Abstract
In mitosis, kinetochores are initially captured by the lateral sides of single microtubules and are subsequently transported toward spindle poles. Mechanisms for kinetochore transport are not yet known. We present two mechanisms involved in microtubule-dependent poleward kinetochore transport in Saccharomyces cerevisiae. First, kinetochores slide along the microtubule lateral surface, which is mainly and probably exclusively driven by Kar3, a kinesin-14 family member that localizes at kinetochores. Second, kinetochores are tethered at the microtubule distal ends and pulled poleward as microtubules shrink (end-on pulling). Kinetochore sliding is often converted to end-on pulling, enabling more processive transport, but the opposite conversion is rare. The establishment of end-on pulling is partly hindered by Kar3, and its progression requires the Dam1 complex. We suggest that the Dam1 complexes, which probably encircle a single microtubule, can convert microtubule depolymerization into the poleward kinetochore-pulling force. Thus, microtubule-dependent poleward kinetochore transport is ensured by at least two distinct mechanisms.
Introduction
The mitotic spindle is composed of microtubules emanating from spindle poles and has pivotal roles in high fidelity chromosome segregation during mitosis (McIntosh et al., 2002; Maiato et al., 2004; Kline-Smith et al., 2005). Kinetochores are initially captured by the lateral surface of spindle microtubules and are transported toward spindle poles during prometaphase (Rieder and Alexander, 1990; K. Tanaka et al., 2005). Subsequently, sister kinetochores are captured by microtubules extending from the opposite spindle poles (called sister kinetochore biorientation or amphitelic attachment) before anaphase onset (T.U. Tanaka et al., 2005). Poleward kinetochore transport during prometaphase is especially crucial when kinetochores are located at a distance from the mitotic spindle.
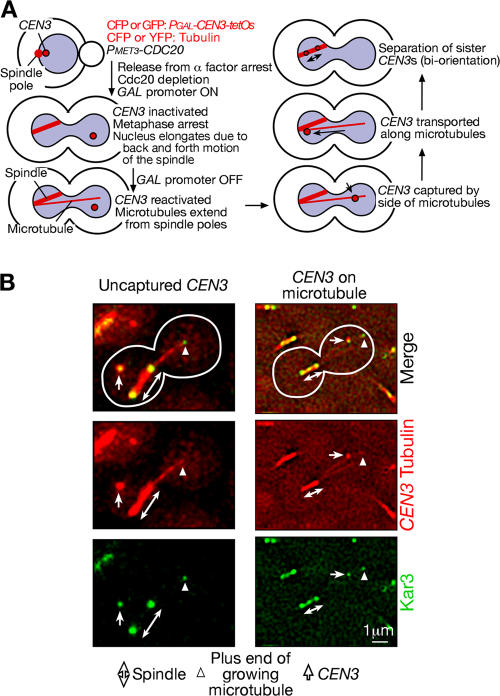
In the budding yeast Saccharomyces cerevisiae, centromeres are tethered to spindle poles by microtubules during most of the cell cycle (Winey and O'Toole, 2001; T.U. Tanaka et al., 2005). Nonetheless, centromeres are released from and recaptured by microtubules during a brief period in S phase, probably as a result of kinetochore disassembly and reassembly upon centromere DNA replication (Pearson et al., 2004; Tanaka, 2005; our unpublished data). We have visualized this process in S phase using electron microscopy and live cell fluorescence microscopy (K. Tanaka et al., 2005). Moreover, to analyze individual kinetochore–microtubule interaction with higher resolution, we displaced a selected centromere (CEN3) from the spindle and other centromeres by conditionally inactivating it (K. Tanaka et al., 2005). Then, during metaphase arrest, we reactivated CEN3, which was subsequently captured by the lateral surface of single microtubules (centromere reactivation system; Fig. 1 A).
Figure 1.
Localization of Kar3 at kinetochores. (A) Experimental system to analyze kinetochore capture and transport by individual microtubules in budding yeast. See details in K. Tanaka et al. (2005) and in the first paragraph of Results. (B) Kar3 localizes at kinetochores before their capture by microtubules (left) and during their transport along microtubules (right). KAR3-4GFP CFP-TUB1 PGAL-CEN3-tetOs TetR-3CFP PMET3-CDC20 cells (T3733) were treated with α factor in methionine drop-out medium with raffinose for 2.5 h and released to YP medium containing galactose, raffinose, and 2 mM methionine. After 3 h, cells were suspended in synthetic complete medium containing glucose and methionine (K. Tanaka et al., 2005). After 3 min, GFP (Kar3; green) and CFP (CEN3 tubulin; red) images were collected. Bidirectional arrows, arrowheads, and arrows indicate metaphase spindle, plus ends of growing microtubules, and CEN3, respectively.
Using this system, we found that Kar3, a kinesin-14 family member and microtubule minus end–directed motor, is involved in poleward kinetochore transport along the lateral surface of microtubules (kinetochore sliding; K. Tanaka et al., 2005). For instance, in the kar3-1 rigor mutant that can bind microtubules but does not have motor activity as a result of an ATP hydrolysis defect (Meluh and Rose, 1990; Maddox et al., 2003), CEN3 was captured by microtubules but frequently was not transported (K. Tanaka et al., 2005). In contrast, KAR3 overexpression accelerated CEN3 transport along microtubules (K. Tanaka et al., 2005). However, it has remained unclear whether Kar3 localizes at kinetochores and directly drives CEN3 sliding along microtubules. Moreover, although Kar3 is apparently involved in kinetochore sliding, CEN3 was still able to reach a spindle pole in the majority of kar3-deleted cells (K. Tanaka et al., 2005). This suggests the involvement of a redundant mechanism for kinetochore transport that still remains to be identified.
The Dam1 complex, which is also called DASH or DDD and is composed of at least 10 proteins, has important roles in ensuring proper kinetochore–microtubule interaction (Cheeseman et al., 2001a; Janke et al., 2002; Li et al., 2002). However, in our CEN3 reactivation system, mutants of the Dam1 complex components did not show substantial defects in CEN3 capture by microtubules or in the subsequent sliding of CEN3 along microtubules (K. Tanaka et al., 2005). It has been recently reported that several Dam1 complexes could gather together and form rings encircling microtubules in vitro (Miranda et al., 2005; Westermann et al., 2005). It is still unclear whether this is the case in vivo or how the complex regulates kinetochore–microtubule interaction.
Here, we studied mechanisms of kinetochore transport by microtubules using our centromere reactivation system (K. Tanaka et al., 2005) as well as in normal cell cycles (i.e., without cell cycle arrest or regulation of centromere activity). We show that poleward movement of kinetochores can occur in two distinct ways: lateral sliding, in which kinetochores move along the side of a microtubule, and end-on pulling, in which the kinetochore is attached to the end of a microtubule and is pulled poleward as the microtubule shrinks. Our study reveals how Kar3 and the Dam1 complex regulate these processes.
Results
Kar3 localizes at kinetochores during their transport along microtubules
To visualize individual kinetochore–microtubule interactions at high resolution in S. cerevisiae, we regulated the activity of a particular centromere (CEN3). CEN3 is displaced from the spindle and other centromeres by conditional inactivation using transcription from the adjacently inserted GAL1-10 promoter (Fig. 1 A; K. Tanaka et al., 2005). Then, during metaphase arrest by Cdc20 depletion, we reactivated CEN3 by turning off the GAL1-10 promoter. CEN3, which was marked with CFP or GFP, was captured by the lateral surface of CFP- or YFP-labeled microtubules.
To address whether Kar3 localizes at kinetochores and directly drives their transport along microtubules, we visualized Kar3 by fusing it with four tandem copies of GFP (Kar3-4GFP). After the CEN3 reactivation, in most cells, Kar3-4GFP was visible at the CFP-labeled CEN3 before its capture by CFP-labeled microtubules and also during its transport along microtubules (Fig. 1 B). Kar3 was also detected at the plus ends of growing microtubules (supplemental note 1; available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1) and at spindle poles as previously reported (Hildebrandt and Hoyt, 2000; Maddox et al., 2003). The amount of Kar3 at kinetochores appeared to decrease after sister kinetochore biorientation (supplemental note 1; Tytell and Sorger, 2006).
Kinetochores attach to microtubule plus ends and are transported poleward as microtubules shrink in the absence of Kar3
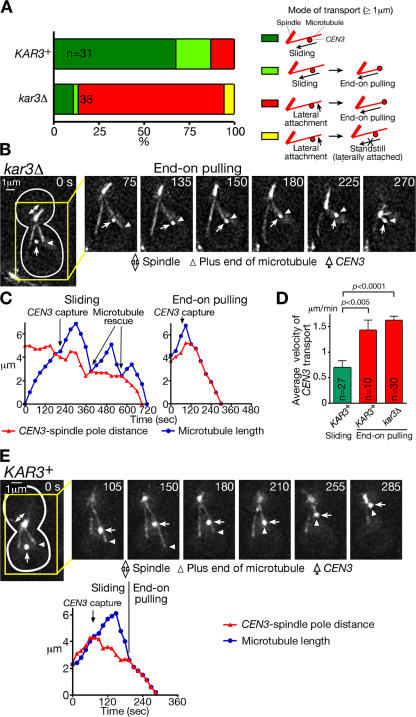
Kar3 is involved in kinetochore transport toward spindle poles (K. Tanaka et al., 2005). However, in the majority of kar3Δ cells, CEN3 still reached spindle poles after being captured by microtubules (K. Tanaka et al., 2005), suggesting the involvement of a redundant mechanism for kinetochore transport. To identify this mechanism, we analyzed poleward kinetochore transport in kar3Δ cells in greater detail. In 68% of KAR3+ wild-type cells, CEN3 reached the spindle by sliding along the lateral surface of microtubules, whereas in kar3Δ cells, this occurred in only 11% of cells (Fig. 2 A, supplemental note 2, and Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1). The sliding observed in kar3Δ cells might depend on other microtubule motors, but, as we discuss below, it probably depends on motor-independent one-dimensional diffusion (supplemental note 3). In many of the kar3Δ cells, after CEN3 capture by the lateral sides of microtubules, CEN3 was tethered at the distal ends of microtubules extending from spindle poles and was pulled poleward as the microtubules shrank (Fig. 2 B, microtubule end-on pulling of kinetochores; and Video 1). 81% of kar3Δ cells showed microtubule end-on pulling of CEN3, whereas this occurred in only 13% of KAR3+ wild-type cells (Fig. 2 A). Thus, in KAR3+ cells, kinetochores were transported mainly by sliding along microtubules, but, in the absence of Kar3, microtubule end-on pulling was the main mode for kinetochore transport.
Figure 2.
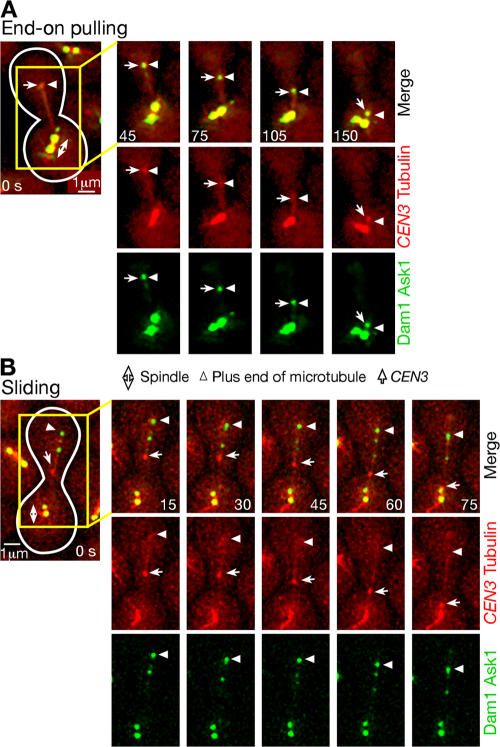
Kinetochores are tethered at the ends of microtubules and are transported poleward as microtubules shrink. KAR3+ (wild-type KAR3; T3531) and kar3Δ (T3860) cells with YFP-TUB1 PGAL-CEN3-tetOs TetR-GFP PMET3-CDC20 were treated as in Fig 1. (A) Percentages of cells in which CEN3 was transported with each mode as colored/illustrated. Sliding and end-on pulling were scored only when CEN3 moved for 1 μm or more by each mode of transport; standstill was scored when CEN3 was at almost the same position on microtubules for considerable time (supplemental note 30). (B) Representative time-lapse images of the microtubule end-on pulling of CEN3. GFP and YFP images were acquired together in T3860 cells. See Video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1). (C) Distance of CEN3 from a spindle pole (red) and length of the microtubule that captured CEN3 (blue) during sliding (left) and end-on pulling (right; images shown in B) of CEN3 in representative KAR3+ and kar3Δ cells, respectively. Sliding in kar3Δ and end-on pulling in KAR3+ also occurred similarly (not depicted). (D) The mean velocity of CEN3 transport by sliding (KAR3+) and end-on pulling (KAR3+ and kar3Δ). Error bars represent SEM. (E) Conversion of CEN3 sliding along a microtubule into microtubule end-on pulling in a representative KAR3+ cell. See Video 2. Such conversion also occurred similarly in kar3Δ cells (not depicted).
In KAR3+ cells, poleward sliding of CEN3 occurred with frequent pausing, and the associated microtubules showed frequent rescue (conversion from shrinkage to growth; Fig. 2 C, left; K. Tanaka et al., 2005). In contrast, in both KAR3+ and kar3Δ cells, microtubule end-on pulling transported CEN3 poleward on average more rapidly than sliding and without pausing (Fig. 2, C [right] and D). During end-on pulling, microtubule rescue was rarely observed (supplemental note 4 and Fig. S1 B). In addition, CEN3 occasionally (4.9%) detached from microtubules during sliding, whereas such detachment was never observed during end-on pulling (supplemental note 5). These differences suggest that the two modes of kinetochore transport are distinctly regulated processes. Nonetheless, CEN3 sliding was sometimes converted to end-on pulling (Fig. 2, A [pale green] and E; and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1), whereas the opposite conversion was seldom, if ever, observed.
Kar3 is the main and probably the sole factor driving kinetochore sliding along microtubules
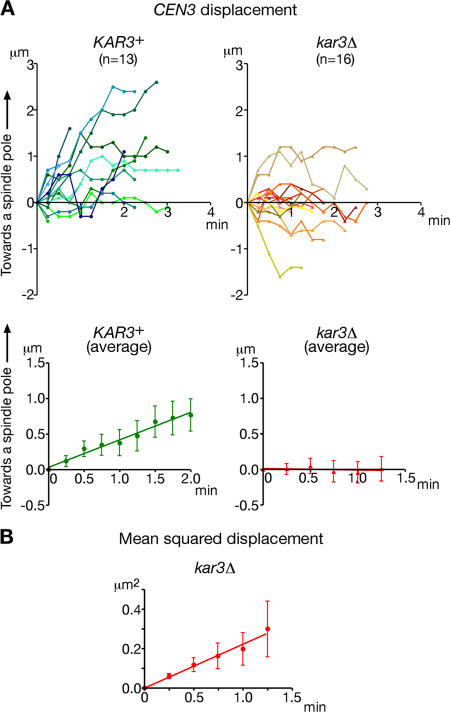
Having established that CEN3 is transported poleward either by sliding or end-on pulling, we were in a position to evaluate the contribution of Kar3 specifically to CEN3 sliding by excluding end-on pulling events from our analysis. To make an unbiased comparison between KAR3+ and kar3Δ cells, we studied CEN3 movement for a short period, during which CEN3 is associated with the microtubule lateral surface but not at the microtubule distal end after its initial capture by the microtubule lateral surface (supplemental note 6). During such a period, in KAR3+ cells, CEN3 travelled preferentially poleward (Fig. 3 A, top left) except in a small number of cells (supplemental note 7). In contrast, in kar3Δ cells, CEN3 moved in both directions apparently equally (Fig. 3 A, top right). Consequently, the mean displacement of CEN3 from its original position (i.e., its position when initially captured by a microtubule) along a microtubule increased with time and was oriented toward a spindle pole in KAR3+ cells at a speed of 0.39 μm/min (Fig. 3 A, bottom left; and supplemental note 6), whereas it remained approximately zero in kar3Δ cells (Fig. 3 A, bottom right).
Figure 3.
Kar3 is the main and probably the sole factor driving kinetochore sliding along microtubules. (A) The position of CEN3 was plotted for a short period, during which it was associated with the microtubule lateral surface but not at the microtubule distal end after its initial capture by a microtubule lateral surface in KAR3+ and kar3Δ cells (supplemental note 6, available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1). T3531 and T3860 cells (see Fig 2) were treated as in Fig 1. (top) Each line represents the time-course trajectory of CEN3 along a microtubule in an individual cell. On the y axis and x axis of the graphs, zero represents the original CEN3 position when initially captured by a microtubule and the time of this capture, respectively; y axis positive values designate CEN3 motion toward a spindle pole along a microtubule. (bottom) The displacement of CEN3 from its original position along a microtubule was averaged among different cells. (B) The MSD, which was averaged among different cells, was plotted against a change of time in kar3Δ cells. Error bars represent SEM.
Given a lack of preferential direction to CEN3 motion in kar3Δ cells, we suspected that CEN3 motility in this mutant might be caused by one-dimensional diffusion along a microtubule. To address this, we plotted the mean squared displacement (MSD) of CEN3 along a microtubule against a change of time (Δt) in kar3Δ cells (Fig. 3 B; Saxton and Jacobson, 1997; Helenius et al., 2006). The MSD increased linearly as Δt increased, which was indeed consistent with the one-dimensional diffusion of CEN3. The diffusion coefficient (D in MSD = 2DΔt) of CEN3 motility along microtubules was calculated as 0.11 μm2/min. This result was consistent with our previous observations that single deletion mutants of other microtubule-dependent motor proteins (Cin8, Kip1, Kip2, Kip3, and Dyn1) had no effect on CEN3 transport (K. Tanaka et al., 2005). We also failed to observe a CEN3 association of these other motor proteins during its transport (supplemental note 8 and Fig. S2, A and B; available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1). These data suggest that Kar3 is the main and probably the sole factor that drives poleward kinetochore sliding along microtubules.
Kar3 partially hinders kinetochores from being tethered at the microtubule plus ends
In most cells that we observed, CEN3 was first captured by a microtubule lateral surface, but, when the microtubule subsequently shrank, its distal end encountered CEN3 (note that the microtubule shrinkage rate exceeds the mean velocity of CEN3 sliding; Fig. 4 A; K. Tanaka et al., 2005) unless either the microtubule was rescued before this or CEN3 reached a spindle pole by sliding. Immediately after the microtubule end encountered CEN3, we observed either of the following two events: (1) microtubules were rescued, and CEN3 remained on the microtubule lateral surface; or (2) CEN3 became tethered at the microtubule end and subsequently was transported poleward by end-on pulling (Fig. 4 A). In such encounters in wild-type KAR3+ cells, we observed microtubule rescue in 59% of cases and the establishment of end-on pulling in 41% of cases (Fig. 4 B).
Figure 4.
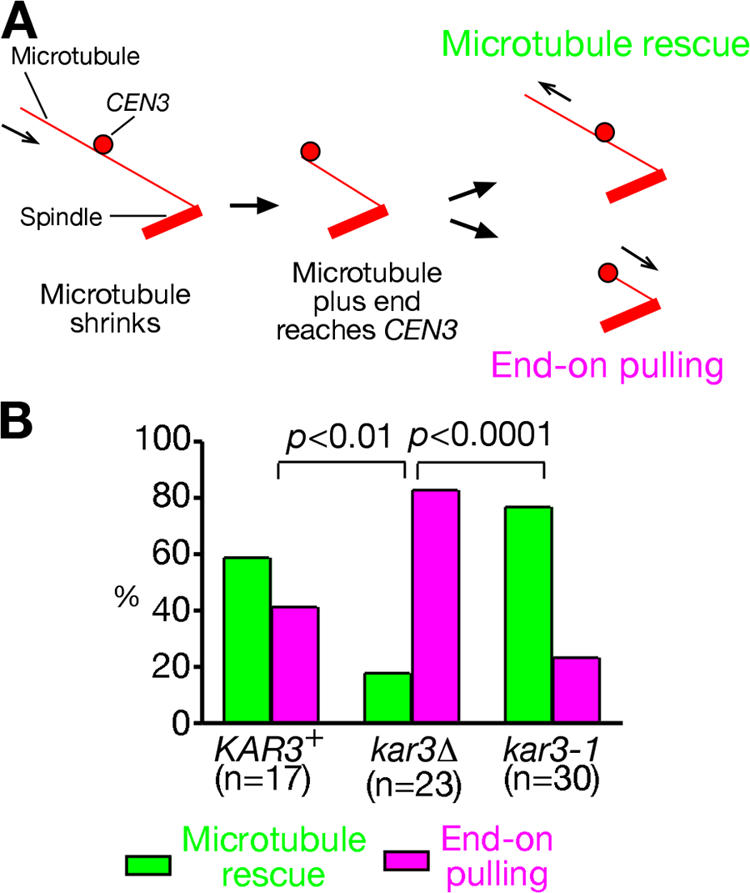
Kar3 partially suppresses the establishment of the microtubule end-on pulling of kinetochores. (A) Immediately after the distal end of a shrinking microtubule encountered CEN3 that had been on its lateral side, the microtubule showed rescue, and CEN3 stayed attached to the lateral surface (microtubule rescue); alternatively, CEN3 became bound to the plus end of the microtubule and was pulled poleward as the microtubule continued to shrink (end-on pulling). (B) Frequency of microtubule rescue (green bars) and end-on pulling (pink bars), as described in A, was scored in KAR3+, kar3Δ, and kar3-1 cells. T3531, T3860 (see Fig. 2). and T3319 (the same genotype as T3531 but kar3-1) cells were treated as in Fig 1.
When microtubule plus ends reach CEN3 after microtubule shrinkage, how do cells choose between these two options? We addressed whether Kar3 affects this choice because end-on pulling was more frequently observed when Kar3 was not functional (Fig. 2 A). For this, we evaluated how frequently each option was chosen when microtubule plus ends reached CEN3 in kar3Δ cells (Fig. 4 B). We found that kar3Δ cells showed a more frequent establishment of end-on pulling than KAR3+ cells, suggesting that Kar3 can suppress the establishment of end-on pulling when microtubule plus ends reached CEN3 (supplemental note 9 and Fig. S2 C).
How does Kar3 partially suppress the establishment of the microtubule end-on pulling of CEN3? We speculated that Kar3 might anchor CEN3 to the microtubule lateral surface (close to microtubule plus ends) and that this might hinder CEN3 from forming a specific attachment required for end-on pulling. To test this idea, we used the kar3-1 mutant, which can bind microtubule lateral surfaces but cannot work as a motor because of an ATP hydrolysis defect (Meluh and Rose, 1990; Maddox et al., 2003; K. Tanaka et al., 2005). In contrast to kar3Δ cells, kar3-1 mutant cells showed a strong reduction in the frequency of establishing end-on pulling (Fig. 4 B, compare kar3-1 with kar3Δ; and supplemental note 10). This result also explains why microtubule-dependent poleward CEN3 transport was more severely delayed in kar3-1 than in kar3Δ (K. Tanaka et al., 2005). The kar3-1 mutant cannot facilitate CEN3 sliding along microtubules as a result of its defective motor activity yet considerably inhibits the establishment of the microtubule end-on pulling of CEN3 in contrast to kar3Δ, thus causing a considerable delay in CEN3 transport (supplemental notes 10 and 11).
Characterizing microtubule depolymerization during end-on pulling
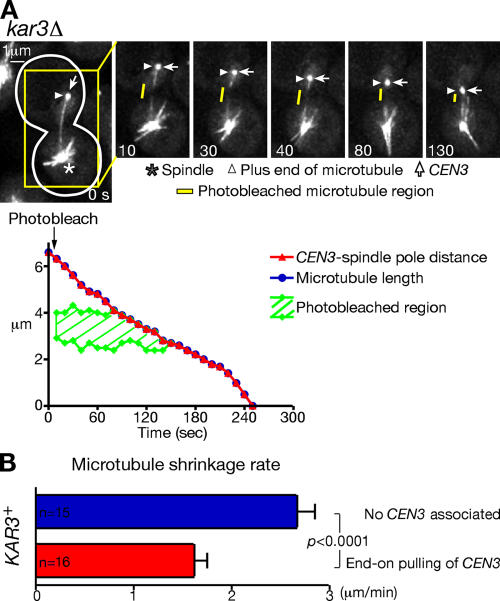
We next addressed which microtubule end underwent depolymerization during the microtubule end-on pulling of CEN3. We marked a microtubule region midway between CEN3 (bound at the microtubule plus end) and a spindle pole by photobleaching the YFP-Tub1 signal (Fig. 5 A). The distance between the photobleached region and a spindle pole did not substantially change as CEN3 was pulled poleward by microtubule shrinkage. Thus, depolymerization occurred at the plus end (i.e., bound to CEN3) but not substantially at the minus end (i.e., at a spindle pole). This is consistent with other results indicating that microtubules are dynamic only at the plus ends in budding yeast (Maddox et al., 2000; K. Tanaka et al., 2005).
Figure 5.
Microtubule dynamics during the end-on pulling of kinetochores. (A) Microtubules depolymerize at their plus ends during the end-on pulling of CEN3. kar3Δ (T3860; see Fig 2) cells were treated as in Fig 1. A microtubule region midway between CEN3 (bound at the microtubule plus end) and a spindle pole was photobleached between time points 0 and 10 s. GFP (CEN3) and YFP (tubulin) signals were acquired together every 10 s. Yellow bars show a photobleached region. The graph shows the distance of CEN3 from a spindle pole (red), length of the microtubule that captured CEN3 (blue), and the photobleached region (green) during the microtubule end-on pulling of CEN3. (B) Microtubule shrinkage rate in the absence of CEN3 associated with the microtubule (blue) and during the microtubule end-on pulling of CEN3 (red). KAR3+ (T3531; see Fig 2) cells were treated as in Fig 1. Error bars represent SEM.
Subsequently, we compared the microtubule depolymerization (shrinkage) rate in the presence of CEN3 associated with the microtubule end (i.e., during end-on pulling) and in the absence of CEN3 association with microtubules (supplemental note 12). The shrinkage rate was significantly slower during the end-on pulling (Fig. 5 B), suggesting that the presence of kinetochores at the microtubule plus ends somehow reduced the rate of microtubule depolymerization at those ends.
In addition, because Kar3 and Kip3 (a kinesin-8 family member) facilitate microtubule depolymerization in vitro (Endow et al., 1994; Chu et al., 2005; Sproul et al., 2005; Gupta et al., 2006; Varga et al., 2006) and foster microtubule disassembly in vivo (Huyett et al., 1998; Hildebrandt and Hoyt, 2000; Severin et al., 2001; West et al., 2001), we addressed whether these motor proteins affect the microtubule shrinkage rate (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1). Deletions of kar3 or kip3 and the kar3 kip3 double mutant did not significantly change the microtubule shrinkage rate either when CEN3 was transported by microtubule end-on pulling or in the absence of CEN3 association. Nonetheless, the mean length of nuclear microtubules was longer in kar3Δ and kip3Δ cells than in wild-type cells (unpublished data) as a result of a reduced frequency of microtubule catastrophe (conversion from growth to shrinkage) in these cells (unpublished data).
The Dam1 complexes along a microtubule are collected at the microtubule plus end as the microtubule depolymerizes
We next studied what factors facilitate the microtubule end-on pulling of kinetochores. The Dam1 complex was a candidate for the following reasons: (1) the Dam1 complex is not required for kinetochore sliding (K. Tanaka et al., 2005) but, nonetheless, becomes associated with kinetochores before sister kinetochores biorient in metaphase (Cheeseman et al., 2001a; Janke et al., 2002; Li et al., 2002); (2) the complex forms a ring encircling microtubules in vitro (Miranda et al., 2005; Westermann et al., 2005) and, if this is the case in vivo, it could tether kinetochores at microtubule plus ends while the complex is pushed poleward as microtubule protofilaments splay out during microtubule shrinkage (Fig. 6 A); and (3) dam1 and kar3 mutants show a synergistic effect on chromosome loss (supplemental note 13), which could be explained if both factors are involved in microtubule-dependent kinetochore transport.
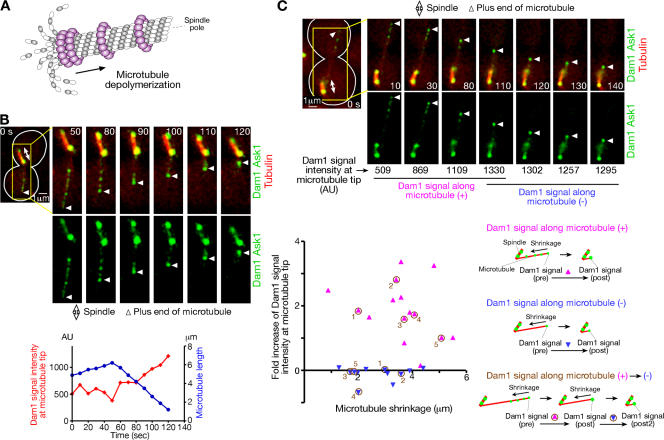
Figure 6.
Localization of the Dam1 complexes at the microtubule plus end. (A) Schematic diagram of microtubule depolymerization and possible localization of the Dam1 complexes. (B) The Dam1 complexes accumulate at the end of a microtubule as it depolymerizes. (top) Representative time-lapse images of Dam1 complex localization during microtubule shrinkage. (bottom) Length of the microtubule (blue) and intensity of Dam1 and Ask1 GFP signals at the microtubule distal end (red). DAM1-4GFP ASK1-4GFP CFP-TUB1 PGAL-CEN3-tetOs TetR-3CFP PMET3-CDC20 cells (T4991) were treated as in Fig. 1 except that glucose-containing media were used throughout the experiment (i.e., CEN3 was continuously active). CFP (CEN3, tubulin; red) and GFP (the Dam1 complex; green) images were acquired every 10 s. AU, arbitrary unit. See Video 3. (C) The Dam1 complexes along a microtubule are collected at its plus end. T4991 cells were treated as in B. The Dam1 complex signal (Dam1-4GFP and Ask1-4GFP) at the microtubule plus end was quantified, and its fold increase during microtubule shrinkage was plotted against the extent of this shrinkage in the presence (pink) and absence (blue) of the Dam1 complexes along the microtubule. Fold increase was calculated by dividing the increase of the signal intensity (pre→post or post→post 2) by pre-signal intensity. Numbers in brown represent each microtubule in which the fold increase was measured consecutively (microtubule #3 is shown in the images). For more information, see supplemental note 31 (available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1).
To gain more clues as to the function of the Dam1 complex, we visualized the complex along microtubules by tagging both Dam1 and Ask1, two components of the complex, with four tandem copies of GFP. The Dam1 complex showed multiple punctate GFP signals along microtubules. Intriguingly, the intensity of the GFP signals at the distal ends of microtubules increased during microtubule shrinkage (Fig. 6 B and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1). We also compared this increase when GFP signals were either present or absent along the extent of microtubule shrinkage (Fig. 6 C). In their presence, the GFP signal intensity at the microtubule tip increased more evidently. These results suggest that GFP spots along microtubules become collected by the microtubule ends. The simplest interpretation is that the Dam1 complexes form a ring encircling a microtubule in vivo and are collected by splaying protofilaments (Fig. 6 A and see Discussion). Moreover, during the microtubule end-on pulling of CEN3, the Dam1 complex continuously colocalized with CEN3 (Fig. 7 A and Video 4). However, such continuous colocalization was not observed during the sliding of CEN3 along microtubules (Fig. 7 B and Video 5), which is consistent with the Dam1 complex not being required for CEN3 sliding along microtubules (K. Tanaka et al., 2005).
Figure 7.
Continuous colocalization of the Dam1 complex with a centromere during end-on puling but not during sliding. (A and B) The Dam1 complexes continuously colocalize with CEN3 during end-on pulling (A) but not during sliding (B). T4991 cells (see Fig. 6) were treated as in Fig. 1, and CFP and GFP images were acquired. See Videos 4 and 5 (available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1).
The Dam1 complex facilitates the microtubule end-on pulling of kinetochores
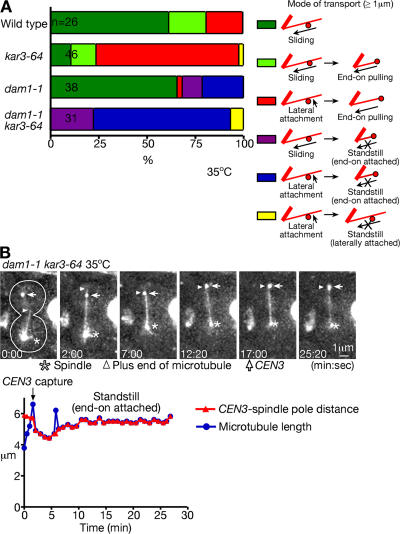
We subsequently investigated the roles of the Dam1 complex in kinetochore transport by impairing the function of the complex. If the Dam1 complex facilitates the microtubule end-on pulling of kinetochores, then this, the main kinetochore transport mode in kar3Δ cells, would be defective when the Dam1 complex is impaired. To test this, we attempted to combine kar3Δ with temperature-sensitive mutants of the Dam1 complex components dam1-1, ask1-3, and spc34-3. However, all of these combinations were lethal even at a permissive temperature for each temperature-sensitive mutant. Therefore, we used the kar3-64 temperature-sensitive mutant instead of kar3Δ (supplemental note 14). Only the dam1-1 kar3-64 double mutant was viable, whereas the other two combinations were lethal.
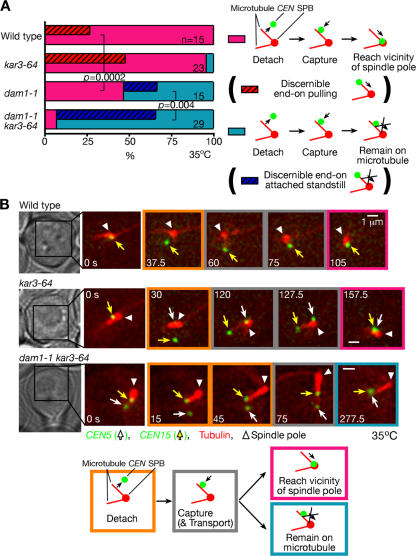
We analyzed CEN3 transport in dam1-1 and kar3-64 single mutants and in the dam1-1 kar3-64 double mutant at 35°C, which is a restrictive temperature for these mutant alleles. In the kar3-64 single mutant, CEN3 was transported toward spindle poles by microtubule end-on pulling in the majority (74%) of cells (Fig. 8 A), which is consistent with the behavior of kar3Δ cells (Fig. 2 A). In the dam1-1 single mutant, lateral sliding was observed in 66% of the cells, similar to the wild type. However, the amount of successful end-on pulling was dramatically reduced to only 3%. The remainder of the dam1-1 cells ended up in a standstill state, with CEN3 at the end of a microtubule that did not shrink any further (similar to the cell in Fig. 8 B; supplemental note 15). This suggests that Dam1 is required for successful end-on pulling. This conclusion was supported by the behavior of the dam1-1 kar3-64 double mutant, in which virtually all cells ended up in the standstill state (Fig. 8 B and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1). These results suggest that although Kar3 is required for lateral sliding, the Dam1 complex has an essential role in the microtubule end-on pulling of kinetochores.
Figure 8.
Both Kar3 and the Dam1 complex are involved in kinetochore transport. DAM1 + KAR3+ wild-type (T3531), dam1-1 (T2897), kar3-64 (T4034), and dam1-1 kar3-64 (T4044) cells with YFP-TUB1 PGAL-CEN3-tetOs TetR-GFP PMET3-CDC20 were treated as in Fig. 1 but with cultures shifted to 35°C 30 min before transfer to glucose-containing medium. Images were acquired every 10 s at 35°C. (A) Percentages of cells in which CEN3 was transported with each mode as colored/illustrated. Sliding, end-on pulling, and standstill were scored as in Fig. 2 A. (B, top) Representative time-lapse GFP and YFP images (acquired together) in T4044 cells. (bottom) CEN3–spindle pole distance (red) and length of the microtubule capturing CEN3 (blue). See Video 6 (available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1).
How could the Dam1 complex facilitate the microtubule end-on pulling of kinetochores? Given that the Dam1 complexes are able to form ring structures, it is likely that during microtubule depolymerization, the Dam1 complex has a key role in converting the splaying out of depolymerizing microtubule protofilaments into the kinetochore pulling force toward a spindle pole (see Fig. 10), as recently suggested by in vitro studies (Asbury et al., 2006; Westermann et al., 2006). A simple model would be that kinetochores are tethered to the Dam1 complex during this process. This model is supported by three pieces of evidence: (1) in the Dam1 complex component mutant spc34-3, 20% of cells showed the detachment of CEN3 from microtubules (Fig. S3 B); (2) once kinetochores were captured by microtubules, subsequent microtubule depolymerization with nocodazole did not abolish association of the Dam1 complex with kinetochores (Li et al., 2002); and (3) the Dam1 complex directly associates with the kinetochore complex Ndc80 (Shang et al., 2003). Nonetheless, only a few dam1-1 cells showed the detachment of CEN3 from microtubules (Fig. S3 B). We suggest that any residual function of the dam1-1 mutant protein was sufficient to maintain CEN3-microtubule attachment. Consistent with this notion, another of the Dam1 complex components still localized at CEN3 during the microtubule end-on attachment of CEN3 in dam1-1 cells (supplemental note 16), and centromeres became detached from microtubules more frequently when dam1-1 was converted into a more defective mutant (supplemental note 16 and Fig. S4 A; available at http://www.jcb.org/cgi/content/full/jcb.200702141).
Figure 10.
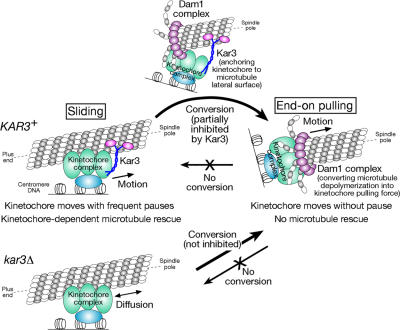
Model for how kinetochores are transported by spindle microtubules. Poleward kinetochore transport occurs in two distinct ways: lateral sliding, in which kinetochores move along the side of a microtubule, and end-on pulling, in which the kinetochore is attached to the end of a microtubule and is pulled poleward as the microtubule shrinks. Kar3 is essential to drive poleward lateral sliding, whereas the Dam1 complex is crucial for end-on pulling. Kar3 partly suppresses the establishment of end-on pulling by anchoring kinetochores to the microtubule lateral surface. In the absence of Kar3 (kar3Δ), kinetochores show diffusion along the microtubule lateral surface.
Kar3 and the Dam1 complex facilitate kinetochore transport by microtubules in normal S phase
To analyze microtubule-dependent kinetochore transport at high resolution, we have so far studied this process using centromere reactivation in metaphase-arrested cells (Fig. 1 A). Next, we wanted to study whether Kar3 and the Dam1 complex also have important roles in the transport of authentic centromeres (i.e., without regulation by an adjacent GAL1-10 promoter) and in normal cell cycles (i.e., without cell cycle arrest).
Recently, we have been able to visualize the transient detachment of GFP-marked centromeres from microtubules in early S phase (supplemental note 17; our unpublished data). This detachment continued for 0.5–1.5 min, during which centromeres typically moved up to 1.4 ± 0.6 μm (mean ± SD) from a spindle pole. After detachment, centromeres were recaptured by microtubules and swiftly (in 0.5–1.0 min) returned to the vicinity of a spindle pole (<0.6 μm from the spindle pole; our unpublished data).
These results prompted us to compare the behavior of centromeres, which detached from microtubules in early S phase in wild-type, dam1-1, and kar3-64 single mutant and dam1-1 kar3-64 double mutant cells. We labeled CEN5, CEN15, and microtubules by CFP, GFP, and YFP, respectively. Before cells started budding (i.e., before entry into S phase), both CENs localized close to a spindle pole in wild-type and kar3-64 cells but often somewhat more distant (>1.0 μm) from a pole while still attached to microtubules in dam1- 1 and dam1-1 kar3-64 cells. Nonetheless, in all four kinds of cells, both CENs equally showed detachment from microtubules when cells started budding (i.e., during S phase; supplemental note 17) and were recaptured by microtubules extending from a spindle pole after a similar time interval (0.5–1.5 min).
In both wild-type and kar3-64, after CENs were recaptured by microtubules, most centromeres promptly (in 0.5–1.0 min) moved to the vicinity of a spindle pole (Fig. 9 A, pink). Microtubule end-on pulling was more frequently discerned in kar3-64 cells than in wild-type cells (Fig. 9, A [red shaded areas] and B; and supplemental note 18), although in other cases, it was difficult in this experimental condition to distinguish CEN transport by sliding along microtubules and by end-on pulling. In dam1-1 and dam1-1 kar3-64, CENs were captured by microtubules but were not transported to the vicinity of a spindle pole in 53% and 93% of cells, respectively (Fig. 9 A, teal). In such cases, we often discerned that CENs attach at microtubule distal ends without being pulled toward a spindle pole (Fig. 9, A [blue shaded area] and B; and supplemental note 18). We also measured the velocity of CEN transport and found that it was distinctly altered by dam1-1 and kar3-64 mutations (supplemental note 19 and Fig. S4 B).
Figure 9.
Kar3 and the Dam1 complex facilitate poleward kinetochore transport in normal S phase. DAM1 + KAR3+ wild-type (T4243), dam1-1 (T5057), kar3-64 (T5058), and dam1-1 kar3-64 (T5056) cells with YFP-TUB1 CEN5-tetOs TetR-3CFP CEN15-lacOs GFP-LacI were treated with α factor and subsequently released to fresh media at 35°C. Images were acquired every 7.5 s. With the applied filter set (JP3 filter; see Materials and methods), CFP/GFP and YFP were separately visualized, and the two CENs could be distinguished, as CEN5-CFP showed lower intensity than CEN15-GFP. The behavior of CEN5 and CEN15 was analyzed when they detached from and were subsequently recaptured by microtubules. (A) Percentage of CENs that did (pink) and did not (teal) swiftly reach (<2 min) the vicinity of the spindle pole (<0.6 μm from the spindle pole). Graphs also indicate the percentage of cells in which we could discern microtubule end-on pulling (red shaded areas) and end-on attached standstill (blue shaded areas) of either CEN5 or CEN15 (supplemental note 17). P-values were obtained by comparing the number of CENs, which are categorized in pink and teal bars. (B) Representative time-lapse images of a wild-type cell that showed swift CEN15 transport, a kar3-64 cell that showed the end-on pulling of CEN15, and a dam1-1 kar3-64 cell that showed CEN5 standstill at the microtubule end. The other CEN was sometimes out of focus and is therefore not indicated. White arrows, yellow arrows, and white arrowheads indicate CEN5, CEN15, and spindle poles, respectively. For more information, see supplemental note 32 (available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1).
Thus, in dam1-1 and kar3-64 single and dam1-1 kar3-64 double mutant cells, we found distinct defects in the microtubule-dependent transport of CENs during normal S phase, which are consistent with our results from the centromere reactivation system. Our data suggest that Kar3 and the Dam1 complex have important roles in the microtubule-dependent poleward transport of authentic centromeres during the normal cell cycle (supplemental notes 20 and 21; Fig. S4 C).
Discussion
We have investigated the mechanisms by which kinetochores are transported toward spindle poles by microtubules in budding yeast. We show that poleward movement can occur in two distinct ways: lateral sliding, in which kinetochores move along the side of a microtubule, and end-on pulling, in which the kinetochore is attached to the end of a microtubule and is pulled poleward as the microtubule shrinks. Kar3 is essential to drive poleward lateral sliding, whereas the Dam1 complex is crucial for end-on pulling (Fig. 10).
It is thought that upon centromere DNA replication in budding yeast, kinetochores are disassembled, causing the release of centromeres from microtubules for a short period (Pearson et al., 2004; Tanaka, 2005; our unpublished data). Soon afterward, kinetochores are reassembled and captured by the lateral sides of microtubules extending from spindle poles (K. Tanaka et al., 2005). Microtubule lateral surfaces can secure initial kinetochore capture by providing a much larger contact surface compared with microtubule tips. The amount of Kar3 loaded at kinetochores probably increases while kinetochores are detached from microtubules (supplemental note 1; Tytell and Sorger, 2006). Kar3 (and the Dam1 complex) is not required for the initial kinetochore capture by microtubules (supplemental note 22; K. Tanaka et al., 2005), but, once kinetochores are captured, the motor activity of Kar3 drives kinetochore sliding along microtubule lateral surfaces toward spindle poles. Kar3 is the main and probably the sole factor driving this kinetochore sliding because in the absence of Kar3, kinetochores only show one-dimensional diffusion along microtubules.
Kinetochore sliding, which is promoted by Kar3, occurs toward a spindle pole with frequent pausing. This is perhaps because the Kar3 molecules loaded on kinetochores do not persistently drive their sliding along microtubules. This is a similar situation to that of Ncd, a putative Kar3 orthologue in Drosophila melanogaster shown to be a nonprocessive motor, whose domain is released from microtubules after each ATPase cycle and must bind microtubules repeatedly to drive motion (Endow, 2003). Probably because of this frequent pausing, the mean velocity of kinetochore sliding is lower than that of microtubule shrinkage (K. Tanaka et al., 2005). Therefore, microtubule plus ends often reach kinetochores unless microtubules are rescued and regrow before this happens, a process involving Stu2 transport from kinetochores (K. Tanaka et al., 2005).
If microtubule plus ends reach kinetochores, cells must choose one of the following two options: (1) microtubules show regrowth (i.e., are rescued), probably facilitated by Stu2 and other factors loaded at kinetochores (K. Tanaka et al., 2005), or (2) kinetochores are tethered at microtubule plus ends, probably as a result of association with the Dam1 complex ring structure, which has been pulled poleward as microtubules shrink. Kar3 reduces the frequency of the second choice (i.e., establishment of microtubule end-on pulling), probably by anchoring kinetochores to the microtubule lateral surface. In addition, the establishment of microtubule end-on pulling seems to be partly affected by stochastic elements (supplemental note 9). In any case, in the first option, kinetochores still remain associated with the lateral surface of microtubules and continue to slide poleward along microtubules. In the second, kinetochores at microtubule ends are continuously pulled poleward (end-on pulling) as the attached microtubules shrink without pausing or rescue. It is currently unclear how microtubule rescue is suppressed during end-on pulling, but it is not solely caused by a lack of Stu2 loaded on kinetochores (supplemental note 4).
The microtubule end-on pulling of kinetochores is facilitated by the Dam1 complex. In vitro experiments suggested that several Dam1 complexes could gather together and form a ring structure encircling a microtubule, which could move along the microtubule (Miranda et al., 2005; Westermann et al., 2005, 2006; Asbury et al., 2006). We found that the Dam1 complexes along a microtubule were collected at the plus ends of depolymerizing microtubules in vivo (supplemental note 23). The simplest interpretation would be that the Dam1 complexes indeed form a ring encircling a microtubule in vivo, which is pushed poleward by splaying protofilaments as the microtubule depolymerizes. However, it cannot be completely ruled out that the Dam1 complexes do not form a ring in vivo (McIntosh, 2005) and that unknown mechanisms collect the complexes along a microtubule at its plus end during microtubule shrinkage. In any case, when Dam1 function was impaired, the microtubule end-on pulling of kinetochores became defective. Our in vivo data support the model (Fig. 10) in which the Dam1 complex tethers kinetochores and plays a crucial role in converting microtubule depolymerization to kinetochore pulling force as initially proposed from the in vitro experiments.
Kinetochore sliding and microtubule end-on pulling are two distinct modes of microtubule-dependent kinetochore transport and seem to use different energy sources to produce the force necessary for kinetochore transport. When microtubules polymerize, the curvature of GDP-bound tubulin dimers is constrained by microtubule geometry so that the polymer lattice stores energy from GTP hydrolysis (Howard and Hyman, 2003). During end-on pulling, the free energy is released and converted to kinetochore pulling force by a power-stroke mechanism as microtubule protofilaments change from a straight to curved form (Grishchuk et al., 2005). The Dam1 complex apparently has an important role in this conversion (Fig. 10). In contrast, kinetochore sliding is driven by the Kar3 motor activity that is dependent on its ATP hydrolysis (i.e., additional energy is consumed; Yun et al., 2001). In spite of this, kinetochore sliding is less processive and achieves less efficient kinetochore transport.
Given these disadvantages, why do cells still use kinetochore sliding for their transport? Kinetochore sliding may have the following merits compared with end-on pulling: (1) for the establishment of microtubule end-on pulling, kinetochores must wait until the associated microtubule shrinks and the microtubule plus end finally reaches the kinetochore. Therefore, depending on the situation, kinetochores may reach a spindle pole earlier by sliding than by end-on pulling. (2) A single microtubule plus end is probably able to attach to only a single kinetochore during end-on pulling (Winey and O'Toole, 2001), but, in contrast, multiple kinetochores could be transported simultaneously by sliding (supplemental note 24). (3) Microtubule rescue, which happens during kinetochore sliding but not during end-on pulling, would increase the chance that kinetochores further afield are also captured by the same microtubule (supplemental note 25).
Because kinetochore sliding is converted into end-on pulling but not vice versa, the population of kinetochores attached to microtubule plus ends increases during poleward kinetochore transport. Both sister kinetochores subsequently interact with microtubules, and the Ipl1 kinase promotes the reorientation of kinetochore–microtubule attachment (T.U. Tanaka et al., 2002, 2005), in which the Dam1 complex is a crucial substrate of the kinase (Cheeseman et al., 2002). Because this reorientation happens in a tension-dependent manner (Nicklas, 1997; Dewar et al., 2004), sister kinetochores eventually attach to microtubules from opposite spindle poles (biorientation). To establish biorientation efficiently, kinetochores must be located within the spindle where microtubules extend from both spindle poles at high density. Because microtubule-dependent transport brings kinetochores close to the spindle, this process should facilitate efficient sister kinetochore biorientation.
The stable maintenance of biorientation crucially requires Dam1 complex function (Janke et al., 2002). Presumably, in metaphase, the Dam1 complex is necessary to pull sister kinetochores toward opposite spindle poles, generating tension across sister kinetochores and, in turn, stabilizing kinetochore–microtubule attachment (Dewar et al., 2004), thus simultaneously avoiding breakage of the attachment when this tension is applied (supplemental note 26). Metaphase is followed by anaphase A (Pearson et al., 2001), in which the kinetochore–spindle pole distance is shortened. We envisage that the Dam1 complex also plays the same role in anaphase A, as we found in prometaphase (i.e., tethering kinetochores at the microtubule plus ends and converting microtubule depolymerization [occurring at kinetochore sides; Maddox et al., 2000] into kinetochore pulling force). Consistent with this notion, we found that the Dam1 complex colocalizes with kinetochores during anaphase A (supplemental note 27; unpublished data).
Recently, the Dam1 complex orthologue was identified in fission yeast (Liu et al., 2005; Sanchez-Perez et al., 2005). In this organism, the Dam1 complex has important roles in sister kinetochore biorientation and kinetochore congression to the spindle midzone, which is consistent with Dam1 complex function in budding yeast. Moreover, kinetochores are still transported poleward in the absence of all of the known microtubule minus end–directed motors (i.e., two kinesin-14s and dynein) in fission yeast (Grishchuk and McIntosh, 2006); thus, perhaps kinetochores are transported by end-on pulling in this organism, as we have shown directly here in budding yeast.
In vertebrate cells, kinetochores are also captured by the lateral sides of single microtubules and are transported toward spindle poles in prometaphase (Rieder and Alexander, 1990). How is the kinetochore transport regulated in vertebrate cells? In contrast to mechanisms in budding yeast (supplemental note 8), vertebrate dynein could be involved in fast and processive kinetochore sliding along microtubules (supplemental note 28; Rieder and Alexander, 1990; King et al., 2000). If this is the case, the depletion of dynein may reveal the microtubule end-on pulling of kinetochores as a possible redundant mechanism for kinetochore transport in vertebrate cells, just as it was revealed by kar3Δ in yeast. Although convincing orthologues of the Dam1 complex components have not yet been identified in vertebrate cells (Meraldi et al., 2006), functional counterparts of the Dam1 complex may have an important role in microtubule end-on pulling. Kinesin-13s (mitotic centromere-associated kinesin, etc.) may be such functional counterparts because they also form rings encircling single microtubules in vitro (Moores et al., 2006; Tan et al., 2006), localize at kinetochores in mitosis (Wordeman, 2005), and act as important substrates of the aurora B kinase in ensuring proper kinetochore–microtubule attachment (Andrews et al., 2004; Lan et al., 2004; Ohi et al., 2004; Sampath et al., 2004).
Kinetochore capture and transport by spindle microtubules is the first crucial step for proper chromosome segregation in all eukaryotic cells. Comparison of kinetochore transport between different organisms will uncover the evolution of regulatory mechanisms for this fundamental cellular process.
Materials and methods
Yeast genetics and molecular biology
The background of yeast strains (W303), methods for yeast culture, and the centromere reactivation system were described previously (Dewar et al., 2004; Amberg et al., 2005; K. Tanaka et al., 2005). All tagging of yeast genes were performed at their C termini at their original gene loci except for the tagging of TUB1. CFP-TUB1 and YFP-TUB1 constructs were integrated at an auxotroph marker locus. Mutant alleles of spc34-3 (Janke et al., 2002), ask1-3 (Li et al., 2002), dam1-1 (Cheeseman et al., 2001b), and kar3-64 (Cottingham et al., 1999) were reported previously. Cells were cultured at 25°C in YP medium containing glucose unless otherwise stated. For more information, see supplemental note 29.
Microscopy
The procedures for time-lapse fluorescence microscopy were described previously (Dewar et al., 2004; K. Tanaka et al., 2005). Time-lapse images were collected every 15 s for 30 min at 23°C (ambient temperature) unless otherwise stated. For image acquisition, we used a microscope (DeltaVision RT; Applied Precision), a UPlanSApo 100× NA 1.40 objective lens (Olympus), a CCD camera (CoolSnap HQ; Photometrics), and SoftWoRx software (Applied Precision). We acquired three to seven (0.7 μm apart) z sections, which were subsequently deconvoluted, projected to two-dimensional images, and analyzed with SoftWoRx and Volocity (Improvision) software. CFP signals were discriminated from either YFP or GFP signals using the JP4 filter set (Chroma Technology Corp.). YFP signals were discriminated from CFP and GFP signals using the JP3 filter set (Chroma Technology Corp.). To collect GFP and YFP signals together without distinguishing the two, the YFP channel of the JP4 filter set was used.
Analyzing the dynamics of kinetochores and microtubules
To evaluate the length of microtubules and position of centromeres, we took account of the distance along the z axis as well as the distance on a projected image. To score modes of kinetochore transport in the CEN3 reactivation system (Figs. 2 A and 8 A), we selected cells in which CEN3 was captured by microtubules (CEN3–spindle pole distance was 2 μm or longer at the initial capture) and subsequently reached spindle poles during a time-lapse observation of 30-min duration. Cells were scored for sliding when CEN3 was transported along microtubules for 1 μm or longer (in most cases toward a spindle pole) and tubulin signal intensity was similar distal and proximal to CEN3 (i.e., CEN3 was not transported with end-on pulling by shorter overlapping microtubules; supplemental note 2). Cells were scored for microtubule end-on pulling when CEN3 was transported poleward for 1 μm or longer with CEN3 attached to the microtubule distal end. Standstill was scored when CEN3 was almost at the same position on microtubules for considerable time (see supplemental note 30 for details). The rate of microtubule growth and shrinkage was evaluated only for approximate linear changes (R2 > 0.85 in linear regression analyses) of microtubule lengths >3 μm. MSD was calculated as described in Helenius et al. (2006). Statistical analyses were performed with the Fisher's exact test (Figs. 4 B, 9 A, and S4 A) or with an unpaired t test (all others) using Prism software (GraphPad) unless otherwise stated. All p-values are two tailed. All error bars in figures represent SEM. For more information, see supplemental note 30.
Online supplemental material
Supplemental notes 1–32 describe more results, discussions, and methods. Fig. S1 A shows kinetochore transport in cells in which microtubule plus ends are labeled, and B shows Stu2 localization at CEN3 during microtubule end-on pulling. Fig. S2 A shows the localization of motor proteins Cin8, Kip1, Kip2, Kip3, and Dyn1, B shows kinetochore transport in kar3 dyn1 and kar3 kip3 double mutants, and C shows Kar3 localization during microtubule end-on pulling. Fig. S3 A shows that Kar3 and Kip3 do not significantly affect the microtubule shrinkage rate, and B shows defects in kinetochore capture and transport in dam1, ask1, and spc34 mutants. Fig. S4 A shows that kinetochores sometimes detach from microtubules in dam1-1-td, B shows CEN transport velocity in normal S phase, and C shows Kar3 localization at kinetochores in normal S phase. Video 1 shows a kar3Δ cell showing the microtubule end-on pulling of CEN3 (video of the cell shown in Fig. 2 B). Video 2 shows the conversion of CEN3 sliding along a microtubule into microtubule end-on pulling (video of the cell shown in Fig. 2 E). Video 3 shows the Dam1 complexes accumulate at a microtubule plus end as it depolymerizes (video of the cell shown in Fig. 6 B). Video 4 shows that the Dam1 complexes continuously colocalize with CEN3 during microtubule end-on pulling (video of the cell shown in Fig. 7 A). Video 5 shows that the Dam1 complexes do not continuously colocalize with CEN3 during sliding along a microtubule (video of the cell shown in Fig. 7 B). Video 6 presents a dam1-1 kar3-64 cell showing the standstill of CEN3 attached at the plus end of a microtubule (video of the cell shown in Fig. 8 B). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200702141/DC1.
Supplementary Material
Acknowledgments
We thank J.J. Blow, L. Clayton, J. Howard, M.J.R. Stark, J.R. Swedlow, and members of the Tanaka laboratory for discussions and reading the manuscript and thank C. Allan and S. Swift for technical help for microscopy/computing. We also thank R. Ciosk, F. Uhlmann, K. Nasmyth, M.A. Hoyt, E. Schiebel, I.M. Cheeseman, G. Barnes, R. Tsien, S.J. Elledge, K. Bloom, J.E. Haber, M.D. Rose, M. Yanagida, A.W. Murray, A.F. Straight, T.N. Davis, EUROSCARF, and the Yeast Resource Centre for reagents.
This work was supported by Cancer Research UK, Wellcome Trust, the Human Frontier Science Program, the Lister Research Institute Prize, and the Association for International Cancer Research. T.U. Tanaka is a Senior Research Fellow of Cancer Research UK.
K. Tanaka's present address is Institute of Development, Aging, and Cancer, Tohoku University, Sendai 980-8575, Japan.
Abbreviation used in this paper: MSD, mean squared displacement.
References
- Amberg, D.C., D.J. Burke, and J.N. Strathern. 2005. Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 230 pp.
- Andrews, P.D., Y. Ovechkina, N. Morrice, M. Wagenbach, K. Duncan, L. Wordeman, and J.R. Swedlow. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268. [DOI] [PubMed] [Google Scholar]
- Asbury, C.L., D.R. Gestaut, A.F. Powers, A.D. Franck, and T.N. Davis. 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA. 103:9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., C. Brew, M. Wolyniak, A. Desai, S. Anderson, N. Muster, J.R. Yates, T.C. Huffaker, D.G. Drubin, and G. Barnes. 2001. a. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155:1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., M. Enquist-Newman, T. Muller-Reichert, D.G. Drubin, and G. Barnes. 2001. b. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152:197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Anderson, M. Jwa, E.M. Green, J. Kang, J.R. Yates III, C.S. Chan, D.G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. [DOI] [PubMed] [Google Scholar]
- Chu, H.M., M. Yun, D.E. Anderson, H. Sage, H.W. Park, and S.A. Endow. 2005. Kar3 interaction with Cik1 alters motor structure and function. EMBO J. 24:3214–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham, F.R., L. Gheber, D.L. Miller, and M.A. Hoyt. 1999. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 147:335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar, H., K. Tanaka, K. Nasmyth, and T.U. Tanaka. 2004. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 428:93–97. [DOI] [PubMed] [Google Scholar]
- Endow, S.A. 2003. Kinesin motors as molecular machines. Bioessays. 25:1212–1219. [DOI] [PubMed] [Google Scholar]
- Endow, S.A., S.J. Kang, L.L. Satterwhite, M.D. Rose, V.P. Skeen, and E.D. Salmon. 1994. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 13:2708–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, E.L., and J.R. McIntosh. 2006. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 25:4888–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk, E.L., M.I. Molodtsov, F.I. Ataullakhanov, and J.R. McIntosh. 2005. Force production by disassembling microtubules. Nature. 438:384–388. [DOI] [PubMed] [Google Scholar]
- Gupta, M.L., Jr., P. Carvalho, D.M. Roof, and D. Pellman. 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8:913–923. [DOI] [PubMed] [Google Scholar]
- Helenius, J., G. Brouhard, Y. Kalaidzidis, S. Diez, and J. Howard. 2006. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 441:115–119. [DOI] [PubMed] [Google Scholar]
- Hildebrandt, E.R., and M.A. Hoyt. 2000. Mitotic motors in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1496:99–116. [DOI] [PubMed] [Google Scholar]
- Howard, J., and A.A. Hyman. 2003. Dynamics and mechanics of the microtubule plus end. Nature. 422:753–758. [DOI] [PubMed] [Google Scholar]
- Huyett, A., J. Kahana, P. Silver, X. Zeng, and W.S. Saunders. 1998. The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J. Cell Sci. 111:295–301. [DOI] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, T.U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J.M., T.S. Hays, and R.B. Nicklas. 2000. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J. Cell Biol. 151:739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith, S.L., S. Sandall, and A. Desai. 2005. Kinetochore-spindle microtubule interactions during mitosis. Curr. Opin. Cell Biol. 17:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, W., X. Zhang, S.L. Kline-Smith, S.E. Rosasco, G.A. Barrett-Wilt, J. Shabanowitz, D.F. Hunt, C.E. Walczak, and P.T. Stukenberg. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286. [DOI] [PubMed] [Google Scholar]
- Li, Y., J. Bachant, A.A. Alcasabas, Y. Wang, J. Qin, and S.J. Elledge. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., I. McLeod, S. Anderson, J.R. Yates, and X. He. 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24:2919–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P.S., K.S. Bloom, and E.D. Salmon. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P.S., J.K. Stemple, L. Satterwhite, E.D. Salmon, and K. Bloom. 2003. The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr. Biol. 13:1423–1428. [DOI] [PubMed] [Google Scholar]
- Maiato, H., J. Deluca, E.D. Salmon, and W.C. Earnshaw. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 117:5461–5477. [DOI] [PubMed] [Google Scholar]
- McIntosh, J.R. 2005. Rings around kinetochore microtubules in yeast. Nat. Struct. Mol. Biol. 12:210–212. [DOI] [PubMed] [Google Scholar]
- McIntosh, J.R., E.L. Grishchuk, and R.R. West. 2002. Chromosome-microtubule interactions during mitosis. Annu. Rev. Cell Dev. Biol. 18:193–219. [DOI] [PubMed] [Google Scholar]
- Meluh, P.B., and M.D. Rose. 1990. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 60:1029–1041. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., A.D. McAinsh, E. Rheinbay, and P.K. Sorger. 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, J.J., P. De Wulf, P.K. Sorger, and S.C. Harrison. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143. [DOI] [PubMed] [Google Scholar]
- Moores, C.A., J. Cooper, M. Wagenbach, Y. Ovechkina, L. Wordeman, and R.A. Milligan. 2006. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 5:1812–1815. [DOI] [PubMed] [Google Scholar]
- Nicklas, R.B. 1997. How cells get the right chromosomes. Science. 275:632–637. [DOI] [PubMed] [Google Scholar]
- Ohi, R., T. Sapra, J. Howard, and T.J. Mitchison. 2004. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B-dependent phosphorylation. Mol. Biol. Cell. 15:2895–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C.G., P.S. Maddox, E.D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C.G., E. Yeh, M. Gardner, D. Odde, E.D. Salmon, and K. Bloom. 2004. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 14:1962–1967. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., and S.P. Alexander. 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 110:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath, S.C., R. Ohi, O. Leismann, A. Salic, A. Pozniakovski, and H. Funabiki. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 118:187–202. [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez, I., S.J. Renwick, K. Crawley, I. Karig, V. Buck, J.C. Meadows, A. Franco-Sanchez, U. Fleig, T. Toda, and J.B. Millar. 2005. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 24:2931–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton, M.J., and K. Jacobson. 1997. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26:373–399. [DOI] [PubMed] [Google Scholar]
- Severin, F., B. Habermann, T. Huffaker, and T. Hyman. 2001. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 153:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, C., T.R. Hazbun, I.M. Cheeseman, J. Aranda, S. Fields, D.G. Drubin, and G. Barnes. 2003. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 14:3342–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul, L.R., D.J. Anderson, A.T. Mackey, W.S. Saunders, and S.P. Gilbert. 2005. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr. Biol. 15:1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, D., A.B. Asenjo, V. Mennella, D.J. Sharp, and H. Sosa. 2006. Kinesin-13s form rings around microtubules. J. Cell Biol. 175:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., N. Mukae, H. Dewar, M. van Breugel, E.K. James, A.R. Prescott, C. Antony, and T.U. Tanaka. 2005. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 434:987–994. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U. 2005. Chromosome bi-orientation on the mitotic spindle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T.U., N. Rachidi, C. Janke, G. Pereira, M. Galova, E. Schiebel, M.J. Stark, and K. Nasmyth. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., M.J. Stark, and K. Tanaka. 2005. Kinetochore capture and bi-orientation on the mitotic spindle. Nat. Rev. Mol. Cell Biol. 6:929–942. [DOI] [PubMed] [Google Scholar]
- Tytell, J.D., and P.K. Sorger. 2006. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172:861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, V., J. Helenius, K. Tanaka, A.A. Hyman, T.U. Tanaka, and J. Howard. 2006. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8:957–962. [DOI] [PubMed] [Google Scholar]
- West, R.R., T. Malmstrom, C.L. Troxell, and J.R. McIntosh. 2001. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell. 12:3919–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, S., A. Avila-Sakar, H.W. Wang, H. Niederstrasser, J. Wong, D.G. Drubin, E. Nogales, and G. Barnes. 2005. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290. [DOI] [PubMed] [Google Scholar]
- Westermann, S., H.W. Wang, A. Avila-Sakar, D.G. Drubin, E. Nogales, and G. Barnes. 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 440:565–569. [DOI] [PubMed] [Google Scholar]
- Winey, M., and E.T. O'Toole. 2001. The spindle cycle in budding yeast. Nat. Cell Biol. 3:E23–E27. [DOI] [PubMed] [Google Scholar]
- Wordeman, L. 2005. Microtubule-depolymerizing kinesins. Curr. Opin. Cell Biol. 17:82–88. [DOI] [PubMed] [Google Scholar]
- Yun, M., X. Zhang, C.G. Park, H.W. Park, and S.A. Endow. 2001. A structural pathway for activation of the kinesin motor ATPase. EMBO J. 20:2611–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.