXT (UXTr) was generated in which silent mutations were introduced into the sequence targeted by siRNA 428 and without changing the amino acid sequence of the proteins expressed. The usefulness of UXTr was confirmed so that siRNA 428 no longer had any effects on UXTr but siRNA 242 could still interfere with UXTr expression (Fig. 6 A). The exogenous wild-type UXT or UXTr was transfected together with siRNA 428 and stimulated with TNF-α as indicated, respectively. Nuclear and cytoplasmic extracts were probed with indicated antibodies. As shown in Fig. 6 B, the expression of UXTr rescued the loss of nuclear p65 caused by endogenous UXT impairment, whereas the expression of wild-type UXT failed to do so. This rescuing effect was also confirmed with EMSA assays (Fig. 6 C). Collectively, these data indicate that the loss of nuclear p65 was directly caused by impairment of UXT function and that there was a direct functional connection between p65 and UXT.
Figure 6.
Rescue of the UXT knockdown effects with a siRNA-resistant form. (A) The HA-tagged wild-type UXT and siRNA-resistant UXT (UXTr) were cotransfected with the indicated siRNAs. After 24 h, cell lysates were subjected to Western blotting for determining exogenous UXT protein levels. (B) 293T cells were transfected with the indicated exogenous UXT and siRNAs. After 48 h, cells were induced by 10 ng/ml TNF-α for the indicated times. Cytoplasmic and nuclear fractions were prepared and immunoblotted with the indicated antibodies, respectively. (C) 293T cells were treated as in B. EMSA was performed to test endogenous NF-κB or sp1 binding to their cognate probes.
UXT is dynamically recruited to the NF-κB enhanceosome upon stimulation in vivo
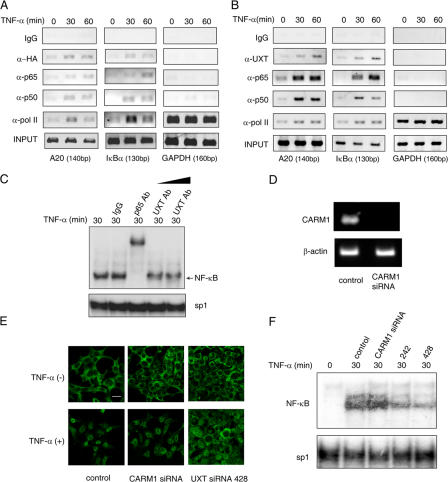
Given that UXT interacted with p65 and was essential to maintain the presence of NF-κB inside the nucleus, we wondered whether UXT was an integral component of the NF-κB transcriptional enhanceosome in vivo. To address this possibility, we transfected HA-UXT into 293T cells and performed systematic ChIP assays on the promoters of A20 and IκBα as described in Materials and methods. It turned out that UXT was indeed present within the NF-κB transcriptional enhanceosome. Notably, its presence became much more prominent upon stimulation, which suggested that UXT was dynamically recruited onto the enhanceosome. In addition, UXT had nothing to do with the transcription complex on the GAPDH promoter, indicating the selectivity of UXT action (Fig. 7 A). We also stimulated 293T cells and performed similar ChIP assays to confirm again that endogenous UXT was recruited onto the NF-κB enhanceosome in response to stimulation (Fig. 7 B).
Figure 7.
UXT forms a dynamic complex with NF-κB and is recruited to the NF-κB enhanceosome upon stimulation. (A and B) 293T cells transfected with (A) or without (B) UXT. After 10 ng/ml TNF-α stimulation, ChIP assays were performed on A20, IκBα, or GAPDH promoters as described. (C) 293T cells were induced by 10 ng/ml TNF-α for 30 min, and nuclear extracts were prepared and incubated with the indicated antibodies to perform EMSA supershift assays. (D) 293T cells were transfected with CARM1 siRNA. 48 h after transfection, the endogenous CARM1 mRNA was shown by RT-PCR. (E) 293T cells were transfected with CARM1 or UXT siRNA and treated with 10 ng/ml TNF-α for 30 min or were left untreated. Cover slides were subjected to immunofluoresence assay with anti-p65 antibody. (F) 293T cells were transfected and treated as in E. Nuclear lysates were used for EMSA to check endogenous NF-κB or sp1 DNA binding activities. Bar, 20 μm.
Alternatively, we performed supershift assay to further substantiate this observation. Stimulation of 293T cells with TNF-α led to a strong NF-κB DNA-binding band composed of κB probe, NF-κB, and its cofactors in EMSA. Interestingly, this EMSA band was markedly diminished when antibody against UXT was introduced into the reaction mixture (Fig. 7 C), which strongly suggests that UXT is an integral component in this band and that it is of importance to foster an NF-κB conformation amenable to its binding. Collectively, these data indicate that UXT forms a dynamic complex with p65 in vivo and is recruited to the NF-κB enhanceosome after stimulation.
The NF-κB enhanceosome consisted of a growing list of transcriptional cofactors. Our current finding that UXT was one of them and that it was able to maintain the stability of the NF-κB enhanceosome led us to ask whether other factors could also accomplish this function. Coactivator-associated arginine methyltransferase 1 (CARM1) is known to be a transcriptional cofactor in the NF-κB enhanceosome. Cells with a reduced expression of CARM1 showed an impaired expression of NF-κB–dependent genes upon stimulation (Covic et al., 2005; Teferedegne et al., 2006). However, immunofluorescence and EMSA experiments indicated that the knockdown of CARM1 did not affect the nuclear presence of p65 (Fig. 7, E and F), which suggested that UXT played a unique role toward NF-κB function.
Knockdown of UXT sensitizes 293T cells to apoptosis induced by TNF-α
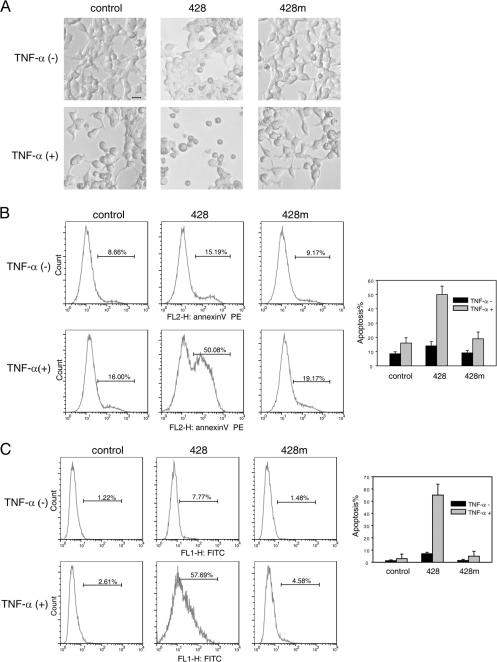
It was well established that NF-κB promotes the survival of most cells through the transcriptional induction of antiapoptotic genes (Barkett and Gilmore, 1999; Zong et al., 1999). Normally, 293T cells would not display apoptotic phenomenon in the presence of TNF-α (<3%). However, loss of NF-κB function would make cells prone to apoptosis. This was confirmed by knocking down UXT upon TNF-α stimulation (∼15%; unpublished data). Cycloheximide (CHX) is a potent protein synthesis inhibitor. However, at the concentration of ≤10 μg/ml, CHX only reduces but does not completely block de novo protein synthesis (Tang et al., 2001). Interestingly, a low concentration of CHX could dramatically augment the apoptotic effects (Yeh et al., 1997; Kelliher et al., 1998). Taking advantage of this cell model, we explored whether UXT was important for cell survival in response to TNF-α treatment. As was shown in Fig. 8, TNF plus 5 μg/ml CHX resulted in ∼10% cell death after 18 h of treatment. More importantly, when siRNA of UXT, which blocked NF-κB activity, was used in combination with TNF and CHX, we detected drastically morphological changes under light microscopy and ∼50% of cells undergoing apoptosis. Alternatively, we used annexin V and TUNEL methods to quantitatively measure the percentages of cells undergoing apoptosis. Consistently, a considerable increase of apoptotic cells was observed in samples with reduced UXT (Fig. 8, B and C). These indicated that the knockdown of UXT sensitizes 293T cells to apoptosis induced by TNF-α.
Figure 8.
Knockdown of UXT sensitizes 293T cells to TNF-α–induced apoptosis. (A) 293T cells were transfected with the indicated siRNAs. 48 h after transfection, cells were treated with 50 ng/ml TNF-α plus 5 μg/ml CHX or were left untreated for 18 h. Representative microscopic images are shown. (B and C) Cells were prepared as in A. After that, cells were analyzed using the annexin V (B) or TUNEL assay (C) to monitor cell apoptosis. The percentages indicate the fractions of positive annexin V cells in total cells. Data represent means ± SD (error bars) of at least three independent experiments. Bar, 10 μm.
Endogenous UXT expression correlates with NF-κB activation in human prostate cancer cell lines
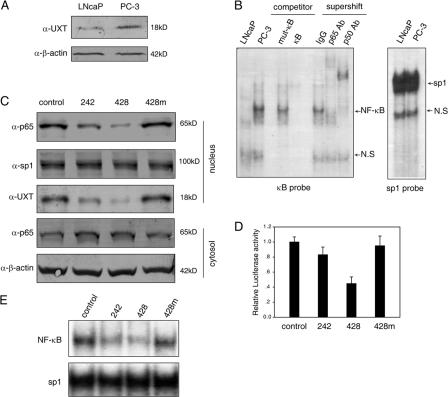
Prostate cancer began as an androgen-dependent tumor and progressed into an androgen-independent tumor. In this progress, NF-κB activity started to behave out of control (Chen and Sawyers, 2002; Zerbini et al., 2003). In androgen-independent PC-3 cells, more NF-κB localized inside the nucleus and constitutively displayed DNA binding activity, whereas it exhibited barely detectable activity in androgen-sensitive LNCaP cells (Palayoor et al., 1999). We confirmed these observations as shown in Fig. 9 B, which indicate that these two cells are ideal models for studying NF-κB regulation within the nucleus.
Figure 9.
UXT protein level correlates with the constitutive NF-κB activity in human prostate cancer cell lines. (A) Equal amounts of whole cell lysates from LNcaP or PC-3 prostate cancer cells were immunoblotted with the indicated antibodies. (B) 10 μg of nuclear extracts from LNCaP or PC-3 were subjected to EMSA with a radiolabeled κB or sp1 probe. NS, nonspecific band. (C) PC-3 cells were transfected with the indicated siRNAs. 48 h after transfection, cytoplasmic and nuclear fractions were prepared and immunoblotted with the indicated antibodies, respectively. (D) PC-3 cells were cotransfected with the indicated siRNAs and 3×κB-Luc. Luciferase assays were performed 48 h after transfection. Data represent means ± SD (error) of at least three independent experiments. (E) PC-3 cells were transfected with the indicated siRNAs. 48 h after transfection, nuclear extracts were prepared. Endogenous NF-κB or sp1 DNA binding activities were examined by EMSA.
In addition, a former study indicated that the UXT mRNA level was considerably elevated in PC-3 as compared with LNCaP cells (Markus et al., 2002). We confirmed this observation in terms of UXT mRNA (not depicted) and also found that this was true for the protein level of endogenous UXT (Fig. 9 A). Given that UXT was recruited onto the NF-κB enhanceosome upon stimulation and that it is essential to maintain the presence of NF-κB inside the nucleus, these results suggest a possible correlation between UXT levels and NF-κB activation.
To further explore this possibility, we determined whether the inhibition of endogenous UXT expression would affect the constitutive amount of nuclear NF-κB in PC-3 cells. As shown in Fig. 9 C, this interference indeed reduced the amount of nuclear NF-κB in this cell, which is consistent with that observed in 293T cells stimulated by TNF-α. In addition, luciferase assay revealed that the decrease of UXT expression suppressed the constitutive transcriptional activity of NF-κB in PC-3 cells (Fig. 9 D). Furthermore, EMSA confirmed that the loss of UXT also suppressed the constitutive DNA binding activity of NF-κB but not the control sp1 (Fig. 9 E). These results indicate that the elevated expression of UXT strongly correlates with constitutive NF-κB activity in prostate cancer cell lines and again substantiates the notion that UXT is essential for NF-κB function in the nucleus.
Discussion
The NF-κB family of transcription factors is crucial for many key cellular processes. Recently, the regulation of NF-κB activity has been cast in the limelight (Dixit and Mak, 2002; Ghosh and Karin, 2002). A handful of transcriptional cofactors were implicated in this process. For example, p300/CBP played a major role in the acetylation of p65 in vivo (Chen et al., 2002). Conversely, acetylated p65 was subjected to deacetylation by HDAC3 (Chen et al., 2001). Recent work also revealed that SIRT1 physically interacted with p65 and promoted p65 deacetylation (Yeung et al., 2004). In addition, NF-κB was reported to be phosphorylated at multiple sites during activation. For example, IKKα was demonstrated to accelerate both the turnover of NF-κB and its removal from proinflammatory gene promoters (Lawrence et al., 2005). An emerging theme is that there are additional important layers of regulation for nuclear NF-κB.
Regulating the duration of the nuclear presence of p65 is another potential mechanism to modulate the NF-κB transcriptional response. In this study, we characterized UXT as a novel and essential cofactor for NF-κB function in its enhanceosome. Several lines of findings support this argument. (1) UXT was shown to interact directly with p65 both in vitro and in vivo. Importantly, this interaction was dependent on external stimuli. Because UXT was almost exclusively present inside the nucleus, we reasoned that only after NF-κB translocation into the nucleus could this interaction take place, which was also substantiated by the observation that NF-κB was constitutively inside the nucleus in PC-3 cells and regulated by UXT as a result. (2) ChIP and EMSA assays demonstrated that UXT localized inside the NF-κB enhanceosome in vivo and was recruited to it upon stimulation. (3) The knockdown of UXT did not influence the molecular events of NF-κB activation outside the nucleus. Instead, it decreased the amount of nuclear p65 and severely impaired NF-κB activation. This decrease could be rescued by a siRNA-resistant exogenous UXT. (4) Overexpression of UXT caused marginal synergic inductions of both A20 and IL-8 in response to TNF-α. We reasoned that there was a sufficient amount of endogenous UXT within the nucleus in 293T cells so that it would be difficult to demonstrate the synergic effect more dramatically. Convincingly, RNAi of UXT resulted in apparent attenuations of NF-κB–responsive reporter expression by various stimuli and, ultimately, the inducible expression of genes tightly regulated by NF-κB. (5) The reduction of endogenous UXT tamed cells prone to apoptosis that was induced by TNF-α, which is a known index for the impairment of NF-κB function. (6) PC-3 cells displayed constitutive NF-κB activity inside the nucleus. This was nicely correlated with the elevated presence of both NF-κB and UXT within the nucleus, which was also consistently substantiated by the loss of NF-κB and its activity when endogenous UXT was diminished in PC-3 cells. Thus, UXT might function to extend the duration of NF-κB or its enhanceosome inside the nucleus.
This function was probably achieved by fostering a favorable conformation for NF-κB in its enhanceosome. UXT was recently predicted as a new member of the α-class PFD family protein. Yeast and human PFDs 1–6 assembled into a hexameric complex, which functioned as a molecular chaperone in protein folding. Our preliminary data also suggested that UXT forms oligomers in vivo (unpublished data). Some members of the PFD family protein were implicated to participate in transcriptional regulation, such as PFDN5 (MM1) in c-myc transcription (Satou et al., 2001) and URI in the rapamycin-sensitive transcription response (Gstaiger et al., 2003). However, the specific mechanisms of their action remain unknown. We have generated several point mutations of UXT (C75A, L32P, L50P, L59P, and L32P/L50P/L59P) and did not reveal any substantial correlation between UXT and p65 interaction. Our speculation is that a three-dimensional juxtaposed motif may be involved in this interaction. Structural analysis of UXT and NF-κB interaction is under way in our laboratory, and hopefully this will shed light on how UXT performs this regulatory role.
Recently, studies suggested that the ubiquitin–proteasomal degradation pathway was also involved in stringent control of the promoter-bound p65 (Ryo et al., 2003; Saccani et al., 2004). An NF-κB coactivator, Pin1, was found to bind p65 and prevent SOCS-1–mediated ubiquitination (Ryo et al., 2003). We have tried to address whether the ubiquitin–proteasome pathway is critical for p65 stability in the context of UXT, but no conclusion can be made as of now. The problem lies in the fact that most proteins are instantly degraded after ubiquitination, which is also the case for p65. To prevent this obstacle, proteasome inhibitors were used to stabilize ubiquitinated proteins. Unfortunately, a critical step during NF-κB activation involved proteasome function (i.e., IκBα degradation), which makes it unpractical to probe endogenous p65 degradation inside the nucleus. The few papers that reported nuclear p65 ubiquitination often used the overexpression of p65 to answer this question, which was a controversial approach. After this method, we did observe traces of p65 ubiquitination when UXT was knocked down (unpublished data), which was comparable with published reports (Ryo et al., 2003). However, we believe it is too early to correlate the loss of UXT with p65 ubiquitination. Substantially, we found that LMB, a specific inhibitor of nuclear export, increased the nuclear accumulation of p65 even though the endogenous UXT was knocked down. More likely, UXT influenced p65 nucleocytoplasmic shuttling.
Given the findings from this study, we favor the notion that UXT is a nuclear chaperone that promotes formation of the NF-κB enhanceosome. Nuclear chaperones usually mediate nucleosome assembly and remodeling (Philpott et al., 2000; Loyola and Almouzni, 2004). Furthermore, they were implicated to directly regulate transcription factors. For example, a nuclear chaperone termed FACT (facilitates chromatin transcription) could facilitate transcript elongation through nucleosomes (Orphanides et al., 1998). Another human nuclear chaperone (bZIP-enhancing factor) promoted the transcriptional activity of bZIP (basic region–leucine zipper DNA-binding domain) proteins (Virbasius et al., 1999). JDP2 was recently demonstrated to be a chaperone for AP1 (Jin et al., 2006). In addition, nuclear chaperones can also negatively regulate transcription. For example, nuclear chaperones p23 and Hsp90 bound to glucocorticoid-induced regulatory complexes and consequently disassembled these complexes (Freeman and Yamamoto, 2002). In conclusion, nuclear chaperones have regulatory functions targeted to their specific client proteins other than the classic functions involved in protein synthesis and maturation. More work needs to be done before the function and mechanism of UXT action are fully understood.
Numerous reports have documented considerable correlations between NF-κB activation and specific types of cancer (for review see Rayet and Gelinas, 1999). By promoting proliferation and inhibiting apoptosis, NF-κB could tip the balance between proliferation and apoptosis toward malignant behavior in tumor cells (for review see Rayet and Gelinas, 1999). Prostate cancer begins as an androgen-dependent tumor and progresses into an androgen-independent tumor. In this progress, NF-κB activity is up-regulated (Chen and Sawyers, 2002; Zerbini et al., 2003). In this study, we provide a new thread of explanation for this correlation and demonstrate that UXT is essential for the constitutive activity of NF-κB in prostate cancer cells.
Collectively, our study reveals that UXT is an integral component of the NF-κB enhanceosome and is essential for its function in the nucleus. UXT may function as a specific molecular chaperone for NF-κB in this process. Future investigations will focus on how UXT interacts with NF-κB and regulates the dynamic processes of NF-κB within the nucleus.
Materials and methods
Reagents
Monoclonal UXT antibodies 6D3 and 105.128 were provided by M.I. Greene (University of Pennsylvania, Philadelphia, PA) and W. Krek (Eidgenössische Technische Hochschule Honggerberg, Zurich, Switzerland), respectively. UXT siRNA duplexes were chemically synthesized by GenePharma. The UXT siRNA sequences were as follows: 242, AGCACUCGGAGUUAUAUAUdTdT; 428, CCAAGGACUCCAUGAAUAUdTdT; and 428M1, CCAACCCCUCCAUGAAUAUdTdT. The control siRNA sequence was UUCUCCGAACGUGUCACGUdTdT, and the CARM1 siRNA sequence was GCAGUCCUUCAUCAUCACCdTdT. Other commercially available reagents and antibodies used were as follows: HA, p65, p65, p50, IκBα, and RNA polymerase II antibodies were purchased from Santa Cruz Biotechnology, Inc. Flag, sp1, and β-actin antibodies were obtained from Sigma-Aldrich. Anti-myc was purchased from Wolwobiotech. rhTNFα and RhIL-1β was purchased from R&D Systems, and lipopolysaccharide was obtained from Sigma-Aldrich.
Plasmids
UXT and its deletion mutants were constructed by PCR from the human thymus library and subsequently cloned into mammalian expression vectors as indicated. The UXT siRNA-resistant form was generated by introducing four silent mutations (373 ACAAAAGATAGC 384) in the siRNA 428 target sequence. Flag-IκBαSR, Flag-TRAF6, and the reporter genes (3×κB-luc and pRL-SV40) have been described previously (Diao et al., 2005). pcDNA3-HA-p65 was a gift from G. Pei (Shanghai Institute of Biochemistry and Cell Biology [SIBCB], Shanghai, China), and its deletion mutants were constructed by PCR. MyD88 was amplified from the human thymus library and subcloned into pcDNA3.1-Flag vector. HA–lymphoid enhancer binding factor 1 was provided by L. Li (SIBCB, Shanghai, China), HA-p50 and mp40-luc were provided by B. Sun (SIBCB, Shanghai, China), myc-cRel was provided by B. Huang (Northeast Normal University, Changchun, China), and A20 was provided by L. Guo (SIBCB, Shanghai, China).
Yeast two-hybrid screening
A cDNA fragment encoding residues 1–312 of human p65 was inserted in frame into the Gal4 DNA-binding domain vector pGBKT7. A human thymus cDNA library (CLONTECH Laboratories, Inc.) was screened according to protocols recommended by the manufacturer.
Cell culture and transfection
293T and RAW264.7 cells were cultured in DME (Invitrogen) supplemented with 10% FBS (Hyclone). PC-3 cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS, and LNCaP cells were cultured in Ham's F12 medium (Invitrogen) supplemented with 10% FBS. All transfections were performed using LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions.
Reporter assays
Cells were seeded in 24-well plates and transfected with 40 pmol siRNA combined with reporters and other constructs as indicated. The total amount of DNA was kept constant by supplementing with pcDNA3. pRL-SV40 (Promega) was cotransfected to normalize transfection efficiency. 48 h after transfection, cells were treated with the indicated reagents or left untreated. Luciferase activity was analyzed with the Dual Luciferase Reporter Assay System (Promega).
In vitro translation
UXT and p65 were in vitro translated and labeled with [35S]methionine using the TNT Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instructions.
Real-time RT-PCR
Total RNA was isolated with TRIzol (Invitrogen) according to the manufacturer's instructions. Reverse transcription of purified RNA was performed using oligonucleotide dT primer. The quantification of gene transcripts was performed by real-time PCR using SYBR green I dye (Invitrogen). Expression values were normalized with control β-actin. The primers used are listed as follows: IL-8, sense (AGGTGCAGTTTTGCCAAGGA) and antisense (TTTCTGTGTTGGCGCAGTGT); IκBα, sense (CTGAGCTCCGAGACTTTCGAGG) and antisense (CACGTGTGGCCATTGTAGTTGG); A20, sense (GCGTTCAGGACACAGACTTG) and antisense (GCAAAGCCCCGTTTCAACAA); UXT, sense (TTTGGGCTGTAACTTCTTCGT) and antisense (ATATTCATGGAGTCCTTGGTG); CARM1, sense (TGCCGACCGCCTATGACT) and antisense (CCCGTGTTGGCTAAAGGAA); β-actin, sense (AAAGACCTGTACGCCAACAC) and antisense (GTCATACTCCTGCTTGCTGAT).
EMSA
EMSAs were performed as described previously (Yang et al., 2006). The probes used are as follows: wild-type κB probe (AGTTGAGGGGACTTTCCCAGGC), mutant κB probe (AGTTGAGGCGACTTTCCCAGGC), and sp1 probe (ATTCGATCGGGGCGGGGCGAGC).
ChIP assay
The ChIP assay kit (Upstate Biotechnology) was used according to the manufacturer's instructions with some variations. Formaldehyde cross-linking was performed at room temperature for 10 min before glycine was added to a final concentration of 125 mM for 5 min. The cells were rapidly collected and lysed in SDS lysis buffer. Suspended chromatin was sheared by sonication to a mean size of 200–1,000 bp, centrifuged to pellet debris, and diluted 10 times with dilution buffer. Extracts were precleared for 2 h with salmon sperm DNA and BSA-saturated protein A/G beads. Immunoprecipitations were performed at 4°C overnight using antibodies as indicated with IgG as a negative control. Immune complexes were collected and washed sequentially with Tris-SDS-EDTA buffer I, II, and III followed by two washes with Tris-EDTA buffer. Immune complexes were then extracted with elution buffer and DNA: protein complexes were disrupted by heating at 65°C overnight. After proteinase K digestion for 1 h, DNA was extracted with phenolchloroform and precipitated in ethanol. About one twentieth of precipitated DNA was used as template in each PCR reaction. The following promoter-specific primers were used: human A20, sense (CAGCCCGACCCAGAGAGTCAC) and antisense (CGGGCTCCAAGCTCGCTT); human GAPDH, sense (AGCTCAGGCCTCAAGACCTT) and antisense (AAGAAGATGCGGCTGACTGT); and human IκBα, sense (TAGTGGCTCATCGCAGGGAG) and antisense (TCAGGCTCGGGGAATTTCC).
Immunofluorescence and microscopy
Cells grown on coverslips were fixed with 4% PFA, permeabilized in 0.1% Triton X-100, blocked by 1% BSA, and stained with the indicated primary antibodies followed by FITC-conjugated anti–mouse IgG (Jackson ImmunoResearch Laboratories). Nuclei were counterstained with DAPI (Sigma-Aldrich). Slides were mounted by Aqua-Poly/Mount (Polysciences). Images were captured at room temperature using a confocal microscope (TCS SP2 ACBS; Leica) with a 63× NA 1.4 oil objective (Leica) except those in Fig. 2 C, which were captured using a camera (DP70; Olympus) on a microscope (BX51; Olympus) with a 10× NA 0.3 objective. The acquiring software was TCS (Leica) or DPcontrol (Olympus).
Apoptosis assay
293T cells were transfected with the indicated siRNA. 48 h after transfection, cells were treated with 50 ng/ml TNF-α and 5 μg/ml CHX or left untreated for 18 h. Floating and adherent cells were collected and analyzed using the annexin V–phycoerythrin Apoptosis Detection kit I (BD Biosciences) and In Situ Cell Death Detection kit (TUNEL; Roche) according to the manufacturer's instructions. The flow cytometer used was a FACSCalibur (BD Biosciences).
Acknowledgments
We thank Dr. Mark I. Greene and Dr. Wilhelm Krek for providing monoclonal UXT antibodies and thank Dr. Baiqu Huang for providing c-Rel plasmid.
C. Wang is a scholar of the Chinese Academy Renovation Program (grant KSCX1-YW-R-06). This study is partly supported by grants from the Ministry of Science and Technology of China (grants 2006CB504301 and 2006AA02Z121) and the National Natural Science Foundation of China (grants 30225013, 30570378, and 30623003).
S. Sun and Y. Tang contributed equally to this paper.
Abbreviations used in this paper: CARM1, coactivator-associated arginine methyltransferase 1; CBP, CREB-binding protein; ChIP, chromatin immunoprecipitation; CHX, cycloheximide; EMSA, electrophoretic mobility shift assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IKK, I κ-B kinase; IL, interleukin; LMB, leptomycin B; NF-κB, nuclear factor κB; PFD, prefoldin; RHD, Rel homology domain; SR, super repressor; UXT, ubiquitously expressed transcript.
References
- Barkett, M., and T.D. Gilmore. 1999. Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene. 18:6910–6924. [DOI] [PubMed] [Google Scholar]
- Chan, H.M., and N.B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363–2373. [DOI] [PubMed] [Google Scholar]
- Chen, C.D., and C.L. Sawyers. 2002. NF-kappa B activates prostate-specific antigen expression and is upregulated in androgen-independent prostate cancer. Mol. Cell. Biol. 22:2862–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F.E., D.B. Huang, Y.Q. Chen, and G. Ghosh. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 391:410–413. [DOI] [PubMed] [Google Scholar]
- Chen, L.F., and W.C. Greene. 2004. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 5:392–401. [DOI] [PubMed] [Google Scholar]
- Chen, L.f., W. Fischle, E. Verdin, and W.C. Greene. 2001. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 293:1653–1657. [DOI] [PubMed] [Google Scholar]
- Chen, L.f., Y. Mu, and W.C. Greene. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 21:6539–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z.J., L. Parent, and T. Maniatis. 1996. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 84:853–862. [DOI] [PubMed] [Google Scholar]
- Covic, M., P.O. Hassa, S. Saccani, C. Buerki, N.I. Meier, C. Lombardi, R. Imhof, M.T. Bedford, G. Natoli, and M.O. Hottiger. 2005. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 24:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, L., C. Wang, E. Spencer, L. Yang, A. Braun, J. You, C. Slaughter, C. Pickart, and Z.J. Chen. 2000. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 103:351–361. [DOI] [PubMed] [Google Scholar]
- Diao, L., B. Zhang, J. Fan, X. Gao, S. Sun, K. Yang, D. Xin, N. Jin, Y. Geng, and C. Wang. 2005. Herpes virus proteins ICP0 and BICP0 can activate NF-kappaB by catalyzing IkappaBalpha ubiquitination. Cell. Signal. 17:217–229. [DOI] [PubMed] [Google Scholar]
- Dixit, V., and T.W. Mak. 2002. NF-kappaB signaling. Many roads lead to madrid. Cell. 111:615–619. [DOI] [PubMed] [Google Scholar]
- Freeman, B.C., and K.R. Yamamoto. 2002. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 296:2232–2235. [DOI] [PubMed] [Google Scholar]
- Gerritsen, M.E., A.J. Williams, A.S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA. 94:2927–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell. 109:S81–S96. [DOI] [PubMed] [Google Scholar]
- Ghosh, S., M.J. May, and E.B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225–260. [DOI] [PubMed] [Google Scholar]
- Gri, G., D. Savio, G. Trinchieri, and X. Ma. 1998. Synergistic regulation of the human interleukin-12 p40 promoter by NFkappaB and Ets transcription factors in Epstein-Barr virus-transformed B cells and macrophages. J. Biol. Chem. 273:6431–6438. [DOI] [PubMed] [Google Scholar]
- Gstaiger, M., B. Luke, D. Hess, E.J. Oakeley, C. Wirbelauer, M. Blondel, M. Vigneron, M. Peter, and W. Krek. 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 302:1208–1212. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, S., M. Kitagawa, K. Nakayama, M. Shirane, M. Matsumoto, K. Hattori, H. Higashi, H. Nakano, K. Okumura, K. Onoe, R.A. Good, and K. Nakayama. 1999. Ubiquitin-dependent degradation of IkappaBalpha is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proc. Natl. Acad. Sci. USA. 96:3859–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden, M.S., and S. Ghosh. 2004. Signaling to NF-kappaB. Genes Dev. 18:2195–2224. [DOI] [PubMed] [Google Scholar]
- Jin, C., K. Kato, T. Chimura, T. Yamasaki, K. Nakade, T. Murata, H. Li, J. Pan, M. Zhao, K. Sun, et al. 2006. Regulation of histone acetylation and nucleosome assembly by transcription factor JDP2. Nat. Struct. Mol. Biol. 13:331–338. [DOI] [PubMed] [Google Scholar]
- Kelliher, M.A., S. Grimm, Y. Ishida, F. Kuo, B.Z. Stanger, and P. Leder. 1998. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 8:297–303. [DOI] [PubMed] [Google Scholar]
- Lawrence, T., M. Bebien, G.Y. Liu, V. Nizet, and M. Karin. 2005. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature. 434:1138–1143. [DOI] [PubMed] [Google Scholar]
- Li, Q., and I.M. Verma. 2002. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2:725–734. [DOI] [PubMed] [Google Scholar]
- Loyola, A., and G. Almouzni. 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta. 1677:3–11. [DOI] [PubMed] [Google Scholar]
- Markus, S.M., S.S. Taneja, S.K. Logan, W. Li, S. Ha, A.B. Hittelman, I. Rogatsky, and M.J. Garabedian. 2002. Identification and characterization of ART-27, a novel coactivator for the androgen receptor N terminus. Mol. Biol. Cell. 13:670–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio, M., J. Ni, P. Feng, and V.M. Dixit. 1997. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 278:1612–1615. [DOI] [PubMed] [Google Scholar]
- Orphanides, G., G. LeRoy, C.H. Chang, D.S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 92:105–116. [DOI] [PubMed] [Google Scholar]
- Palayoor, S.T., M.Y. Youmell, S.K. Calderwood, C.N. Coleman, and B.D. Price. 1999. Constitutive activation of IkappaB kinase alpha and NF-kappaB in prostate cancer cells is inhibited by ibuprofen. Oncogene. 18:7389–7394. [DOI] [PubMed] [Google Scholar]
- Philpott, A., T. Krude, and R.A. Laskey. 2000. Nuclear chaperones. Semin. Cell Dev. Biol. 11:7–14. [DOI] [PubMed] [Google Scholar]
- Rayet, B., and C. Gelinas. 1999. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 18:6938–6947. [DOI] [PubMed] [Google Scholar]
- Ryo, A., F. Suizu, Y. Yoshida, K. Perrem, Y.C. Liou, G. Wulf, R. Rottapel, S. Yamaoka, and K.P. Lu. 2003. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell. 12:1413–1426. [DOI] [PubMed] [Google Scholar]
- Saccani, S., I. Marazzi, A.A. Beg, and G. Natoli. 2004. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor κB response. J. Exp. Med. 200:107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou, A., T. Taira, S.M. Iguchi-Ariga, and H. Ariga. 2001. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J. Biol. Chem. 276:46562–46567. [DOI] [PubMed] [Google Scholar]
- Schroer, A., S. Schneider, H. Ropers, and H. Nothwang. 1999. Cloning and characterization of UXT, a novel gene in human Xp11, which is widely and abundantly expressed in tumor tissue. Genomics. 56:340–343. [DOI] [PubMed] [Google Scholar]
- Senftleben, U., Y. Cao, G. Xiao, F.R. Greten, G. Krahn, G. Bonizzi, Y. Chen, Y. Hu, A. Fong, S.C. Sun, and M. Karin. 2001. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 293:1495–1499. [DOI] [PubMed] [Google Scholar]
- Siegert, R., M.R. Leroux, C. Scheufler, F.U. Hartl, and I. Moarefi. 2000. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell. 103:621–632. [DOI] [PubMed] [Google Scholar]
- Spencer, E., J. Jiang, and Z.J. Chen. 1999. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 13:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja, S.S., S. Ha, N.K. Swenson, I.P. Torra, S. Rome, P.D. Walden, H.Y. Huang, E. Shapiro, M.J. Garabedian, and S.K. Logan. 2004. ART-27, an androgen receptor coactivator regulated in prostate development and cancer. J. Biol. Chem. 279:13944–13952. [DOI] [PubMed] [Google Scholar]
- Tang, G., J. Yang, Y. Minemoto, and A. Lin. 2001. Blocking caspase-3-mediated proteolysis of IKKbeta suppresses TNF-alpha-induced apoptosis. Mol. Cell. 8:1005–1016. [DOI] [PubMed] [Google Scholar]
- Teferedegne, B., M.R. Green, Z. Guo, and J.M. Boss. 2006. Mechanism of action of a distal NF-kappaB-dependent enhancer. Mol. Cell. Biol. 26:5759–5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman, K.S., M.A. Powers, and D.J. Forbes. 1997. Nuclear export receptors: from importin to exportin. Cell. 90:967–970. [DOI] [PubMed] [Google Scholar]
- Vainberg, I.E., S.A. Lewis, H. Rommelaere, C. Ampe, J. Vandekerckhove, H.L. Klein, and N.J. Cowan. 1998. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 93:863–873. [DOI] [PubMed] [Google Scholar]
- Virbasius, C.M., S. Wagner, and M.R. Green. 1999. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell. 4:219–228. [DOI] [PubMed] [Google Scholar]
- Wertz, I.E., K.M. O'Rourke, H. Zhou, M. Eby, L. Aravind, S. Seshagiri, P. Wu, C. Wiesmann, R. Baker, D.L. Boone, et al. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 430:694–699. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y., U.N. Verma, S. Prajapati, Y.T. Kwak, and R.B. Gaynor. 2003. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 423:655–659. [DOI] [PubMed] [Google Scholar]
- Yang, K., H. Shi, R. Qi, S. Sun, Y. Tang, B. Zhang, and C. Wang. 2006. Hsp90 regulates activation of interferon regulatory factor 3 and TBK-1 stabilization in Sendai virus-infected cells. Mol. Biol. Cell. 17:1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, W.C., A. Shahinian, D. Speiser, J. Kraunus, F. Billia, A. Wakeham, J.L. de la Pompa, D. Ferrick, B. Hum, N. Iscove, et al. 1997. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 7:715–725. [DOI] [PubMed] [Google Scholar]
- Yeung, F., J.E. Hoberg, C.S. Ramsey, M.D. Keller, D.R. Jones, R.A. Frye, and M.W. Mayo. 2004. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23:2369–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbini, L.F., Y. Wang, J.Y. Cho, and T.A. Libermann. 2003. Constitutive activation of nuclear factor kappaB p50/p65 and Fra-1 and JunD is essential for deregulated interleukin 6 expression in prostate cancer. Cancer Res. 63:2206–2215. [PubMed] [Google Scholar]
- Zhao, H., Q. Wang, H. Zhang, Q. Liu, X. Du, M. Richter, and M.I. Greene. 2005. UXT is a novel centrosomal protein essential for cell viability. Mol. Biol. Cell. 16:5857–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, W.X., L.C. Edelstein, C. Chen, J. Bash, and C. Gelinas. 1999. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 13:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]