Abstract
Transforming growth factor β1 (TGFβ1), an important regulator of cell behavior, is secreted as a large latent complex (LLC) in which it is bound to its cleaved prodomain (latency-associated peptide [LAP]) and, via LAP, to latent TGFβ-binding proteins (LTBPs). The latter target LLCs to the extracellular matrix (ECM). Bone morphogenetic protein 1 (BMP1)–like metalloproteinases play key roles in ECM formation, by converting precursors into mature functional proteins, and in morphogenetic patterning, by cleaving the antagonist Chordin to activate BMP2/4. We provide in vitro and in vivo evidence that BMP1 cleaves LTBP1 at two specific sites, thus liberating LLC from ECM and resulting in consequent activation of TGFβ1 via cleavage of LAP by non–BMP1-like proteinases. In mouse embryo fibroblasts, LAP cleavage is shown to be predominantly matrix metalloproteinase 2 dependent. TGFβ1 is a potent inducer of ECM formation and of BMP1 expression. Thus, a role for BMP1-like proteinases in TGFβ1 activation completes a novel fast-forward loop in vertebrate tissue remodeling.
Introduction
TGFβs are cytokines with manifold key roles in modulating cell proliferation and differentiation, apoptosis, immune responses, tissue repair, and ECM formation (Massague, 1990; Border and Ruoslahti, 1992; Letterio and Roberts, 1998; Derynck et al., 2001). TGFβ1, the first of 3 isoforms (TGFβ1–3) to be identified, is the prototype of a superfamily of cytokines, related in domain structure and sequence homology, that is divisible into subfamilies based on degree of sequence homology (Ducy and Karsenty, 2000). The TGFβ1–3 subfamily is closely related to the activin subfamily and to growth and differentiation factors (GDFs) 8 (aka myostatin) and 11 (Ducy and Karsenty, 2000). All superfamily members act by binding cell surface type I and II receptor heterotetramers, with signal transduction to the nucleus via Smad-dependent or -independent pathways (Massague et al., 2005).
Signaling by TGFβ-like ligands is tightly controlled by various intracellular, cell surface, and extracellular inhibitory proteins (Massague et al., 2005; Zacchigna et al., 2006). Signaling by TGFβ1–3 and GDF8 and -11 is also blocked by formation of a noncovalent latent complex between the functional ligand and its cleaved prodomain (Massague, 1990; Annes et al., 2003; Wolfman et al., 2003; Ge et al., 2005), designated latency-associated peptide (LAP) for TGFβ1–3 (Annes et al., 2003). TGFβ1–3 are produced as large latent complexes (LLCs) in which LAP is disulfide bonded to a latent TGFβ-binding protein (LTBP), of which mammals have four (Annes et al., 2003). It remains unclear which processes activate TGFβ latent complexes in vivo, although cleavage within LAP sequences by plasmin and/or matrix metalloproteinases (MMPs; Lyons et al., 1988; Sato and Rifkin, 1989; Yu and Stamenkovic, 2000; D'Angelo et al., 2001; Maeda et al., 2001; Mu et al., 2002) and/or nonproteolytic dissociation of LAP from active TGFβ, caused by interactions with thrombospondin (Crawford et al., 1998) or integrin αVβ6 (Munger et al., 1999), may be involved. Although overall in vivo roles of LTBPs remain obscure, they serve to bind LLCs to ECM, perhaps via covalent linkage of LTBPs to ECM components (Nunes et al., 1997). Ability of LTBP1 to fix LLC to ECM appears involved in its ability to promote αVβ6-mediated TGFβ activation, whereas the apparent ability of LTBPs to facilitate TGFβ activity in other situations is by as-yet-unclear mechanisms (Flaumenhaft et al., 1993; Nakajima et al., 1997; Gualandris et al., 2000).
Bone morphogenetic protein 1 (BMP1)–like proteinases affect morphogenesis by biosynthetic processing of precursors to mature functional proteins necessary to ECM formation. Such proteins include collagens I–III (Kessler et al., 1996), V, VII, and XI; proteoglycans biglycan and osteoglycin; basement component laminin 5; and the cross-linking enzyme lysyl oxidase (Ge and Greenspan, 2006). BMP1-like proteinases also affect morphogenesis by activating BMP2 and -4 via cleaving the extracellular antagonist Chordin and thus regulating patterning (Blader et al., 1997; Piccolo et al., 1997; Pappano et al., 2003). In addition, BMP1-like proteinases activate GDF8 and -11 by cleaving within noncovalently bound prodomains to release mature GDF8 and -11 from latent complexes (Wolfman et al., 2003; Ge et al., 2005), thus affecting negative growth control of skeletal muscle and neural tissues, for which GDF8 and -11, respectively, are responsible (McPherron et al., 1997; Wu et al., 2003).
Here, we use in vitro and in vivo approaches to demonstrate that BMP1-like proteinases serve to activate TGFβ by direct cleavage of LTBP1, resulting in liberation of LLCs from ECM and consequent MMP-dependent LAP cleavage. Because BMP1 is potently induced by TGFβ1 (Lee et al., 1997), its role in TGFβ1 activation completes a novel amplification loop in vertebrate tissue remodeling. Moreover, BMP1-like proteinases are identified as regulators capable of orchestrating TGFβ signaling with ECM deposition, patterning, and negative feedback control of muscle and neural tissue growth.
Results
BMP1 cleaves LTBP1 but not LAP
To determine whether BMP1 might play a role in proteolytic activation of TGFβ1, BMP1 was incubated with TGFβ1 LLC. A Western blot probed with antibody to a C-terminal His tag demonstrated dose-dependent cleavage of an ∼17-kD fragment from the C-terminal portion of ∼190-kD LTBP1 (Fig. 1 A). A similar blot probed with anti-LTBP1 (Fig. 1 B) demonstrated a corresponding reduction in size of the remainder of LTBP1 to produce a doublet or, at higher BMP1 concentrations, a single band of ∼150 kD. N-terminal amino acid sequencing of the 150- and 17-kD LTBP1 fragments demonstrated that cleavages had occurred at peptide bonds 485L-D486 and 1285Q-D1286, respectively. The former of these sites occurs between the most N-terminal 8-Cys motif and the second most N-terminal non-Ca2+ binding EGF-like domain, within the flexible hinge region of LTBP1 (Fig. 1 C). The latter cleavage occurs between the most C-terminal 8-Cys motif and the most C-terminal non-Ca2+ binding EGF-like domain. Both sites have P1′ Asp residues and other features characteristic of cleavage sites of previously identified BMP1 substrates (Fig. 1 D). Control experiments showed both sites to be efficiently cleaved, with cleavage at the N- and C-terminal sites occurring within 10 and 30 min of incubation with BMP1, respectively (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200606058/DC1). Control experiments also demonstrated that BMP1 must be catalytically active for LTBP1 cleavage to occur, as even prolonged incubation with catalytically inactive BMP1, in which the protease domain active site Glu94 had been substituted for with Ala, did not alter LTBP1 electrophoretic mobility (Fig. S1).
Figure 1.
BMP1 cleaves LTBP-1, but not LAP, in vitro. TGFβ1 LLC was incubated in the absence (−) or presence of low (25 ng) or high (250 ng) amounts of BMP1, and cleavage monitored by Western blots was probed with antibody to the C-terminal His tag of LTBP1 (A) or with anti-LTBP1 antibody (B). (C) A schematic is shown of LTBP1. H, hybrid domain; C1–3, 8-cysteine domains 1–3. Green boxes indicate non-Ca2+ binding EGF-like repeats, light orange boxes indicate Ca2+ binding EGF-like repeats, and the scissors indicate BMP1 cleavage sites. LAP (blue) is shown disulfide bonded to the C2 domain, and active TGFβ (dark orange) is shown noncovalently bound to LAP. (D) Alignment of LTBP1 cleavage sites with cleavage sites of known substrates of BMP1-like proteinases. Aspartates present at the P1′ sites of most cleavage sites of BMP1-like proteinases are in red, whereas other residues enriched near cleavage sites of known substrates of BMP1-like proteinases are in green. (E and F) TGFβ1 LLC was incubated in the absence (−) or presence (+) of BMP1, with samples analyzed by SDS-PAGE under reducing (E) or nonreducing (F) conditions, followed by Western blot analysis with anti-LAP antibodies. Numbers to the sides of blots correspond to the positions and approximate sizes in kD of molecular mass markers or known protein bands. In some cases, the molecular masses of known proteins are noted parenthetically next to the name of the protein. SM, starting material.
In contrast to its ability to cleave LTBP1, active BMP1 did not cleave ∼37-kD LAP to a smaller size, nor did it cleave within the ∼50-kD precursor molecule consisting of LAP covalently bound to the mature portion of TGFβ1 (Fig. 1 E). Under nonreducing conditions, an immunoblot analysis with anti-LAP antibodies showed ∼290-kD intact LLC to be cleaved to an ∼245-kD form by BMP1 (Fig. 1 F). Interestingly, mobility corresponding to 245 kD, rather than the 37 kD expected for dissociated LAP, indicates that LAP remains associated with LTBP1 subsequent to cleavage of the latter by BMP1 (Fig. 1 F).
BMP1-like proteinases are necessary for direct cleavage of LTBP1 and indirect cleavage of LAP by cells
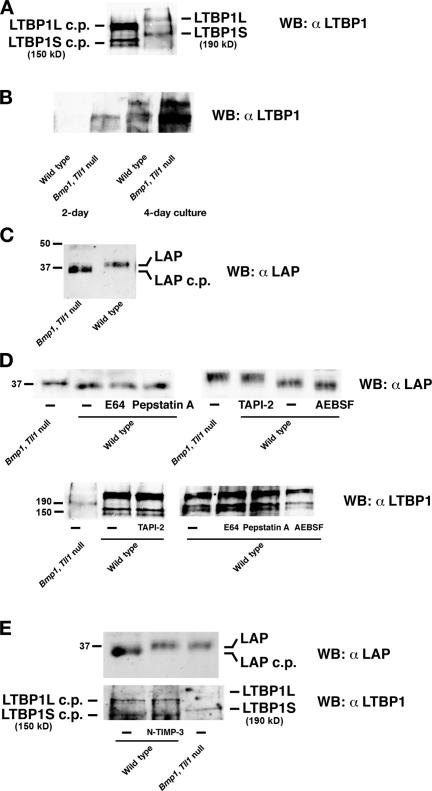
To determine whether BMP1-like proteinases are used for in vivo LTBP1 cleavage, we compared processing of LTBP1 by wild-type mouse embryo fibroblasts (MEFs) to processing of LTBP1 by MEFs derived from embryos doubly homozygous null for the Bmp1 gene, which encodes alternatively spliced mRNAs for proteinases BMP1 and mammalian tolloid (Ge and Greenspan, 2006), and the Tll1 gene, which encodes the BMP1-like proteinase mammalian tolloid–like 1 (Ge and Greenspan, 2006). Simultaneous ablation of the functionally redundant products of Bmp1 and Tll1 has previously been shown to remove detectable processing activity for various substrates of BMP1-like proteinases (Pappano et al., 2003). Cells produce short and long forms of LTBP1 (LTBP1S and -L, respectively), resulting from alternative splicing and multiple transcriptional promoters (Koski et al., 1999), and both LTBP1S and -L were found to differ in electrophoretic mobility in wild-type and Bmp1/Tll1 doubly null MEF media samples (Fig. 2 A). Moreover, sizes of LTBP1S in wild-type (150 kD) and Bmp1/Tll1-null (190 kD) samples corresponded, respectively, to sizes of BMP1-cleaved and uncleaved recombinant LTPB1S (Fig. 1 B).
Figure 2.
LLCs are not proteolytically processed in cultures of MEFs doubly homozygous null for the Bmp1 and Tll1 genes. Western blot analysis with anti-LTPB1 antibodies was used to monitor proteolytic processing of LTBP1 to LTBP1 cleavage products (cp) in conditioned media from 4-d cultures of wild-type or Bmp1/Tll1 doubly null MEFs (A) or monitor amounts of LTBP1 released by plasmin from ECM of 2- or 4-d wild-type or Bmp1/Tll1 doubly null MEF cultures (B). (C) Western blot analysis with anti-LAP antibodies was used to monitor LAP proteolytic processing in conditioned media from 4-d wild-type or Bmp1/Tll1 doubly null MEF cultures. (D) Western blot analyses were used to monitor proteolytic processing of LAP and LTBP1 in conditioned media of wild-type MEFs cultured in the presence of inhibitors of metallo- (TAPI-2), cysteine- (E64), aspartic- (pepstatin A), and serine- (AEBSF) proteinases. (E) Western blot analyses with anti-LAP and anti-LTBP1 antibodies shows that the inhibitory N-terminal domain of TIMP-3 (N-TIMP-3) inhibits processing of LAP, but not LTBP1, in wild-type MEF cultures. Numbers to the left of blots correspond to the positions and approximate sizes in kD of molecular mass markers or known protein bands. In some cases, the molecular masses of known proteins are noted parenthetically next to the name of the protein.
It has been reported that removal of sequences N-terminal to the hinge region is sufficient to release LTBP1 from transglutaminase-dependent covalent association with ECM (Taipale et al., 1994; Nunes et al., 1997). Because such sequences are N-terminal to the 485L-D486 BMP1 cleavage site demonstrated here, the decreased amounts of LTBP1S and -1L in Bmp1/Tll1-null, compared with wild-type MEF medium (Fig. 2 A), is consistent with the probability that cells use BMP1-like proteinases to process the LTBP1 hinge region and that this is sufficient to release TGFβ1 LLC from ECM. Buttressing this probability was the observation that markedly greater amounts of LTBP1 fragments are released by plasmin from Bmp1/Tll1-null than from wild-type MEF ECM (Fig. 2 B).
Surprisingly, although BMP1 does not cleave LAP in vitro (Fig. 1 E), the electrophoretic mobility of LAP was increased in wild-type compared with Bmp1/Tll1-null MEF media (Fig. 2 C). Because the mobility of LAP from Bmp1/Tll1-null MEF media is equal to that of full-length LAP, these results are consistent with the probability that LAP is cleaved in wild-type, but not Bmp1/Tll1-null, MEF cultures. To determine the type of proteinases responsible for LAP cleavage, wild-type MEFs were cultured in the presence of inhibitors of metallo- (TNF-α processing inhibitor 2 [TAPI-2]), cysteine- (E64), aspartic- (pepstatin A), and serine- (4-[2-aminoethyl]-benzenesulfonyl fluoride [AEBSF]) proteinases. Only TAPI-2, which inhibits metalloproteinase sheddases and a range of MMPs, inhibited LAP processing in MEF cultures, whereas none of the inhibitors interfered with LTBP1S or -L processing (Fig. 2 D). Importantly, BMP1 is not inhibited by even 200 μM TAPI-2 in vitro (unpublished data), consistent with the interpretation that LAP in wild-type MEF cultures is cleaved by metalloproteinases not of the BMP1 subgroup.
Tissue inhibitor of metalloproteinase 3 (TIMP-3) has inhibitory activity toward all MMPs and against some members of the ADAM (a disintegrin and metalloproteinase) family of proteinases (Visse and Nagase, 2003) but has no inhibitory activity against BMP1 (Wang et al., 2006). When wild-type MEFs were cultured in the presence of the N-terminal inhibitory domain of TIMP-3, which has the same inhibitory activity as the full-length protein (Kashiwagi et al., 2001), processing of LAP but not LTBP1 was inhibited (Fig. 2 E), consistent with the conclusion that LAP is cleaved by non–BMP1-like metalloproteinases.
LAP cleavage is MMP2 dependent
MMP2, - 9, and -14 have previously been identified in vitro as metalloproteinase activators of TGFβ (Sato and Rifkin, 1989; Yu and Stamenkovic, 2000; Mu et al., 2002). Thus, RNAi knockdown was performed in wild-type MEFs, to test whether any of these might play a role in cleaving LAP in LLCs released from ECM by BMP1. RNAi knock down of each MMP was effective at RNA (Fig. 3 A) and protein (Fig. 3 B) levels. Interestingly, MMP9 RNA and protein levels were also reduced upon knock down of MMP2.
Figure 3.
LAP cleavage is MMP2 dependent. (A) Levels of MMP2, MMP9, MMP14, or GAPDH RNA were compared by RT-PCR. Sizes of cDNA bands in bp are noted in brackets to the right of the panel. Western blot analysis using anti-proMMP2, anti-MMP9, or anti-PCOLCE1 antibodies (B) or anti–phospho-Smad2/3 or anti-tubulin antibodies (D) was performed on samples from wild-type MEFs subjected to RNAi knock down of MMP2, -9, or -14, or using a scrambled RNAi duplex control. (C) Western blot analysis with anti-LAP or anti-LTBP1 antibodies was performed on samples from Bmp1/Tll1 doubly null MEFs or from wild-type MEFs subjected to RNAi knock down of MMP2, -9, or -14, or using a scrambled RNAi duplex control. Western blot analysis with anti-LAP antibodies was performed on SLC (E) or on LLC (F and G) incubated with (+) or without (−) MMP2. In F and G, MMP2 cleavage was subsequent to incubation in the presence (+) or absence (−) of BMP1. The numbers beneath lanes indicate signal intensity relative to signal in control lane (B and D). The numbers to the left of blots correspond to the positions and approximate sizes in kD of molecular mass markers or known protein bands. In some cases, the molecular masses of known proteins are noted parenthetically next to the name of the protein.
Analysis of LAP from conditioned media showed that knock down of MMP2, but not of the other two MMPs, resulted in inhibition of LAP cleavage, such that electrophoretic mobility was the same as for LAP isolated from Bmp1/Tll1-null MEF medium (Fig. 3 C). Consistent with the latter results, Smad2/3 phosphorylation, a direct downstream indicator of TGFβ-specific signaling (Massague et al., 2005), was much reduced in MMP2 RNAi-treated MEFs, compared with untreated MEFs or MEFs treated with MMP9 or -14 RNAi (Fig. 3 D). Thus, MMP2 is necessary for LAP cleavage and TGFβ activation in MEFs, subsequent to BMP1 liberation of LLC from ECM. MMP2-dependent TGFβ activation probably explains the decrease in MMP9 RNA/protein levels upon MMP2 RNAi treatment, as MMP9 RNA/protein levels are elevated by TGFβ activity (Sehgal and Thompson, 1999). In Fig. 3 E, it is demonstrated that MMP2 is capable of in vitro cleavage of LAP in small latent complexes (SLC), yielding a fragment similar in size to the LAP fragment in wild-type MEF media (Figs. 2, C and D; Fig. 3 C). In contrast, MMP2 cleavage of LLC-associated LAP was extremely inefficient, without prior incubation of LLC with BMP1 (Fig. 3 F). Thus, in vitro, as in vivo, prior LLC processing by BMP1 greatly facilitates subsequent MMP2 cleavage of LAP. Under nonreducing conditions, immunoblot analysis with anti-LAP antibodies showed LLC to be cleaved to an ∼230-kD form upon incubation with BMP1 and MMP2 (Fig. 3 G). Mobility corresponding to a 230-kD form, rather than the 35 kD expected for cleaved and dissociated LAP, indicates that LAP remains associated with LTBP1 subsequent to BMP1 cleavage of LTBP1 and MMP2 cleavage of LAP.
It should be noted that the aforementioned studies do not formally rule out roles for MMP9 and -14 in TGFβ activation in MEFs, as RNAi knock down of MMP2 also resulted in a knock down of MMP9 protein that was more dramatic than that achieved with MMP9 RNAi (Fig. 3 B), whereas levels of MMP14 protein could not be ascertained in knockdowns because of the unavailability of antibodies capable of detecting murine MMP14 via immunoblotting. Indeed, the role of MMP14 in activating MMP2 (Visse and Nagase, 2003) ensures some level of contribution of MMP14 to TGFβ activation in MEFs.
Cell cultures and tissues have increased levels of LTBP1 deposition and reduced TGFβ activity in the absence of BMP1-like proteinases
It has previously been shown that LTBP1-containing LLCs can be deposited in ECM as extracellular fibrillar structures associated with fibrillin microfibrils (Dallas et al., 2000; Isogai et al., 2003). To determine whether ablation of Bmp1 and Tll1 affects the appearance of LTBP1 in ECM, anti-LTBP1 immunofluorescence was performed for comparison of wild-type and Bmp1/Tll1-null MEF cell layers. Consistent with the results of Fig. 2 B, in which levels of ECM-associated LTBP1 were markedly increased in Bmp1/Tll1-null MEF cell layers compared with wild type, detectable fibrillar structures containing LTBP1 were greatly increased in number and thickness in Bmp1/Tll1-null MEF cell layers compared with wild type (Fig. 4 A). In contrast, fibrillin 1 microfibrils were similar in numbers and thickness in mutant and wild-type cell layers.
Figure 4.
Comparison of fibrillar LTBP1 deposition, TGFβ activity, and MMP levels in Bmp1/Tll1 doubly null and wild-type MEF cultures. (A) Wild-type (WT) and Bmp1/Tll1 doubly null MEF cell layers were subjected to immunofluorescence staining, using polyclonal anti-LTBP1 or anti–fibrillin 1 (FBN1) antibodies. Images were deconvoluted and contrast was adjusted with the same parameters. Photomicrograph exposure times were identical per given antibody (1/8 and 1/15 s for anti-LTBP1 and anti–fibrillin 1, respectively) to ensure comparability of signal levels for wild-type and mutant tissues. Bar, 20 μm. (B–D) Bmp1/Tll1 doubly null MEF culture media contain similar levels of TGFβ but markedly lower levels of active TGFβ than wild-type culture media. Conditioned media from Bmp1/Tll1 doubly null and wild-type MEFs were separately added to a reporter gene assay without (B) or with (C) heat activation, for detection of active or total TGFβ, respectively. Aliquots of 2.5, 5, 10, 20, and 50 μl were added, as indicated. (D) In a control experiment, conditioned media from Bmp1/Tll1 doubly null and wild-type MEFs were incubated 1 h at 4°C in the absence (−) or presence of either anti-TGFβ1 (αTGFβ1) or pan-specific (α pan TGFβ) anti-TGFβ antibody before addition to the reporter gene assay. (E) MEFs were cocultured with T-MLEC reporter cells, followed by scraping of cell layers for determination of luciferase levels. (B–E) Numbers on the ordinate axis represent fold increases in levels of luciferase activity. (F–H) Levels of TGFβ-induced MMPs and CYR61 are notably higher in wild-type than in Bmp1/Tll1 doubly null MEF cultures. (F) 25, 30, and 35 cycles of RT-PCR were performed on RNA from wild-type and Bmp1/Tll1 doubly null MEFs. Sizes of cDNA bands in bp are noted in brackets to the right of the panel. (G and H) Western blot analyses were performed on media (G) and cell layers (H) of wild-type or Bmp1/Tll1 doubly null MEFs using antibodies to MMP2 (Chemicon), MMP9 (Abcam), CYR61 (Santa Cruz Biotechnology, Inc.), and p-Smad2/3, and against PCOLCE1 (G; Lee et al., 1997) and tubulin (H; Oncogene) as loading controls. Molecular masses of known proteins are noted parenthetically next to the name of the protein.
Cleavage of LAP in wild-type but not Bmp1/Tll1-null MEF cultures suggested that TGFβ activity might be reduced in the latter. To test this, levels of active TGFβ in wild-type and Bmp1/Tll1-null MEF cultures were compared by adding conditioned media to a reporter gene assay consisting of mink lung epithelial cells stably transfected with a luciferase gene driven by TGFβ-responsive plasminogen activator inhibitor 1 promoter sequences (T-MLEC [transfected mink lung epithelial cell]). Assay results clearly demonstrated Bmp1/Tll1-null MEF media to have lower TGFβ activity levels than wild type (Fig. 4 B), even though the two types of media had similar levels of TGFβ protein, as demonstrated by the similar levels of activity obtained upon heat activation (Fig. 4 C). Thus, although the two types of MEF culture media have similar levels of TGFβ, much more is in a latent form in Bmp1/Tll1-null than in wild-type medium. In a control experiment, the use of either of two different TGFβ-neutralizing antibodies (one specific for TGFβ1 and the other a pan-specific TGFβ antibody that reacts with TGFβ1–5) was capable of reducing both wild- type and Bmp1/Tll1-null MEF media activity levels in the T-MLEC reporter gene assay to baseline levels (Fig. 4 D), thereby demonstrating such activity to be due predominantly to TGFβ1, rather than to other factors that act through the Smad2/3 signaling pathway. In another control experiment, to ensure that latent TGFβ complexes in harvested wild-type and Bmp1/Tll1-null MEF conditioned media were not differentially activated during in vitro handling before addition to T-MLEC cultures; MEFs were cocultured with T-MLECs, directly followed by harvesting of monolayers and determination of amounts of luciferase activity. As shown in Fig. 4 E, coculturing confirmed wild-type MEFs to have intrinsically higher levels of TGFβ signaling than Bmp1/Tll1-null MEFs. We have found T-MLECs to secrete active BMP1 (unpublished data), which may release some LLC from ECM during coculturing, thus accounting for the somewhat smaller difference in TGFβ activity levels between wild-type and Bmp1/Tll1-null MEFs in the coculturing experiment (Fig. 4 E), compared with differences found in experiments in which conditioned media were added to T-MLEC cultures (Figs. 4, B and D).
We next assayed for evidence of effects of decreased TGFβ activity on Bmp1/Tll1-null MEF cells. It has been reported that TGFβ1 induces increased levels of MMP2 and -9 via transcriptional, posttranscriptional, and posttranslational mechanisms (Brown et al., 1990; Overall et al., 1991; Marti et al., 1994; Sehgal and Thompson, 1999). Thus, we compared MMP2 and -9 levels in Bmp1/Tll1-null and wild-type MEF conditioned media. Results showed levels of MMP2 RNA (Fig. 4 F) and protein (Fig. 4 G) to be markedly higher in wild-type than in Bmp1/Tll1-null MEF cultures and showed differences in MMP9 RNA and protein levels to be even more pronounced. Similarly, levels of CYR61, an established marker for TGFβ activity (Brunner et al., 1991), were markedly higher in wild-type than in Bmp1/Tll1-null MEF cultures. In contrast, levels of the protein PCOLCE1, expression of which has been shown not to be affected by TGFβ1 (Lee et al., 1997), were similar in Bmp1/Tll1-null and wild-type MEF media (Fig. 4 G). Next, levels of phosphorylated/activated Smad2/3 were compared in Bmp1/Tll1-null and wild-type MEFs. As shown in Fig. 4 H, phosphorylated Smad2/3, readily detectable in wild-type MEFs, was difficult to detect in Bmp1/Tll1-null MEFs under the same conditions. Thus, Bmp1/Tll1-null MEFs bear multiple hallmarks of cells with reduced levels of TGFβ signaling.
We next assayed for differences in levels of detectable LTBP1 and levels of TGFβ signaling in tissues of 13.5-d post conception (dpc) wild-type and Bmp1/Tll1-null embryos. As previously reported (Isogai et al., 2003), labeling of wild-type tissues with anti–LTBP-1 was sparse. However, markedly higher levels of LTBP1 fibrillar networks were detected in Bmp1/Tll1-null tissues (Fig. 5 A). In contrast, wild-type tissues had markedly higher levels of phosphorylated Smad2/3 than did Bmp1/Tll1-null tissues, consistent with markedly higher levels of TGFβ signaling (Fig. 5 A). In contrast to both these results, levels and distributions of fibrillin 1 microfibrils were similar in the two types of tissues. That the overabundant LTBP1 in Bmp1/Tll1-null tissues, which predominantly colocalizes with fibrillin fibrils (unpublished data), does not alter the appearance of fibrillin fibrils compared with wild type is consistent with previous biochemical evidence that LTBP1 is a microfibril-associated protein, rather than an integral structural component of microfibrils (Isogai et al., 2003).
Figure 5.
Increased LTBP1 accumulation and decreased TGFβ signaling are found in Bmp1/Tll1 doubly null tissues and accompany developmental failure of the frontal bone in Bmp1−/−/Tll1+/− embryos. (A) Bmp1/Tll1 doubly null tissues show decreased TGFβ signaling and increased LTBP1 accumulation compared with wild type (WT). Serial parasagittal sections of tissues of 13.5-dpc wild-type and Bmp1/Tll1 doubly null embryos were subjected to hematoxylin/eosin (H&E) staining (top) and immunofluorescent staining with polyclonal antibodies to LTBP1, phosphorylated Smad2/3, and fibrillin 1 (FBN1). Shown are cross sections of presumptive rib. (B) Increased LTBP1 accumulation and decreased TGFβ signaling accompany developmental failure of the frontal bone in Bmp1 −/−/Tll1 +/− embryos. Serial coronal sections of 17.5-dpc wild-type and Bmp1 −/−/Tll1 +/− embryos were subjected to hematoxylin/eosin staining (top) or immunofluorescent staining. Arrowheads mark the boundaries of ossified and nonossified mesenchymal portions of presumptive frontal bones. Insets in the top panels of B correspond to the hematoxylin/eosin-stained sections immediately below. Note that although both margins of ossified frontal bone are visible near the midline in wild type (B), only a single Bmp1 −/−/Tll1 +/− margin is shown because of the wide gap separating the two margins in retarded mutant frontal bone. For immunofluorescence, photomicrograph exposure times were identical per given antibody (1/8, 1/4, and 1/30 s for anti-LTBP1, anti–p-Smad2/3, and anti-FBN1, respectively) to ensure comparability of signal levels for wild-type and mutant tissues. Bars: (B, top) 80 μm; (A and B) 20 μm.
Interestingly, it has previously been shown that impairment of TGFβ signaling (Sasaki et al., 2006) and ablation of the Bmp1 gene (Suzuki et al., 1996) both result in retardation in formation of the intramembranous frontal bone of the skull. Bmp1/Tll1 doubly homozygous null embryos die at 13.5 dpc (Pappano et al., 2003), before formation of the frontal bone. Thus, we assayed for possible differences in LTBP1 accumulation and TGFβ signaling in frontal bones of 17.5-dpc Bmp1 −/−/Tll1 +/− and wild-type embryos. As previously reported for Bmp1 −/− 17.5-dpc embryos (Suzuki et al., 1996), the gap between ossified portions of the forming frontal bones is greatly increased in Bmp1 −/−/Tll1 +/− compared with wild-type 17.5-dpc embryos (Fig. 5 B). Immunofluorescent staining showed fibrillar structures containing LTBP1 to be greatly increased in number and thickness in Bmp1 −/−/Tll1 +/− compared with wild-type frontal bone, with localization predominantly to the periosteum lining the ossified bone and nonossified mesenchyme of the frontal bone primordium (Fig. 5 B). In contrast, markedly higher levels of signal for phosphorylated Smad2/3 were found in wild-type presumptive frontal bone and adjoining tissues than in corresponding Bmp1 −/−/Tll1 +/− tissues. Levels of fibrillin 1 microfibrils were similar in wild-type and mutant tissues.
Discussion
Much of the TGFβ in tissues is in the form of LLCs linked to ECM via LTBPs. As such, they constitute a reservoir of key cytokines that can be rapidly mobilized in response to tissue perturbations. Interestingly, identified activators of TGFβ may all represent readouts of ECM perturbation (Annes et al., 2003). For example, MMP2 and -9 and plasmin play central roles in ECM degradation, whereas thrombospondin-1 expression is induced in response to tissue damage and is involved in the early stages of ECM regeneration (Bornstein et al., 2004). Thus, in many cases, ECM perturbation may be an important signal causally preceding activation of TGFβ in ECM-bound LLCs. Indeed, ablation of fibrillin1, the protein via which LTBP1 binds LLCs to ECM, is sufficient to yield raised TGFβ activity in tissues (Neptune et al., 2003); similarly, overexpression of truncated LTBP1 that is capable of binding SLC but incapable of binding ECM also yields raised TGFβ activity (Mazzieri et al., 2005). Both results support the concept that activation of LLC-associated TGFβ is greatly enhanced by, and may depend on, prior release from ECM.
Here, we describe a novel mechanism for TGFβ activation, involving LLC liberation from ECM via specific LTBP1 cleavage by BMP1-like proteinases. This cleavage does not free LAP from LTBP1, nor does it free active TGFβ from LAP. Rather, the effect of this cleavage in both cell cultures and tissues appears to be consequent cleavage of LAP and activation of TGFβ by non–BMP1-like metalloproteinases. In MEF cultures, we have demonstrated this consequent cleavage to be dependent on MMP2 and perhaps on other non–BMP1-like metalloproteinases as well and have demonstrated that prior BMP1 cleavage of LLC in vitro results in greatly enhanced susceptibility of LLC LAP to cleavage by MMP2. In other cell types, additional proteases may be involved in LAP cleavage, subsequent to BMP1 liberation of LLCs from ECM.
TGFβ potently induces net ECM formation by effecting decreased production of some ECM-degrading proteases and increased production of (1) endogenous inhibitors for such proteases, (2) ECM structural proteins, (3) enzymes that stabilize ECM via cross-link formation (i.e., lysyl oxidase; Massague, 1990), and (4) metalloproteinases necessary for biosynthetic processing of ECM structural proteins and lysyl oxidase to their mature functional forms (e.g., the BMP1-like proteinases; Lee et al., 1997; Wang et al., 2003). Thus, demonstration here that BMP1-like proteinases effect activation of TGFβ completes a novel feed-forward loop for net ECM deposition. This loop, illustrated in Fig. 6, is likely to feature in various morphogenetic events in which both BMP1-like proteinases and TGFβ and have been implicated as key players, including development, wound repair, angiogenesis (St Croix et al., 2000; Bertolino et al., 2005), and synaptic plasticity (Zhang et al., 1997). Data presented herein are consistent with the possibility that this loop is necessary, in a nonredundant way, for normal formation of the frontal bone of the skull.
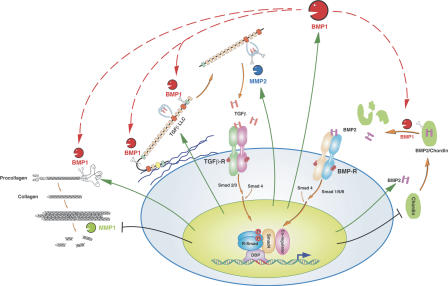
Figure 6.
Manifold roles for BMP1-like proteinases and the BMP1/TGFβ feed-forward loop for tissue remodeling. BMP1-like proteinases biosynthetically process ECM precursors (e.g., procollagen) to mature functional ECM components. They also activate TGFβ by processing LTBP1 to release truncated LLC from ECM, leading to consequent activation via LAP cleavage by metalloproteinases such as MMP2. Activated TGFβ induces activation of R-Smad2 and -3, which combine with Smad4 for translocation to the nucleus and up-regulation (vertical arrows) of BMP1, ECM precursors (e.g., procollagen), MMP2, and TGFβ itself. TGFβ also down-regulates some MMPs that degrade ECM (e.g., MMP1). BMP1-like proteinases also activate BMP2/4 by cleaving the antagonist Chordin, thus inducing activation of R-Smad1, -5, and -8. The latter may compete with R-Smad2 and -3 for limiting amounts of Smad4. Conceivably, Chordin competes with ECM precursors and LTBP1 for BMP1-like proteinases. Thus, Smad4 and BMP1-like proteinases may represent two levels at which cross talk between TGFβ and BMP signaling pathways orchestrates tissue remodeling with patterning.
MMP2 and -9 and various other MMPs are capable of playing key roles in the tissue remodeling associated with the growth, angiogenesis, and invasiveness of tumors (Yu and Stamenkovic, 2000; Sternlicht and Werb, 2001). The same is true for TGFβ (Yu and Stamenkovic, 2000; Derynck et al., 2001), whereas high throughput screens have identified BMP1 RNA sequences as among the most up-regulated in activated endothelia associated with tumor angiogenesis (St Croix et al., 2000). Thus, the fast-forward loop involving activation of TGFβ by TGFβ-inducible BMP1, with subsequent roles played by MMPs (Fig. 6), is of potential importance to the tissue remodeling associated with morphogenesis and to the pathogenesis of cancers as well.
Clearly, mammals possess a variety of molecular mechanisms for activating latent TGFβ, each of which may be suitable to a limited set of circumstances (Annes et al., 2003). Thus, BMP1 activation of TGFβ may be limited to some subset of cell types, to responses to only certain stimuli, and/or to the etiology of only some pathologies involving TGFβ activity. The range of cells and situations in which BMP1 participates in TGFβ activation in vivo remains to be determined.
Finally, BMP1-like proteinases are also responsible for activating BMP2 and -4 via cleavage of Chordin (Pappano et al., 2003) and GDF8 and -11 via cleavage of prodomain sequences (Wolfman et al., 2003; Ge et al., 2005). Thus, they may serve to orchestrate signaling by these different morphogenetic TGFβ superfamily ligands and perhaps contribute to coordination between R-Smad2/3 and R-Smad1/5/8 signaling pathways, used by TGFβ/GDF8/GDF11 and BMP2/4, respectively (Massague et al., 2005). They may even contribute to antagonism between the two pathways if, as in the case of Smad4 (Candia et al., 1997), these proteinases occur in limiting amounts. Importantly, the previously characterized roles of the BMP1-like proteinases in ECM formation (Ge and Greenspan, 2006) and their newly identified roles as activators of TGFβ, make these proteinases potential targets for anti-fibrotic therapeutic interventions.
Materials and methods
Production of recombinant proteins
Human LTBP1S sequences were PCR amplified from placenta cDNA in two fragments. Primers were as follows: fragment 1 (amino acids 22–637), 5′-GATCGCTAGCACACACTGGCCGCATCAAGGT-3′ (forward) and 5′-CGTCCGGCCTCAGGCATT-3′ (reverse); and fragment 2 (amino acids 637–1396), 5′-CGAATGCCTGAGGCCGGA-3′ (forward) and 5′-GATCGCGGCCGCTAATGGTGATGGTGATGATGCTCCAGGTCACTGTCTTTCTCT-3′ (reverse). Amplicons were joined at a shared Bsu36I site and joined via an NheI site to BM40 signal peptide sequences. The insert was placed between HindIII and NotI sites of tetracycline-inducible vector pcDNA4/TO (Invitrogen). The resulting construct (pcDNA4/LTBP1S) expresses C-terminal His-tagged LTBP1S with BM40 signal peptide sequences for optimization of secretion. Human TGFβ1 sequences were PCR amplified from placenta cDNA using primers 5′-GATCGCTAGCAGACTACAAAGACGATGACGACAAGCTATCCACCTGCAAGACTATC-3′ (forward) and 5′-GATCGCGGCCGCTTAGCTGCACTTGCAGGAGCGCA-3′ (reverse). The amplicon, joined at an NheI site to BM40 signal peptide sequences, was subcloned between HindIII and NotI sites of tetracycline-inducible vector pcDNA5/TO (Invitrogen). The resulting construct (pcDNA5/TGFβ1) expresses TGFβ1 in which the native signal peptide is replaced by the BM40 signal peptide and, upon cleavage of the signal peptide, a FLAG epitope remains at the LAP N terminus.
293 T-REx cells (Invitrogen) were maintained in DME, 5 μg/ml blasticidin, and 10% FBS. 80% confluent cells were cotransfected with pcDNA4/LTBP-1S and pcDNA5/TGFβ1, using Lipofectamine (Invitrogen). After 48 h, cells were selected in the same type of medium containing 200 μg/ml Zeocin and 250 μg/ml hygromycin B. Ring-cloned colonies producing the highest tetracycline-induced levels of secreted LTBP1S and TGFβ1 were used for LLC production.
Confluent cells were washed twice with PBS and incubated for 15 min in serum-free DME at 37°C. Cells were then washed once with PBS and placed in serum-free DME/1 μg/ml tetracycline/40 μg/ml soybean trypsin inhibitor. Conditioned medium was harvested every 24 h, and protease inhibitors were added to final concentrations of 1 mM phenylmethylsulfonyl fluoride, 1 mM N-ethylmaleimide, and 1 mM p-aminobenzonic acid. Conditioned medium was centrifuged to remove debris, and LLC was purified by sequential affinity purification on Ni-NTA (QIAGEN) and anti-FLAG M2 matrix (Sigma-Aldrich).
In vitro cleavage assays
500 ng LLC was incubated for 3 h alone or with 25 or 250 ng Flag-tagged BMP1 in 20 μl 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 10 mM CaCl2 at 37°C. An additional 25 or 250 ng BMP1 was then added and the reaction continued another 3 h. Reactions were stopped with 2× SDS sample buffer/1% β-mercaptoethanol and boiling for 5 min. For MMP2 cleavage, 50 ng SLC or 150 ng LLC preincubated with/without BMP1 was incubated 16 h with/without 60 ng p-aminophenylmercuric acetate–activated proMMP2 (R&D Systems) in 20 μl 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 10 mM CaCl2 at 37°C. Reactions were stopped with 2× SDS sample buffer/1% β-mercaptoethanol and boiling for 5 min.
Amino acid sequence analysis
2 μg purified recombinant LLC were cleaved as above. Products were resolved on a 12% polyacrylamide SDS-PAGE gel, electrotransferred to Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad Laboratories), and stained with 0.025% Coomassie brilliant blue R-250. N-terminal amino acid sequencing was done by Edman degradation at the Harvard Microchemistry Facility.
MEFs
Confluent MEFs, isolated from 13.5-dpc embryos and immortalized as described previously (Steiglitz et al., 2004), were washed twice with PBS, incubated in serum-free DME for 15 min at 37°C, and incubated for 24 h in serum-free DME and 40 μg/ml soybean trypsin inhibitor. Conditioned media were harvested, and protease inhibitors were added to final concentrations of 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM N-ethylmaleimide, and 1 mM p-aminobenzonic acid for media used for Western blots. Inhibitors were not added to media for reporter gene assays. All media were centrifuged to remove debris. For inhibition profiles, 80% confluent wild-type or Bmp1/Tll1 doubly null MEFs were cultured 24 h in DME/10% FBS with/without 20 μM E-64, 20 μM pepstatin A, 100 μM AEBSF (Sigma-Aldrich), or 200 μM TAPI-2 (BIOMOL Research Laboratories, Inc.). Cells were then switched to inhibitor-containing serum-free DME, and conditioned media were harvested 24 h later. For TIMP-3 inhibition, 90% confluent wild-type MEFs were cultured for 24 h in serum-free DME with/without 100 ng/ml recombinant N-terminal inhibitor domain of TIMP-3, which has inhibitory activity comparable to that of full-length TIMP-3 (Kashiwagi et al., 2001). Plasmin extraction of cell-associated ECM was as described previously (Koli et al., 2005).
Reporter gene assay
1.6 × 104 T-MLECs, stably transfected with a luciferase gene driven by TGFβ-responsive plasminogen activator inhibitor-1 promoter sequences (Abe et al., 1994; a gift from D. Rifkin, New York University, New York, NY), were allowed to attach for 3 h in 96-well plates. MEF conditioned medium (for active TGFβ estimation) or heat-treated conditioned medium (for total TGFβ estimation) were harvested and immediately (without storage) added to the T-MLECs, which were harvested 24 h later. Coculture experiments were performed as previously described (Jenkins et al., 2006). 4 × 104 wild-type or Bmp1/Tll1 doubly null MEFs were cultured for 16 h in DME/10% FBS in 96-well plates to allow adhesion and cell spreading. 1.6 × 104 T-MLECs were then added and allowed to attach for 3 h. Cells were then switched to serum-free medium containing 0.1% BSA. After 16 h, TGFβ activity was assessed by measuring luciferase activity in coculture cell layer lysates. Luciferase activity was measured using the Luciferase assay system (Promega).
Immunohistochemistry and immunocytochemistry
13.5-dpc embryos were harvested, embedded in OCT (Tissue-Tek), frozen, and cut at −30°C into 10-μm sections. Frozen sections were thawed, fixed for 20 min at 4°C with 2% paraformaldehyde in PBS, and washed for 15 min at 4°C with PBS. Fixed cryosections were blocked for 4 h with 10% goat serum in PBS at room temperature and incubated overnight at 4°C with primary antibodies diluted in blocking solution. Rabbit polyclonal anti–phospho-Smad2/3 (Santa Cruz Biotechnology, Inc.), Ab39 anti-LTBP1 (Kanzaki et al., 1990; a gift from C.-H. Heldin, Ludwig Institute for Cancer Research, Uppsala, Sweden), and 9543 anti–fibrillin 1 (Isogai et al., 2003; a gift from L. Sakai, Shriners Hospital for Children, Portland, OR) primary antibodies were diluted 1:1,000. Sections were washed with PBS and incubated for 1 h at room temperature with Alexa Fluor 555 goat anti–rabbit secondary antibodies diluted in blocking solution. After washing with PBS, sections were mounted in Immu-Mount (Thermo Electron Corporation), and viewed with a microscope (Axiophot 2; Carl Zeiss MicroImaging, Inc.) with a Plan-NEOFLUAR 40× objective/0.75 aperture. Images were captured with a digital camera (ZVS-3C75DE; Carl Zeiss MicroImaging, Inc.) and Digital Acquire software (DEI 750; Optronics).
4.5 × 104 MEFs were cultured 2 d in Lab-Tek chambers, washed once with PBS, fixed for 10 min with methanol at –20°C, rinsed with PBS, blocked for 4 h with 3% BSA/PBS at room temperature, and incubated overnight at 4°C with primary antibodies diluted in blocking solution. Cells were washed with PBS, incubated for 1 h at room temperature with Alexa Fluor 555 goat anti–rabbit secondary antibody diluted in blocking solution, washed with PBS, and mounted in Immu-Mount. Images were deconvoluted using AutoDeblur and AutoVisualize version 9.3 (AutoQuant Imaging, Inc.), and contrast was adjusted using Photoshop version 7.0 (Adobe) with the same parameters.
RNAi
5 × 105 wild-type MEFs/well on a 6-well plate were transfected with 250 pmol Stealth RNAi duplexes for MMP2 (sense, 5′-UAUUCCCGACCGUUGAACAGGAAGG-3′), MMP9 (sense, 5′-UAUACAGCGGGUACAUGAGCGCUUC-3′), MMP14 (sense, 5′-AAACUUAUCCGGAACACCACAGCGA-3′), or Stealth medium GC RNAi negative control (Invitrogen), using Lipofectamine. After 6 h, cells were placed in DME/10% FBS, and 18 h later, they were changed into serum-free medium. Conditioned media were harvested after 24 h, as above. Cell layers were washed twice with ice-cold PBS and scraped into hot SDS sample buffer for Western blotting. For RT-PCR, RNA was isolated with TRIzol (Invitrogen), and cDNA was synthesized using 1 μg RNA, random primers, and Super-Script II reverse transcriptase (Invitrogen). PCR was performed at 95°C/3 min, followed by 25, 30, or 35 cycles of 95°C/1 min, 60°C/1 min, and 72°C/1 min, and final extension at 72°C/10 min. Primers were as follows: MMP2, 5′-ACCCATTTGATGGCAAGGAT-3′ (forward) and 5′-TTGTTGCCCAGGAAAGTGAA-3′ (reverse); MMP9, 5′-GGAGAAGGCAAACCCTGTGT-3′ (forward) and 5′-AGGCTGTACCCTTGGTCTGG-3′ (reverse); MMP14, 5′-TCCTGGCTCATGCCTACTTC-3′ (forward) and 5′-GGTGTCAAAGTTCCCGTCAC-3′ (reverse); CYR61, 5′-TCACCCTTCTCCACTTGACC-3′ (forward) and 5′-AGGGTCTGCCTTCTGACTGA-3′ (reverse); GAPDH, 5′-TGGCCAAGGTCATCCATGAC-3′ (forward) and 5′-ATGTAGGCCATGAGGTCCAC -3′ (reverse).
Online supplemental material
Fig. S1 presents a control showing that LTBP1 cleavage occurs only with a form of BMP1 that is catalytically active and demonstrates that the LTBP1 N-terminal site is somewhat more efficiently cleaved than the C-terminal site. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200606058/DC1.
Supplementary Material
Acknowledgments
The authors are grateful for the generous gifts of anti-LTBP1 and anti-fibrillin antibodies and of T-MLECs from Carl-Henrik Heldin, Lynn Sakai, and Dan Rifkin, respectively.
This work was supported by National Institutes of Health grant R01 GM71679 (to D.S. Greenspan).
The authors have no financial conflicts of interest in this work.
Abbreviations used in this paper: AEBSF, 4-(2-aminoethyl)-benzenesulfonyl fluoride; BMP, bone morphogenetic protein; dpc, days post conception; GDF, growth and differentiation factor; LAP, latency-associated peptide; LLC, large latent complex; LTBP, latent TGFβ-binding protein; MEF, mouse embryo fibroblast; MMP, matrix metalloproteinase; SLC, small latent complex; TAPI-2, TNF-α processing inhibitor 2; TIMP, tissue inhibitor of metalloproteinase; T-MLEC, transfected mink lung epithelial cell.
References
- Abe, M., J.G. Harpel, C.N. Metz, I. Nunes, D.J. Loskutoff, and D.B. Rifkin. 1994. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 216:276–284. [DOI] [PubMed] [Google Scholar]
- Annes, J.P., J.S. Munger, and D.B. Rifkin. 2003. Making sense of latent TGF β activation. J. Cell Sci. 116:217–224. [DOI] [PubMed] [Google Scholar]
- Bertolino, P., M. Deckers, F. Lebrin, and P. ten Dijke. 2005. Transforming growth factor-β signal transduction in angiogenesis and vascular disorders. Chest. 128:585S–590S. [DOI] [PubMed] [Google Scholar]
- Blader, P., S. Rastegar, N. Fischer, and U. Strahle. 1997. Cleavage of the BMP-4 antagonist chordin by zebrafish tolloid. Science. 278:1937–1940. [DOI] [PubMed] [Google Scholar]
- Border, W.A., and E. Ruoslahti. 1992. Transforming growth factor-β in disease: the dark side of tissue repair. J. Clin. Invest. 90:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein, P., A. Agah, and T.R. Kyriakides. 2004. The role of thrombospondins 1 and 2 in the regulation of cell-matrix interactions, collagen fibril formation, and the response to injury. Int. J. Biochem. Cell Biol. 36:1115–1125. [DOI] [PubMed] [Google Scholar]
- Brown, P.D., A.T. Levy, I.M. Margulies, L.A. Liotta, and W.G. Stetler-Stevenson. 1990. Independent expression and cellular processing of Mr 72,000 type IV collagenase and interstitial collagenase in human tumorigenic cell lines. Cancer Res. 50:6184–6191. [PubMed] [Google Scholar]
- Brunner, A., J. Chinn, M. Neubauer, and A.F. Purchio. 1991. Identification of a gene family regulated by transforming growth factor-β. DNA Cell Biol. 10:293–300. [DOI] [PubMed] [Google Scholar]
- Candia, A.F., T. Watabe, S.H. Hawley, D. Onichtchouk, Y. Zhang, R. Derynck, C. Niehrs, and K.W. Cho. 1997. Cellular interpretation of multiple TGF-β signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 124:4467–4480. [DOI] [PubMed] [Google Scholar]
- Crawford, S.E., V. Stellmach, J.E. Murphy-Ullrich, S.M. Ribeiro, J. Lawler, R.O. Hynes, G.P. Boivin, and N. Bouck. 1998. Thrombospondin-1 is a major activator of TGF-β 1 in vivo. Cell. 93:1159–1170. [DOI] [PubMed] [Google Scholar]
- D'Angelo, M., P.C. Billings, M. Pacifici, P.S. Leboy, and T. Kirsch. 2001. Authentic matrix vesicles contain active metalloproteases (MMP): a role for matrix vesicle-associated MMP-13 in activation of transforming growth factor-β. J. Biol. Chem. 276:11347–11353. [DOI] [PubMed] [Google Scholar]
- Dallas, S.L., D.R. Keene, S.P. Bruder, J. Saharinen, L.Y. Sakai, G.R. Mundy, and L.F. Bonewald. 2000. Role of the latent transforming growth factor β binding protein 1 in fibrillin-containing microfibrils in bone cells in vitro and in vivo. J. Bone Miner. Res. 15:68–81. [DOI] [PubMed] [Google Scholar]
- Derynck, R., R.J. Akhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29:117–129. [DOI] [PubMed] [Google Scholar]
- Ducy, P., and G. Karsenty. 2000. The family of bone morphogenetic proteins. Kidney Int. 57:2207–2214. [DOI] [PubMed] [Google Scholar]
- Flaumenhaft, R., M. Abe, Y. Sato, K. Miyazono, J. Harpel, C.H. Heldin, and D.B. Rifkin. 1993. Role of the latent TGF-β binding protein in the activation of latent TGF-β by co-cultures of endothelial and smooth muscle cells. J. Cell Biol. 120:995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, G., and D.S. Greenspan. 2006. Developmental roles of the BMP1/TLD metalloproteinases. Birth Defects Res. C Embryo Today. 78:47–68. [DOI] [PubMed] [Google Scholar]
- Ge, G., D.R. Hopkins, W.B. Ho, and D.S. Greenspan. 2005. GDF11 forms a bone morphogenetic protein 1-activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol. Cell. Biol. 25:5846–5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualandris, A., J.P. Annes, M. Arese, I. Noguera, V. Jurukovski, and D.B. Rifkin. 2000. The latent transforming growth factor-β-binding protein-1 promotes in vitro differentiation of embryonic stem cells into endothelium. Mol. Biol. Cell. 11:4295–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai, Z., R.N. Ono, S. Ushiro, D.R. Keene, Y. Chen, R. Mazzieri, N.L. Charbonneau, D.P. Reinhardt, D.B. Rifkin, and L.Y. Sakai. 2003. Latent transforming growth factor β-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J. Biol. Chem. 278:2750–2757. [DOI] [PubMed] [Google Scholar]
- Jenkins, R.G., X. Su, G. Su, C.J. Scotton, E. Camerer, G.J. Laurent, G.E. Davis, R.C. Chambers, M.A. Matthay, and D. Sheppard. 2006. Ligation of protease-activated receptor 1 enhances α(v) β 6 integrin-dependent TGF-β activation and promotes acute lung injury. J. Clin. Invest. 116:1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki, T., A. Olofsson, A. Moren, C. Wernstedt, U. Hellman, K. Miyazono, L. Claesson-Welsh, and C.H. Heldin. 1990. TGF-β 1 binding protein: a component of the large latent complex of TGF-β 1 with multiple repeat sequences. Cell. 61:1051–1061. [DOI] [PubMed] [Google Scholar]
- Kashiwagi, M., M. Tortorella, H. Nagase, and K. Brew. 2001. TIMP-3 is a potent inhibitor of aggrecanase 1 (ADAM-TS4) and aggrecanase 2 (ADAM-TS5). J. Biol. Chem. 276:12501–12504. [DOI] [PubMed] [Google Scholar]
- Kessler, E., K. Takahara, L. Biniaminov, M. Brusel, and D.S. Greenspan. 1996. Bone morphogenetic protein-1: the type I procollagen C-proteinase. Science. 271:360–362. [DOI] [PubMed] [Google Scholar]
- Koli, K., M. Hyytiainen, M.J. Ryynanen, and J. Keski-Oja. 2005. Sequential deposition of latent TGF-β binding proteins (LTBPs) during formation of the extracellular matrix in human lung fibroblasts. Exp. Cell Res. 310:370–382. [DOI] [PubMed] [Google Scholar]
- Koski, C., J. Saharinen, and J. Keski-Oja. 1999. Independent promoters regulate the expression of two amino terminally distinct forms of latent transforming growth factor-β binding protein-1 (LTBP-1) in a cell type-specific manner. J. Biol. Chem. 274:32619–32630. [DOI] [PubMed] [Google Scholar]
- Lee, S., D.E. Solow-Cordero, E. Kessler, K. Takahara, and D.S. Greenspan. 1997. Transforming growth factor-β regulation of bone morphogenetic protein-1/procollagen C-proteinase and related proteins in fibrogenic cells and keratinocytes. J. Biol. Chem. 272:19059–19066. [DOI] [PubMed] [Google Scholar]
- Letterio, J.J., and A.B. Roberts. 1998. Regulation of immune responses by TGF-β. Annu. Rev. Immunol. 16:137–161. [DOI] [PubMed] [Google Scholar]
- Lyons, R.M., J. Keski-Oja, and H.L. Moses. 1988. Proteolytic activation of latent transforming growth factor-β from fibroblast-conditioned medium. J. Cell Biol. 106:1659–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, S., D.D. Dean, I. Gay, Z. Schwartz, and B.D. Boyan. 2001. Activation of latent transforming growth factor β1 by stromelysin 1 in extracts of growth plate chondrocyte-derived matrix vesicles. J. Bone Miner. Res. 16:1281–1290. [DOI] [PubMed] [Google Scholar]
- Marti, H.P., L. Lee, M. Kashgarian, and D.H. Lovett. 1994. Transforming growth factor-β 1 stimulates glomerular mesangial cell synthesis of the 72-kd type IV collagenase. Am. J. Pathol. 144:82–94. [PMC free article] [PubMed] [Google Scholar]
- Massague, J. 1990. The transforming growth factor-β family. Annu. Rev. Cell Biol. 6:597–641. [DOI] [PubMed] [Google Scholar]
- Massague, J., J. Seoane, and D. Wotton. 2005. Smad transcription factors. Genes Dev. 19:2783–2810. [DOI] [PubMed] [Google Scholar]
- Mazzieri, R., V. Jurukovski, H. Obata, J. Sung, A. Platt, E. Annes, N. Karaman-Jurukovska, P.E. Gleizes, and D.B. Rifkin. 2005. Expression of truncated latent TGF-β-binding protein modulates TGF-β signaling. J. Cell Sci. 118:2177–2187. [DOI] [PubMed] [Google Scholar]
- McPherron, A.C., A.M. Lawler, and S.J. Lee. 1997. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature. 387:83–90. [DOI] [PubMed] [Google Scholar]
- Mu, D., S. Cambier, L. Fjellbirkeland, J.L. Baron, J.S. Munger, H. Kawakatsu, D. Sheppard, V.C. Broaddus, and S.L. Nishimura. 2002. The integrin αvβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-β1. J. Cell Biol. 157:493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger, J.S., X. Huang, H. Kawakatsu, M.J. Griffiths, S.L. Dalton, J. Wu, J.F. Pittet, N. Kaminski, C. Garat, M.A. Matthay, et al. 1999. The integrin αv β6 binds and activates latent TGF β1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 96:319–328. [DOI] [PubMed] [Google Scholar]
- Nakajima, Y., K. Miyazono, M. Kato, M. Takase, T. Yamagishi, and H. Nakamura. 1997. Extracellular fibrillar structure of latent TGF β binding protein-1: role in TGF β-dependent endothelial-mesenchymal transformation during endocardial cushion tissue formation in mouse embryonic heart. J. Cell Biol. 136:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neptune, E.R., P.A. Frischmeyer, D.E. Arking, L. Myers, T.E. Bunton, B. Gayraud, F. Ramirez, L.Y. Sakai, and H.C. Dietz. 2003. Dysregulation of TGF-β activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 33:407–411. [DOI] [PubMed] [Google Scholar]
- Nunes, I., P.E. Gleizes, C.N. Metz, and D.B. Rifkin. 1997. Latent transforming growth factor-β binding protein domains involved in activation and transglutaminase-dependent cross-linking of latent transforming growth factor-β. J. Cell Biol. 136:1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall, C.M., J.L. Wrana, and J. Sodek. 1991. Transcriptional and post-transcriptional regulation of 72-kDa gelatinase/type IV collagenase by transforming growth factor-β 1 in human fibroblasts. Comparisons with collagenase and tissue inhibitor of matrix metalloproteinase gene expression. J. Biol. Chem. 266:14064–14071. [PubMed] [Google Scholar]
- Pappano, W.N., B.M. Steiglitz, I.C. Scott, D.R. Keene, and D.S. Greenspan. 2003. Use of Bmp1/Tll1 doubly homozygous null mice and proteomics to identify and validate in vivo substrates of bone morphogenetic protein 1/tolloid-like metalloproteinases. Mol. Cell. Biol. 23:4428–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo, S., E. Agius, B. Lu, S. Goodman, L. Dale, and E.M. De Robertis. 1997. Cleavage of Chordin by Xolloid metalloprotease suggests a role for proteolytic processing in the regulation of Spemann organizer activity. Cell. 91:407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, T., Y. Ito, P. Bringas Jr., S. Chou, M.M. Urata, H. Slavkin, and Y. Chai. 2006. TGFβ-mediated FGF signaling is crucial for regulating cranial neural crest cell proliferation during frontal bone development. Development. 133:371–381. [DOI] [PubMed] [Google Scholar]
- Sato, Y., and D.B. Rifkin. 1989. Inhibition of endothelial cell movement by pericytes and smooth muscle cells: activation of a latent transforming growth factor-β1-like molecule by plasmin during co-culture. J. Cell Biol. 109:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal, I., and T.C. Thompson. 1999. Novel regulation of type IV collagenase (matrix metalloproteinase-9 and -2) activities by transforming growth factor-β1 in human prostate cancer cell lines. Mol. Biol. Cell. 10:407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Croix, B., C. Rago, V. Velculescu, G. Traverso, K.E. Romans, E. Montgomery, A. Lal, G.J. Riggins, C. Lengauer, B. Vogelstein, and K.W. Kinzler. 2000. Genes expressed in human tumor endothelium. Science. 289:1197–1202. [DOI] [PubMed] [Google Scholar]
- Steiglitz, B.M., M. Ayala, K. Narayanan, A. George, and D.S. Greenspan. 2004. Bone morphogenetic protein-1/Tolloid-like proteinases process dentin matrix protein-1. J. Biol. Chem. 279:980–986. [DOI] [PubMed] [Google Scholar]
- Sternlicht, M.D., and Z. Werb. 2001. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 17:463–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N., P.A. Labosky, Y. Furuta, L. Hargett, R. Dunn, A.B. Fogo, K. Takahara, D.M. Peters, D.S. Greenspan, and B.L. Hogan. 1996. Failure of ventral body wall closure in mouse embryos lacking a procollagen C-proteinase encoded by Bmp1, a mammalian gene related to Drosophila tolloid. Development. 122:3587–3595. [DOI] [PubMed] [Google Scholar]
- Taipale, J., K. Miyazono, C.H. Heldin, and J. Keski-Oja. 1994. Latent transforming growth factor-β1 associates to fibroblast extracellular matrix via latent TGF-β binding protein. J. Cell Biol. 124:171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visse, R., and H. Nagase. 2003. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ. Res. 92:827–839. [DOI] [PubMed] [Google Scholar]
- Wang, W.M., S. Lee, B.M. Steiglitz, I.C. Scott, C.C. Lebares, M.L. Allen, M.C. Brenner, K. Takahara, and D.S. Greenspan. 2003. Transforming growth factor-β induces secretion of activated ADAMTS-2. A procollagen III N-proteinase. J. Biol. Chem. 278:19549–19557. [DOI] [PubMed] [Google Scholar]
- Wang, W.M., G. Ge, N.H. Lim, H. Nagase, and D.S. Greenspan. 2006. TIMP-3 inhibits the procollagen N-proteinase ADAMTS-2. Biochem. J. In press. [DOI] [PMC free article] [PubMed]
- Wolfman, N.M., A.C. McPherron, W.N. Pappano, M.V. Davies, K. Song, K.N. Tomkinson, J.F. Wright, L. Zhao, S.M. Sebald, D.S. Greenspan, and S.J. Lee. 2003. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc. Natl. Acad. Sci. USA. 100:15842–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H.H., S. Ivkovic, R.C. Murray, S. Jaramillo, K.M. Lyons, J.E. Johnson, and A.L. Calof. 2003. Autoregulation of neurogenesis by GDF11. Neuron. 37:197–207. [DOI] [PubMed] [Google Scholar]
- Yu, Q., and I. Stamenkovic. 2000. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev. 14:163–176. [PMC free article] [PubMed] [Google Scholar]
- Zacchigna, L., C. Vecchione, A. Notte, M. Cordenonsi, S. Dupont, S. Maretto, G. Cifelli, A. Ferrari, A. Maffei, C. Fabbro, et al. 2006. Emilin1 links TGF-β maturation to blood pressure homeostasis. Cell. 124:929–942. [DOI] [PubMed] [Google Scholar]
- Zhang, F., S. Endo, L.J. Cleary, A. Eskin, and J.H. Byrne. 1997. Role of transforming growth factor-β in long-term synaptic facilitation in Aplysia. Science. 275:1318–1320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.