Abstract
We report the interactions amongst 20 proteins that specify their assembly to the centromere–kinetochore complex in human cells. Centromere protein (CENP)-A is at the top of a hierarchy that directs three major pathways, which are specified by CENP-C, -I, and Aurora B. Each pathway consists of branches that intersect to form nodes that may coordinate the assembly process. Complementary EM studies found that the formation of kinetochore trilaminar plates depends on the CENP-I/NUF2 branch, whereas CENP-C and Aurora B affect the size, shape, and structural integrity of the plates. We found that hMis12 is not constitutively localized at kinetochores, and that it is not essential for recruiting CENP-I. Our studies also revealed that kinetochores in HeLa cells contain an excess of CENP-A, of which ∼10% is sufficient to promote the assembly of normal levels of kinetochore proteins. We elaborate on a previous model that suggested kinetochores are assembled from repetitive modules (Zinkowski, R.P., J. Meyne, and B.R. Brinkley. 1991. J. Cell Biol. 113:1091–110).
Introduction
Faithful segregation of chromosomes during mitosis requires a dynamic interaction between spindle microtubules and the kinetochore, which is a macromolecular complex that localizes at the centromere of mitotic chromosomes. The kinetochore was originally identified by EM as a trilaminar stack of plates that formed along the outer surface of the centromere region of each sister chromatid. As part of the centromere–kinetochore complex, the inner centromere is defined as the region between the inner plates of apposing sister kinetochores, and it is occupied largely by centromeric heterochromatin. Extending away from the surface of the outer plate is an electron-dense cloud that is termed the fibrous corona (Cleveland et al., 2003; Maiato et al., 2004). Early EM studies revealed that trilaminar kinetochore structure is only visible from late prophase until the end of mitosis, suggesting that the kinetochore undergoes an assembly/disassembly cycle during each mitosis (Brenner et al., 1981; He and Brinkley, 1996).
Molecular studies over the past two decades have confirmed the notion that the centromere–kinetochore complex undergoes cell cycle–dependent changes. Currently, >100 proteins, many of which are evolutionarily conserved, have been reported to associate with the centromere–kinetochore complex in human cells (Chan et al., 2005; Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006). Some of the proteins are constitutively associated with centromeres throughout the cell cycle, whereas others are only transiently detected at the centromere–kinetochore complex from late G2 to telophase. The cell cycle–dependent localization of proteins to the kinetochore is consistent with the EM data that showed kinetochores are assembled and disassembled during mitosis. Moreover, observations made in human cells showing that different proteins exhibit a distinct temporal order of appearance at the kinetochore suggested that the trilaminar plates may be assembled in a stepwise fashion. In human cells, CENP-A, -C, -H, and -I (hMis6) are constitutive centromere proteins that become part of the inner plate of the mature kinetochore. These proteins are therefore likely to participate in the earliest steps of kinetochore assembly, which is thought to initiate during G2, after centromeric chromatin have replicated. Although kinetochore structures are not visible at this time, proteins such as hZwint-1, BUB1, Aurora B, MCAK, and CENP-F begin to accumulate at the nascent centromere–kinetochore complex. Immediately after nuclear envelope breakdown, when trilaminar plates are first visible, a host of proteins that are important for microtubule binding and checkpoint control, including the dynein–dynactin complex, CENP-E, CDC20, MAD1, MAD2, BUBR1, hMPS1, hZW10, and hROD assemble onto the mature kinetochore (Maiato et al., 2004; Chan et al., 2005).
Despite the fact that proteins exhibit a temporal order of assembly at kinetochores, the cumulative data does not support a single linear assembly pathway. It appears that kinetochore assembly in human cells, as well as in a variety of model organisms, is complex (Blower and Karpen, 2001; McAinsh et al., 2003; Amor et al., 2004; Cheeseman et al., 2004; Hauf and Watanabe, 2004; Maiato et al., 2004; Vigneron et al., 2004; Chan et al., 2005). For example, CENP-I is a constitutive protein that specifies the assembly of CENP-F, MAD1, and MAD2, but not BUB1, BUBR1, hZW10, and hROD (Liu et al., 2003b). Interestingly, kinetochore localizations of CENP-F and MAD2 (as with CENP-E and BUBR1) were reported to depend on BUB1 (Johnson et al., 2004), although BUB1 localization is independent of CENP-I. This suggests that multiple pathways may be necessary to recruit some proteins to the kinetochore.
Although there are numerous studies describing the relationships amongst selected kinetochore proteins, our current understanding of how proteins come together to construct the kinetochore trilaminar structure remains fragmentary (Tomkiel et al., 1994; Salina et al., 2003; Deluca et al., 2005). No single study of a large set of proteins has been conducted to achieve a global view of kinetochore assembly. To integrate findings reported by different labs and to advance our understanding of how kinetochores are assembled, we examined the relationships amongst twenty human centromere–kinetochore proteins and their contributions toward their organization at the ultrastructural level in HeLa cells.
Results
Overview of the kinetochore assembly pathways
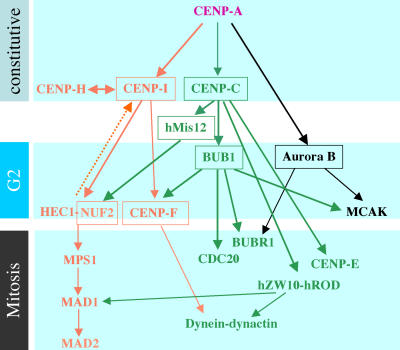
We chose to examine the relationships amongst twenty of the best-characterized proteins as the first step toward mapping the pathways for kinetochore assembly. These proteins include the constitutive centromere proteins CENP-A, -B, -C, -H, and -I; the inner centromere protein Aurora B; the microtubule-interacting proteins dynein–dynactin complex (represented by p150glued); CENP-E, -F (Feng et al., 2006), and MCAK; and the mitotic checkpoint proteins BUB1, BUBR1, MAD1, MAD2, hMPS1, hZW10, and hROD, as well as HEC1, NUF2, and hMis12. From this group, Aurora B was classified as a “chromosomal passenger protein” or “inner centromere protein” (Adams et al., 2001), but its role in kinetochore–microtubule attachment, mitotic checkpoint signaling, and recruitment of kinetochore proteins (Murata-Hori and Wang, 2002; Ditchfield et al., 2003; Hauf et al., 2003; Andrews et al., 2004; Lan et al., 2004) led us to include it in our analysis.
The relationships between the aforementioned proteins were established by depleting specific proteins by siRNA and examining the effects on the localization of other proteins. Representative results of RNAi-induced knockdown of specific proteins and the specificities of several new antibodies developed during this study are shown in Fig. S1 (available at http://www.jcb.org/cgi/content/full/jcb.200606020/DC1). Table I summarizes our epistasis analysis of the 20 kinetochore proteins. A genetic interaction map (Fig. 1) was constructed based on Table I and other studies (Martin-Lluesma et al., 2002; Ditchfield et al., 2003; Hauf et al., 2003; Liu et al., 2003b; Andrews et al., 2004; Kops et al., 2005). The proteins are organized based on their relative temporal order of appearance (top to bottom), as documented in previous studies (Jablonski et al., 1998; Chan et al., 2000; Johnson et al., 2004) or uncovered in this study; BUB1 and Aurora B, HEC1, and CENP-F, respectively, appear at prekinetochores around the same time during late G2 (unpublished data). The reader should refer to this map as we present the details of our experiments. A more comprehensive map may be obtained upon request.
Table I.
Defining the dependency among proteins for kinetochore assembly
| siRNA target |
Effects on kinetochore localizationa | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CENP-A | CENP-B | CENP-C | CENP-E | CENP-F | CENP-H | CENP-I | hMIS12 | Aurora B | MCAK | BUB1 | BUBR1 | MAD1 | MAD2 | hMPS1 | hZW10 | hROD | p150glued | HEC1 | Nuf2 | CDC20 | |
| CENP-A | + | + | + | + | + | ||||||||||||||||
| CENP-B | + | − | |||||||||||||||||||
| CENP-C | + | + | − | + | − | + | + | + | + | + | + | + | + | ||||||||
| CENP-H | − | + | + | − | |||||||||||||||||
| CENP-I | − | − | − | − | + | + | + | − | − | − | − | − | + | + | + | − | − | + | + | + | |
| hMIS12 | − | − | − | + | − | − | + | − | + | ||||||||||||
| Aurora B | − | − | − | − | + | + | − | + | − | − | − | − | − | − | |||||||
| hMPS1 | − | − | − | − | + | + | + | − | − | − | |||||||||||
| BUB1 | − | + | − | − | + | + | + | − | + | − | − | − | − | − | − | + | |||||
Kinetochore localization was examined by immunofluorescence and compared between control and target siRNA–transfected cells. + and – stand for the dependency or nondependency, respectively, between two proteins. Not all possible combinations were tested (blanks) because they were either already reported or because they can be inferred based on the assembly pathway.
bRefer to the main text for the complex hMis12–CENP-I relationship.
Figure 1.
A network of intersecting pathways that specify formation of a kinetochore module. Inner kinetochore proteins are positioned at the top. Proteins are also arranged (top to bottom) with respect to their relative temporal order of appearance at kinetochores (the temporal sequence for mitotic kinetochore proteins is not very clear, except that BUBR1 appears before CENP-E). Thick solid arrows show connections examined in this study; thinner arrows indicate previous published interactions that were not elaborated on in this study. The dashed arrow denotes a potential feedback mechanism between CENP-I and the HEC1–NUF2 complex. Boxes denote proteins whose roles in kinetochore assembly were examined by EM.
Our map shows that the centromere-specific histone H3 variant CENP-A occupies the top of a hierarchy that directs three major assembly pathways that are specified by CENP-C, -I, and Aurora B. The pathways are not linear, but contain multiple branches that intersect to form a network that defines the spatial and temporal relationships amongst the proteins that were examined. No direct physical interaction between proteins is implied, although that is certainly possible, as in the cases of CENP-H–HEC1–NUF2 and hMis12–HEC1–NUF2 (Cheeseman et al., 2004; Obuse et al., 2004; Mikami et al., 2005). The contribution of seven proteins to the ultrastructure of the kinetochore was examined by EM (Table II and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200606020/DC1).
Table II.
Defects in kinetochore ultrastructure resulting from depletion of specific kinetochore proteinsa
| siRNA | Kinetochores with triplate and >4 associated mts (normal) |
Triplate with few (<4) or no associated mts |
Partial plates and/or pulled out plates and/or fibrillar structure at outer plates |
Fuzzy ball | Small kinetochore plates |
Thin and/or punctate plates |
Subjacent chromatin decondense and/or C-shaped plates |
Total number of kinetochores counted |
|---|---|---|---|---|---|---|---|---|
| CENP-I | (no mts) 5 | (<4 mt) 10 | (no mts) 12 | 38 | ||||
| (<4 mt) 3 | (≥4 mts) 5 | (ama) 1 | ||||||
| (ama) 2 | ||||||||
| NUF2 | 1 | (no mts)( 2 (<4 mts) 6 | (<4 mt) 27 (≥4 mts) 4 | (no mts) 37 (<4 mts) 5 (ama) 1 | 83 | |||
| NUF2 | 1 | (no mts) 2 | (<4 mt) 27 | (no mts) 37 | 83 | |||
| (<4 mts) 6 | (≥4 mts) 4 | (<4 mts) 5 | ||||||
| (ama) 1 | ||||||||
| CENP-F | 4 | (<4 mt) 4 | (ama) 3 | 12 | ||||
| (≥4 mts) 1 | ||||||||
| CENP-C | 2 | (no mts) 4 | (<4 mt) 5 | (no mts) 1 | (<4 mts) 4 | (no mts) 7 | 50 | |
| (<4 mts) 9 | (≥4 mts) 1 | (<4 mts) 5 | (<4 mts) 1 | |||||
| (ama) 2 | (≥4 mts) 7 | |||||||
| (ama) 2 | ||||||||
| BUB1 | 9 | (no mts) 2 | (<4 mt) 1 | (no mts) 6 | 22 | |||
| (<4 mts) 1 | (ama) 1 | (<4 mts) 1 | ||||||
| (ama) 1 | ||||||||
| hMis12 | 3 | (no mts) 2 | (<4 mt) 2 | (no mts) 3 | 18 | |||
| (<4 mts) 1 | (<4 mts) 5 | |||||||
| (ama) 2 | ||||||||
| Aurora B | (no mts) 7 | (<4 mt) 3 | (no mts) 3 | 67 | ||||
| (<4 mts) 3 | (ama) 3 | (<4 mts) 24 | ||||||
| (≥4 mts) 10 | ||||||||
| (ama) 14 | ||||||||
| Examples | Fig. 5 F, d | Fig. 2 G | Fig. 2, D–F | Fig. 5 F, a | Fig. 5 F, c and e | Fig. 7 F, a | ||
| Fig. 5 F, b |
Microtubule (Mts) counts are based on a single section. A section with >4 microtubules is considered normal. ama, aberrant Mts association.
The CENP-I assembly pathway includes HEC1, NUF2, and hMPS1
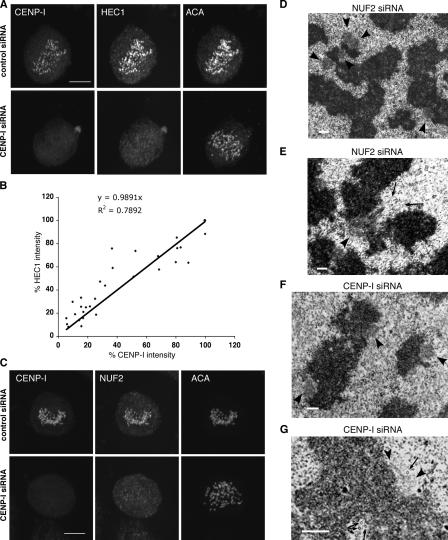
CENP-I was previously shown to specify the assembly of CENP-F, MAD1, and MAD2, but not the localization of BUB1, BUBR1, hZW10, and hROD (Liu et al., 2003b). As the localization of MAD1, MAD2, and hMPS1 to kinetochores was reported to depend on HEC1 (Martin-Lluesma et al., 2002), we tested and found that HEC1 localization depends on CENP-I (Fig. 2 A). Examination of cells that differed in the amounts of CENP-I depletion showed that the fluorescence intensities between CENP-I and HEC1 exhibited a relationship near 1:1, which was held over a 20-fold range (Fig. 2 B). Consistent with the fact that HEC1 is part of a complex with NUF2 (Bharadwaj et al., 2004; McCleland et al., 2004), we found that the localization of NUF2 was also sensitive to CENP-I level (Fig. 2 C). These results are consistent with those reported by Hori et al. (2003), who showed in chicken DT-40 cells that the localization of HEC1-GFP and NUF2-GFP at kinetochores requires CENP-I.
Figure 2.
Localization of HEC1 complex depends on CENP-I. (A and C) Control or CENP-I–depleted HeLa cells (top and bottom rows, respectively) were costained with CENP-I, anticentromere antibody (ACA), and HEC1 (A) or NUF2 (C) antibodies. Images are presented as maximum projections of z series. (B) HEC1 fluorescence intensities on kinetochores with different levels of CENP-I (31 kinetochores in 10 cells) were quantitated, normalized to the brightest signal (100%), and plotted against corresponding CENP-I intensities. (D–G) Representative EM images of kinetochores in NUF2 (D–E) and CENP-I (F–G) siRNA-transfected cells. (D and F) Fuzzy ball–shaped kinetochores (arrowheads) lacking microtubules are commonly seen. (E) Microtubules (arrows) interact laterally with a fuzzy kinetochore (arrowhead). (G) A kinetochore with half plate (left arrowhead) and half fuzzy ball (right arrowhead) structure. Some microtubules (arrows) pass perpendicularly to the section plane. Bars: (A and C) 10 μm; (D–G) 400 nm.
We also found that the localization of hMPS1 at kinetochores was dependent on CENP-I (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200606020/DC1). Thus, CENP-I specifies the following assembly pathway: CENP-I → HEC1–NUF2 complex → hMPS1 → MAD1 → MAD2, which links the inner kinetochore with the outer kinetochore. The position of the HEC1–NUF2 complex in this pathway is consistent with our observation that HEC1 was first detected at kinetochores in late G2, before the appearance of hMPS1, MAD1, and MAD2 (unpublished data). Although we previously showed that the localization of CENP-F and p150glued also depend on CENP-I (Fig. S2 B; Liu et al., 2003b), our studies and those reported by others (DeLuca et al., 2002; Holt et al., 2005) indicated that neither belongs to HEC1–NUF2 pathway. Nonetheless, CENP-F may regulate the kinetochore localization of the dynein–dynactin complex (Yang et al., 2005). Thus, CENP-I specifies at least two separate assembly branches for recruiting CENP-F and the HEC1–NUF2 complex.
Kinetochore ultrastructure in NUF2- and CENP-I–depleted cells
EM studies showed that depletion of NUF2 from HeLa cells resulted in disorganized kinetochores with poorly defined outer plates (Deluca et al., 2005). We made similar observations, as >50% of the kinetochores (43 out of 83) examined in NUF2-depleted cells lacked trilaminar plates, but instead displayed a “fuzzy ball”–shaped structure with few, if any, bound microtubules (Fig. 2 D and Table II). Consistent with Deluca et al.'s (2005) proposition that NUF2 may contribute to the end-on attachment of microtubules to kinetochore, we observed microtubules in longitudinal profile associated with the surface of the fuzzy ball–shaped kinetochore masses in NUF2-depleted cells (Fig. 2 E).
Importantly, similar fuzzy ball structures or expanded kinetochore structures were detected in 30 out of 38 kinetochores scored in CENP-I siRNA–transfected cells (Fig. 2, F– G, Table II). Kinetochores with defects of different extent were observed even in the same cell (Table S1), which was attributed to the variability in the extent or timing of protein depletion mediated by siRNA. These EM data nonetheless provide further confirmation that CENP-I and NUF2 lie in the same assembly pathway and contribute to the formation of the higher order trilaminar plate structure.
We also examined kinetochore structures in CENP-F–depleted cells (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200606020/DC1) to see if this portion of the CENP-I pathway contributed toward trilaminar plate formation. Consistent with light microscopy data from recent CENP-F siRNA studies (Bomont et al., 2005; Holt et al., 2005; Yang et al., 2005; Feng et al., 2006), microtubules were attached to most of these kinetochores. Kinetochores displayed a variety of morphologies, including an uncondensed fuzzy ball or pulled out and fibrillar appearance (Fig. S3 A, a), a trilaminar structure (Fig. S3 A, b), and a trilaminar structure associated with strands of material emanating from the kinetochore region and intertwined with the kinetochore-associated microtubules (Fig. S3 A, c). These observations suggest that plate development can proceed further without CENP-F than without NUF2, but is nevertheless compromised.
CENP-I localization requires only a fraction of the normal level of CENP-A
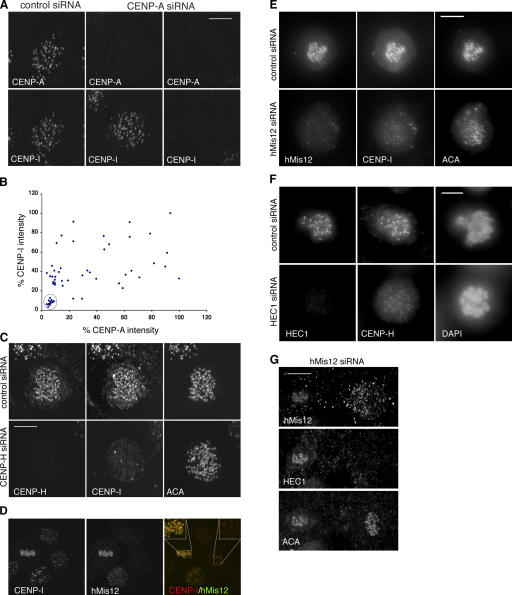
CENP-I localization at kinetochores was reported to depend on CENP-A (Goshima et al., 2003). However, we found that cells with significantly diminished levels of CENP-A still displayed bright CENP-I staining (compare the left and middle columns in Fig. 3 A). Therefore, we quantitated the intensity of CENP-A signals in interphase cells that were transfected with CENP-A siRNA and then compared them with the intensity of CENP-I staining at the same kinetochores. To eliminate experiment-to-experiment variations in absolute staining intensities, we normalized the signal intensity to the brightest CENP-A signal in mock-transfected cells. We observed that when the reduction in CENP-A was <10-fold, CENP-I intensities were clustered at 37 and 75% of the brightest CENP-I signal (Fig. 3, A [middle column] and B). The twofold difference in signal intensity probably reflects the relative abundance of CENP-I in cells before and after replication of their centromeres. We independently confirmed that cells in G2 with duplicated, but unseparated, centromeres exhibited a twofold higher intensity of CENP-I staining compared with cells with unreplicated centromeres (unpublished data). Near-normal levels of CENP-I were also seen in mitotic cells whose CENP-A levels were reduced by <10-fold (unpublished data).
Figure 3.
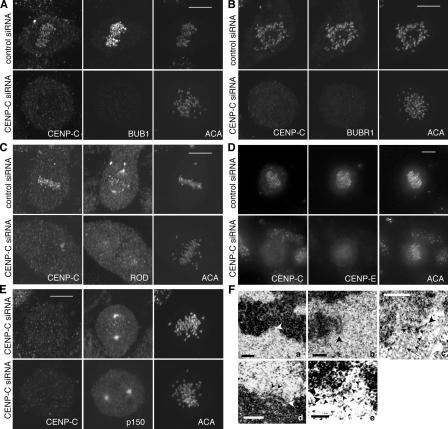
Localization of CENP-I depends on CENP-A and -H, but not hMis12. (A) Interphase cells with different levels of CENP-A were costained with CENP-I. Exposure times were identical for the same protein. (B) Comparison of the normalized fluorescence intensities of CENP-A and -I at 54 centromeres from 12 interphase cells transfected with CENP-A siRNA. The intensity of each protein at the brightest kinetochore is taken as 100%. (C) Maximum projections of a control (top) and a CENP-H–depleted mitotic cell (bottom) stained with antibodies against CENP-H, -I, and ACA. (D) Maximum projections of deconvolved images of normal HeLa cells in mitosis (center left) and early G1 (center right) that were stained with anti–CENP-I and anti-hMis12 antibodies. (E) Maximum projections of deconvolved images of a control (top) and a hMis12-depleted mitotic cell (bottom) stained with antibodies against hMis12, CENP-I, and ACA. (F) Comparison of HEC1 and CENP-H intensities in a control (top) and HEC1 siRNA–transfected (bottom) cells. Single optical plane is shown here. (G) A hMis12-reduced and a hMis12-depleted cell in the same field were stained with ACA and antibodies against hMis12 and HEC1. Bar, 10 μm.
A dramatic drop in CENP-I levels was seen only when CENP-A was reduced by >10-fold (Fig. 3, A [right column] and B [circled datapoints]). Fisher's exact test showed that CENP-I levels at kinetochores with CENP-A intensity below or above 10% of normal level are significantly different (P = 0.00029). When CENP-A is reduced by ∼20-fold, CENP-I levels can be reduced to as low as 3% of controls. These cells were difficult to find, and the precise level of reduction was difficult to determine, as we were approaching the limits of detection. In addition, the loss of other centromere markers (i.e., ACA staining) in cells with extremely low levels of CENP-A made it more difficult to identify kinetochores with certainty. Thus, we can only estimate that substantial reduction of CENP-I was achieved when there was a 10–20-fold reduction in CENP-A. We conclude that only 10% of the normal level of CENP-A is sufficient to assemble near normal levels of CENP-I to the kinetochore.
Effects on CENP-I localization by CENP-H, hMis12, and HEC1
In addition to the dependence on CENP-A, we found that the localization of CENP-I depended on CENP-H (Fig. 3 C), as reported for chicken CENP-I (Nishihashi et al., 2002). The localization of CENP-H also depended on CENP-I (Fig. S2 C), which is consistent with a recent study that found that they can form a complex with several other proteins (Okada et al., 2006).
hMis12 was reported to be a constitutive kinetochore protein that specified CENP-I localization independently of CENP-A (Goshima et al., 2003). However, examination of HeLa and immortalized normal hTERT-RPE1 cells showed that hMis12 is not constitutively localized to kinetochores, as its signals started to decline during late anaphase and were not detectable (<10% of metaphase signals) in late telophase or early G1 cells (Fig. 3 D). This pattern was confirmed by real-time analysis of cells expressing GFP-hMis12 (unpublished data). Thus, hMis12 localization to kinetochores is cell cycle dependent, although its pattern is unique amongst the transient kinetochore proteins. Detailed characterization of hMis12 dynamics will be reported elsewhere. Of relevance to this study, we found normal levels of CENP-I at kinetochores that were devoid of hMis12 (Fig. 3 D). This observation demonstrates that the presence of hMis12 is not a prerequisite for localization of CENP-I to interphase centromeres.
In mitotic cells, kinetochores depleted of hMis12 did exhibit a slight reduction in the level of CENP-I staining (Fig. 3 E). Quantitative analysis showed that even when hMis12 levels were reduced by 10-fold, there was only a twofold reduction in the level of CENP-I at kinetochores. The relationship was puzzling until we discovered connections between hMis12, HEC1, and CENP-I. When HEC1 was depleted from kinetochores by 10-fold, CENP-H and -I levels were reduced by ∼40% (Fig. 3 F and not depicted). This suggests a negative feedback loop between HEC1 and CENP-H/-I. As depletion of hMis12 was shown to reduce HEC1 at kinetochores (Fig. 3 G; Kline et al., 2006), the twofold reduction of CENP-I in hMis12-depleted cells is likely to be indirectly caused by the loss of HEC1 from kinetochores. Regardless, the combined data suggest that hMis12 is not essential for kinetochore localization of CENP-I in interphase or mitosis.
The CENP-C branch specifies localization of hMis12, BUB1, BUBR1, CENP-E, hZW10, and hROD
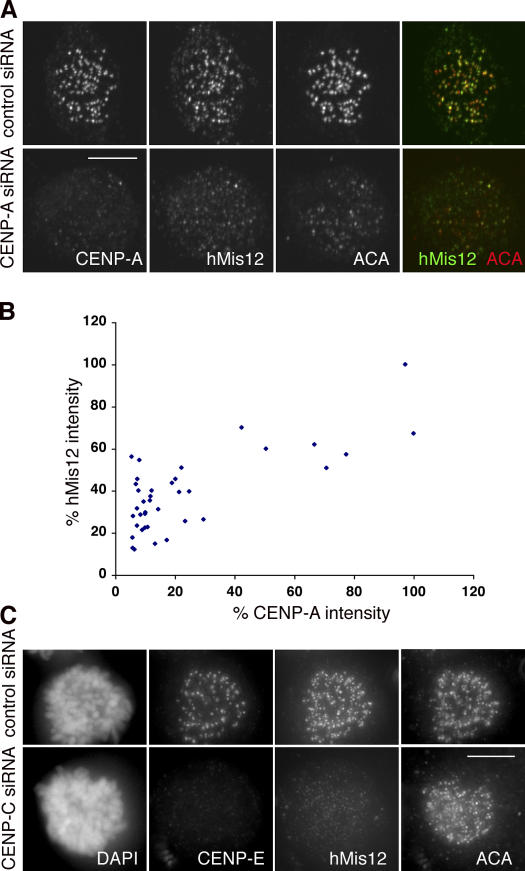
The localization of hMis12 to kinetochores in both Schizosaccharomyces pombe and human cells was reported to be independent of CENP-A (Takahashi et al., 2000; Goshima et al., 2003). In contrast, Caenorhabditis elegans Mis12 has been placed downstream of CENP-A (Cheeseman et al., 2004). Our finding that hMis12 is not constitutively localized to kinetochores led us to reexamine its relationship with CENP-A. As shown in Fig. 4 A, localization of hMis12 was clearly affected by the loss of CENP-A. Quantitative analysis showed an ∼2–3-fold reduction of hMis12 when CENP-A levels were reduced to between 5- and 10-fold (Fig. 4 B). This moderate reduction may be caused by the possibility that there is a separate pool of hMis12 whose localization at centromeres is specified by HP1α and HP1γ (Obuse et al., 2004; Chan et al., 2005). It is also possible that kinetochores in HeLa cells can tolerate a 10-fold reduction in CENP-A, and thus, a >10-fold depletion of CENP-A would be required to see significant loss of downstream proteins. As CENP-C lies downstream of CENP-A (Goshima et al., 2003), we tested and found that depletion of CENP-C resulted in a greater than fivefold reduction of hMis12 from kinetochores (Fig. 4 C). Thus, hMis12 localization can be specified by CENP-A and lies downstream of CENP-C.
Figure 4.
hMis12 localization depends on CENP-A and CENP-C. (A) Maximum projections of a control (top) and a CENP-A–depleted mitotic cell (bottom) stained with CENP-A, hMis12, and ACA. Note the weaker hMis12 and ACA signals in the CENP-A–depleted cell. (B) Comparison of the normalized fluorescence intensities of CENP-A and hMis12 at 38 kinetochores from 13 mitotic cells transfected with CENP-A siRNA. The intensity of each protein at the brightest kinetochore is taken as 100%. (C) Maximum projections of a control (top) and a CENP-C–depleted mitotic cell (bottom) stained with CENP-E, hMis12, and ACA antibodies. CENP-E is used as a readout for depletion of CENP-C. Bars, 10 μm.
We next examined other proteins whose assembly at kinetochores depended on CENP-C. As CENP-C and -I specify separate pathways (Goshima et al., 2003), we focused on proteins that did not depend on CENP-I (Liu et al., 2003b). Cells depleted of CENP-C exhibited severe chromosome missegregation and micronucleation that are fully consistent with those reported for antibody injection experiments (Tomkiel et al., 1994). Kinetochores depleted of CENP-C were found to lack nearly all the transiently localized kinetochore proteins except the inner centromere protein Aurora B. Thus, BUB1, BUBR1, hROD, hZW10, and CENP-E all failed to assemble onto kinetochores that lacked CENP-C (Fig. 5, A–D; Fig. S4 A, a, available at http://www.jcb.org/cgi/content/full/jcb.200606020/DC1; and Table I). HEC1 and p150glued, whose localization at kinetochores depends on CENP-I, were also found to depend on CENP-C (Fig. 5 E; Fig. S4 B).
Figure 5.
Kinetochore localization of BUB1, BUBR1, hROD, CENP-E, and p150glued depends on CENP-C. (A–E) Maximum projections of control cells (A–D) or cells with reduced CENP-C (E; top rows) and CENP-C–depleted mitotic cells (bottom rows) costained with CENP-C, ACA, and BUB1 (A), BUBR1 (B), hROD (C), CENP-E (D), p150glued (E). Bar, 10 μm. (F) Representative EM images of kinetochores in CENP-C siRNA–transfected cells. Big arrowheads indicate the positions of kinetochores. (a) A kinetochore with short plates and a few attached microtubules. (b–c) Kinetochores pulled out of the surface of the chromosome. A few attached microtubules are indicated by small arrows in c. They are absent in b. (d) A kinetochore with no microtubule binding showing some fibrils extending (small arrowheads) from the outer plate. (c and e) Some kinetochores show a distinct beaded structure in their outer plates (small arrowheads). Bars: (a–d) 400 nm; (e) 200 nm.
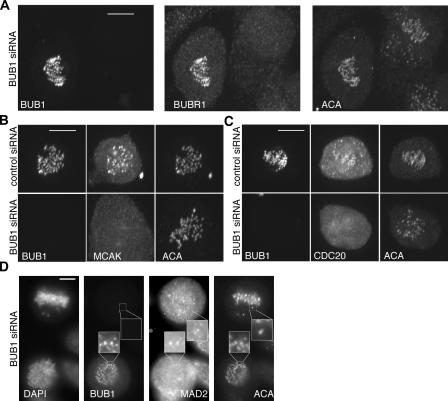
We next examined the relationships amongst the proteins that lie downstream of CENP-C. BUB1 was given special attention, as there were conflicting reports about its checkpoint and recruitment functions (Johnson et al., 2004; Tang et al., 2004; Meraldi and Sorger, 2005). We confirmed that BUB1 is, indeed, essential for the mitotic checkpoint, as microinjection of BUB1 antibodies or transfection of BUB1 siRNA prevented HeLa cells from delaying mitosis in response to nocodazole (unpublished data). As for its role in recruiting proteins to kinetochores, we found that BUBR1, MCAK, CENP-F, and CDC20 depended on BUB1 for localization, whereas hMis12, hROD, HEC1, hMPS1, MAD1, MAD2, CENP-E, p150glued, and Aurora B did not (Table I; Fig. 6, A–D; and Fig. S4, C–G). Within this latter group, we found that hMis12 specified kinetochore localization of HEC1 (Fig. 3 G and Fig. S4 B). However, hROD and CENP-E were not dependent on either BUB1 or hMis12 for their kinetochore localization (Fig. S4 E and not depicted). The combined data suggest that CENP-C specifies three or four subbranches (Fig. 1).
Figure 6.
Kinetochore localization of BUBR1, MCAK, and CDC20, but not MAD2, depends on BUB1. Maximum projections of untransfected cells (A, left cell; D, bottom cell) or control cells (B–C, top rows) and BUB1-depleted mitotic cells (A, right cell; B–C bottom rows; D, top cell) costained with BUB1, ACA, and BUBR1 (A), MCAK (B), CDC20 (C), MAD2 (D). Bar, 10 μm.
EM analysis showed that all cells depleted of CENP-C (n = 6) contained kinetochores with discernible laminar plate structure (Table II and Table S1). However, they were usually deformed, as they appeared smaller (4/50; Fig. 5 F, a), pulled away from the underlying centromere heterochromatin (8/50; Fig. 5 F, b and c), or exhibited thinner outer plates that often displayed a beaded appearance (17/50; Fig. 5 F, c and e). Microtubules were usually absent from these kinetochores, and prominent fibrillar extensions were sometimes observed extending from the outer plates (Fig. 5 F, d). Both the kinetochore morphology and microtubule pattern observed in these cells were comparable to those seen in cells injected with antibodies to CENP-C (Tomkiel et al., 1994), but were distinct from kinetochores depleted of CENP-I. EM analysis of kinetochores in BUB1- or hMis12-depleted cells did not recapitulate all the ultrastructural defects found in CENP-C–depleted cells. For example, we did not find kinetochores with smaller plates. However, kinetochores with plate structure but no microtubule binding and kinetochores with pulled out, thinner, or punctate plates were observed (Fig. S3, B–C, and Table II). In some cells depleted of BUB1 or hMis12, we noticed undercondensed chromatin, either in centromere regions or elsewhere along the chromosomes (Fig. S3, B [a] and C [b]). The reasons for this are unclear, but we rarely observed such changes in CENP-C– depleted cells. Despite these discrepancies (see Discussion), our observations suggest that some of the ultrastructural anomalies resulting from the loss of CENP-C may be attributed to the disruption of the BUB1 and hMis12 branches.
CENP-A specifies the localization of Aurora B and MCAK at the inner centromere
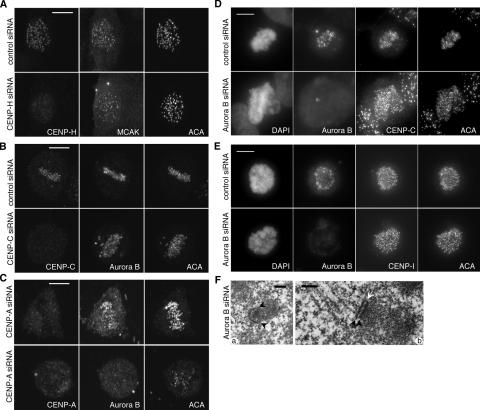
Aurora B and the microtubule depolymerase MCAK are both concentrated at the inner centromere from late G2 until metaphase (Andrews et al., 2004). Both the localization and enzymatic activity of MCAK are regulated by Aurora B kinase (Andrews et al., 2004; Lan et al., 2004). Although MCAK was reported to interact with CENP-H in vitro (Sugata et al., 2000), neither Aurora B nor MCAK localization was affected when CENP-H or -I was depleted from cells (Fig. 7 A and not depicted). Furthermore, Aurora B or MCAK localization was not dependent on CENP-C (Fig. 7 B and not depicted). Conversely, the localization of CENP-I and -C also did not depend on Aurora B (Fig. 7, D–E). We found, however, that the localization of Aurora B was dependent on CENP-A (Fig. 7 C). Therefore, CENP-A specifies a third assembly branch that specifies the localization of Aurora B and MCAK to the inner centromere (Fig. 1).
Figure 7.
Localization of Aurora B depends on CENP-A. (A) A control (top) and a CENP-H–depleted (bottom) late G2/early prophase cell costained for CENP-H, MCAK, and ACA. (B and C) Maximum projections of cells costained for Aurora B, ACA, and CENP-C (B) or -A (C). The bottom rows show the CENP-C– (B) or CENP-A–depleted (C) cells. The top row in B shows a control mitotic cell. The top row in C shows a cell with less reduced CENP-A and nearly normal levels of Aurora B and ACA. (D–E) Maximum projections of control and Aurora B–depleted cells (top and bottom rows, respectively) costained with Aurora B, ACA, and CENP-C (D) or -I (E). (F) Under EM, the kinetochores in Aurora B–depleted cells maintain the plate structures, but either expand and extend significantly to a “C” shape without microtubule binding (a) or display “hairpin” structure, with outer and inner plates seemingly fused at one end (b).
Consistent with its reported roles in chromatin condensation and kinetochore–microtubule interactions (Murata-Hori and Wang, 2002; Ditchfield et al., 2003; Hauf et al., 2003; Andrews et al., 2004; Johnson et al., 2004; Lan et al., 2004), two major types of kinetochore defects were observed in cells depleted of Aurora B (Table II). The first type showed an expanded, C-shaped outer plate accompanied by undercondensed subjacent chromatin and no discernable inner plate (Fig. 7 F, a). The second type of defect displayed both inner and outer plates that appeared more electron dense, and the outer and inner plates often appeared to be fused at one end (Fig. 7 F, b). The chromosomes in these cells also appeared undercondensed, similar to those seen in BUB1-depleted cells (Fig. S3 B; Table S1).
Discussion
Mapping the pathways that specify kinetochore assembly
We have performed an epistasis analysis of 20 kinetochore proteins with respect to their assembly at the centromere–kinetochore complex and constructed a genetic interaction map based on our analysis and previously published data (Fig. 1). The map shows CENP-A occupying the top of a hierarchy that consists of three major branches that are specified by CENP-C, -I, and Aurora B. Each of the three branches form subbranches that intersect with one another to form a network that we believe describes the temporal and spatial relationship of these proteins. If kinetochores are assembled from repetitive units (Zinkowski et al., 1991), this map depicts the organization of a single unit that, in concert with other similar units, would specify the structure that is seen by EM. Depletion of specific components of the epistasis groups differentially affects the integrity of the unit and the unit–unit interactions.
Each of the three major branches appears to play distinct roles in the formation of the trilaminar kinetochore structure. The CENP-I branch is clearly essential for trilaminar plate formation because, in its absence, we saw a structure that is reminiscent of the fuzzy balls that were described early on as prekinetochores (Brenner et al., 1981; He and Brinkley, 1996; Roos, 1973). However, the molecular composition of the fuzzy balls described in this paper differs from prekinetochores because they contain proteins (e.g., CENP-E and BUBR1) that normally assemble after nuclear envelope breakdown. Fully consistent with the fact that NUF2 belongs to the CENP-I pathway, its depletion also produced the fuzzy balls. However, the CENP-F subbranch does not participate in this organizational step.
The fuzzy ball structure is also reminiscent of the structures seen when cells were depleted of the nuclear pore protein Nup358/RanBP2 (Salina et al., 2003). Interestingly, depletion of HEC1 and NUF2 are known to affect the kinetochore targeting of Nup358/RanBP2, suggesting the latter may contribute to the maturation of kinetochore plates (Joseph et al., 2004). Consistent with this, depletion of Nup358/RanBP2, indeed, affected the localization of several kinetochore proteins (Joseph et al., 2004). However, Nup358/RanBP2 seems not to depend on CENP-I for its kinetochore targeting, and it is unlikely that Nup358/RanBP2 acts as a scaffold for assembly because its own localization at kinetochores depends on microtubule attachments, whereas the proteins that rely on Nup358/RanBP2 do not (Joseph et al., 2004; Joseph et al., 2002). Given that Nup358/RanBP2 has SUMO-conjugating activity (Pichler et al., 2002), it may act independently of kinetochores by modifying proteins to facilitate their assembly into kinetochores.
Disruption of the CENP-C and Aurora B pathways affected the structure of the trilaminar plate in a qualitative way. The CENP-C pathway appeared to specify the compaction and dimensions of the kinetochore plates, as its loss resulted in small-sized kinetochores and kinetochores with a thin or expanded and beaded outer plate. As CENP-C is involved in recruiting a substantial number of proteins, its loss would reduce the number of fully assembled unit modules. As siRNA depletes CENP-C to different extents in any given cell (Fig. 5 E), sufficient modules might be available to form a trilaminar plate, albeit one of reduced dimension (Fig. 5 F). Depletion of proteins downstream of CENP-C, such as BUB1 and hMis12, partially recapitulates the thin plates, but does not produce small-sized kinetochores that were seen in cells depleted of CENP-C (Fig. S3, B–C). As the localization of proteins like hROD and CENP-E were affected by CENP-C, but not by BUB1 or hMis12, it is likely there are unidentified proteins that are downstream of CENP-C and contribute to this aspect of trilaminar plate organization. Two candidates may be the human homologues of KNL-1 and -3, which are two proteins in C. elegans that lie downstream of CENP-C. The human counterpart of KNL-1, AF15q14, has already been shown to be a kinetochore protein (Cheeseman et al., 2004).
Defects associated with the Aurora B pathway seem to affect the relationship between the kinetochore and the subjacent chromatin, as seen by the formation of C-shaped kinetochores and kinetochores with seemingly continuous outer and inner plates. There is no easy explanation for how the inner and outer plates become fused, but C-shaped kinetochores have been seen in prematurely condensed chromosomes induced by either cell fusion or caffeine treatment (Rattner and Wang, 1992; Wise and Brinkley, 1997). In both situations, the condensation state of the underlying heterochromatin may be altered, thus, impacting the relationship between the kinetochore and the chromosome. Despite the impact of Aurora B on the inner regions of the kinetochore, proteins such as CENP-C and -I, which normally localize to the inner plate, are still retained at kinetochores after depletion of Aurora B. This suggests that neither of these proteins are able to form or maintain a distinct inner kinetochore plate ultrastructure in the absence of Aurora B. As Aurora B is also present throughout the cell, its actions on kinetochores do not have to be restricted to the pool that is localized at the inner centromere. Indeed, the undercondensation of chromosomes in both Aurora B– and BUB1-depleted cells may reflect additional roles of these kinases that are not linked to the kinetochore.
Evolutionary conservation of kinetochore assembly
Many of the centromere–kinetochore proteins examined here are evolutionarily conserved, and thus, similarity in their assembly amongst other organisms was expected (Cleveland et al., 2003; Maiato et al., 2004; Chan et al., 2005). However, we would like to point out that there are distinctive features of human kinetochore assembly that distinguish it from some model organisms. For example, both budding and fission yeasts perform closed mitosis, and thus, most if not all of the kinetochore proteins are present at kinetochores throughout the cell cycle. This is in contrast to the distinct temporal order of assembly for a large number of human kinetochore proteins. In addition, there are important human centromere–kinetochore components that are missing in some other species and vice versa. For instance, CENP-I and -H homologues are not found in C. elegans and Drosophila melanogaster, and ZW10 or ROD homologues are absent from fungal genomes (Chan et al., 2000; Okada et al., 2006). These differences suggest some flexibility in how the conserved proteins are assembled.
Nodes within the kinetochore assembly network
At least six proteins or protein complexes were found to occupy nodes (HEC1–NUF2, MAD1, CENP-F, BUBR1, dynein–dynactin, and MCAK) that linked various branches within the network that is depicted in Fig. 1. We believe that these intersections reflect coordination among different assembly pathways so that no single pathway outpaces the other during the construction of a functional kinetochore. It is interesting that BUB1 occupies a position that links three pathways. Although earlier siRNA results on BUB1 challenged its role as a bona fide checkpoint protein (Johnson et al., 2004; Tang et al., 2004), a recent study confirmed its status as a functional mitotic checkpoint kinase and proposed a new role for BUB1 to resolve improper lateral attachment and mediate the bi-orientation of sister chromatids (Meraldi and Sorger, 2005). Along this line, we found BUB1 controls the localization of BUBR1 and CDC20, two essential components of the mitotic checkpoint. Previous works claimed that MAD2 localization at kinetochores depends on BUB1 (Johnson et al., 2004; Meraldi and Sorger, 2005). We found no evidence that MAD2, or any of its upstream components (MAD1, hMPS1, and HEC1), to depend on BUB1 (Fig. 6 D; Fig. S4, D and G). In regard to its role in correcting the improper kinetochore–microtubule attachment, we have shown that BUB1 is required for the localization of MCAK, the microtubule depolymerase that, together with Aurora B kinase, is involved in correcting syntelic attachment (Andrews et al., 2004; Lan et al., 2004). Considering recent reports that BUB1 also affects centromeric cohesion through Sgo1 (Tang et al., 2004; Kitajima et al., 2005; McGuinness et al., 2005), it will be interesting to understand why a single protein is assigned to coordinate these diverse functions.
A critical amount of CENP-A in maintaining centromere–kinetochore structure
We were initially surprised to learn that only a fraction (<10% by intensity) of the CENP-A that is normally present at kinetochores is sufficient to maintain near-normal levels of CENP-I (Fig. 3, A–B). Further studies showed that this relationship was extended to include CENP-C, hMis12, and Aurora B (Fig. 4, A–B, and Fig. 7 E; unpublished data). Although we did not directly test whether 10% levels of CENP-A supported normal kinetochore functions, we routinely observed normal metaphase cells whose kinetochores exhibited 10% levels of CENP-A. It remains to be tested whether this level of CENP-A can support a normal mitotic checkpoint response. We suggest that kinetochores in HeLa cells contain an amount of CENP-A that is in excess to that required for kinetochore assembly. Examination of extended chromatin fibers in D. melanogaster and human cells has revealed that CENP-A (CID) and histone H3 are interspersed along the chromatin fiber. However, 3D reconstruction of their localization at kinetochores shows that domains of CENP-A and histone H3 are spatially separated (Blower et al., 2002). This suggests that the CENP-A nucleosomes that are interspersed along the chromatin fiber can contact one another so that the intervening H3-containing chromatin loops out to form a spatially separated domain (Blower et al., 2002). Although we cannot say with certainty the minimal number of CENP-A nucleosomes that are required for a normal kinetochore structure, our data in HeLa cells would suggest that this higher order structure can still be established when the level of CENP-A at kinetochores is reduced by 10-fold. Below this level, the density of CENP-A along the chromatin may be too low to be reliably organized into a domain that can promote the assembly of the subsequent layer of proteins that include Aurora B, CENP-C, and -I.
In conclusion, we show that CENP-A plays a central role in kinetochore structure, as it specifies the assembly of proteins that form not only the trilaminar plates but also components of the inner centromere. Three major assembly pathways that contribute to different aspects of the higher order organization of the kinetochore were defined. A network of intersecting branches suggests some level of coordination between the assembly pathways. The biochemical interactions that link the assembly steps remain to be determined. The recent isolation of discrete kinetochore protein complexes consisting of hMis12, CENP-A, -I, and -H have revealed a constellation of novel kinetochore proteins whose characterization will provide insights into this question (Cheeseman et al., 2004; Obuse et al., 2004; Foltz et al., 2006; Izuta et al., 2006; Okada et al., 2006). As new kinetochore proteins are identified, they will be assigned to our map to see how they are related to the pathways that we have identified here. The website address to an interactive map is available upon request.
Materials and methods
Cell culture
HeLa cells were grown in a humidified 37°C incubator with 5% CO2 level in DME supplemented with 10% fetal bovine serum, nonessential amino acids, and 2 mM l-glutamine.
siRNA sequences and transfection
All siRNAs (synthesized by Dharmacon; sequences can be provided on request) were transfected at the final concentration of 100 nM using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Usually, HeLa cells were grown on coverslips in 24-well plates and transfected at 50% confluency. Coverslips were processed 4 d later, except those transfected with BUB1, Aurora B, HEC1, or hMPS1 siRNA, which were fixed 48 h after transfection. Fresh medium was added or cells were split when necessary. For BUB1 and Aurora B, double-thymidine synchronization was used to increase the number of mitotic cells at the harvesting time point; cells were incubated with 2 mM thymidine for 15 h after siRNA transfection, released for 8 h, and blocked again for 15 h with 2 mM thymidine. They were harvested 10–11 h after release, usually after 1 h of treatment of MG132 at 1 μM final concentration. In some early experiments, CENP-I siRNA1 was transfected as previously described (Liu et al., 2003b). Usually, several different siRNAs against the same target were used to validate the knockdown results.
Antibodies
Rabbit, rat, or mouse monoclonal MAD1 antibody, Rabbit polyclonal MAD2 antibody, and rat or mouse monoclonal antibodies against CENP-A, -I, -E, -F, BUB1, BUBR1, hZW10, and hROD have been previously described (Liu et al., 2003b). Other antibodies used include monoclonal CENP-B antibody 2D8D8 (H. Masumoto, Nagoya University, Nagoya, Japan; Suzuki et al., 2004), rabbit anti-hMPS1 antibody Ag3 (Liu et al., 2003a), rabbit anti-MCAK antibody (a gift from Linda Wordeman, University of Washington, Seattle, WA), anti-p150 Glued (subunit of dynactin) rabbit polyclonal (from Linda Wordeman), and mouse monoclonal antibody (BD Biosciences), mouse monoclonal HEC1 antibody (BD Biosciences), anti-Aurora B monoclonal antibody (BD Biosciences), and rabbit polyclonal antibody (Zymed). Anticentromere antibody (ACA) serum was provided by J.B. Rattner.
Rat polyclonal and monoclonal anti–CENP-H antibodies, rabbit polyclonal anti-hMis12 antibody, rabbit anti–CENP-C antibody, and rabbit or rat anti-Nuf2 polyclonal antibodies were made against GST-CENP-H, GST-hMis12, GST-CENP-C, and GST-Nuf2, and the antisera were affinity purified through immobilized antigen columns.
Immunofluorescence staining and microscopy
Cells were plated onto No. 1.5 glass coverslips, fixed for 7 min in freshly prepared 3.5% paraformaldehyde (in PBS), pH 7.0, extracted in KB (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, and 0.1% BSA) plus 0.2% Triton X-100 for 5 min at room temperature, and rinsed in KB. For optimal staining of hMis12 at kinetochores, cells were preextracted for 1 min in microtubule stabilization buffer (MTSB: 4.0 M glycerol, 0.1 M Pipes, pH 6.9, and 1 mM EGTA) plus 0.5% Triton X-100, incubated with MTSB for 2 min, fixed in –20°C methanol for 7 min, and then equilibrated with KB. Primary and secondary antibodies were diluted in KB and added to coverslips for 30–60 min at 37°C in a humidified chamber. All the secondary antibodies were conjugated to Alexa Fluor dyes (488, 555, 568, or 647). DAPI was used to counterstain DNA at 5 ng/ml final concentration. The coverslips were mounted onto slides using mounting medium containing 90% glycerol and 0.1% n-propylgallate made in 1×PBS.
Images were captured with a spinning disc confocal microscope (Ultraview; Perkin-Elmer) that consisted of a microscope (Eclipse TE2000S; Nikon) and a charge-coupled device camera (ORCA-ERG; Hamamatsu). Some images were taken on an inverted microscope (Eclipse TE2000 E; Nikon) equipped with a Cascade 512F camera (Roper) and controlled by Metavue (Molecular Devices) software. Images were visualized with either 60× or 100×, 1.4 NA, objectives.
Image processing and fluorescence intensity measurement were performed basically as previously described using ImagePro Plus 5.0 software (Media Cybernetics) and 12-or 16-bit raw image stacks obtained from the aforementioned microscopes (Liu et al., 2003b). To eliminate variations of staining between experiments and coverslips, we normalized the absolute intensity values to the intensity of the brightest stained kinetochore, which was set at 100%. The relative intensities at kinetochores of different proteins were plotted to examine the relationship. AutoDeblur software (Media Cybernetics) was used to perform image deconvolution.
EM
HeLa cells were grown in 35-mm cell culture dishes and transfected with control and target siRNAs. 2–4 d after transfection, the cells were fixed in 3% glutaraldehyde and 0.2% tannic acid in 200 mM sodium cacodylate buffer for 1 h at room temperature. Post fixation was in 2% OsO4 for 20 min. The cells were dehydrated in ethanol, and then infiltrated with Polybed 812 resin. Polymerization was performed at 60°C for 24 h. Silver-gray sections were cut with an ultramicrotome (Leica) equipped with a diamond knife (Diatome), and sections were stained with uranyl acetate and lead citrate and examined in an EM (H-7000; Hitachi).
Online supplemental material
Fig. S1 shows representative results of siRNA knockdown and specificity test of several antibodies. Fig. S2 shows that CENP-I is required for kinetochore localization of hMPS1, p150glued, and CENP-H. Fig. S3 shows representative EM images of chromosomes or kinetochores in cells depleted of CENP-F, BUB1 or hMis12. Fig. S4 shows the effects of CENP-C and BUB1 depletion on the localization of other kinetochore proteins. Table S1 shows the details of EM examination on kinetochore structures in cells transfected with siRNAs targeting CENP-I, NUF2, CENP-F, -C, BUB1, hMis12, and Aurora B. Online supplemental materials are available at http://www.jcb.org/cgi/content/full/jcb.200606020/DC1.
Supplementary Material
Acknowledgments
We thank Jim Hittle and Beatrice Conner for technical support, Jie Feng for discussion, and the animal, hybridoma, and biostatistics facilities (Tianyu Li) at the Fox Chase Cancer Center for help.
J.B. Rattner is supported by a grant from the National Science and Engineering Council of Canada. T.J. Yen is supported by National Institutes of Health grants GM44762, CA99423, and CA75138, core grant CA06927, and an appropriation from the Commonwealth of Pennsylvania.
Abbreviation used in this paper: ACA, anticentromere antibody.
References
- Adams, R.R., M. Carmena, and W.C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49–54. [DOI] [PubMed] [Google Scholar]
- Amor, D.J., P. Kalitsis, H. Sumer, and K.H. Choo. 2004. Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol. 14:359–368. [DOI] [PubMed] [Google Scholar]
- Andrews, P.D., Y. Ovechkina, N. Morrice, M. Wagenbach, K. Duncan, L. Wordeman, and J.R. Swedlow. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268. [DOI] [PubMed] [Google Scholar]
- Bharadwaj, R., W. Qi, and H. Yu. 2004. Identification of two novel components of the human NDC80 kinetochore complex. J. Biol. Chem. 279:13076–13085. [DOI] [PubMed] [Google Scholar]
- Blower, M.D., and G.H. Karpen. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3:730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M.D., B.A. Sullivan, and G.H. Karpen. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomont, P., P. Maddox, J.V. Shah, A.B. Desai, and D.W. Cleveland. 2005. Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. EMBO J. 24:3927–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., D. Pepper, M.W. Berns, E. Tan, and B.R. Brinkley. 1981. Kinetochore structure, duplication, and distribution in mammalian cells: analysis by human autoantibodies from scleroderma patients. J. Cell Biol. 91:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G.K., S.A. Jablonski, D.A. Starr, M.L. Goldberg, and T.J. Yen. 2000. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2:944–947. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., S.T. Liu, and T.J. Yen. 2005. Kinetochore structure and function. Trends Cell Biol. 15:589–598. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Niessen, S. Anderson, F. Hyndman, J.R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D.W., Y. Mao, and K.F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., B. Moree, J.M. Hickey, J.V. Kilmartin, and E.D. Salmon. 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca, J.G., Y. Dong, P. Hergert, J. Strauss, J.M. Hickey, E.D. Salmon, and B.F. McEwen. 2005. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol. Biol. Cell. 16:519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield, C., V.L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S.S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J., H. Huang, and T.J. Yen. 2006. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 115:320–329. [DOI] [PubMed] [Google Scholar]
- Foltz, D.R., L.E. Jansen, B.E. Black, A.O. Bailey, J.R. Yates III, and D.W. Cleveland. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469. [DOI] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf, S., and Y. Watanabe. 2004. Kinetochore orientation in mitosis and meiosis. Cell. 119:317–327. [DOI] [PubMed] [Google Scholar]
- Hauf, S., R.W. Cole, S. LaTerra, C. Zimmer, G. Schnapp, R. Walter, A. Heckel, J. van Meel, C.L. Rieder, and J.M. Peters. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, D., and B.R. Brinkley. 1996. Structure and dynamic organization of centromeres/prekinetochores in the nucleus of mammalian cells. J. Cell Sci. 109:2693–2704. [DOI] [PubMed] [Google Scholar]
- Holt, S.V., M.A. Vergnolle, D. Hussein, M.J. Wozniak, V.J. Allan, and S.S. Taylor. 2005. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J. Cell Sci. 118:4889–4900. [DOI] [PubMed] [Google Scholar]
- Hori, T., T. Haraguchi, Y. Hiraoka, H. Kimura, and T. Fukagawa. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347–3362. [DOI] [PubMed] [Google Scholar]
- Izuta, H., M. Ikeno, N. Suzuki, T. Tomonaga, N. Nozaki, C. Obuse, Y. Kisu, N. Goshima, F. Nomura, N. Nomura, and K. Yoda. 2006. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes Cells. 11:673–684. [DOI] [PubMed] [Google Scholar]
- Jablonski, S.A., G.K. Chan, C.A. Cooke, W.C. Earnshaw, and T.J. Yen. 1998. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 107:386–396. [DOI] [PubMed] [Google Scholar]
- Johnson, V.L., M.I. Scott, S.V. Holt, D. Hussein, and S.S. Taylor. 2004. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117:1577–1589. [DOI] [PubMed] [Google Scholar]
- Joseph, J., S.H. Tan, T.S. Karpova, J.G. McNally, and M. Dasso. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J., S.T. Liu, S.A. Jablonski, T.J. Yen, and M. Dasso. 2004. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14:611–617. [DOI] [PubMed] [Google Scholar]
- Kitajima, T.S., S. Hauf, M. Ohsugi, T. Yamamoto, and Y. Watanabe. 2005. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr. Biol. 15:353–359. [DOI] [PubMed] [Google Scholar]
- Kline, S.L., I.M. Cheeseman, T. Hori, T. Fukagawa, and A. Desai. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops, G.J., Y. Kim, B.A. Weaver, Y. Mao, I. McLeod, J.R. Yates III, M. Tagaya, and D.W. Cleveland. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, W., X. Zhang, S.L. Kline-Smith, S.E. Rosasco, G.A. Barrett-Wilt, J. Shabanowitz, D.F. Hunt, C.E. Walczak, and P.T. Stukenberg. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286. [DOI] [PubMed] [Google Scholar]
- Liu, S.T., G.K. Chan, J.C. Hittle, G. Fujii, E. Lees, and T.J. Yen. 2003. a. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol. Biol. Cell. 14:1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.T., J.C. Hittle, S.A. Jablonski, M.S. Campbell, K. Yoda, and T.J. Yen. 2003. b. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 5:341–345. [DOI] [PubMed] [Google Scholar]
- Maiato, H., J. DeLuca, E.D. Salmon, and W.C. Earnshaw. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 117:5461–5477. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma, S., V.M. Stucke, and E.A. Nigg. 2002. Role of hec1 in spindle checkpoint signaling and kinetochore recruitment of mad1/mad2. Science. 297:2267–2270. [DOI] [PubMed] [Google Scholar]
- McAinsh, A.D., J.D. Tytell, and P.K. Sorger. 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19:519–539. [DOI] [PubMed] [Google Scholar]
- McCleland, M.L., M.J. Kallio, G.A. Barrett-Wilt, C.A. Kestner, J. Shabanowitz, D.F. Hunt, G.J. Gorbsky, and P.T. Stukenberg. 2004. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr. Biol. 14:131–137. [DOI] [PubMed] [Google Scholar]
- McGuinness, B.E., T. Hirota, N.R. Kudo, J.M. Peters, and K. Nasmyth. 2005. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., and P.K. Sorger. 2005. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 24:1621–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami, Y., T. Hori, H. Kimura, and T. Fukagawa. 2005. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol. Cell. Biol. 25:1958–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori, M., and Y.L. Wang. 2002. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12:894–899. [DOI] [PubMed] [Google Scholar]
- Nishihashi, A., T. Haraguchi, Y. Hiraoka, T. Ikemura, V. Regnier, H. Dodson, W.C. Earnshaw, and T. Fukagawa. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2:463–476. [DOI] [PubMed] [Google Scholar]
- Obuse, C., O. Iwasaki, T. Kiyomitsu, G. Goshima, Y. Toyoda, and M. Yanagida. 2004. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 6:1135–1141. [DOI] [PubMed] [Google Scholar]
- Okada, M., I.M. Cheeseman, T. Hori, K. Okawa, I.X. McLeod, J.R. Yates III, A. Desai, and T. Fukagawa. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457. [DOI] [PubMed] [Google Scholar]
- Pichler, A., A. Gast, J.S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 108:109–120. [DOI] [PubMed] [Google Scholar]
- Rattner, J.B., and T. Wang. 1992. Kinetochore formation and behaviour following premature chromosome condensation. J. Cell Sci. 103:1039–1045. [DOI] [PubMed] [Google Scholar]
- Roos, U.P. 1973. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma. 41:195–220. [DOI] [PubMed] [Google Scholar]
- Salina, D., P. Enarson, J.B. Rattner, and B. Burke. 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162:991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugata, N., S. Li, W.C. Earnshaw, T.J. Yen, K. Yoda, H. Masumoto, E. Munekata, P.E. Warburton, and K. Todokoro. 2000. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere–kinetochore complexes. Hum. Mol. Genet. 9:2919–2926. [DOI] [PubMed] [Google Scholar]
- Suzuki, N., M. Nakano, N. Nozaki, S. Egashira, T. Okazaki, and H. Masumoto. 2004. CENP-B interacts with CENP-C domains containing Mif2 regions responsible for centromere localization. J. Biol. Chem. 279:5934–5946. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., E.S. Chen, and M. Yanagida. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 288:2215–2219. [DOI] [PubMed] [Google Scholar]
- Tang, Z., Y. Sun, S.E. Harley, H. Zou, and H. Yu. 2004. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. USA. 101:18012–18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiel, J., C.A. Cooke, H. Saitoh, R.L. Bernat, and W.C. Earnshaw. 1994. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol. 125:531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneron, S., S. Prieto, C. Bernis, J.C. Labbe, A. Castro, and T. Lorca. 2004. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol. Biol. Cell. 15:4584–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, D.A., and B.R. Brinkley. 1997. Mitosis in cells with unreplicated genomes (MUGs): spindle assembly and behavior of centromere fragments. Cell Motil. Cytoskeleton. 36:291–302. [DOI] [PubMed] [Google Scholar]
- Yang, Z., J. Guo, Q. Chen, C. Ding, J. Du, and X. Zhu. 2005. Silencing mitosin induces misaligned chromosomes, premature chromosome decondensation before anaphase onset, and mitotic cell death. Mol. Cell. Biol. 25:4062–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski, R.P., J. Meyne, and B.R. Brinkley. 1991. The centromere–kinetochore complex: a repeat subunit model. J. Cell Biol. 113:1091–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.