Abstract
Terminal differentiation of distinct cell types requires the transcriptional activation of differentiation-specific genes and the suppression of genes associated with the precursor cell. For example, the expression of utrophin (Utrn) is suppressed during skeletal muscle differentiation, and it is replaced at the sarcolemma by the related dystrophin protein. The MyoD transcription factor directly activates the expression of a large number of skeletal muscle genes, but also suppresses the expression of many genes. To characterize a mechanism of MyoD-mediated suppression of gene expression, we investigated two genes that are suppressed in fibroblasts converted to skeletal muscle by MyoD, follistatin-like 1 (Fstl1) and Utrn. MyoD directly activates the expression of a muscle-specific microRNA (miRNA), miR-206, which targets sequences in the Fstl1 and Utrn RNA, and these sequences are sufficient to suppress gene expression in the presence of miR-206. These findings demonstrate that MyoD, in addition to activating muscle-specific genes, induces miRNAs that repress gene expression during skeletal muscle differentiation.
Introduction
Cell commitment and differentiation involves the carefully regulated expression of genes associated with terminal differentiation, and many different biological systems have been used to identify the factors and networks that regulate these genes. Skeletal myogenesis represents a particularly well-studied model system (Tapscott, 2005). Vertebrate skeletal myogenesis is regulated by a family of four related basic helix-loop-helix transcription factors: Myf5, MyoD, Myog, and Mrf4 (Molkentin and Olson, 1996; Puri and Sartorelli, 2000; Pownall et al., 2002; Buckingham et al., 2003). Recent studies indicate that MyoD, and perhaps by analogy Myf5, binds to promoters of genes expressed throughout the program of myogenesis, establishing temporal patterning through feed-forward mechanisms with other regulatory factors (Bergstrom et al., 2002; Penn et al., 2004; Cao et al., 2006). An interesting and relatively unstudied feature of the MyoD-mediated differentiation program, however, is the suppression of the nonmuscle phenotype in cells that are converted to skeletal muscle by MyoD. For example, expression of MyoD in melanocytes, adipocytes, and chondrocytes results not only in the positive regulation of skeletal muscle genes, but also in the suppression of molecular phenotypes specific to the melanocyte, adipocyte, and chondrocyte, respectively (Weintraub et al., 1989; Choi et al., 1990). In this regard, conversion of a cell to skeletal muscle by MyoD is accompanied by the suppression of the nonmuscle phenotype of that cell. This is a specific example of a general feature of cell differentiation; establishing a new cellular phenotype requires suppression of features associated with the prior phenotype or other related phenotypes.
Although the activation of skeletal muscle gene expression by MyoD has been intensively studied, very little is known about the ability of MyoD to suppress gene expression. Several studies have indicated that MyoD can form repressive complexes, and these complexes have been suggested to be a mechanism of directly recruiting transcriptional repressors to specific promoters. For example, MyoD has been shown to recruit HDAC1 to the Myog promoter in myoblasts with an associated hypoacetylation of regional histones (Mal et al., 2001; Mal and Harter, 2003), and a similar MyoD-mediated recruitment of the Sir2 HDAC to suppress gene expression has been previously described (Fulco et al., 2003). When cells differentiate, MyoD has been shown to recruit histone acetylases and chromatin-remodeling complexes to some of the same promoters shown to be suppressed by HDAC recruitment in myoblasts (de la Serna et al., 2001, 2005; Bergstrom et al., 2002), suggesting that a switch between a repressive complex and an activating complex occurs at the initiation of terminal differentiation. In addition, components of the repressive Polycomb complex have been shown to be associated with repressed muscle genes before differentiation and replaced by MyoD and other activators during differentiation (Caretti et al., 2004). Therefore, there is precedent for a regulated transition from repressive promoter complexes to activating complexes during skeletal muscle differentiation, and some evidence that MyoD can recruit either activators or repressors to the promoter regions, depending on cellular context, i.e., whether a cell is a replicating myoblast or differentiating myotube.
These developmental transitions can account for a general switch from repression to activation, but do not necessarily account for the simultaneous activation and repression of sets of genes during MyoD-mediated myogenesis. Our expression array study with an inducible MyoD in fibroblasts showed that MyoD activates the expression of several distinct temporal clusters of genes and simultaneously suppresses the expression of other gene sets (Bergstrom et al., 2002). In our current study, we use this model system of MyoD-mediated myogenesis to determine how a single transcription factor simultaneously activates and suppresses different sets of genes during myogenic differentiation. We demonstrate that MyoD directly regulates the transcription of microRNA (miRNA) expression that suppresses specific targets during myogenic differentiation. In addition, our demonstration that a MyoD-induced miRNA targets the Utrn RNA and can posttranscriptionally suppress expression through this sequence suggests that therapies of Duchenne muscular dystrophy based on increasing Utrn expression should include modulation of these posttranscriptional mechanisms.
Results
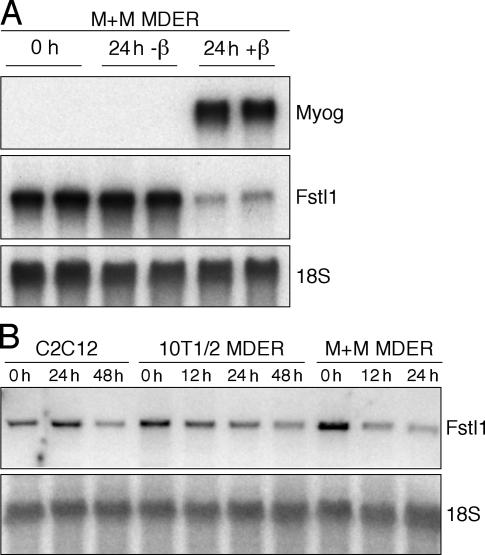
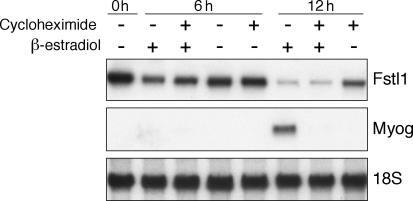
Previously, we have used microarray studies to show that MyoD both induces expression of a large number of genes and simultaneously elicits a reduction in the RNA abundance of a subset of genes (Bergstrom et al., 2002). To study the mechanism of decreased RNA abundance by MyoD, we focused on follistatin-like 1 (Fstl1). Although Fstl1 does not have a defined biological role in muscle physiology, we chose to study this gene because it showed a robust decrease in its message during the first 24 h of MyoD induction. As in our previous studies, we used mouse embryonic fibroblasts (MEFs) derived from mice with both the endogenous MyoD and Myf5 genes disrupted (M+M cells) that contain a constitutively expressed fusion protein between mouse MyoD and the hormone-binding domain of the estrogen receptor (MyoD estrogen receptor [MDER]; Hollenberg et al., 1993; Bergstrom et al., 2002). Transition of the cells to low-serum differentiation medium (DM; see Materials and methods) with the addition of β-estradiol synchronously induces MyoD activity and initiates differentiation in these cells (Bergstrom et al., 2002). Consistent with our previous findings, Fstl1 RNA decreased several fold at 24 h after the induction of MyoD activity by β-estradiol in the M+M cells, whereas the abundance of Myog, which is a downstream transcriptional target of MyoD, increased substantially (Fig. 1 A). The decreased abundance of Fstl1 RNA in response to MyoD induction and myogenic differentiation was also evident in the myoblast cell line C2C12 that expresses endogenous MyoD and in the 10T1/2 MEF cell line that constitutively expresses MDER (Fig. 1 B). Notably, the abundance of Fstl1 RNA decreases when MyoD is induced in the presence of the protein synthesis inhibitor cycloheximide, indicating that MyoD causes a decrease in this RNA in the absence of new protein synthesis (Fig. 2).
Figure 1.
Fstl1 is suppressed in response to MyoD expression. Northern blot analysis of Myog and Fstl1 expression at various times after the induction of MDER expression. RNA was harvested from cells maintained in growth medium (0 h) or induced in DM (+ β-estradiol) for the durations indicated. 18S RNA levels were analyzed as a loading control. (A) Analysis of Myog and Fstl1 levels in uninduced (0 h, 24 h –β) versus induced (24 h + β) M+M MDER cells. (B) Analysis of Fstl1 levels in C2C12, 10T1/2 MDER, and M+M MDER cells.
Figure 2.
Fstl1 is suppressed in the presence of MyoD and cycloheximide. Northern blot analysis of Fstl1 expression after MDER induction. MDER expression was induced by switching to DM with β-estradiol in the absence or presence of the protein synthesis inhibitor cycloheximide, as indicated. RNA was harvested at 6 or 12 h after treatment with β-estradiol and/or cycloheximide, as indicated. Fstl1 expression was also analyzed in the absence of cycloheximide and β-estradiol (0 h). Myog RNA levels were analyzed as a positive control for MDER activity and cycloheximide function. 18S RNA levels were analyzed as a loading control.
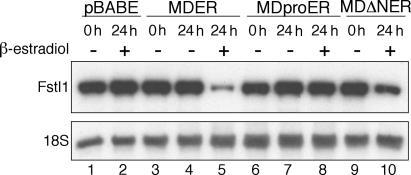
MyoD is a transcription factor with a well-defined DNA-binding domain and an N-terminal acidic activation domain. To determine whether the DNA-binding or activation functions of MyoD are necessary to suppress the abundance of Fstl1, we tested the activity of two MDER mutant proteins. MDproER has an inactivating point mutation (A114P) in the MyoD DNA-binding domain (Davis et al., 1990), whereas MDΔNER has a deletion of the MyoD activation domain (aa 3–56; Tapscott et al., 1988). These mutant proteins are expressed at levels comparable to wild-type MDER in M+M cells (unpublished data). Consistent with prior studies showing that the activation functions of E-protein heterodimers and other recruited factors can partially compensate for the absence of the MyoD activation domain on many promoters (Berkes et al., 2004), we found that MDΔNER had a modest but discernable effect on Fstl1 mRNA levels (Fig. 3, compare lanes 9 and 10), whereas the induction of the DNA binding–deficient MDproER mutant did not affect the abundance of Fstl1 (Fig. 3, compare lanes 7 and 8). Together, these data suggest that DNA binding by MyoD is required to diminish the abundance of Fstl1.
Figure 3.
MyoD DNA-binding and activation activities are required for repression of Fstl1. Northern blot analysis of Fstl1 expression in M+M cells either lacking MyoD expression (pBABE), or expressing MDER (wild type), MDproER (DNA-binding domain mutant), or MDΔNER (activation domain mutant). RNA was harvested from cells maintained either in growth medium (0 h), or in DM for 24 h with or without the addition of β-estradiol, as indicated. 18S RNA levels were analyzed as a loading control.
We next used chromatin immunoprecipitation (ChIP) to determine whether MyoD was down-regulating Fstl1 through direct binding to regulatory regions. We identified conserved regions within 5 kb of the promoter and used primers targeted to these regions (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200603039/DC1); however, because of the size of the fragmented DNA for the ChIPs it is likely that we effectively screened the entire region 2.5 kb upstream of the promoter. Using antisera to the MyoD protein, we looked for enrichment of MyoD at the Fstl1 regulatory regions, compared with negative (IgH) and positive (Myog) internal controls where we have previously determined MyoD binding (Bergstrom et al., 2002; Berkes et al., 2004; Penn et al., 2004). ChIP assays showed robust MyoD binding at the Myog regulatory region at 24 h after MyoD induction, but none of the regions in the vicinity of the Fstl1 promoter showed enrichment relative to the IgH internal negative control (Fig. S1), indicating that MyoD was not directly binding near the promoter of the Fstl1 gene.
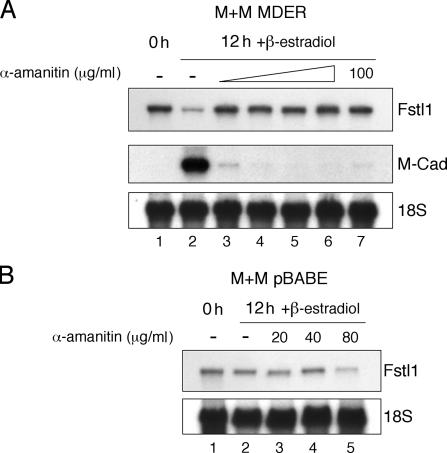
To determine whether RNA transcription is necessary for MyoD to decrease the abundance of Fstl1 RNA, we treated cells with α-amanitin, which, at low concentrations, is an RNA polymerase II inhibitor (Wieland and Faulstich, 1978). At doses of α-amanitin that are sufficient to prevent the transcription of MyoD target genes, we found that Fstl1 RNA was no longer down-regulated upon MyoD induction (Fig. 4).
Figure 4.
Suppression of Fstl1 in response to MDER induction requires transcription. Northern blot analysis of Fstl1 expression in the absence or presence of the transcriptional inhibitor α-amanitin. RNA was harvested from cells maintained either in growth medium (0 h) or in DM with β-estradiol for 12 h, in the presence of 0, 20, 40, 80, or 100 μg/ml α-amanitin, as indicated. 18S RNA levels were analyzed as a loading control. (A) Analysis of Fstl1 and M-cadherin expression in M+M MDER cells. α-Amanitin was added to the cells 15 min before the addition of β-estradiol, except in lane 7, where the reagents were added simultaneously. M-cadherin expression was analyzed as a positive control for MDER activity and α-amanitin function. (B) Analysis of Fstl1 expression in M+M pBABE cells, which do not express MDER.
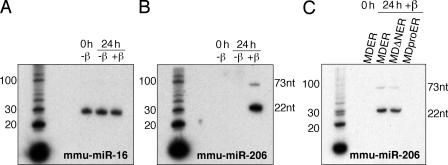
These results indicate that MyoD decreases the abundance of Fstl1 in a manner that requires DNA binding and transcription, but not protein translation, and is consistent with MyoD-regulated expression of a regulatory RNA, such as a miRNA or siRNA, which posttranscriptionally regulates the abundance of the Fstl1 mRNA. Although in many cases miRNAs block translation, RNA degradation can also be induced by miRNA (for review see Ambros, 2004; Yekta et al., 2004). We used the miRanda algorithm to identify several miRNAs that are predicted to potentially bind sites in Fstl1 (John et al., 2004). Within this candidate set of miRNAs, Northern blot analysis demonstrated that the abundance of miR-206 was dramatically up-regulated by the induction of MyoD activity (Fig. 5). Up-regulation of miR-206 was dependent on active MyoD, as cells that did not express MDER did not show miR-206 expression (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200603039/DC1). These data are consistent with recently published studies indicating an up-regulation of miR-206 in differentiated C2C12 myoblasts (Chen et al., 2006; Rao et al., 2006). In addition, miR-206 was not induced by the MDproER mutation in the DNA-binding domain (Fig. 5 C).
Figure 5.
miR-206 is up-regulated in response to MyoD expression. Northern blot analysis of miRNA expression. Total RNA was harvested from M+M cells that were maintained either in growth medium (0 h), or in DM with β-estradiol for 24 h, as indicated. M+M cells expressed wild-type MDER, MDΔNER, or MDproER, as indicated. miRNA was detected using radioactive end-labeled oligonucleotide probes complementary to the mature miRNA sequence. The sizes of the mature (22 nt) and pre-miRNA (73 nt) transcripts are indicated. A and B consist of the same RNA probed sequentially for miR-206 and -16. Radiolabeled 10-bp DNA ladder is shown as a size standard. (A) Analysis of miR-16 expression in M+M MDER cells. (B) Analysis of miR-206 expression in M+M MDER cells. (C) Analysis of miR-206 expression in M+M MDER, M+M MDΔNER, and M+M MDproER cells.
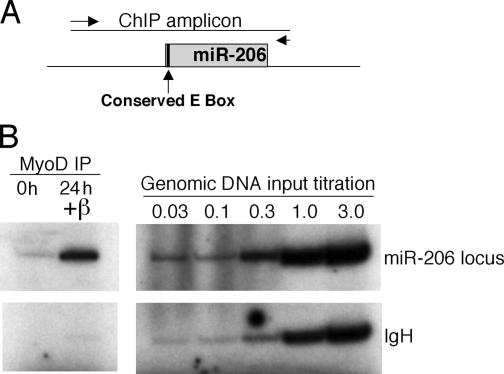
The locus encoding mmu-miR-206 is on chromosome 1 and the 5′ end of the pre–miR-206 closely coincides with the 5′ end of a longer transcript (AK132542). A conserved E box is near the predicted transcription start of the putative miR-206 hairpin and the AK132542 transcript. ChIP assays show robust MyoD binding to this putative regulatory region of miR-206 (Fig. 6). (In addition, while this manuscript was under review, Rao et al., 2006 identified miR-206 as a MyoD target by ChIP.) Northern and RT-PCR analysis demonstrate that the longer AK132542 transcript is also induced by MyoD (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200603039/DC1), suggesting that a larger RNA might be transcribed and processed to produce miR-206. It is interesting to note that the miR-133b sequence is also contained in the AK132542 transcript. miR-133 has recently been shown to be induced during differentiation of C2C12 muscle cells (Chen et al., 2006; Rao et al., 2006), and miR-133 is induced by MyoD together with miR-206 in M+M MDER cells (unpublished data). Therefore, it is possible that the induction of AK132542 by MyoD might lead to the production of both miR-206 and -133. Our data indicate, therefore, that MyoD directly binds the promoter region of the miR-206 precursor, and the binding is correlated with transcription of a longer precursor RNA and the appearance of the mature miR-206.
Figure 6.
MyoD binding is enriched at the miR-206 locus. ChIP analysis of MyoD binding. (A) Schematic representation of the genomic location of the putative miR-206 hairpin and chromatin IP amplicon. Predicted miR-206 hairpin is represented as a gray rectangle. The conserved E box that is the putative MyoD-binding site in the region is indicated. Positions of the primers used to produce the ChIP amplicon are represented as arrows. (B) ChIP assay. Chromatin from cells either maintained in growth medium (0 h) or in DM for 24 h (24 h +β) was immunoprecipitated with an antibody against MyoD and interrogated by duplex PCR. A genomic DNA titration was performed to indicate relative PCR efficiencies based on equivalent template amounts.
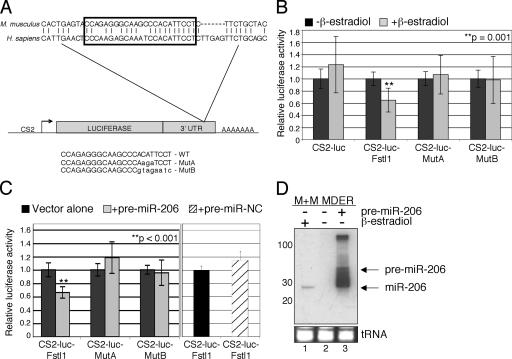
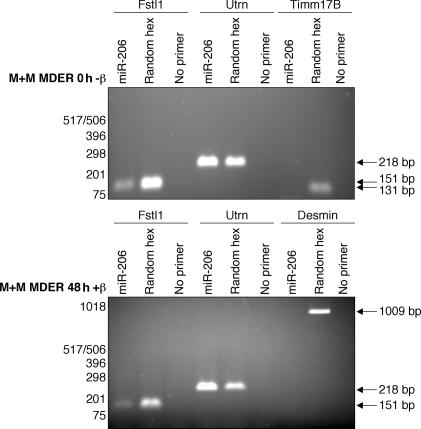
To determine whether the miR-206 can target the 3′UTR of the Fstl1 gene, we cloned three copies of the putative target sequence (Fig. 7 A) into the 3′ UTR of a luciferase reporter gene (CS2-luc-Fstl1) and, as a control, we cloned similar multimers of the target sequence containing mutations that should disrupt miR-206 binding (MutA and MutB). Compared with the parent vector, the expression of the CS2-luc-Fstl1 reporter was significantly suppressed by MyoD induction, whereas the MutA and MutB were not suppressed by MyoD (Fig. 7 B). These data suggest that MyoD activation results in specific repression targeted to the Fstl1 3′UTR. To show a direct effect of miR-206 on the CS2-luc-Fstl1 reporter, we cotransfected cells with a miR-206 hairpin precursor (pre–miR-206), which is processed by the M+M MDER cells to produce the mature, 22 nucleotide miR-206 (Fig. 7, C and D). Consistent with the effect of MDER induction, we see a decrease in CS2-luc-Fstl1 reporter activity, but not in MutA or MutB reporter activity, when cells are cotransfected with pre–miR-206. Together, these data demonstrate that the conserved sequence motif in the 3′-UTR of Fstl1 is targeted by miR-206. To confirm that miR-206 can target the Fstl1 mRNA, and to develop a method to identify additional miR-206 targets, we used a miR-206 oligonucleotide as a primer in a reverse transcriptase reaction to see if it would prime the Fstl1 mRNA, as detected by subsequent PCR. An oligonucleotide of the miR-206 sequence primed first strand synthesis from the Fstl1 mRNA, whereas neither desmin nor Timm17b (a constitutively expressed control RNA) were primed by miR-206 (Fig. 8). Cloning and sequencing the miR-206–primed cDNA confirmed that miR-206 binds to the predicted target site in Fstl1 and to an additional site further 3′ in the RNA, suggesting that multiple miR-206–binding sites might exist in the Fstl1 3′ UTR (unpublished data). In considering the aforementioned data, we conclude that MyoD down-regulates Fstl1 by transcriptionally activating the miR-206 gene, which posttranscriptionally regulates Fstl1 through the conserved motif in its 3′UTR sequence.
Figure 7.
miR-206 targets the Fstl1 3′UTR. (A) Schematic representation of the CS2-luciferase constructs containing multimers of the putative miR-206-binding site in the 3′ UTR of Fstl1. The nucleotides targeted by miR-206 are indicated by a boxed area. The sequences from mouse and human Fstl1 are shown to indicate the evolutionary conservation of this region. The murine sequence was multimerized and inserted into the 3′UTR of the luciferase gene. The mutations in the miR-206 target sequence that distinguish the MutA and MutB constructs are shown. (B) Induction of MyoD-ER expression represses CS2-luc-Fstl1 activity. Cells were transfected with CS2-luc, CS2-luc-Fstl1 (WT), CS2-luc-MutA, or CS2-luc-MutB. CS2-β-gal was cotransfected as an internal control. After transfection, cells were switched to DM with β-estradiol. Control cells were not treated with β-estradiol. (C) Exogenous pre–miR-206 targets CS2-luc-Fstl1. Cells were cotransfected with pre–miR-206 or pre–miR-Negative Control (NC) and the reporter constructs (MutA and MutB were not tested with pre–miR-NC, as they showed no response to pre–miR-206). Vector alone cells were not cotransfected with pre–miRs. (D) Exogenous pre–miR-206 is processed to produce mature miR-206 in M+M MDER cells. MiRNA Northern blot analysis of miR-206. Total RNA from M+M MDER cells transfected with 25 nM pre–miR-206 (lane 3) was compared with endogenous miR-206 from nontransfected cells that had been maintained either in growth medium (lane 2) or in DM with β-estradiol (lane 1). The positions of the unprocessed pre–miR-206 and the mature miR-206 are indicated. An ethidium bromide–stained image of the tRNA contained in each lane serves as a loading control. A radiolabeled 10-bp DNA ladder was used as a size standard (not depicted).
Figure 8.
Fstl1 and Utrn mRNA contain miR-206 target sites. RT-PCR analysis of Fstl1 and Utrn using a miR-206 oligo to prime first-strand synthesis. After collection of RNA from M+M MDER cells either maintained in growth medium (0 h −β) or cultured in DM with β-estradiol (48 h +β), reverse transcription was performed in which either a DNA oligo corresponding to miR-206, a random hexamer oligo mixture (positive control), or no primer (negative control) was used. PCR was subsequently performed with primers specific for Fstl1, Utrn, Timm17b, and desmin, as indicated. Timm17b and Desmin are not predicted targets of miR-206 and are used to demonstrate the specificity of miR-206 oligo priming.
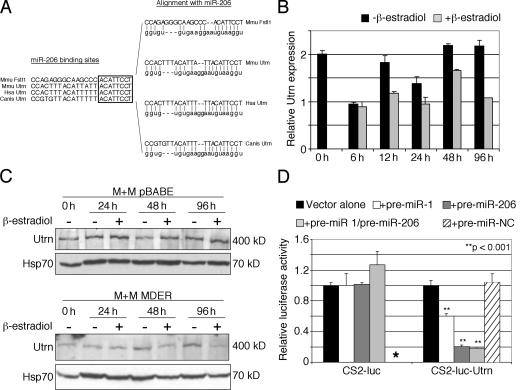
Although little is known about the potential function of Fstl1 in myogenesis, other predicted targets of miR-206 have been extensively studied. For example, Utrn is a predicted target of miR-206 (http://microrna.sanger.ac.uk/). Consistent with the predicted interaction, a miR-206 oligonucleotide can prime the Utrn mRNA in a reverse transcriptase reaction (Fig. 8), and cloning and sequencing of the miR-206–primed cDNA confirmed that the miR-206 oligonucleotide binds to the predicted target site in Utrn (not depicted). The predicted miR-206–binding site in Utrn is highly conserved in mouse, human, and dog and shares the same “seed” sequence as the miR-206 site in Fstl1 (Fig. 9 A). In our model system of MyoD-induced myogenesis, there is a small decrease in the abundance of the Utrn mRNA after 12 h of MyoD induction (Fig. 9 B) and a more marked decrease in the abundance of the protein at 48 and 96 h of MyoD induction, which is dependent on the presence of an active MyoD (Fig. 9 C, compare the pBABE control cells with the MDER cells). A CS2-luciferase reporter containing a multimer of the putative miR-206–binding site from the Utrn mRNA shows substantial inhibition when cotransfected with the pre–miR-206 construct (Fig. 9 D), demonstrating that this site is indeed targeted by miR-206. Another muscle-specific miRNA, miR-1, has high sequence homology to miR-206, and it is also induced in the M+M MDER cells (unpublished data). Transfection of pre–miR-1 had a significant, but more modest, suppression of the CS2–luc–Utrn construct, and cotransfection of both pre–miR-206 and -1 did not show synergistic activity. Therefore, we conclude that the induction of miR-206 by MyoD can alter the stability and/or the translation of subpopulations of RNAs, some of which are biologically relevant to the process of terminal myogenic differentiation. Furthermore, the demonstration that at least a portion of the suppression of Utrn expression occurs posttranscriptionally suggests specific therapies aimed at increasing or maintaining Utrn expression in Duchenne muscular dystrophy.
Figure 9.
miR-206 targets the Utrn 3′UTR. (A) The 3′UTR of Utrophin contains an evolutionarily conserved miR-206 target sequence. (left) Alignment between the miR-206 target sites of Mus musculus Fstl1 and the miR-206 target sites of Utrophin from M. musculus, Homo sapiens, and Canis familiaris. The highly conserved, 8-bp “seed” sequence for miR-206 target recognition is indicated by a box. (right) Alignment between the respective miR-206 target sites and miR-206. (B) Real-time PCR analysis of utrophin expression in M+M MDER cells. RNA was harvested from cells incubated for up to 96 h in DM in the presence or absence of β-estradiol, as indicated, and the relative abundance of utrophin transcripts was measured using Timm17b transcript abundance as a reference. (C) Western blot analysis of utrophin in M+M MDER cells. Lysates were prepared from M+M pBABE cells (lacking MDER) or M+M MDER cells that had been cultured for up to 96 h in DM in the presence or absence of β-estradiol, as indicated. Each sample contained 70 μg total protein, as determined by BCA assay. Samples were subjected to SDS-PAGE and immunoblotted with anti-utrophin antibody. The samples were subsequently immunoblotted with anti-Hsp70 antibody as a loading control. (D) Exogenous pre–miR-206 targets the CS2–luc–Utrn reporter in the absence of MDER expression. The miR-206 target sequence in the 3′UTR of Utrn was multimerized and inserted into the 3′UTR of the luciferase gene, as shown. Cells were cotransfected with pre–miR-206, -1 (alone and in combination with pre–miR-206), or -Negative Control (NC) reagent and the reporter constructs, as indicated. Vector alone cells were not cotransfected with pre–miR reagent. Asterisk indicates that the CS2-luc construct was not tested with the pre–miR-NC reagent, as it did not respond to pre–miR-1 or pre–miR-206.
Discussion
Expression of MyoD in a wide variety of cell types is sufficient to activate the program of skeletal muscle differentiation (Tapscott, 2005). Although different sets of genes are expressed in specific temporal patterns during muscle differentiation, we have shown that MyoD directly binds to the regulatory regions of genes expressed both early and late in the program (Bergstrom et al., 2002). Temporal patterning is achieved through a feed-forward mechanism, where transcription factors activated by MyoD early in the myogenic program feed-forward to cooperate with MyoD to activate genes expressed later in the program (Penn et al., 2004). In this manner, a single transcription factor can directly orchestrate the temporal program of gene activation during skeletal muscle differentiation. Our findings in this study demonstrate that the same transcription factor that has a central role in activating muscle-specific genes simultaneously induces miRNAs that repress gene expression during myogenic differentiation. Specifically, we demonstrate that MyoD acts as a transcriptional activator of the miR-206 pre–miRNA transcript and the induced high levels of the mature miR-206 result in the down-regulation of specific target genes, such as Fstl1 and Utrn.
We chose to investigate the regulation of Fstl1 because it was identified as a suppressed RNA in our time-course study of MyoD-regulated genes (Bergstrom et al., 2002). Fstl1 was originally identified as TSC-36 (TGF-β stimulated clone 36) in a screen for genes induced by TGF-β in mouse osteoblasts, and was observed to have some amino acid homology to Fst (Shibanuma et al., 1993). It is not currently known whether Fstl1 has overlapping or distinct functions from Fst, however, the different developmental expression patterns of these two genes (de Groot et al., 2000) suggest distinct biological functions. Additionally, a functional domain important for Fst activity, the activin-binding domain (Wang et al., 2000), is not conserved in Fstl1. Therefore, although Fst has been shown to antagonize activin and myostatin during myogenesis (Amthor et al., 2004), the role of Fstl1 in this process remains unknown.
In contrast, the decreased expression of Utrn protein in skeletal muscle is thought to be an important process in differentiation. During muscle differentiation, dystrophin, which is a Utrn paralog, replaces the Utrn protein in the dystrophin-associated glycoprotein complex. Importantly, the loss of dystrophin protein is the etiologic basis for Duchenne muscular dystrophy. The demonstration that constitutive expression of Utrn protein in the skeletal muscle of transgenic mice (Tinsley et al., 1996) can partially compensate for the loss of dystrophin has led to attempts to transcriptionally induce Utrn gene expression in mature muscle cells, in the hope that Utrn up-regulation might prove therapeutic to DMD patients. Our demonstration that miR-206 can suppress expression posttranscriptionally through a sequence in the Utrn RNA suggests that therapies based on posttranscriptional regulation of the Utrn RNA should also be explored.
Since the initial discovery that lin-4 acts as a regulatory RNA in Caenorhabditis elegans (Lee et al., 1993), there has been growing interest in understanding how miRNAs, acting posttranscriptionally, interface with well-characterized transcriptional regulatory networks to enforce precise regulation of gene expression programs in animals (Ambros, 2004). As a result, we are beginning to learn that miRNAs play important roles in embryonic development and cell fate (for review see Pasquinelli et al., 2005). Several groups have demonstrated tissue-specific distribution of miRNAs in developing embryos and adult animals (Lagos-Quintana et al., 2002; Mansfield et al., 2004; Hornstein et al., 2005). Further, it seems that several miRNAs act in the specification of cell lineage. For example, miR-181, -223, and -142 are preferentially expressed in mouse hematopoietic tissues, where mir-181 specifies the B cell lineage and miR-223 promotes granulocyte differentiation (Chen et al., 2004; Fazi et al., 2005). Recently, the role of miRNAs in skeletal and cardiac muscle biology has been the focus of intense interest.
To date, three muscle-specific miRNAs have been identified: miR-1, -133, and -206. miR-1 expression is strictly limited to cardiac and skeletal muscle in Drosophila melanogaster, zebrafish, and mouse, although species-specific variations in expression patterns have been noted (Biemar, et al., 2005; Brennecke et al., 2005; Kwon et al., 2005; Sokol and Ambros, 2005; Wienholds and Plasterk, 2005; Zhao et al., 2005). Expression of miR-1 is up-regulated in response to differentiating signals in cardiac and skeletal muscle in vivo and differentiated myoblasts in vitro, and it has been shown to be activated by several factors, including SRF, Twist, myocardin, and Mef2 (Kwon et al., 2005; Sokol and Ambros, 2005; Zhao et al., 2005). Another muscle-specific miRNA, miR-133, has been shown to promote proliferation of myoblasts by antagonizing SRF (Chen et al., 2006). Recently, a role for miR- 181 in muscle differentiation and regeneration was also described (Naguibneva et al., 2006). We now show that miR-206 is a direct transcriptional target of MyoD and functions to suppress targets during muscle differentiation.
Although our study has focused on a single miRNA and its targets, we assume that multiple miRNAs will be similarly regulated by MyoD. Similarly, it is likely that the related myogenic bHLH proteins (Myf5, Myog, and Mrf4) will also induce miR-206. Indeed, our recent ChIP study identified four additional miRNA promoter regions bound by MyoD: miR-100, -138-2, -191, and -22 (Cao et al., 2006), and a study published while this paper was under review showed that MyoD and Myog bind the putative regulatory regions of miR-206, -1, and -133 (Rao et al., 2006). Because many of the targets of miRNAs can be translationally inhibited in the absence of RNA degradation, the expression array studies we used as the basis for investigating the regulation of Fstl1 are likely underestimating the extent of genes suppressed during myogenesis. Additionally, because it has been demonstrated that transfection of a single miRNA (e.g., miR-1) into HeLa cells decreases nearly 100 mRNA transcripts (Lim et al., 2005), it seems likely that future studies will show multiple additional genes to be targeted by miRNAs in response to MyoD.
Materials and methods
Cell culture
MEFs null for both MyoD and Myf5 (M+M cells) were infected with a retrovirus containing the pBABE vector expressing a puromycin-resistance gene and MDER (Hollenberg et al., 1993). Mutant MDER proteins (MDproER and MDΔNER) were constructed in the same vector context. Control cells (M+M pBABE) were infected with virus containing only vector and selectable marker. Cells were maintained in growth medium, which consisted of DME containing 1% l-glutamine and 10% bovine calf serum (Hyclone). Infected cells were selected in 1.2 μg/ml puromycin. To induce expression of MDER, cells were switched to DM, which consisted of DME containing 1% l-glutamine, 0.5% horse serum, 10 μg/ml insulin, 10 μg/ml transferrin, and 10−7 M β-estradiol.
Transient transfection and luciferase assay
All transient transfection/luciferase assays were performed in triplicate on 35-mm tissue culture dishes (Corning). For transfections, each plate was seeded with 105 cells and incubated overnight at 37°C. Cells were transfected using Superfect reagent (QIAGEN) as specified by the manufacturer. The constructs used for transfections contained the indicated regions of Fstl1 or Utrn 3′UTR sequence inserted 3′ of the firefly luciferase gene, under the control of the CMV promoter in the CS2 vector background. Each plate was transfected with 100 ng of CS2-luc reporter vector and 2 μg of empty CS2 vector. Where indicated, cells were cotransfected with pre–miR-206, -1, or -Negative Control #1 (Ambion) to a final concentration of 25 nM, and luciferase activity was measured 24 h after transfection. Where indicated, MDER activity was induced in transfected cells at 16–24 h after transfection by switching cells to DM with 10−7 M β-estradiol (uninduced cells were switched to DM without β-estradiol), and luciferase activity was measured at 24 h after induction. For all experiments, cells were cotransfected with 200 ng of CS2-β-gal, and assayed for β-galactosidase activity as an internal control for transfection efficiency using the MUG assay. Luciferase assays and MUG assays were performed as previously described (Bergstrom and Tapscott, 2001), using AutoLumat LB 953 (BERTHOLD TECHNOLOGIES) and MicroFluor (Dynatech) instrumentation.
Northern blot analysis
Cultured cells were harvested by scraping, and RNA was prepared using the RNEasy kit (QIAGEN). Northern blot analysis was performed according to standard techniques, which were previously described (Bergstrom et al., 2002). Probes were generated by PCR amplification of cDNA reverse transcribed from total M+M MDER RNA using an oligo-dT primer and SuperScript II (Invitrogen), and were radioactively labeled using Ready-to-Go DNA labeling beads (GE Healthcare). Images were captured on film, digitized, and if needed, minor linear adjustments in contrast were made using Adobe Photoshop software.
miRNA northern blot analysis
Total RNA was prepared using Trizol (Invitrogen) extraction, per the manufacturer's instructions. Trizol purification was followed by acid phenol extraction to remove any DNA contamination. Each sample contained 20 μg RNA. Samples were separated electrophoretically in a 20% polyacrylamide/8 M urea/1× TBE gel. RNA was electroblotted onto Nytran SPC nylon membrane in 1× TBE at 250 mA for 45 min, and was fixed to the membrane by UV cross-linking. Blots were hybridized overnight at 35°C in Ultrahybe Oligo buffer (Ambion) with radiolabeled oligonucleotide probes complementary to the mature miRNA sequences of interest. Images were captured on film, digitized, and if needed, minor linear adjustments in contrast were made using Photoshop software (Adobe).
ChIP assay
ChIP analysis was performed as previously described (Filippova et al., 2001; Penn et al., 2004). Precipitations from 750–1,000 μg M+M MDER cell lysate were incubated overnight at 4°C with 5 μl anti-MyoD antiserum (Tapscott et al., 1988). Duplex PCR was performed by coamplifying test control regions from MyoD target genes with an internal control region from the IgH enhancer. Amplification reactions included 32P-dCTP for incorporation into PCR product that was then detected and quantified by PhosporImager analysis using ImageQuant software (Molecular Dynamics). For input samples, a titration of 0.03–3.00 ng of genomic DNA was used as a template for duplex PCR to establish relative ratios of PCR product in the absence of any asymmetry in target abundance. For MyoD IPs, 5% of the IP sample was used per reaction and amplified over 32 PCR cycles. Results were analyzed for enrichment at the muscle-specific target sequence relative to the target sequence/control sequence ration established using input DNA sample. PCR linearity was confirmed by titrating the input DNA over a 30-fold range. Efficiency of MyoD IP was confirmed by analysis of MyoD enrichment at the Myog promoter, as described previously (Bergstrom et al., 2002).
Primer extension
Total RNA was collected from M+M MDER cells cultured in either growth medium (0 h) or DM with β-estradiol (48 h) using the RNEasy kit (QIAGEN). RT-PCR was performed by standard methodology using 0.5 ug of total RNA per sample. For reverse transcription, a DNA oligonucleotide corresponding to mir-206 (5′-TGGAATGTAAGGAAGTGTGTGG-3′) was used as a primer, a sample primed with random hexamer was used as a positive control, and an unprimed sample was used as a negative control. Thermocycler conditions for reverse transcription with random hexamer primers were as follows: 45°C/15 min, 50°C/15 min, 55°C/15 min, and 75°C/15 min. For the miR-206–primed samples, higher temperatures were used to achieve higher target specificity as follows: 50°C/45 min, 55°/15 min, 60°C/15 min, and 75°C/15 min. The thermocycler conditions for the unprimed negative control samples were the same as for the miR-206–primed samples. After reverse transcription, all samples were treated with RNaseH for 1 h at 37°C before performing PCR. For the detection of cDNAs by PCR, 28 cycles were performed for Fstl1 and Utrn; 26 cycles for desmin and 29 cycles for Timm17b. For detection of Fstl1, the primer sequences were as follows: Fstl1-F, 5′-TCACAGCAGCAATGCCATCATCAA-3′; Fstl1-R, 5′-GATTGGCCAACAGACACTGCAGCTA-3′. For the detection of Utrn, the primer sequences were as follows: Utrn-F, 5′-TGC CAATCCCAAGACCCATTCAAC; and Utrn-R, 5′-TCAGTGACAAATGCTTTACCACCTCCA-3′.
For the detection of desmin, the primer sequences used were as follows: desmin-F: 5′-CTCGAGCAGGCTTCGGTACC-3′; desmin-R: 5′-CTTGG- CGCAGCGCATCGTTG-3′.
For the detection of Timm17b, the same primer set was used as for real-time PCR (see next section). PCR products were resolved on 1% agarose gels and stained with SYBR gold (Invitrogen).
Quantitative real-time PCR
Cultured cells were harvested by scraping and RNA was prepared using the RNEasy kit (QIAGEN). Real-time PCR was performed using TaqMan universal PCR mix reagent, according to the manufacturer's instructions (Applied Biosystems). For detection of utrophin, the primers and probe sequences were as follows: Utrn-F primer, 5′-GGCAGAACGAATTCAGTGAC-3′; Utrn-R primer, 5′-ATCACTGATGGGTGGTTTCC-3′; and Utrn probe, 5′-CCAAATGGATAAACGCTCGATTTTCCA-3′.
For detection of Timm17b, the primers and probe sequences were as follows: Timm17b-F primer, 5′-TGTCATTGGTGGTGGAGTCT-3′; Timm17b-R primer, 5′-ACTGCAAAGCTTCCTCCAAT-3′; and Timm17b probe, 5′-TGCTGTGAGGATCCGGGCAC-3′.
Real-time PCR was performed on an Sequence Detection System instrument (ABI Prism 7900HT; Applied Biosystems), and expression levels were quantitated using SDS 2.1 software (Applied Biosystems). Each sample was assayed in triplicate; data represents the mean and SD for the triplicates. The relative expression levels of utrophin were normalized to those of Timm17b in the same samples.
Western blot
Cultured cells were harvested by scraping, resuspended in RIPA lysis buffer with 5% SDS (150 mM NaCl, 10 mM Tris, pH 7.2, 1% Triton X-100, 1% deoxycholate, and 5 mM EDTA), and homogenized by repeated manipulation through a 22-gauge needle. Protein concentration for each lysate preparation was determined by BCA assay. Samples were concentrated by methanol-chloroform extraction (Wessel and Flugge, 1984) and resuspended in loading buffer. Each sample analyzed contained 70 μg total protein. Samples were electrophoresed on a 6% SDS-PAGE and transferred to nitrocellulose for 3.5 h at 500 mA. Utrophin was detected using goat anti-utrophin (E-16) polyclonal antibody (Santa Cruz Biotechnology, Inc.); Hsp70 was detected using mouse anti-Hsp70 monoclonal antibody (Stressgen). Images were captured on film, digitized, and if needed, minor linear adjustments in contrast were made using Photoshop software.
Online supplemental material
Fig. S1 shows ChIP analysis of MyoD binding at the Fstl1 promoter, where MyoD binding was not detected. Fig. S2 shows that M+M pBABE cells (M+M cells that do not express MDER) do not express miR-206 upon switching to DM. Fig. S3 shows that MyoD induces the expression of the AK132542 transcript. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200603039/DC1.
Supplementary Material
Acknowledgments
We acknowledge Bennett Penn, Sarah Mahoney, Erica Jacobs, and Stavros Lomvardas for helpful discussions and technical advice.
This work was supported by National Institutes of Health National Institute of Arthritis and Musculoskeletal Diseases (NIH NIAMS) grant AR045113 and NIH National Institute of Neurological Disorders and Stroke grant NS046788 (to S.J. Tapscott); NIH Chromosome Metabolism and Cancer Training grant T32 CA09657 (to M.I. Rosenberg); and NIH Chromosome Metabolism and Cancer Training grant T32 CA09657-14 and NIH NIAMS grant F32 AR052581 (S.A. Georges).
M.I. Rosenberg and S.A. Georges contributed equally to this paper.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; DM, differentiation medium; Fstl1, follistatin-like 1; MDER, MyoD estrogen receptor; MEF, mouse embryonic fibroblast; miRNA, microRNA.
References
- Ambros, V. 2004. The functions of animal microRNAs. Nature. 431:350–355. [DOI] [PubMed] [Google Scholar]
- Amthor, H., G. Nicholas, I. McKinnell, C.F. Kemp, M. Sharma, R. Kambadur, and K. Patel. 2004. Follistatin complexes Myostatin and antagonises Myostatin-mediated inhibition of myogenesis. Dev. Biol. 270:19–30. [DOI] [PubMed] [Google Scholar]
- Bergstrom, D.A., and S.J. Tapscott. 2001. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol. Cell. Biol. 21:2404–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom, D.A., B.H. Penn, A. Strand, R.L. Perry, M.A. Rudnicki, and S.J. Tapscott. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 9:587–600. [DOI] [PubMed] [Google Scholar]
- Berkes, C.A., D.A. Bergstrom, B.H. Penn, K.S. Seaver, P.S. Knoepfler, and S.J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 14:465–477. [DOI] [PubMed] [Google Scholar]
- Biemar, F., R. Zinzen, M. Ronshaugen, V. Sementchenko, J.R. Manak, and M.S. Levine. 2005. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 102:15907–15911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke, J., A. Stark, and S.M. Cohen. 2005. Not miR-ly muscular: microRNAs and muscle development. Genes Dev. 19:2261–2264. [DOI] [PubMed] [Google Scholar]
- Buckingham, M., L. Bajard, T. Chang, P. Daubas, J. Hadchouel, S. Meilhac, D. Montarras, D. Rocancourt, and F. Relaix. 2003. The formation of skeletal muscle: from somite to limb. J. Anat. 202:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y., R.M. Kumar, B.H. Penn, C.A. Berkes, C. Kooperberg, L.A. Boyer, R.A. Young, and S.J. Tapscott. 2006. Global and gene-specific analyses show distinct roles for MyoD and Myog at a common set of promoters. EMBO J. 25:502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti, G., M. Di Padova, B. Micales, G.E. Lyons, and V. Sartorelli. 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18:2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Z., L. Li, H.F. Lodish, and D.P. Bartel. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science. 303:83–86. [DOI] [PubMed] [Google Scholar]
- Chen, J.F., E.M. Mandel, J.M. Thomson, Q. Wu, T.E. Callis, S.M. Hammond, F.L. Conlon, and D.Z. Wang. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38:228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J., M.L. Costa, C.S. Mermelstein, C. Chagas, S. Holtzer, and H. Holtzer. 1990. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc. Natl. Acad. Sci. USA. 87:7988–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R.L., P.F. Cheng, A.B. Lassar, and H. Weintraub. 1990. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 60:733–746. [DOI] [PubMed] [Google Scholar]
- de Groot, E., A. Feijen, D. Eib, A. Zwijsen, H. Sugino, G. Martens, and A.J. Van Den Eijnden-Van Raaij. 2000. Expression patterns of follistatin and two follistatin-related proteins during mouse development. Int. J. Dev. Biol. 44:327–330. [PubMed] [Google Scholar]
- de la Serna, I.L., K.A. Carlson, and A.N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187–190. [DOI] [PubMed] [Google Scholar]
- de La Serna, I.L., Y. Ohkawa, C.A. Berkes, D.A. Bergstrom, C.S. Dacwar, S.J. Tapscott, and A.N. Imbalzano. 2005. MyoD targets chromatin complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25:3997–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi, F., A. Rosa, A. Fatica, V. Gelmetti, M.L. De Marchis, C. Nervi, and I. Bozzoni. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 123:819–831. [DOI] [PubMed] [Google Scholar]
- Filippova, G.N., C.P. Thienes, B.H. Penn, D.H. Cho, Y.J. Hu, J.M. Moore, T.R. Klesert, V.V. Lobanenkov, and S.J. Tapscott. 2001. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 28:335–343. [DOI] [PubMed] [Google Scholar]
- Fulco, M., R.L. Schiltz, S. Iezzi, M.T. King, P. Zhao, Y. Kashiwaya, E. Hoffman, R.L. Veech, and V. Sartorelli. 2003. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 12:51–62. [DOI] [PubMed] [Google Scholar]
- Hollenberg, S.M., P.F. Cheng, and H. Weintraub. 1993. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc. Natl. Acad. Sci. USA. 90:8028–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein, E., J.H. Mansfield, S. Yekta, J.K. Hu, B.D. Harfe, M.T. McManus, S. Baskerville, D.P. Bartel, and C.J. Tabin. 2005. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 438:671–674. [DOI] [PubMed] [Google Scholar]
- John, B., A.J. Enright, A. Aravin, T. Tuschl, C. Sander, and D.S. Marks. 2004. Human MicroRNA targets. PLoS Biol. 2:E363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C., Z. Han, E.N. Olson, and D. Srivastava. 2005. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl. Acad. Sci. USA. 102:18986–18991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735–739. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., R.L. Feinbaum, and V. Ambros. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75:843–854. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., N.C. Lau, P. Garrett-Engele, A. Grimson, J.M. Schelter, J. Castle, D.P. Bartel, P.S. Linsley, and J.M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773. [DOI] [PubMed] [Google Scholar]
- Mal, A., and M.L. Harter. 2003. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl. Acad. Sci. USA. 100:1735–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal, A., M. Sturniolo, R.L. Schiltz, M. Ghosh, and M.L. Harter. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20:1739–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, J.H., B.D. Harfe, R. Nissen, J. Obenauer, J. Srineel, A. Chaudhuri, R. Farzan-Kashani, M. Zuker, A.E. Pasquinelli, G. Ruvkun, et al. 2004. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 36:1079–1083. [DOI] [PubMed] [Google Scholar]
- Molkentin, J.D., and E.N. Olson. 1996. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6:445–453. [DOI] [PubMed] [Google Scholar]
- Naguibneva, I., M. Ameyar-Zazoua, A. Polesskaya, S. Ait-Si-Ali, R. Groisman, M. Souidi, S. Cuvellier, and A. Harel-Bellan. 2006. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 8:278–284. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, A.E., S. Hunter, and J. Bracht. 2005. MicroRNAs: a developing story. Curr. Opin. Genet. Dev. 15:200–205. [DOI] [PubMed] [Google Scholar]
- Penn, B.H., D.A. Bergstrom, F.J. Dilworth, E. Bengal, and S.J. Tapscott. 2004. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev. 18:2348–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pownall, M.E., M.K. Gustafsson, and C.P. Emerson Jr. 2002. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 18:747–783. [DOI] [PubMed] [Google Scholar]
- Puri, P.L., and V. Sartorelli. 2000. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J. Cell. Physiol. 185:155–173. [DOI] [PubMed] [Google Scholar]
- Rao, P.K., R.M. Kumar, M. Farkhondeh, S. Baskerville, and H.F. Lodish. 2006. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA. 103:8721–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibanuma, M., J. Mashimo, A. Mita, T. Kuroki, and K. Nose. 1993. Cloning from a mouse osteoblastic cell line of a set of transforming-growth- factor-beta 1-regulated genes, one of which seems to encode a follistatin-related polypeptide. Eur. J. Biochem. 217:13–19. [DOI] [PubMed] [Google Scholar]
- Sokol, N.S., and V. Ambros. 2005. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 19:2343–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott, S.J. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 132:2685–2695. [DOI] [PubMed] [Google Scholar]
- Tapscott, S.J., R.L. Davis, M.J. Thayer, P.F. Cheng, H. Weintraub, and A.B. Lassar. 1988. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 242:405–411. [DOI] [PubMed] [Google Scholar]
- Tinsley, J.M., A.C. Potter, S.R. Phelps, R. Fisher, J.I. Trickett, and K.E. Davies. 1996. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. Nature. 384:349–353. [DOI] [PubMed] [Google Scholar]
- Wang, Q., H.T. Keutmann, A.L. Schneyer, and P.M. Sluss. 2000. Analysis of human follistatin structure: identification of two discontinuous N-terminal sequences coding for activin A binding and structural consequences of activin binding to native proteins. Endocrinology. 141:3183–3193. [DOI] [PubMed] [Google Scholar]
- Weintraub, H., S.J. Tapscott, R.L. Davis, M.J. Thayer, M.A. Adam, A.B. Lassar, and A.D. Miller. 1989. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA. 86:5434–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel, D., and U.I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141–143. [DOI] [PubMed] [Google Scholar]
- Wieland, T., and H. Faulstich. 1978. Amatoxins, phallotoxins, phallolysin, and antamanide: the biologically active components of poisonous Amanita mushrooms. CRC Crit. Rev. Biochem. 5:185–260. [DOI] [PubMed] [Google Scholar]
- Wienholds, E., and R.H. Plasterk. 2005. MicroRNA function in animal development. FEBS Lett. 579:5911–5922. [DOI] [PubMed] [Google Scholar]
- Yekta, S., I.H. Shih, and D.P. Bartel. 2004. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., E. Samal, and D. Srivastava. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 436:214–220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.