Abstract
Skeletal muscle side population (SP) cells are thought to be “stem”-like cells. Despite reports confirming the ability of muscle SP cells to give rise to differentiated progeny in vitro and in vivo, the molecular mechanisms defining their phenotype remain unclear. In this study, gene expression analyses of human fetal skeletal muscle demonstrate that bone morphogenetic protein 4 (BMP4) is highly expressed in SP cells but not in main population (MP) mononuclear muscle-derived cells. Functional studies revealed that BMP4 specifically induces proliferation of BMP receptor 1a–positive MP cells but has no effect on SP cells, which are BMPR1a-negative. In contrast, the BMP4 antagonist Gremlin, specifically up-regulated in MP cells, counteracts the stimulatory effects of BMP4 and inhibits proliferation of BMPR1a-positive muscle cells. In vivo, BMP4-positive cells can be found in the proximity of BMPR1a-positive cells in the interstitial spaces between myofibers. Gremlin is expressed by mature myofibers and interstitial cells, which are separate from BMP4-expressing cells. Together, these studies propose that BMP4 and Gremlin, which are highly expressed by human fetal skeletal muscle SP and MP cells, respectively, are regulators of myogenic progenitor proliferation.
Introduction
In skeletal muscle, satellite cells have long been described as “reserve” or “stem” cells. They are located between the basal lamina and the sarcolemma of myofibers and are able to self-renew and differentiate into mature muscle (Mauro, 1961; Armand et al., 1983; Zammit and Beauchamp, 2001). Satellite cells are the most efficient cell type in skeletal muscle repair after acute injury (Schultz et al., 1985; Darr and Schultz, 1987; Appell et al., 1988; Grounds and Yablonka-Reuveni, 1993; Partridge, 2004; Sherwood et al., 2004; Collins et al., 2005). However, contribution of other stem-like cells to muscle regeneration has been reported (Shi and Garry, 2006). One of these additional cell populations is the so-called side population (SP), which has been isolated from skeletal muscle using the FACS based on its greater ability to efflux the fluorescent dye Hoechst 33342, when compared with the main population (MP; Gussoni et al., 1999; Jackson et al., 1999; Asakura et al., 2002; Montanaro et al., 2004). In vivo, mouse muscle SP cells can fuse to dystrophic myofibers after systemic delivery (Gussoni et al., 1999) and can give rise to Myf5-positive myogenic cells after intramuscular injection in acutely injured muscle (Asakura et al., 2002). In vitro, muscle SP cells can adapt myogenic specification after coculture with C2C12 (Asakura et al., 2002) or are able to express the myogenic marker Pax7 after intravenous injections into mdx5cv mice (Bachrach et al., 2004). Despite studies supporting the ability of SP cells to give rise to differentiated progeny in vitro and in vivo, the molecular pathways that define their phenotype remain unclear. We hypothesized that the specific molecular networks responsible for the phenotype of SP cells could be identified by global gene expression analysis.
During embryonic development, a morphogen gradient of bone morphogenetic protein 4 (BMP4) and its antagonists plays important roles in mesoderm induction, establishment of dorso-ventral polarity, ectodermal differentiation, somite formation, and myogenesis induction (Hogan et al., 1994; Sasai and De Robertis, 1997; Dale and Jones, 1999; Giudice, 2001; Wang and Ferguson, 2005). In the paraxial mesoderm, local variation of BMP4 concentrations created by interaction between BMP4 and its antagonists are known to differentially affect the induction of Pax3 and MyoD (Reshef et al., 1998). In developing somites, BMP4 is expressed by ventral cells that give rise to the sclerotome, whereas BMP4 antagonists, such as Noggin, Chordin, Gremlin, and Follistatin are expressed in the dorsal part, which gives rise to the dermomyotome. A depletion of the BMP antagonists Noggin, Chordin, and Follistatin leads to a catastrophic loss of dorsal structures in Xenopus laevis (Khokha et al., 2005).
In the present study, microarray analysis revealed that BMP4, a known repressor of myogenic differentiation (Dahlqvist et al., 2003; Liu and Harland, 2003), is highly expressed in muscle SP cells, whereas its antagonist, Gremlin (Hsu et al., 1998; Merino et al., 1999; Zuniga et al., 1999; Topol et al., 2000), is up-regulated in MP cells. Functional studies demonstrate that BMP4 expressed by muscle SP cells induces proliferation of BMP receptor 1a (BMPR1a)–positive MP cells, and this effect can be reversed by Gremlin. Detection of BMP4+ and BMPR1a+ cells by immunohistochemistry in human fetal skeletal muscle revealed that BMP4+ cells are located near BMPR1a+ cells in the interstitial spaces, supporting the hypothesis that interactions between these cells occur in vivo. Gremlin is expressed by mature myofibers and interstitial muscle cells, which are separate from BMP4-expressing cells. Our results propose a functional role for BMP4 and Gremlin, which are expressed by muscle SP and MP cells, respectively, as regulators of proliferation and differentiation of myogenic progenitors in human fetal skeletal muscle.
Results
SP cells are present in human skeletal muscle
SP cells have been isolated from multiple tissues (Goodell et al., 1996, 1997), including murine skeletal muscle (Gussoni et al., 1999; Jackson et al., 1999; Asakura et al., 2002; Montanaro et al., 2004). To identify SP cells in human skeletal muscle, dissociated mononuclear cells from discarded muscle samples of individuals aged 14 gestational weeks to 63 yr were stained with the vital DNA dye Hoechst 33342. The dye concentration used for each sample was individually optimized because of observed interindividual variability in sensitivity to Hoechst 33342 dye (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200511036/DC1). After the initial optimization, independent SP cell isolations from the same individual demonstrated minimal variability (Fig. S1, C–F). For fetal samples, the optimal Hoechst dye concentration ranged from 3–9 μg/ml, whereas for adults it ranged from 7.5 to 12.5 μg/ml (Fig. S1, G and H). A control sample stained in the presence of reserpine allowed definition of the appropriate SP gate. Because fetal samples contained the highest proportion of muscle SP cells (Fig. S1), all subsequent studies were performed on human fetal muscle SP cells.
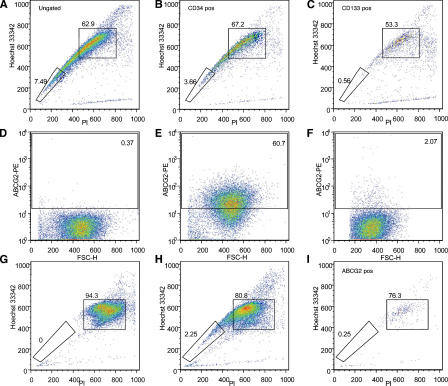
To identify muscle SP-specific cell surface markers, the expression of two antigens present on stem cells from other tissues, CD34 (Krause et al., 1996; Asakura et al., 2002) and CD133 (Miraglia et al., 1997; Yin et al., 1997), was analyzed. As shown in Fig. 1 (B and C), only 3.6% of CD34-positive cells and 0.56% CD133-positive cells were detected within the SP gate. The high dye efflux ability of SP cells has been previously attributed to the function of the ABCG2 transporter, which is expressed in multiple tissues, including skeletal muscle (Zhou et al., 2001; Martin et al., 2004; Meeson et al., 2004). In our experiments, the ABCG2 transporter was detected in 2% of the cells in human fetal muscle (Fig. 1 F), with 76% of these cells present within the MP gate and only 0.25% in the muscle SP gate (Fig. 1 I). In control samples of mouse 3T3 cells transfected with either a mock vector (Fig. 1 D) or with a vector encoding the human ABCG2 cDNA (Fig. 1 E), 0.37 and 60.7% of ABCG2-positive cells were detected, respectively. Thus, our studies indicate that human fetal skeletal muscle–derived SP cells are largely ABCG2-negative.
Figure 1.
Expression of cell surface antigens in human fetal skeletal mononuclear cells. (A–C) Flow cytometry analysis of 18-wk human fetal skeletal muscle cells. Cells were costained with 3 μg/ml Hoechst 33342, anti–CD34-FITC, anti–CD133-PE, and PI. (A) Cells stained with Hoechst 33342. 7.49% of the cells are detected in the SP gate and 62.9% in the MP gate. (B) CD34-positive cells stained with Hoechst 33342. 3.66% of the cells are detected in the SP gate and 67.2% in the MP. (C) CD133-positive cells stained with Hoechst 33342. 0.56% of the cells are detected in the SP gate and 53.3% in the MP gate. (D–I) Expression of ABCG2 (D) 3T3 cells transfected with a mock vector. 99.6% of the cells are ABCG2-negative. (E) 3T3 cells transfected with human ABCG2. 60.7% of the cells are ABCG2-positive. (F–I) Three-color flow cytometry (Hoechst 33342, PI, and ABCG2-PE) staining profiles of 18-wk-old human fetal skeletal muscle. Cells were costained with 3 μg/ml Hoechst 33342, anti–ABCG2-FITC, and PI. (F) 2% of human fetal skeletal myoblasts express ABCG2. (G–H) Hoechst/PI profiles of all the cells stained in the presence (G) or absence (H) of reserpine. 2.25% of cells were detected as SP. Hoechst/PI profile of ABCG2-positive cells (I) showed only one ABCG2-positive cell detected in the SP gate.
Previous studies reported that a variable percentage of murine skeletal muscle SP cells are of hematopoietic origin and express the pan-hematopoietic marker CD45 (McKinney-Freeman et al., 2002). We found that only 0.44% of human fetal skeletal muscle SP cells expressed CD45, suggesting that >99% of these cells are not of hematopoietic origin (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200511036/DC1).
Differential expression of BMP4 and Gremlin in human skeletal muscle SP and MP cells
To characterize the repertoire of expressed genes and identify a developmental hierarchy among mononuclear cells within human muscle, microarray analyses were performed on sorted, noncultured human fetal skeletal muscle SP (n = 10) and MP (n = 9) cells. A detailed description of the samples used in the microarray studies is provided in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200511036/DC1). The percentage of SP cells in fetal muscle samples derived from 15 different individuals ranged between 0 and 6.57%, with no significant correlation to gestational age or gender.
A geometric fold change analysis of the microarray data (Kho et al., 2004) identified 222 unique genes/ESTs as differentially regulated between human fetal skeletal muscle SP and MP cells (Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200511036/DC1). Of these genes, 162 known genes and 4 ESTs were significantly overexpressed in SP cells compared with MP cells and were called SP genes, whereas 60 genes were significantly underexpressed in the SP population and were called MP genes (Table S2). Gene ontology analysis of the differentially expressed genes using DAVID (http://niaid.abcc.ncifcrf.gov/) indicated that SP cells express high levels of transcriptional repressors and negative cell-cycle regulators, whereas MP cells are enriched for genes involved in metabolism, DNA replication, and cell surface membrane proteins (Table I).
Table I.
Genes differentially expressed by human fetal skeletal muscle SP/MP cells
| Functional category | Genes up-regulated in SP | Genes up-regulated in MP |
|---|---|---|
| Transcription | CCR5, RLF, ZNF44, SIRT1, ZNF297B, TLE4, HBP1, HBXAP, DDIT3, MYST4, MECP2, NFATC1, NSBP1, EGR3, ZHX2, NFIB, ELF2, NFX1, HEY2, BLZF1, ZNF667, TBX19, SIRT4, ZNF304, FLJ11011, SIX1, ZNF230, JARID1A, MBD2, ZNF222, MLL, ZNF451, NR3C2, CIR, ARID4A, TMF1, CAMTA1, PER1, ZNF20, NR4A2, ZNF211, MAF, MLLT3, TNRC9 |
LMO4, MCM4, POLR3K, PARP1, TFDP1 |
| Development | SIRT1, CDC2L5, MYLIP, MPZ, EGR3, DGCR2, HEY2, BLZF1, TBX19, LECT1, SIRT4, WDR33, SIX1, RAI2, MLL, AVIL, GAS2, CCNA1, BMP4, IGFBP2, EDN3, CALC |
GREM1, TPM1, DIRAS3, NRP1, LMO4, TPM4, ACTA2, SRI |
| Cell cycle | CDKN1B, RIOK3, PLK2, CDC2L5, BCAR3, DDIT3, RAD52, NFIB, ABCF2, PPM1D, MLL, GAS2, CCNA1, RBBP6 |
DIRAS3, RRM2, RFC4, MCM4, TFDP1, HUS1, MCTS1, PRIM1 |
| Biogenesis | SIRT1, MYST4, BLZF1, SIRT4, H2AFJ, AVIL, ARID4A, GAS2, HIST1H2AE, IGFBP2, PEX1 |
RTCD1 |
| Membrane proteins | CCR5, MYLIP, MPZ, PCNX, SLC39A9, CNR1, DGCR2, AQP8, KCNAB1, POMT1, LECT1, SYT17, SCN3B, SEL1L, ABCF2, LGALS4, PLA2G4C, ITGAM, PEX1, CALCR |
CD47, PLAUR, NRP1, COQ2, CD59, SSR1, ENTPD6, ICMT, DSCR2, MSN, TSPAN6, PIGF, ALG8, ATP5G3 |
Bold type indicates genes that were further investigated for function in the current study.
Two members of TGFβ signaling pathway, BMP4 and ID4, were found among the transcriptional repressors up-regulated in human muscle SP cells. Intriguingly, muscle MP cells expressed significantly higher levels of Gremlin, a known antagonist of BMP4 signaling (Hsu et al., 1998; Merino et al., 1999; Zuniga et al., 1999). The presence of high levels of BMP4 in SP cells and its inhibitor, Gremlin, in MP cells led to the hypothesis that these secreted factors could create antagonistic effects in the cellular environment, regulating the proliferation and differentiation of mononuclear cells in human fetal skeletal muscle.
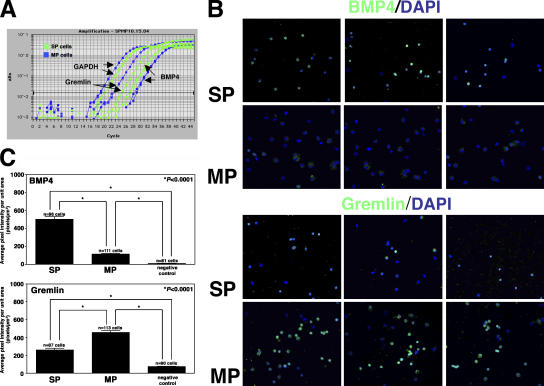
To test this hypothesis, we first confirmed the differential expression of BMP4 (overexpressed in SP cells) and Gremlin (overexpressed in MP cells) by quantitative real-time RT-PCR using total RNA extracted from SP and MP cells (Fig. 2 A and Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200511036/DC1). The results confirmed that SP cells express eightfold higher levels of BMP4 compared with MP cells. Expression of Gremlin mRNA was twofold higher in MP cells compared with SP cells (Fig. 2 A), whereas other known BMP4 antagonists such as Chordin, Noggin, and Follistatin were not up-regulated in MP compared with SP cells at the mRNA level (not depicted).
Figure 2.
Expression of BMP4 and Gremlin in human fetal skeletal muscle SP and MP cells. (A) Quantitative real-time PCR of BMP4 and Gremlin mRNA expression in SP (green) and MP (blue) cells. mRNA expression was measured as Ct values. Ct values for each gene were normalized to the respective values for GAPDH. (B) Expression of BMP4 (top) and Gremlin (bottom) proteins in SP and MP cells. BMP4- and Gremlin-positive cells are stained in green (Alexa Fluor 488), and nuclei are stained in blue (DAPI). Three representative fields are shown for each population. (C) Quantitative immunofluorescence of BMP4 (top) and Gremlin (bottom) protein expression in SP and MP cells, measured as mean pixel intensity per unit area. For BMP4, the mean pixel intensity per unit area for SP cells (n = 96) is 506 ± 30, versus 113 ± 8 pixels/μm2 for MP cells (n = 111; mean ± SEM). Gremlin expression is significantly higher in human MP cells (n = 113; 455 ± 22 pixels/μm2 [mean ± SEM]) than in human SP cells (n = 87; 261 ± 17 pixels/μm2 [mean ± SEM]). *, P < 0.0001
Next, BMP4 and Gremlin protein expression was assessed by immunohistochemistry on cytospins of SP and MP cells (Fig. 2, B and C). All images were acquired at the same exposure time to enable a quantitative analysis of protein expression levels. This analysis demonstrated that SP cells expressed significantly higher levels of BMP4 protein than MP cells, with the mean pixel intensity per unit area for SP cells measured at 506 ± 30 versus 113 ± 8 pixels/μm2 for MP cells (mean ± SEM; P < 0.0001; Fig. 2 C). Gremlin was significantly more abundant in MP cells (455 ± 22 pixels/μm2) compared with SP cells (261 ± 17 pixels/μm2 [mean ± SEM]; P < 0.0001; Fig. 2 C), confirming the results obtained by quantitative real-time RT-PCR.
Effects of BMP4 and Gremlin on human fetal SP and MP cells
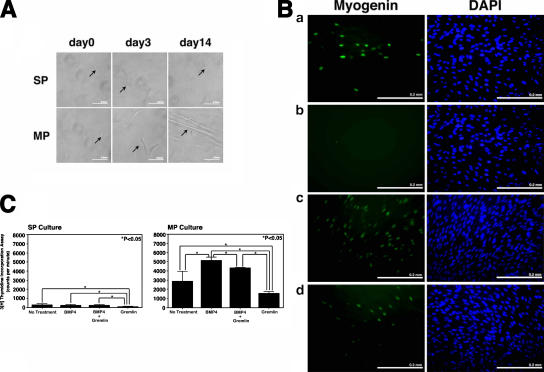
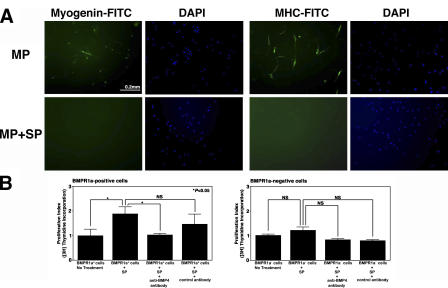
Because both RNA and protein studies demonstrated that human fetal skeletal muscle SP cells express high levels of BMP4, whereas MP cells express high levels of Gremlin, it was important to investigate the functional effects of these factors on purified human muscle SP and MP cells. Both BMP4 and Gremlin are secreted morphogens and can act in the immediate cellular environment as regulators of cellular proliferation or differentiation. To study these effects, purified human fetal muscle SP and MP cells were cultured for 14 d under the following four conditions: (1) control (no factors), (2) in the presence of BMP4, (3) in the presence of BMP4 and Gremlin, and (4) in the presence of Gremlin (Fig. 3). The factors were added at the beginning of the culture and exchanged every 48 h. BMP4 was added at 25 ng/ml according to the previously reported data (Dahlqvist et al., 2003), and Gremlin was added at 2 μg/ml based on ED50 inhibitory concentration reported by the manufacturer (R&D Systems). Although MP cells differentiated and gave rise to myotubes, SP cells maintained their undifferentiated phenotype under all four conditions, indicating that BMP4 and/or Gremlin do not induce differentiation of SP cells (Fig. 3 A). The degree of myogenic differentiation of MP cells was measured by assessing the percentage of myogenin-positive cells (Fig. 3 B). When assessed by expression of myogenin at day 7 of culture in proliferation medium, control MP cultures had 9.07 ± 2.92% (mean ± SD) of myogenin-positive cells (Fig. 3 B a), whereas in the presence of BMP4, the percentage of myogenin-positive nuclei decreased to 0.1 ± 0.31% (mean ± SD; P < 0.001; Fig. 3 B b). Addition of Gremlin to BMP4 reversed expression of myogenin in 14 ± 2.93% (mean ± SD) of cells (P < 0.001; Fig. 3 B c). Addition of Gremlin alone did not show a significant difference with control cultures, as 13.91 ± 3.55% (mean ± SD) of cells were myogenin-positive (Fig. 3 Bd ). Therefore, BMP4 had a strong inhibitory effect on the differentiation of MP cells, whereas addition of Gremlin led to a complete loss of this inhibitory effect.
Figure 3.
Effects of BMP4 and Gremlin on the differentiation and proliferation of human SP and MP cells. (A) Brightfield photographs of cultured SP and MP cells. Under all conditions, muscle SP cells were nonadherent and morphologically round throughout the culture period, whereas MP cells differentiated into myotubes. Arrows point to mononuclear cells (SP and MP) and myotubes (MP, day 14). (B) Effects of BMP4 and Gremlin on human fetal muscle MP cell differentiation. Myogenic differentiation was visualized by immunostaining with anti–myogenin-FITC (green). (a) MP cells cultured under normal differentiation conditions without addition of factors. (b) MP cells cultured in the presence of 25 ng/ml BMP4. (c) MP cells cultured in the presence of 25 ng/ml BMP4 and 2 μg/ml Gremlin. (d) MP cells cultured in the presence of 2 μg/ml Gremlin. Nuclei are stained in blue with DAPI. (C) [3H]thymidine incorporation by SP cells (left) and MP cells (right) cultured as described above, performed at day 7 of culture. Addition of BMP4 to MP cells resulted in a significant increase of [3H]thymidine incorporation compared with nontreated controls, whereas it had no effect on proliferation of SP cells. Addition of Gremlin to BMP4-treated MP cells significantly decreased their proliferation rate compared with MP cells treated with BMP4 alone. Gremlin alone inhibited proliferation of MP and SP cells compared with untreated control cultures. Error bars indicate SD. *, P < 0.05. Bars, 0.2 mm.
The effect of BMP4 and Gremlin on proliferation of SP and MP cells was studied by [3H]thymidine incorporation assay (Fig. 3 C). In cultured SP cells, exposure to Gremlin had a significant inhibitory effect on their proliferation (Fig. 3 C, left) compared with untreated control SP cell cultures (100 ± 62 vs. 300 ± 103 [mean ± SD] cpm; P < 0.05), and this effect was reversed by the addition of BMP4. BMP4 alone, however, had no effect on proliferation of SP cells compared with untreated control cultures (252 ± 79 vs. 300 ± 10 [mean ± SD] cpm; NS). In contrast, addition of BMP4 to MP cultures (Fig. 3 C, right) resulted in a significant increase in their proliferation compared with untreated controls (5,174 ± 340 vs. 2,875 ± 1,088 [mean ± SD] cpm; P < 0.05). This effect was reversed by the addition of Gremlin (4,336 ± 46 vs. 5,174 ± 340 mean ± SD] cpm; P < 0.05), and Gremlin alone inhibited proliferation of MP cells compared with the untreated control (1,579 ± 178 vs. 2,875 ± 1,088 [mean ± SD] cpm; P < 0.05).
These results demonstrate that addition of BMP4 inhibits differentiation and induces proliferation of MP cells but has no effect on SP cells, and that addition of Gremlin can reverse these effects. Gremlin appears to inhibit proliferation of SP and MP cells, whereas it has no effect on their differentiation.
BMP4 induces proliferation of BMPR1a+ cells in human skeletal muscle
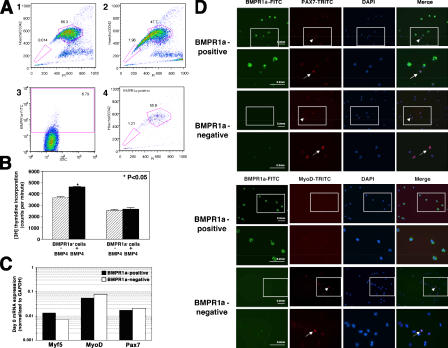
To identify the population within muscle MP cells responsive to BMP4-induced proliferation, the expression of the BMP4-specific receptor BMPR1a (Massague et al., 1994; Wrana et al., 1994a) was studied by flow cytometry (Fig. 4). 4–11% of human muscle cells expressed BMPR1a (Fig. 4 A 3; n = 5), with >98% of BMPR1a+ cells located outside the SP gate (Fig. 4 A 4). To study the molecular mechanism of BMP4-induced proliferation, BMPR1a+ and BMPR1a− cells were cultured in growth medium alone or in the presence of BMP4, and [3H]thymidine incorporation assay was performed on culture day 4 (Fig. 4 B). Results demonstrated that addition of BMP4 to BMPR1a+ cultures resulted in a significant increase in their proliferation compared with untreated controls (4,620 ± 63 vs. 3,672 ± 124 [mean ± SD] cpm; P < 0.05). Addition of BMP4 to BMPR1a− cells had no effect on their proliferation compared with untreated cells. Untreated BMPR1a+ cells demonstrated significantly higher proliferation than BMPR1a− cells (3,672 ± 124 vs. 2,553 ± 161 [mean ± SD] cpm; P < 0.05). These results demonstrate that BMP4 induces proliferation of BMPR1a+ cells, whereas it has no effect on BMPR1a− cells, suggesting that the proliferation induced by BMP4 is mediated via BMPR1a. Further, BMPR1a+ cells have a higher intrinsic proliferative capacity than BMPR1a− cells.
Figure 4.
Effect of BMP4 stimulation on BMPR1a-expressing cells. (A) Expression of BMPR1a in human fetal skeletal myoblasts. Three-color flow cytometry (Hoechst 33342, PI, and BMPR1a-FITC) staining profiles of an 18-wk gestation sample. Cells were costained with 3 μg/ml Hoechst 33342, anti–BMPR1a- FITC, and PI. (1 and 2) Cells stained with Hoechst 33342 in the presence (1) or absence (2) of reserpine. 1.96% of the cells are detected in the SP gate. (3) 6.79% of cells are BMPR1a+. (4) Approximately 99% of BMPR1a+ cells are located outside of the SP gate. (B) [3H]thymidine incorporation by BMPR1a+ and BMPR1a− cells cultured with and without BMP4 for 4 d. Addition of BMP4 to BMPR1a+ cultures resulted in a significant increase of [3H]thymidine incorporation compared with nontreated controls. BMP4 had no effect on the proliferation of BMPR1a− cells. *, P < 0.05. Error bars indicate SD. (C) Expression of Myf5, MyoD, and Pax7 mRNA in freshly isolated BMPR1a+ and BMPR1a− cells. mRNA expression levels were normalized to GAPDH for each condition. Bars represent on a logarithmic scale the level of expression of each myogenic factor in BMPR1a+ and BMPR1a− relative to the expression of GAPDH, which is arbitrarily considered to be 1. The baseline expression of Myf5 is twofold higher in BMPR1a+ cells than in BMPR1a− cells, whereas MyoD and Pax7 mRNA were expressed 1.5-fold higher in BMPR1a− cells. (D) Expression of Pax7 and MyoD on cytospins of purified human fetal skeletal muscle BMPR1a+ and BMPR1a− cells. By total cell count, 1.04% of 192 analyzed BMPR1a+ cells and 12.9% of 743 analyzed BMPR1a− cells expressed Pax7. None of 205 analyzed BMPR1a+ cells and 7.4% of 594 analyzed BMPR1a− cells expressed MyoD. BMPR1a+ cells are stained in green (FITC), Pax7- and MyoD-positive cells are stained in red (TRITC), and nuclei are stained in blue with DAPI. The arrows indicate Pax7- and MyoD-positive cells. The arrowheads in the insets in the top panels highlight the same Pax7- and MyoD-positive cells in the bottom panels, which are marked by arrows. Bars, 0.2 mm.
The activation of myogenic cells is a highly regulated process that includes down-regulation of the satellite cell–specific marker Pax7 (Zammit et al., 2004; Kuang et al., 2006) and the up-regulation of the myogenic-specific factors Myf5 and MyoD (Rudnicki et al., 1993; Weintraub, 1993; Rudnicki and Jaenisch, 1995; Molkentin and Olson, 1996). Muscle cells obtained from Myf5−/− mice show severe proliferation deficiency and undergo premature differentiation (Montarras et al., 2000). Thus, Myf5, which is normally expressed by satellite cells and activated myoblasts, is important for muscle cell proliferation (Sabourin et al., 1999; Yablonka-Reuveni et al., 1999; Cornelison et al., 2000). Expression of Myf5, MyoD, and Pax7 was assayed in BMPR1a+ and BMPR1a− cells by real-time quantitative RT-PCR (Fig. 4 C) and by immunofluorescence on cytospins (Fig. 4 D). The baseline expression of Myf5 was twofold higher in BMPR1a+ cells than in BMPR1a− cells, whereas MyoD and Pax7 were expressed at higher level in BMPR1a− cells (Fig. 4 C). Pax7 and MyoD protein expression assessed by cytospins demonstrated that significantly more BMPR1a− than BMPR1a+ cells expressed Pax7 and MyoD proteins (Fig. 4 D). By total cell count, 1.04% of BMPR1a+ cells (n = 192) and 12.9% of BMPR1a− cells (n = 743) expressed Pax7. None of BMPR1a+ cells (n = 205) and 7.4% of BMPR1a− cells (n = 594) expressed MyoD. These results define BMPR1a+ cells as a Myf5 high, MyoD low, and Pax7 low cell population, and may provide an explanation for their increased proliferation activity.
Effects of muscle SP cells on proliferation and differentiation of MP, BMPR1a+, or BMPR1a− cells
Our studies using synthetic morphogenic factors showed that addition of BMP4 induced proliferation and delayed differentiation of muscle MP and BMPR1a+ cells. To investigate whether BMP4-expressing SP cells also inhibit differentiation of MP cells, freshly purified SP cells were irradiated at 25Gy (Mozdziak et al., 1996) and cocultured with an equal number of MP cells. Myogenic differentiation was assessed by immunostaining for myogenin or myosin heavy chain (MHC) at culture day 7 (Fig. 5 A). In control MP cultures (MP cells alone), 22.9 ± 4.4% (mean ± SD) myogenin-positive cells and 22.4 ± 6.9% MHC-positive cells were detected, whereas in cocultures of MP cells with irradiated SP cells, no myogenin or MHC expression was observed (Fig. 5 A). These results demonstrate that BMP4-expressing SP cells inhibited differentiation of muscle MP cells.
Figure 5.
Effect of SP cells on myogenic differentiation. (A) Purified human fetal muscle MP cells were cocultured with irradiated SP cells. Myogenic differentiation was visualized by immunostaining for myogenin (green) and MHC (green) at day 7 of culture. Control MP cultures had 22.9 ± 4.4% (mean ± SD) of myogenin-positive cells and 22.4 ± 6.9% (mean ± SD) of MHC-positive cells, whereas MP cells cocultured with irradiated SP cells demonstrated no myogenin or MHC expression. (B) Effect of purified muscle SP cells on proliferation of BMPR1a+ (left) and BMPR1a− (right) cells. BMPR1a+ and BMPR1a− cells were cultured with no treatment, cocultured with irradiated SP cells, cocultured with irradiated SP cells and BMP4 blocking antibody, or cocultured with irradiated SP cells and IgG2b isotype control antibody. Proliferation was assessed using a [3H]thymidine incorporation assay after 5 d of culture, and proliferation indexes were established by calculating the ratios of thymidine incorporation in treated versus untreated cells. Error bars indicate SD.
To investigate whether SP cells exert a similar proliferative effect as recombinant BMP4 on BMPR1a+ cells and whether this effect can be abrogated by specific BMP4 blockade, BMPR1a+ and BMPR1a− cells were cultured for 5 d under the following conditions: (1) no treatment, (2) cocultured with irradiated SP cells, (3) cocultured with irradiated SP cells and 2 μg/ml BMP4 blocking antibody, and (4) cocultured with irradiated SP cells and 2 μg/ml isotype control antibody (Fig. 5 B). Proliferation was assessed using a [3H]thymidine incorporation assay, and proliferation indexes were established by calculating the ratios of thymidine incorporation in treated versus untreated cells. BMPR1a+ cells cocultured with irradiated SP cells proliferated significantly more than control BMPR1a+ cells (proliferation index: 1.89 ± 0.27 [mean ± SEM]; P < 0.05). This effect was significantly reversed when 2 μg/ml of BMP4 blocking antibody were added (proliferation index: 1.03 ± 0.05 [mean ± SEM]; P < 0.05). IgG2b isotype control antibody had no significant inhibitory effect on the proliferation of BMPR1a+ cells cocultured with SP cells (proliferation index: 1.47 ± 0.4 [mean ± SEM]; NS). No significant effects were observed on proliferation of BMPR1a− cells compared with untreated controls by coculture with irradiated SP cells (proliferation index: 1.22 ± 0.28 [mean ± SEM]; NS), coculture with irradiated SP cells in the presence of BMP4 blocking antibody (proliferation index: 0.843 ± 0.10 [mean ± SEM]; NS), or coculture with irradiated SP cells in the presence of IgG2b isotype control antibody (proliferation index: 0.802 ± 0.08 [mean ± SEM]; NS). These results demonstrate that SP-induced proliferation of BMPR1a+ cells is mediated primarily by BMP4 signaling, as it is abrogated by specific BMP4 blockade.
Tissue localization of cells expressing BMP4, BMPR1a, or Gremlin
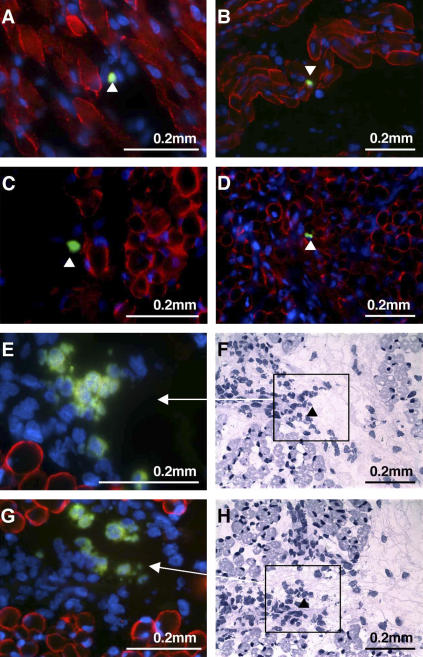
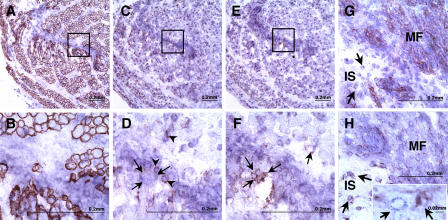
As BMP4 expression was found significantly elevated in the majority of human muscle SP cells compared with MP cells, immunostaining of human fetal skeletal muscle tissue sections obtained from six individuals was performed using an anti-BMP4 antibody in an attempt to localize muscle SP cells in vivo. Dystrophin staining was performed simultaneously to detect the sarcolemma of myofibers (Fig. 6). Rare interstitial BMP4+ cells were observed in muscle tissue sections (Fig. 6, A–D). Small clusters of BMP4+ cells were also found in areas distinct from myofibers (Fig. 6, E–H). In eight random microscopic fields obtained from different samples, the frequency of BMP4+ cells was estimated to be 1 for every 58 nuclei, corresponding to 1.8% of the cells. To define the spatial relationships between BMP4- and BMPR1a-expressing cells, immunohistochemistry was performed on sequential sections of a 20-wk gestation fetal skeletal muscle sample (Fig. 7, A–F). BMP4+ cells (Fig. 7, C and D) were found in the vicinity of BMPR1a+ cells (Fig. 7, E and F) in the interstitial spaces surrounded by myofibers (Fig. 7, A and B). BMP4-reactive cells are rounded in contours and have smaller cross-sectional diameters than BMP4− skeletal muscle fibers. They focally rim the interstitial spaces formed by mature muscle fibers. Occasional cells within the interstitial spaces show variable membrane reactivity for BMPR1a, where they can be also found as clusters surrounded by BMP4+ cells. Furthermore, staining for BMP4 and Gremlin (Fig. 7, G–H) was mutually exclusive. Gremlin was highly expressed in mature myofibers (Fig. 7 G) and scattered rounded cells in the interstitium (Fig. 7 G, arrows), whereas staining for BMP4 was present only in rounded cells that are most prominent in the interstitium (Fig. 7 H, arrows). Double labeling for BMP4 and Gremlin (Fig. 7 H) confirmed the reciprocal staining pattern of interstitial BMP4- or Gremlin-expressing cells.
Figure 6.
Expression of BMP4 in rare interstitial human fetal muscle cells. (A–D) Arrowheads point to single BMP4-positive cells (green, FITC) located between dystrophin positive myofibers (stained in red, Texas red). Nuclei are stained in blue (DAPI). (E and G) Clusters of interstitial BMP4-positive cells with corresponding hematoxylin/eosin staining (F and H) at lower magnification. Black squares in F and H are shown at higher magnification in E and G, respectively.
Figure 7.
Immunohistochemistry of human embryonic skeletal muscle, 20-wk gestation. A–F represent consecutive sections. Insets in A, C, and E correspond precisely to the high magnifications (1,000×) portrayed in B, D, and F, respectively. (A and B) Dystrophin staining defines mature muscle fibers, represented in cross section. Inset denotes interstitial space containing dystrophin-negative nucleated cells. (C and D) BMP4-reactive cells are rounded in contours and have smaller cross-sectional diameters than mature BMP4-negative skeletal muscle fibers. BMP4-positive cells (arrowheads) focally rim interstitial spaces formed by mature muscle fibers. Black arrows (D) define the area of BMPR1a+ cells shown in F. (E and F) Occasional cells within interstitial spaces show variable membrane reactivity for BMPR1a (arrows), where they are found as clusters surrounded by BMP4-positive cells (D and F). (G and H) Sequential sections of 20-wk fetal skeletal muscle stained for BMP4 and Gremlin. (G) Staining for Gremlin (brown; 400×); (H) staining for BMP4 (brown; 400×). Nuclei are stained violet. Note Gremlin staining in mature myofibers (MF) and scattered rounded cells in the interstitium (IS; G, arrows). Staining for BMP4 is present only in rounded cells that are most prominent in the interstitium (H, arrows). Inset (1,000×) is double labeled (nuclei are unstained), showing mutually exclusive staining for BMP4 (blue) and Gremlin (brown) by interstitial cells. Bars: (A–H) 0.2 mm; (H, inset) 0.02 mm.
Discussion
Skeletal muscle SP cells represent a primitive stem cell population capable of myogenic differentiation in vitro and in vivo (Gussoni et al., 1999; Jackson et al., 1999; Asakura et al., 2002). Our data show that human fetal skeletal muscle SP cells express high levels of BMP4, whereas MP cells express high levels of the BMP4 antagonist Gremlin.
As a member of the TGFβ signaling family, BMP4 induces a variety of cellular responses, initiated by binding to its specific cellular receptor, BMPR1a (Massague et al., 1994; Wrana et al., 1994a,b). We found by flow cytometry that >98% of BMPR1a+ cells are located outside of the SP gate. Our data demonstrate that BMP4 specifically stimulates proliferation of BMPR1a+ cells, whereas it has no effect on BMPR1a− cells, suggesting that BMP4-induced proliferation is likely mediated by the BMPR1a receptor. BMP4 has been shown to inhibit transcription of MyoD (Dahlqvist et al., 2003), a muscle-specific transcription factor associated with withdrawal from the cell cycle and terminal differentiation. The myogenic transcription factor Myf5 is thought to be developmentally “upstream” of MyoD (Sabourin et al., 1999; Seale et al., 2000, 2004), and it is expressed in actively proliferating myogenic cells (Sabourin et al., 1999; Yablonka-Reuveni et al., 1999; Cornelison et al., 2000). Our studies demonstrated a twofold increase in expression of Myf5 in uncultured, untreated BMPR1a+ cells compared with BMPR1a− cells, suggesting that these cells may have intrinsic high proliferative potential. Coculture of irradiated, BMP4-secreting muscle SP cells with BMPR1a+ cells significantly induced proliferation of the latter compared with control BMPR1a+ cells cultured alone, and this effect was specifically abrogated by BMP4 blockade. In contrast, BMPR1a− cells did not increase proliferation when cultured in the presence of irradiated SP cells. These results suggest that muscle SP cells are able to induce proliferation of other myogenic precursors and that this effect is mediated via BMP4 signaling. In support of this hypothesis, BMP4+ and BMPR1a+ cells were found in vivo in close proximity of each other, with BMP4+ cells surrounding small clusters of BMPR1a+ cells, located in the interstitial spaces between myofibers.
From the current study, muscle MP cells express high levels of the BMP4 inhibitor Gremlin (Hsu et al., 1998), which is known to bind BMP4, thus reducing BMP4 concentration in the cellular microenvironment. In vivo data indicate that Gremlin is highly expressed by mature myofibers and interstitial cells, which are distinct from BMP4-expressing cells. The observed Gremlin-induced growth inhibition of SP and MP cultures may play an important role in counteracting the stimulatory effects of BMP4 and in regulating the proliferation of BMPR1a+ MP cells. In addition to its function as a BMP4 antagonist, Gremlin has been shown to suppress tumor growth by mechanisms that are independent from BMP4, which involve both up-regulation of p21Cip1 and down-regulation of p42/44 MAPK (Chen et al., 2002). Remarkably, in our studies, only Gremlin is differentially expressed between SP and MP cells, whereas other BMP4 antagonists, such as Chordin, Noggin, and Follistatin, are not up-regulated in MP cells. We therefore hypothesize that secretion of BMP4 by SP cells and of Gremlin by MP cells generate antagonistic effects, influencing the rate of proliferation and differentiation of myogenic cells in human muscle.
High concentrations of BMP4 have been shown to induce osteogenic differentiation of muscle progenitor cells (Katagiri et al., 1994; Wright et al., 2002) and are counteracted by BMP4 inhibitors under normal conditions (Pereira et al., 2000). Overexpression of BMP4 and the inability to mount an appropriate antagonist (including Gremlin) response have been implicated in the pathogenesis of a human disease of heterotopic osteogenesis, fibrodysplasia ossificans progressiva (FOP; Kan et al., 2004). The process of ectopic ossification in this disease is often precipitated by muscle injury, i.e., biopsy or trauma, when progenitor cells are being activated for muscle repair. It is conceivable that the BMP4high SP cells participate in muscle repair after acute injury in response to trauma, creating an unopposed local increase in BMP4, which may contribute to the formation of islands of heterotopically ossified muscle tissue in patients afflicted by FOP.
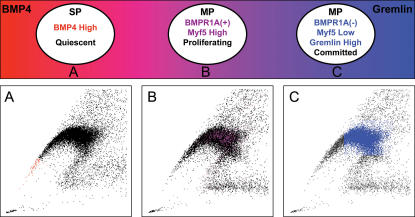
The data presented here suggest that SP cells regulate proliferation of more committed cells in a paracrine fashion by secreting BMP4, which is antagonized by its inhibitor Gremlin, secreted by MP cells. The functional counteraction of BMP4 signaling by Gremlin may play a role in preventing uncontrolled proliferation, thus maintaining the appropriate number of progenitors within the tissue. Based on our in vitro findings, we propose that expression and secretion of BMP4 by quiescent muscle SP cells (Fig. 8 A) induces proliferation of BMPR1a+ Myf5high muscle MP cells (Fig. 8 B). This proliferation is counteracted by secretion of Gremlin by BMPR1a− Myf5low committed MP cells (Fig. 8 C). The demonstrated tissue localization of BMP4+ and BMPR1a+ cells in close proximity of each other and the mutually exclusive staining pattern of BMP4 and Gremlin suggest that these interactions also occur in vivo. This study provides additional insights into the complex functional hierarchy among the mononuclear cells in skeletal muscle and leads to important correlations with other stem cell niches, such as the skin, bone marrow, and intestine, where stem cells use BMP4 signaling to regulate the proliferation of their daughter cells, which in turn differentiate into committed cells and tissues (Kobielak et al., 2003; Zhang et al., 2003; Haramis et al., 2004).
Figure 8.
Schematic illustration of the proposed cellular hierarchy within skeletal muscle. BMP4high quiescent SP cells (A, red) induce the proliferation of BMPR1a+ Myf5high MP cells (B, purple) via BMP4 secretion, and BMPR1a− Myf5low Gremlinhigh committed MP cells (C, blue) may inhibit the stimulatory effect of BMP4 by secreting Gremlin. Cells are displayed schematically according to their respective positions in the Hoechst/PI profile.
Materials and methods
Tissue preparation, Hoechst 33342 staining, and flow cytometry analyses
Mononuclear cells were isolated from discarded human fetal skeletal muscle of gestational age 14–18 wk (Advanced Bioscience Resources). Tissues were collected in sterile HBSS with 1% glucose and 1% penicillin/streptomycin, finely minced using sterile scalpels, and digested using dispase (0.6 U/ml) and collagenase (0.5 mg/ml; Worthington) diluted in PBS. Digested samples were filtered through a 40-μm cell strainer before staining and analysis (Pavlath and Gussoni, 2005).
Mononuclear cells were centrifuged at 365 g for 10 min and resuspended at a concentration of 106 cells/ml in prewarmed (37°C) PBS/0.5% BSA. Hoechst 33342 dye (Sigma-Aldrich) was added at a final concentration of 3–9 μg/ml for fetal samples and 7.5–12.5 μg/ml for adult samples. In parallel, as a negative control for SP cell gating, 5 × 105 muscle cells were stained with Hoechst 33342 in the presence of 5 μM reserpine (Sigma-Aldrich). Cells were incubated for 60 min at 37°C, protected from light in a waterbath, and washed by adding 3.5–5 volumes of ice-cold PBS/0.5% BSA.
For labeling with specific antibodies, after Hoechst 33342 staining, samples were pelleted, resuspended at a concentration of 107–108 cells/ml, and incubated with the following antibodies: anti-human CD34-FITC (BD Biosciences), anti-human CD133-PE (Miltenyi Biotec), anti-CD45-PE (Miltenyi Biotec), rat IgG-PE (BD Biosciences), anti-ABCG2 (Stem Cell Technologies), or anti-BMPR1a (Santa Cruz Biotechnology, Inc.). Control samples of 3T3 cells transfected with either mock- or human ABCG2–encoding vectors were provided by B. Sorrentino (St. Jude Children's Research Hospital, Memphis, TN). Cells were incubated for 15 min on ice, washed, and resuspended in ice-cold PBS/0.5% BSA containing 2 μg/ml propidium iodide (PI). For detection of ABCG2 and BMPR1a+ cells, incubation with FITC-conjugated secondary anti-mouse (BD Biosciences) or anti-rabbit (Jackson ImmunoResearch Laboratories) antibody was performed for 10 min on ice. Subsequently, the cells were washed with cold PBS/0.5% BSA and resuspended in ice-cold 1× PBS/0.5% BSA containing 2 μg/ml PI. Flow cytometric analyses and cell sorting were performed on a dual-laser FACSVantage SE flow cytometer (Becton Dickinson) equipped with two lasers: one with 200 MW power (488 nm) and one with 150 MW power (UV). Both the Hoechst and PI dyes were excited at 350 nm, and their fluorescence was measured at 400 and 600 long pass, respectively, with a 550 short pass dichroic mirror. For each sample analyzed, 20,000 total cell counts were acquired using CellQuest software version 3.3 (Becton Dickinson).
Sample preparation for microarray analysis
Total RNA was extracted from sorted human fetal muscle SP and MP cells using the RNeasy kit (QIAGEN). Total RNA (100 ng) from 10 SP and 9 MP samples were subjected to two rounds of linear amplification using a MessageAmp kit (Ambion). After amplification, 20 μg of fragmented, biotin-labeled cRNA was prepared and hybridized onto the Affymetrix human HG-U133A GeneChip (as per the manufacturer's instructions). Microarrays were optically scanned, and images were processed using the Affymetrix GeneChip MAS5.0 analysis software, which were then summarized in a CHP file. Each surveyed gene transcript was given a mean fluorescence intensity value (signal) indicating the amount of the transcript detected and a call (present, absent, or marginal) indicating the robustness of its detection.
Microarray data analysis
Each Affymetrix HG-U133A microarray contains 22,283 individual probes for gene/RNA transcripts. Using a geometric logarithmic fold analysis (Haslett et al., 2003; Kho et al., 2004), 244 probes had a statistically significant differential reading between the SP (n = 10) and MP (n = 9) groups. Two criteria had to be satisfied for significant differential expression: (1) AbsoluteValue(AvgLF) − StdLF > 0.4 × Local_Fold_Noise and (2) AbsoluteValue(AvgLF) > max(Local_Fold_Noise, Global_Fold_Noise). AvgLF and StdLF denote the mean and standard deviation of the logarithmic fold change of a probe in the SP relative to the MP group, respectively. For each probe, Local_Fold_Noise is the largest logarithmic fold change for the probe within the MP or the SP groups. Global_Fold_Noise is the mean of Local_Fold_Noise over all 22,283 probes. The false discovery rate (FDR) for this approach was <1% after a permutation analysis with 10,000 independent shuffling iterations. In each iteration, the 19 sample labels (MP or SP) of each of the 22,283 probes were first randomly shuffled. Then, the number of probes from this shuffled dataset that came up as significant (i.e., false positives) using the two criteria above were counted. The FDR is the median of these 10,000 false positive counts over 244, the number of probes called significantly differentially expressed in the original unshuffled dataset. These 244 probes (corresponding to 222 unique genes) were functionally annotated using the web-based Database for Annotation, Visualization, and Integrated Discovery 2.0 (DAVID 2.0; http://niaid.abcc.ncifcrf.gov/; Dennis et al., 2003).
Real-time quantitative PCR analysis
Total RNA (100 ng) isolated from freshly sorted human fetal skeletal muscle SP and MP cells or from BMPR1a+ and BMPR1a− cells was reverse transcribed using the Taqman reverse transcription kit (Applied Biosystems) in a 100 μl reaction volume according to the manufacturer's protocol. Approximately 20 pcg of cDNA (1 μl of RT reaction product) was used for PCR amplification. Samples were assayed using Sybergreen chemistry and kinetic PCR (ABI 7700 Sequence Detector; Applied Biosystems). Samples were amplified using the Sybergreen PCR reagent kit (Applied Biosystems) according to the manufacturer's protocol. Sense and antisense primers were used at a final concentration of 300 nM. The primer sequences for the genes tested are listed in Table S3. The cDNA samples were amplified under “default” conditions: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of amplification at 94°C for 15 s and 60°C for 1 min. For data analysis, the cycle threshold (Ct) number was computed for each sample using the Sequence Detection Software (Applied Biosystems). The Ct baseline was preset at 10 standard deviations above the fluorescence background, which was determined in the first 3–15 cycles. All quantitations were normalized to the endogenous control gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to account for variability in the initial concentration and quality of the total RNA, and the efficiency of the reverse transcription reaction. Differential Ct values (Ct for the gene of interest minus Ct for GAPDH) were calculated for each amplified gene. Logarithmic fold change of gene expression between samples was calculated by subtracting gene-specific differential Ct values.
Immunohistochemistry
For cytospin preparation, human fetal skeletal muscle SP, MP, BMPR1a+, and BMPR1a− cells were purified by flow cytometry. Cells (1,000 cells/slide) were placed on glass slides by centrifugation in a cytospin centrifuge (Thermo Electron Corp.). Slides were fixed in absolute methanol for 3 min at room temperature, blocked for 30 min in blocking solution (10% fetal bovine serum and 0.1% Triton X-100), and incubated with mouse anti-Pax7 monoclonal antibody (diluted 1:15; Developmental Studies Hybridoma Bank) or mouse anti-MyoD monoclonal antibody (diluted at 1:50; BD Biosciences) overnight at 4°C. Slides were washed in 1× PBS/0.1% Triton X-100 and incubated with goat anti–mouse or goat anti–rabbit FITC-conjugated secondary antibody (diluted 1:100; Jackson ImmunoResearch Laboratories) for 1 h at room temperature. For BMP4 and Gremlin staining, cytospin slides were fixed in 4% paraformaldehyde for 20 min at room temperature, blocked for 30 min in 10% fetal bovine serum and 0.1% Triton X-100, and incubated with mouse anti-BMP4 monoclonal antibody (diluted 1: 20; Novocastra) or rabbit anti-Gremlin polyclonal antibody (diluted 1:50; Abgent) overnight at 4°C. Slides were washed in 1× PBS/0.1% Triton X-100 and incubated with goat anti–mouse Alexa Fluor 488 or goat anti–rabbit Alexa Fluor 488 secondary antibody (diluted 1:200; Invitrogen) for 1 h at room temperature and washed in 1× PBS/0.1% Triton X-100. Slides were then mounted in Vectashield (Vector Laboratories) supplemented with 100 ng/ml DAPI to visualize nuclei. Images from Pax7- and MyoD-stained BMPR1a+ and BMPR1a− cells were acquired in five random microscopic fields for each sample using 200 and 400 magnification objective on a microscope (E1000; Nikon), photographed using a camera (Orca ER; Hamamatsu), and processed using OpenLab software version 3.1.5 (Improvision). For quantitative analysis, images from BMP4- and Gremlin-stained SP and MP cells were acquired in 10 random microscopic fields for each sample on an automated upright fluorescent microscope (Eclipse 90I; Nikon) using 200 magnification objective and processed using MetaMorph 7.0 software (Molecular Devices). Images were photographed using a camera (Orca II; Hamamatsu). The intensity of fluorescence labeling between the SP and MP cells was compared using the mean integrated intensity per unit area, to correct for differences in cell size. Using MetaMorph software, a region of interest was drawn around each cell to be measured and the integrated intensity of the region was calculated. To correct for local changes in background intensity, the region was then moved to a background area of the field of view close to the cell of interest and integrated intensity was recorded. The integrated intensity of the background was subtracted from the integrated intensity of the cell being measured. Integrated pixel intensity was calculated for each cell in the field and then divided by the area of the cell to calculate mean pixel intensity per unit area. Mean pixel intensities for SP and MP cells were then statistically compared using the nonparametric Wilcoxon test.
For immunofluorescence analysis of cultured human MP cells, cells seeded in 96-well plates were washed three times with PBS, fixed in absolute methanol, blocked for 30 min in 10% fetal bovine serum/0.1% Triton X-100 in PBS, and incubated with mouse anti-MHC monoclonal antibody (MF20; diluted 1:15; Developmental Studies Hybridoma Bank) or anti-myogenin mouse monoclonal (diluted 1:50; DakoCytomation) overnight at 4°C. Wells were washed, incubated with goat anti–mouse FITC-conjugated secondary antibody (diluted 1:100; Jackson ImmunoResearch Laboratories), and processed as described for cytospins. Plates were visualized with a 200 magnification objective on a microscope (Eclipse TE2000-S; Nikon) and photographed using an Orca ER camera, and images were processed using OpenLab software version 3.1.5.
For immunofluorescence studies on human fetal skeletal muscle, 10-μm tissue sections were collected on sialanazed slides (Polysciences, Inc.), fixed in absolute methanol, and blocked as described for cytospins before incubation with mouse anti-BMP4 (diluted 1:20; Novocastra) and anti-dystrophin CAP6-10 polyclonal antibody diluted at 1:500 in PBS (Byers et al., 1993; Gussoni et al., 1999). Slides were subsequently processed as described for cytospins.
For immunohistochemistry, 5-μm-thick sequentially cut muscle cryosections were fixed in −20°C acetone for 5 min. Air-dried sections were incubated with anti-dystrophin CAP6-10 polyclonal antibody diluted at 1:1,000, 2.5 μg/ml anti-BMP4 mouse mAb (R&D Systems) at 4°C overnight, or 0.01 μg/ml anti-BMPR1a rabbit polyclonal antibody (Orbigen, Inc.) for 1 h at room temperature or a mix of Gremlin polyclonal antibody (diluted 1:50; Abgent) and 2.5 μg/ml anti-BMP4 mouse mAb. As negative controls, 10 μg/ml mouse IgG and 2.5 μg/ml rabbit IgG were used. Sections were washed with PBS 3× for 5 min and incubated with 1:200 horseradish peroxidase–conjugated goat anti–rabbit secondary antibody for dystrophin and BMPR1a staining and 1:200 horseradish peroxidase–conjugated horse anti–mouse secondary antibody or 1:200 alkaline phosphatase–conjugated horse anti–mouse secondary antibody for BMP4 staining, and 1:200 biotinylated goat anti–rabbit secondary antibody for 30 min at room temperature (Vector Laboratories) for Gremlin staining, followed by ABC complex incubation at room temperature for 30 min. Immunoreactivity was detected using NovaRed substrate (Vector Laboratories). After washing, the sections were mounted with Vectashield mounting medium. Immunohistochemistry reactivity was visualized with a 200, 400, and 1,000 magnification objective on a microscope (Eclipse 80I; Nikon) coupled to a Cytovision system (Applied Imaging) and photographed using an Orca ER camera.
Cell culture and proliferation assays
Sorted human fetal muscle BMPR1a+ and BMPR1a− cells, as well as SP and MP cells, were seeded in 96-well plates at 3 × 103 cells/well in triplicates and incubated in growth medium consisting of DME with 4% glucose, 20% fetal bovine serum (vol/vol), 1% (vol/vol) penicillin-streptomycin (10,000 UI/ml–10,000 μg/ml; Invitrogen) for 4–14 d. For BMP4 and Gremlin stimulation, 25 ng/ml human recombinant BMP4 (R&D Systems) or 2 μg/ml mouse recombinant Gremlin (R&D Systems) were exchanged every 48 h. Control wells contained sorted SP and MP cells maintained in growth medium without the addition of BMP4 or Gremlin. For coculture experiments, muscle SP cells were irradiated at 25 Gy and seeded together with MP, BMPR1a+, or BMPR1a− cells at 1:1 ratios. Blocking anti-BMP4 antibody (R&D Systems) and isotype control IgG2b antibody (R&D Systems) were added to cocultured SP with BMPR1a+ or BMPR1a− cells at 2 μg/ml every 48 h of culture.
Cell proliferation was assessed for SP/MP, BMPR1a+, and BMPR1a− cells after a culture period of 4–7 d by [3H]thymidine incorporation (1 μCi/well) for the last 18 h of culture (Frank et al., 2001). Cells were harvested using an automated cell harvester, and incorporated radioactivity was assessed by a BetaMax counter (Beckman Coulter). The results of proliferation assays were compared using the Kruskal-Wallis test for comparing populations with unknown distributions (results are shown as p-values). Differences with P < 0.05 were considered statistically significant.
Online supplemental materials
Fig. S1 illustrates Hoechst/PI profiles of human skeletal muscle cells obtained from different individuals. Fig. S2 illustrates expression of CD45 by human fetal skeletal muscle SP cells. Table S1 describes SP and MP samples subjected to microarray studies. Table S2 lists 222 genes differentially expressed between human fetal skeletal muscle SP and MP cells. Table S3 provides sequences for primers used in real-time PCR reactions. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200511036/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank the Mental Retardation and Developmental Disabilities Research Center (DDRC) Cell sorting and Microarray Core facilities (supported by P01 HD18655). The authors would also like to thank all the members of the Kunkel Laboratory and Drs. Regina Sohn and Federica Montanaro for helpful discussions and critical review of this manuscript.
This work was supported by grants from the National Institutes of Health (NIH 1PO1NS40828 and NIH 5R01 NS047727 to E. Gussoni and NIH K08 NS051349 to N.Y. Frank), the Muscular Dystrophy Association (MDA3589 to E. Gussoni), and the National Cancer Institute/National Institutes of Health (1RO1CA113796-01A1 to M.H. Frank).
Abbreviations used in this paper: BMP4, bone morphogenetic protein 4; BMPR1a, BMP receptor 1a; Ct, cycle threshold; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MHC, myosin heavy chain; MP, main population; PI, propidium iodide; SP, side population.
References
- Appell, H.J., S. Forsberg, and W. Hollmann. 1988. Satellite cell activation in human skeletal muscle after training: evidence for muscle fiber neoformation. Int. J. Sports Med. 9:297–299. [DOI] [PubMed] [Google Scholar]
- Armand, O., A.M. Boutineau, A. Mauger, M.P. Pautou, and M. Kieny. 1983. Origin of satellite cells in avian skeletal muscles. Arch. Anat. Microsc. Morphol. Exp. 72:163–181. [PubMed] [Google Scholar]
- Asakura, A., P. Seale, A. Girgis-Gabardo, and M.A. Rudnicki. 2002. Myogenic specification of side population cells in skeletal muscle. J. Cell Biol. 159:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrach, E., S. Li, A.L. Perez, J. Schienda, K. Liadaki, J. Volinski, A. Flint, J. Chamberlain, and L.M. Kunkel. 2004. Systemic delivery of human microdystrophin to regenerating mouse dystrophic muscle by muscle progenitor cells. Proc. Natl. Acad. Sci. USA. 101:3581–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, T.J., H.G. Lidov, and L.M. Kunkel. 1993. An alternative dystrophin transcript specific to peripheral nerve. Nat. Genet. 4:77–81. [DOI] [PubMed] [Google Scholar]
- Chen, B., M. Athanasiou, Q. Gu, and D.G. Blair. 2002. Drm/Gremlin transcriptionally activates p21(Cip1) via a novel mechanism and inhibits neoplastic transformation. Biochem. Biophys. Res. Commun. 295:1135–1141. [DOI] [PubMed] [Google Scholar]
- Collins, C.A., I. Olsen, P.S. Zammit, L. Heslop, A. Petrie, T.A. Partridge, and J.E. Morgan. 2005. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 122:289–301. [DOI] [PubMed] [Google Scholar]
- Cornelison, D.D., B.B. Olwin, M.A. Rudnicki, and B.J. Wold. 2000. MyoD(−/−) satellite cells in single-fiber culture are differentiation defective and MRF4 deficient. Dev. Biol. 224:122–137. [DOI] [PubMed] [Google Scholar]
- Dahlqvist, C., A. Blokzijl, G. Chapman, A. Falk, K. Dannaeus, C.F. Ibanez, and U. Lendahl. 2003. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development. 130:6089–6099. [DOI] [PubMed] [Google Scholar]
- Dale, L., and C.M. Jones. 1999. BMP signalling in early Xenopus development. Bioessays. 21:751–760. [DOI] [PubMed] [Google Scholar]
- Darr, K.C., and E. Schultz. 1987. Exercise-induced satellite cell activation in growing and mature skeletal muscle. J. Appl. Physiol. 63:1816–1821. [DOI] [PubMed] [Google Scholar]
- Dennis, G., Jr., B.T. Sherman, D.A. Hosack, J. Yang, W. Gao, H.C. Lane, and R.A. Lempicki. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:P3. [PubMed] [Google Scholar]
- Frank, M.H., M.D. Denton, S.I. Alexander, S.J. Khoury, M.H. Sayegh, and D.M. Briscoe. 2001. Specific MDR1 P-glycoprotein blockade inhibits human alloimmune T cell activation in vitro. J. Immunol. 166:2451–2459. [DOI] [PubMed] [Google Scholar]
- Giudice, G. 2001. Conserved cellular and molecular mechanisms in development. Cell Biol. Int. 25:1081–1090. [DOI] [PubMed] [Google Scholar]
- Goodell, M.A., K. Brose, G. Paradis, A.S. Conner, and R.C. Mulligan. 1996. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183:1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodell, M.A., M. Rosenzweig, H. Kim, D.F. Marks, M. DeMaria, G. Paradis, S.A. Grupp, C.A. Sieff, R.C. Mulligan, and R.P. Johnson. 1997. Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat. Med. 3:1337–1345. [DOI] [PubMed] [Google Scholar]
- Grounds, M.D., and Z. Yablonka-Reuveni. 1993. Molecular and cell biology of skeletal muscle regeneration. Mol. Cell Biol. Hum. Dis. Ser. 3:210–256. [DOI] [PubMed] [Google Scholar]
- Gussoni, E., Y. Soneoka, C.D. Strickland, E.A. Buzney, M.K. Khan, A.F. Flint, L.M. Kunkel, and R.C. Mulligan. 1999. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 401:390–394. [DOI] [PubMed] [Google Scholar]
- Haramis, A.P., H. Begthel, M. van den Born, J. van Es, S. Jonkheer, G.J. Offerhaus, and H. Clevers. 2004. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 303:1684–1686. [DOI] [PubMed] [Google Scholar]
- Haslett, J.N., D. Sanoudou, A.T. Kho, M. Han, R.R. Bennett, I.S. Kohane, A.H. Beggs, and L.M. Kunkel. 2003. Gene expression profiling of Duchenne muscular dystrophy skeletal muscle. Neurogenetics. 4:163–171. [DOI] [PubMed] [Google Scholar]
- Hogan, B.L., M. Blessing, G.E. Winnier, N. Suzuki, and C.M. Jones. 1994. Growth factors in development: the role of TGF-beta related polypeptide signalling molecules in embryogenesis. Dev. Suppl. 53–60. [PubMed]
- Hsu, D.R., A.N. Economides, X. Wang, P.M. Eimon, and R.M. Harland. 1998. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell. 1:673–683. [DOI] [PubMed] [Google Scholar]
- Jackson, K.A., T. Mi, and M.A. Goodell. 1999. Hematopoietic potential of stem cells isolated from murine skeletal muscle. Proc. Natl. Acad. Sci. USA. 96:14482–14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, L., M. Hu, W.A. Gomes, and J.A. Kessler. 2004. Transgenic mice overexpressing BMP4 develop a fibrodysplasia ossificans progressiva (FOP)-like phenotype. Am. J. Pathol. 165:1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri, T., A. Yamaguchi, M. Komaki, E. Abe, N. Takahashi, T. Ikeda, V. Rosen, J.M. Wozney, A. Fujisawa-Sehara, and T. Suda. 1994. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 127:1755–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho, A.T., Q. Zhao, Z. Cai, A.J. Butte, J.Y. Kim, S.L. Pomeroy, D.H. Rowitch, and I.S. Kohane. 2004. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 18:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha, M.K., J. Yeh, T.C. Grammer, and R.M. Harland. 2005. Depletion of three BMP antagonists from Spemann's organizer leads to a catastrophic loss of dorsal structures. Dev. Cell. 8:401–411. [DOI] [PubMed] [Google Scholar]
- Kobielak, K., H.A. Pasolli, L. Alonso, L. Polak, and E. Fuchs. 2003. Defining BMP functions in the hair follicle by conditional ablation of BMP receptor IA. J. Cell Biol. 163:609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, D.S., M.J. Fackler, C.I. Civin, and W.S. May. 1996. CD34: structure, biology, and clinical utility. Blood. 87:1–13. [PubMed] [Google Scholar]
- Kuang, S., S.B. Charge, P. Seale, M. Huh, and M.A. Rudnicki. 2006. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 172:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K.J., and R.M. Harland. 2003. Cloning and characterization of Xenopus Id4 reveals differing roles for Id genes. Dev. Biol. 264:339–351. [DOI] [PubMed] [Google Scholar]
- Martin, C.M., A.P. Meeson, S.M. Robertson, T.J. Hawke, J.A. Richardson, S. Bates, S.C. Goetsch, T.D. Gallardo, and D.J. Garry. 2004. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev. Biol. 265:262–275. [DOI] [PubMed] [Google Scholar]
- Massague, J., L. Attisano, and J.L. Wrana. 1994. The TGF-beta family and its composite receptors. Trends Cell Biol. 4:172–178. [DOI] [PubMed] [Google Scholar]
- Mauro, A. 1961. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9:493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney-Freeman, S.L., K.A. Jackson, F.D. Camargo, G. Ferrari, F. Mavilio, and M.A. Goodell. 2002. Muscle-derived hematopoietic stem cells are hematopoietic in origin. Proc. Natl. Acad. Sci. USA. 99:1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeson, A.P., T.J. Hawke, S. Graham, N. Jiang, J. Elterman, K. Hutcheson, J.M. Dimaio, T.D. Gallardo, and D.J. Garry. 2004. Cellular and molecular regulation of skeletal muscle side population cells. Stem Cells. 22:1305–1320. [DOI] [PubMed] [Google Scholar]
- Merino, R., J. Rodriguez-Leon, D. Macias, Y. Ganan, A.N. Economides, and J.M. Hurle. 1999. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 126:5515–5522. [DOI] [PubMed] [Google Scholar]
- Miraglia, S., W. Godfrey, A.H. Yin, K. Atkins, R. Warnke, J.T. Holden, R.A. Bray, E.K. Waller, and D.W. Buck. 1997. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 90:5013–5021. [PubMed] [Google Scholar]
- Molkentin, J.D., and E.N. Olson. 1996. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Dev. 6:445–453. [DOI] [PubMed] [Google Scholar]
- Montanaro, F., K. Liadaki, J. Schienda, A. Flint, E. Gussoni, and L.M. Kunkel. 2004. Demystifying SP cell purification: viability, yield, and phenotype are defined by isolation parameters. Exp. Cell Res. 298:144–154. [DOI] [PubMed] [Google Scholar]
- Montarras, D., C. Lindon, C. Pinset, and P. Domeyne. 2000. Cultured myf5 null and myoD null muscle precursor cells display distinct growth defects. Biol. Cell. 92:565–572. [DOI] [PubMed] [Google Scholar]
- Mozdziak, P.E., E. Schultz, and R.G. Cassens. 1996. The effect of in vivo and in vitro irradiation (25 Gy) on the subsequent in vitro growth of satellite cells. Cell Tissue Res. 283:203–208. [DOI] [PubMed] [Google Scholar]
- Partridge, T. 2004. Reenthronement of the muscle satellite cell. Cell. 119:447–448. [DOI] [PubMed] [Google Scholar]
- Pavlath, G.K., and E. Gussoni. 2005. Human myoblasts and muscle-derived SP cells. Methods Mol. Med. 107:97–110. [DOI] [PubMed] [Google Scholar]
- Pereira, R.C., A.N. Economides, and E. Canalis. 2000. Bone morphogenetic proteins induce gremlin, a protein that limits their activity in osteoblasts. Endocrinology. 141:4558–4563. [DOI] [PubMed] [Google Scholar]
- Reshef, R., M. Maroto, and A.B. Lassar. 1998. Regulation of dorsal somitic cell fates: BMPs and Noggin control the timing and pattern of myogenic regulator expression. Genes Dev. 12:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki, M.A., and R. Jaenisch. 1995. The MyoD family of transcription factors and skeletal myogenesis. Bioessays. 17:203–209. [DOI] [PubMed] [Google Scholar]
- Rudnicki, M.A., P.N. Schnegelsberg, R.H. Stead, T. Braun, H.H. Arnold, and R. Jaenisch. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 75:1351–1359. [DOI] [PubMed] [Google Scholar]
- Sabourin, L.A., A. Girgis-Gabardo, P. Seale, A. Asakura, and M.A. Rudnicki. 1999. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J. Cell Biol. 144:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai, Y., and E.M. De Robertis. 1997. Ectodermal patterning in vertebrate embryos. Dev. Biol. 182:5–20. [DOI] [PubMed] [Google Scholar]
- Schultz, E., D.L. Jaryszak, and C.R. Valliere. 1985. Response of satellite cells to focal skeletal muscle injury. Muscle Nerve. 8:217–222. [DOI] [PubMed] [Google Scholar]
- Seale, P., L.A. Sabourin, A. Girgis-Gabardo, A. Mansouri, P. Gruss, and M.A. Rudnicki. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell. 102:777–786. [DOI] [PubMed] [Google Scholar]
- Seale, P., J. Ishibashi, C. Holterman, and M.A. Rudnicki. 2004. Muscle satellite cell-specific genes identified by genetic profiling of MyoD-deficient myogenic cell. Dev. Biol. 275:287–300. [DOI] [PubMed] [Google Scholar]
- Sherwood, R.I., J.L. Christensen, I.M. Conboy, M.J. Conboy, T.A. Rando, I.L. Weissman, and A.J. Wagers. 2004. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 119:543–554. [DOI] [PubMed] [Google Scholar]
- Shi, X., and D.J. Garry. 2006. Muscle stem cells in development, regeneration, and disease. Genes Dev. 20:1692–1708. [DOI] [PubMed] [Google Scholar]
- Topol, L.Z., W.S. Modi, S. Koochekpour, and D.G. Blair. 2000. DRM/GREMLIN (CKTSF1B1) maps to human chromosome 15 and is highly expressed in adult and fetal brain. Cytogenet. Cell Genet. 89:79–84. [DOI] [PubMed] [Google Scholar]
- Wang, Y.C., and E.L. Ferguson. 2005. Spatial bistability of Dpp-receptor interactions during Drosophila dorsal-ventral patterning. Nature. 434:229–234. [DOI] [PubMed] [Google Scholar]
- Weintraub, H. 1993. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 75:1241–1244. [DOI] [PubMed] [Google Scholar]
- Wrana, J.L., L. Attisano, R. Wieser, F. Ventura, and J. Massague. 1994. a. Mechanism of activation of the TGF-beta receptor. Nature. 370:341–347. [DOI] [PubMed] [Google Scholar]
- Wrana, J.L., H. Tran, L. Attisano, K. Arora, S.R. Childs, J. Massague, and M.B. O'Connor. 1994. b. Two distinct transmembrane serine/threonine kinases from Drosophila melanogaster form an activin receptor complex. Mol. Cell. Biol. 14:944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, V., H. Peng, A. Usas, B. Young, B. Gearhart, J. Cummins, and J. Huard. 2002. BMP4-expressing muscle-derived stem cells differentiate into osteogenic lineage and improve bone healing in immunocompetent mice. Mol. Ther. 6:169–178. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni, Z., M.A. Rudnicki, A.J. Rivera, M. Primig, J.E. Anderson, and P. Natanson. 1999. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev. Biol. 210:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, A.H., S. Miraglia, E.D. Zanjani, G. Almeida-Porada, M. Ogawa, A.G. Leary, J. Olweus, J. Kearney, and D.W. Buck. 1997. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 90:5002–5012. [PubMed] [Google Scholar]
- Zammit, P., and J. Beauchamp. 2001. The skeletal muscle satellite cell: stem cell or son of stem cell? Differentiation. 68:193–204. [DOI] [PubMed] [Google Scholar]
- Zammit, P.S., J.P. Golding, Y. Nagata, V. Hudon, T.A. Partridge, and J.R. Beauchamp. 2004. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J. Cell Biol. 166:347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., C. Niu, L. Ye, H. Huang, X. He, W.G. Tong, J. Ross, J. Haug, T. Johnson, J.Q. Feng, et al. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 425:836–841. [DOI] [PubMed] [Google Scholar]
- Zhou, S., J.D. Schuetz, K.D. Bunting, A.M. Colapietro, J. Sampath, J.J. Morris, I. Lagutina, G.C. Grosveld, M. Osawa, H. Nakauchi, and B.P. Sorrentino. 2001. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 7:1028–1034. [DOI] [PubMed] [Google Scholar]
- Zuniga, A., A.P. Haramis, A.P. McMahon, and R. Zeller. 1999. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 401:598–602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.