Abstract
Focal adhesion kinase family interacting protein of 200 kD (FIP200) has been shown to regulate diverse cellular functions such as cell size, proliferation, and migration in vitro. However, the function of FIP200 in vivo has not been investigated. We show that targeted deletion of FIP200 in the mouse led to embryonic death at mid/late gestation associated with heart failure and liver degeneration. We found that FIP200 knockout (KO) embryos show reduced S6 kinase activation and cell size as a result of increased tuberous sclerosis complex function. Furthermore, FIP200 KO embryos exhibited significant apoptosis in heart and liver. Consistent with this, FIP200 KO mouse embryo fibroblasts and liver cells showed increased apoptosis and reduced c-Jun N-terminal kinase phosphorylation in response to tumor necrosis factor (TNF) α stimulation, which might be mediated by FIP200 interaction with apoptosis signal–regulating kinase 1 (ASK1) and TNF receptor–associated factor 2 (TRAF2), regulation of TRAF2–ASK1 interaction, and ASK1 phosphorylation. Together, our results reveal that FIP200 functions as a regulatory node to couple two important signaling pathways to regulate cell growth and survival during mouse embryogenesis.
Introduction
FAK-family interacting protein of 200 kD (FIP200; also known as RB1CC1, or RB1-inducible coiled-coil 1) encodes a 200-kD protein (1591 aa) characterized by a large coiled-coil region (CC; residues 860–1391) containing a leucine zipper motif (residues 1371–1391; Ueda et al., 2000; Chano et al., 2002b). FIP200 was originally identified by us as a Pyk2 interacting protein through a yeast two-hybrid screen (Ueda et al., 2000). It was shown to interact with Pyk2 kinase domain and inhibit its kinase activity in in vitro kinase assays (Ueda et al., 2000). FIP200 was subsequently also shown to interact with FAK and function to inhibit FAK activity and autophosphorylation as well as FAK-promoted cellular functions, including cell spreading, cell migration, and cell cycle progression (Abbi et al., 2002). Shortly after these studies, FIP200 was also independently identified by Chano et al. (2002a) as a potential regulator of the RB1 gene.
Recent studies suggest that the tuberous sclerosis complex (TSC)–mammalian target of rapamycin (mTOR) signaling network plays an essential role in the regulation of cell growth (Kwiatkowski, 2003; Fingar and Blenis, 2004; Hay and Sonenberg, 2004). TSC1 and -2 are both tumor suppressor genes responsible for tuberous sclerosis, which is characterized by the formation of hamartomas in a wide range of tissues. TSC1 and -2 can form a physical and functional complex in vivo (Kwiatkowski, 2003) and function as potent negative regulators of cell growth mainly by their inhibition of mTOR and its targets ribosomal S6 kinase (S6K) and eukaryotic initiation factor 4E binding protein 1 (4EBP1), which play essential roles in the regulation of protein synthesis and cell size. Recent studies suggested that TSC2 functions as the GTPase-activating protein of the small G protein Rheb, an upstream activator of mTOR, and that the TSC1–TSC2 complex antagonizes the mTOR signaling pathway via stimulation of GTP hydrolysis of Rheb (Manning and Cantley, 2003; Inoki et al., 2005). Interestingly, we have recently found a potentially novel function for FIP200 in the regulation of cell growth through its interaction with TSC1 and inhibition of TSC1–TSC2 complex function (Gan et al., 2005).
During embryonic development, cell survival/death is tightly regulated by both intrinsic and extrinsic factors. The intrinsic death pathway is activated by the release of cytochrome c from mitochondria in response to various stress and developmental death cues, whereas the extrinsic death pathway is mainly activated by the binding of death receptors of the TNF receptor (TNFR) superfamily to their ligands. One of the ligands of death receptors is TNFα. The binding of TNFα to its receptor TNFR1 triggers several intracellular events that regulate both cell survival and cell death. TNFα-induced cell death is mainly mediated by the activation of caspase-8, whereas cell survival effect of TNFα is mainly mediated by the NF-κB pathway (Chen and Goeddel, 2002; Ghosh and Karin, 2002). TNFα stimulation can also activate JNK through TNFR1–TNFR-associated factor 2 (TRAF2)–apoptosis signal–regulating kinase 1 (ASK1)–MAPK kinase (MKK) 4/7–JNK signaling cascade (Nishitoh et al., 1998; Davis, 2000). However, the exact role of JNK in TNFα-stimulated cell death signaling is complicated, as JNK has been found to play both antiapoptotic and proapoptotic roles in TNFα signaling in different cellular contexts. A recent study showed that JNK1 and -2 double-knockout (KO) mouse embryo fibroblasts (MEFs) exhibited increased TNFα-stimulated apoptosis, suggesting, at least in MEFs, that JNK could mediate a survival response in TNFα signaling (Lamb et al., 2003). Mice KO studies highlight the important role of TNFα signaling in the regulation of cell survival/death during embryonic development. Deletion of some of the genes involved in TNFα signaling, such as Rel A (a subunit of NF-κB), IκB kinase β, and IκB kinase γ, leads to mid/late gestational lethality associated with increased apoptosis in liver, indicating the role of TNFα signaling in the regulation of cell survival and death in the liver development during embryogenesis (Beg et al., 1995; Li et al., 1999; Rudolph et al., 2000).
FIP200 is widely expressed in various human tissues (Bamba et al., 2004) and is an evolutionarily conserved protein present in human, mouse, rat, Xenopus laevis, Drosophila melanogaster, and Caenorhabditis elegans. The high degree of conservation during evolution suggests that FIP200 plays important functions in vivo. Despite these recent studies suggesting a role of FIP200 in the regulation of a variety of cellular functions in vitro, however, the function of FIP200 in vivo remains totally unknown. In the present study, we generated FIP200 KO mouse to study the physiological role of FIP200 in vivo. The analyses of FIP200 KO embryos and isolated cells reveal the important function of FIP200 in cell size control and cell survival during embryogenesis and highlight a previously unappreciated role of FIP200 in TNFα-JNK signaling.
Results
Ablation of FIP200 in mouse leads to embryonic lethality at mid/late gestation
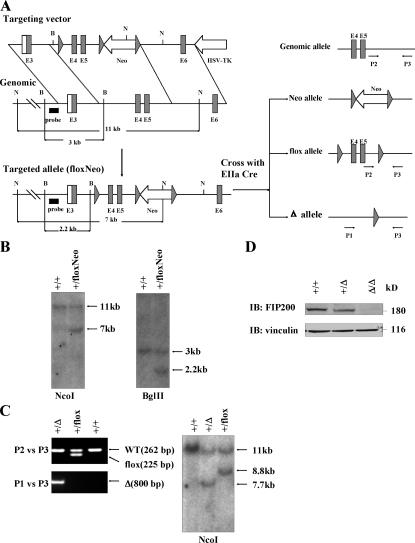
To study the potential function of FIP200 in vivo, we generated a mouse strain that contains three loxP sequences that flank the exons 4 and 5 of mouse FIP200 gene and a neo cassette (FIP200 floxNeo/+) via a standard homologous recombination technique (Fig. 1, A and B). The FIP200 floxNeo/+ mice were crossed with EIIa-Cre mice (Lakso et al., 1996), which led to the generation of three types of offspring: flox allele with neo cassette deleted (FIP200 flox/+), neo allele with exons 4 and 5 deleted (FIP200 Neo/+), and total deletion allele with both exons 4 and 5 and neo cassette deleted (FIP200 Δ/+; Fig. 1 A). Cre-mediated deletion of exons 4 and 5 leads to a frame-shift mutation because of direct splicing from exon 3 (containing ATG codon) to exon 6, producing a small truncated and nonfunctional peptide. The FIP200 Δ/+ and FIP200 flox/+ mice were identified by PCR analysis of tail DNA (Fig. 1 C, left), and the PCR results were confirmed by Southern blotting (Fig. 1 C, right).
Figure 1.
Generation of FIP200 KO mice. (A) Schematic representation of the FIP200 targeting vector, mouse FIP200 genomic structure, and the loxP modified FIP200 loci. Large solid triangles represent loxP sites. The relevant restriction sites (B, BglII; N, NcoI), position of probe (thick line) for Southern blotting, and primers (P1, P2, and P3) for PCR genotyping are indicated. Crosses of the mice with targeted allele with EIIa Cre mice result in three possible outcomes at the FIP200 loci (Neo, flox, and Δ allele). (B) Southern blotting analysis of the DNA extracted from ES cells after digestion with corresponding restriction enzymes as indicated. (C) Genomic DNA was extracted from mouse tail and analyzed by PCR (left) and Southern blotting (right), as indicated, to distinguish different types of alleles resulting from Cre-mediated recombination. (D) Western blotting analysis of protein extracts from whole embryos with various genotypes by anti-FIP200 or anti-vinculin, as indicated.
FIP200 Δ/+ mice are normal and fertile and were intercrossed to generate FIP200 KO mice. Genotyping of >300 weaning-age pups identified no homozygous mice, whereas a Mendelian ratio of nearly 1:2 was found for wild-type (WT) and heterozygous pups, suggesting that deletion of the FIP200 gene results in embryonic lethality. Mating between heterozygous FIP200 flox/+ mice yielded homozygous floxed FIP200 mice (FIP200flox/flox) at the expected Mendelian ratio, and FIP200flox/flox mice were viable and fertile and did not show any gross or histological abnormalities (unpublished data), further confirming that the embryonic lethality for FIP200 Δ/Δ mice is caused by FIP200 gene ablation.
Extensive timed matings were then performed to determine when FIP200 KO embryos die and to characterize phenotypic defects in the KO embryos. Embryos were analyzed from embryonic day (E) 9.5 to birth (Table I). The normal Mendelian ratio of 1:2:1 of WT, heterozygous, and homozygous embryos was observed until E13.5. Approximately 25 and 60% of homozygous embryos were found dead at E14.5 and E15.5, respectively, and the total number of homozygous embryos (including both alive and dead embryos) at each day were very close to the number of WT embryos. No live FIP200 KO embryos were identified at E16.5 and thereafter. The small size and autolysis of the dead KO embryos recovered after E16.5 are compatible with embryonic lethality between E13.5 and E16.5. Analysis of extracts from whole embryos verified the absence of FIP200 in homozygous KO embryos and reduced expression of FIP200 in the heterozygotes (Fig. 1 D). Together, these results show that homozygous deletion of FIP200 leads to embryonic lethality at mid/late gestation.
Table I.
Genotypes of progeny from FIP200+/Δ heterozygous intercrosses
| Embryonic day | +/+ | +/Δ | Δ/Δ (alive) | Δ/Δ (dead) | Total | Litter no. |
|---|---|---|---|---|---|---|
| E9.5 | 6 | 11 | 8 | 0 | 25 | 3 |
| E10.5 | 8 | 18 | 9 | 0 | 35 | 4 |
| E11.5 | 7 | 17 | 7 | 0 | 31 | 4 |
| E12.5 | 7 | 18 | 8 | 0 | 33 | 4 |
| E13.5 | 15 | 29 | 16 | 0 | 60 | 8 |
| E14.5 | 44 | 86 | 31 | 10 | 171 | 22 |
| E15.5 | 22 | 32 | 9 | 12 | 79 | 10 |
| E16.5 | 8 | 13 | 0 | 5 | 26 | 3 |
| E17.5 | 7 | 9 | 0 | 5 | 21 | 3 |
| E18.5 | 4 | 7 | 0 | 4 | 15 | 2 |
| Postnatal | 101 | 205 | 0 | 0 | 306 | 51 |
Defective heart and liver development in FIP200 KO embryos
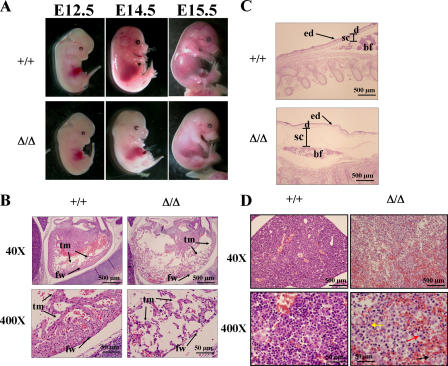
Gross examination of the embryos showed normal morphology for FIP200 KO embryos until E12.5 (Fig. 2 A). At E13.5, ∼20% of the FIP200 KO embryos were pale, and at E14.5 and E15.5 the majority of them were pale compared with WT littermates. Histological studies of the FIP200 KO embryos showed severe cardiac abnormalities characterized by ventricular dilation, sparsely cellular myocardium (Fig. 2 B), and generalized edema (Fig. 2 C) in a majority of the FIP200 KO embryos analyzed at E14.5 and E15.5. The heart ventricular wall in KO embryos has lost the normal trabecular and external compact myocytes, and the wall was significantly thinner and contained fewer cells when compared with the WT littermates (Fig. 2 B). In the most severely affected mutants, the myocardium appears to be composed almost entirely of the trabecular layer with no appreciable compact layer underneath the epicardium. In a majority of the FIP200 KO embryos analyzed at E14.5 and E15.5, we also observed liver lesions characterized by loosely arranged hepatocytes mixed with numerous red blood cells (Fig. 2 D). Hepatocytes were separated from each other with disrupted architecture because of dissecting hemorrhage in between the hepatic cords (Fig. 2 D). Histological examination of FIP200 KO embryos at E14.5 and E15.5 showed apparently normal morphogenesis of the other major organs as well as extraembryonal tissues, including the placenta (unpublished data). Together, these results suggest that the significant defects in the formation and development of the myocardium and liver are the most likely cause of the embryonic lethality observed in FIP200 KO embryos.
Figure 2.
Defective heart and liver development in FIP200 KO embryos. (A) Gross examination of whole mount FIP200 WT (+/+) and KO (Δ/Δ) embryos at E12.5, E14.5, and E15.5. Note the paleness of FIP200 KO embryos compared with WT littermates at E14.5 and E15.5. (B) Histological sections from the heart of FIP200 WT (+/+) and KO (Δ/Δ) embryos at E14.5. The heart in the KO embryo shows marked left ventricular dilation and a sparsely cellular thin wall that is composed of wisps of thin trabecular myocardium and is devoid of the compact subepicardial myocardium. fw, left ventricular free wall; tm, trabecular myocardium. (C) Histological sections of skin from the dorsum of FIP200 WT (+/+) and KO (Δ/Δ) embryos at E14.5. The marked expansion of the subcutis, the mild expansion of the dermis, and the thin, undulating epidermis are indicative of acute edema. ed, epidermis; d, dermis; sc, subcutis; bf, brown fat. (D) Histological sections of liver FIP200 WT (+/+) and KO (Δ/Δ) embryos at E14.5. Liver from KO embryo showed disrupted architecture with loss of hepatocytes and multifocal variable sized hemorrhages. Also note that many hepatocytes have condensed nuclei (karyopyknosis; red arrow) or fragmented nuclei (karyorrhexis; yellow arrow) or lost their nuclei (karyolysis; black arrow) in the KO embryo (bottom right).
FIP200 deletion leads to increased TSC function and corresponding reduction of cell size in vivo
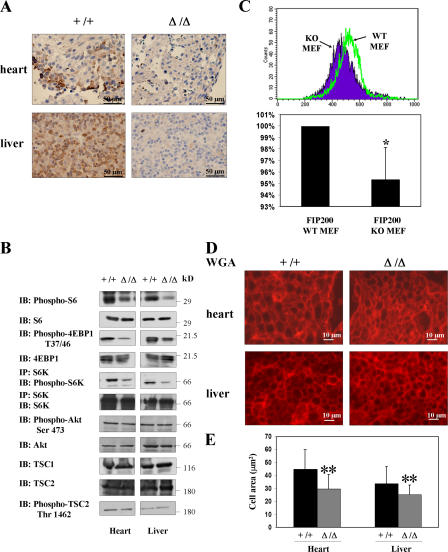
Our previous studies showed FIP200 interaction with TSC1 and inhibition of TSC function in cell size control in vitro (Gan et al., 2005). Interestingly, TSC1 KO embryos also exhibited defects in both heart and liver, with thickened ventricular walls (Kobayashi et al., 2001; Kwiatkowski et al., 2002). Therefore, deletion of a putative inhibitor of TSC, FIP200, could lead to the thin ventricular walls in the FIP200 KO embryos. To test this possibility and investigate the cellular and molecular mechanisms of FIP200 function in embryogenesis, we examined the effect of FIP200 deletion on TSC functions and cell size regulation in vivo. Embryo sections were analyzed for the activity of S6K, a major target of the TSC–mTOR signaling pathway, by immunohistochemical staining with antibodies against phosphorylated S6. We found that FIP200 KO embryos showed generally weaker staining for phospho-S6 compared with WT littermate embryos (unpublished data). In particular, significantly decreased staining for phospho-S6 was found in heart and liver (Fig. 3 A). Western blotting analysis of protein extracts from heart and liver confirmed reduced phosphorylation of S6, S6K, and 4EBP1 in FIP200 KO embryos compared with the WT littermates (Fig. 3 B). Furthermore, we did not observe any change in the expression levels of TSC1 or -2 or the phosphorylation of Akt or TSC2 in the heart and liver from FIP200 KO embryos compared with that from the WT embryos (Fig. 3 B).
Figure 3.
Reduced S6K activity and cell size in FIP200 KO embryos. (A) Immunohistochemical staining of heart and liver sections from FIP200 WT (+/+) and KO (Δ/Δ) embryos at E12.5 with Ser240/244 phospho-S6 antibody. Note that phospho-S6 staining signal is reduced in these organs in the FIP200 KO embryos. (B) Heart and liver protein extracts prepared from FIP200 WT (+/+) and KO (Δ/Δ) embryos were analyzed by Western blotting using various antibodies, as indicated. Alternatively, they were immunoprecipitated by anti-S6K and analyzed by Western blotting using antibody against Thr389 phospho-S6K and S6K. (C, top) Histograms of mean FSC-H comparing primary FIP200 WT and KO MEFs. (bottom) Relative mean FSC-H + SEM for three independent experiments (results are normalized to FIP200 WT MEFs). *, P < 0.05. (D and E) Heart and liver sections from FIP200 WT (+/+) and KO (Δ/Δ) embryos at E12.5 were stained with WGA-TRITC. (D) Representative fields for each section. (E) Mean + SEM of calculated area of cross sections of cells from three independent experiments in multiple fields. **, P < 0.001.
Our previous study showed that FIP200 KO MEFs exhibited decreased S6K activation (Gan et al., 2005). Consistent with this, we also found decreased cell size in FIP200 KO MEFs compared with WT MEFs (Fig. 3 C). We then examined the cell size in FIP200 KO embryos by staining with wheat germ agglutinin (WGA)–tetramethyl rhodamine isothiocyanate (TRITC), which outlines the cell membranes in the tissue sections (Bueno et al., 2000). Fig. 3 D shows a decreased cell size in both heart and liver in FIP200 KO embryos compared with WT littermates. Quantitative analysis of multiple samples suggested an ∼33 and 25% reduction of mean cell size in heart and liver, respectively (Fig. 3 E). The decreased S6 phosphorylation and cell size were found in FIP200 KO embryos at E14.5 as well as E12.5 and E13.5 when there was no apparent histological defect in FIP200 KO embryos. Therefore, the decreased S6 phosphorylation and cell size were unlikely to be an indirect consequence of other defects (such as apoptosis; see the following paragraph) in FIP200 KO embryos. Together, these results suggested that, consistent with our previous findings in vitro (Gan et al., 2005), FIP200 functions as an inhibitor for TSC, and deletion of FIP200 could lead to increased TSC activity and corresponding decreases in S6K activation and cell size in the embryos.
Role of FIP200 in the regulation of apoptosis and cell survival in embryogenesis
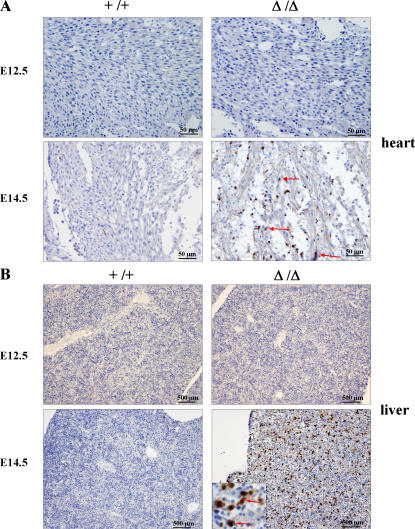
Although the decreased cell size could contribute to the thin ventricular wall defect in the FIP200 KO embryos, we also observed a reduced number of cells in the mutant embryo heart that could not be explained by reduction in cell size. This could be caused by either a decreased cell proliferation or an increased apoptosis or both in the FIP200 KO embryos. Analysis of heart and liver sections from FIP200 KO embryos at E13.5–15.5 by immunohistochemical staining of proliferating cell nuclear antigen showed no apparent difference from WT littermate samples (unpublished data). In contrast, significantly increased apoptosis was found in the mutant heart as measured by immunohistochemical staining for cleaved caspase-3 at E14.5, whereas no significant signals were detected at E12.5 or the WT littermates at E12.5 or E14.5 (Fig. 4 A).
Figure 4.
Increased apoptosis in FIP200 KO embryos. Immunohistochemical staining of heart (A) and liver (B) sections from FIP200 WT (+/+) and KO (Δ/Δ) embryos at E12.5 or E14.5 with antibody against cleaved caspase-3. There is no immunostaining observed in heart or liver sections from either the KO or WT embryo section at E12.5 or in the WT embryo at E14.5. In contrast, in the KO embryo at E14.5, selected myocardial cells (arrows) show intense cytoplasmic and nuclear staining. Many hepatocytes also show intense cytoplasmic and nuclear staining. In the high-magnification inset, selected hepatocytes are marked with arrows.
Consistent with the increased apoptosis in the mutant heart, signs of cell death were also observed in the liver of FIP200 KO embryos at both E14.5 and E15.5 (Fig. 2 D). Immunohistochemical staining for cleaved caspase-3 of liver sections confirmed significantly increased apoptosis in FIP200 KO embryos at E14.5 (Fig. 4 B) and E15.5 (not depicted) compared with WT littermates. The majority of the apoptotic cells were identified as hepatocytes based on their larger size with large eosinophilic cytoplasm and large, vesicular, single, round nucleus possessing a single, prominent, central nucleolus, compared with hematopoietic cells present in the liver. Interestingly, the increased apoptosis in the mutant embryos appeared to be restricted to the liver and heart, as other organs of the FIP200 KO embryos did not show an obvious increase in cleaved caspase-3 staining (unpublished data). As observed in the heart, no increased apoptosis was found for the liver of mutant embryos at E12.5. Together, these results suggest that increased apoptosis, rather than decreased proliferation, is responsible for the decreased cell number of the thin ventricular walls as well as liver lesions in the FIP200 KO embryos.
FIP200 deletion leads to increased susceptibility to TNFα-induced apoptosis
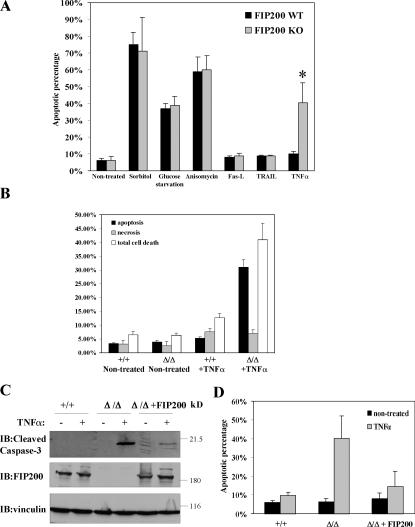
To understand the mechanism of regulation of apoptosis by FIP200, we isolated primary MEFs from FIP200 KO and WT littermate embryos. Under normal culture conditions, FIP200 KO MEFs did not show increased apoptosis compared with the WT MEFs. Furthermore, FIP200 KO and WT MEFs showed a similar extent of apoptosis upon several apoptotic stimuli, such as glucose depletion and sorbitol and anisomycin treatment. In contrast, FIP200 KO MEFs showed an elevated sensitivity to TNFα-induced apoptosis compared with WT MEFs (Fig. 5 A). Consistent with previous studies (Hoeflich et al., 2000; Rudolph et al., 2000; Lamb et al., 2003), TNFα treatment did not cause significant apoptosis in WT MEFs (Fig. 5 A). Interestingly, it induced significant apoptosis in the FIP200 KO MEFs compared with the untreated cells, suggesting an increased susceptibility to TNFα-induced apoptosis. In contrast, deletion of FIP200 did not sensitize cells to FAS-L– and TRAIL-induced apoptosis in MEFs (Fig. 5 A), suggesting that FIP200 plays a specific role in TNFα-induced apoptosis. Furthermore, it appears that TNFα-induced necrosis was not increased in FIP200 KO MEFs (Fig. 5 B). To verify that the increased cell death was caused by apoptosis, lysates from FIP200 KO and WT MEFs that had been treated with TNFα or untreated cells were analyzed by Western blotting for cleaved caspase-3, a marker of apoptotic cell death. Fig. 5 C shows that TNFα treatment induced an increased amount of cleaved caspase-3 in FIP200 KO but not WT MEFs. Furthermore, reexpression of FIP200 in FIP200 KO MEFs significantly abolished increased caspase-3 cleavage and apoptosis by TNFα treatment in these cells (Fig. 5, C and D). Together, these results suggest that FIP200 functions as a suppressor of TNFα-induced apoptosis, and increased susceptibility to TNFα-induced apoptosis may play an important role in the heart and liver defects and embryonic lethality in FIP200 KO mice.
Figure 5.
Increased susceptibility of FIP200 KO MEFs to TNFα-induced apoptosis. (A) Primary FIP200 KO and WT MEFs were cultured in DME supplemented with 10% FBS. The cells were left untreated; treated for 1 d with 200 mM sorbitol, 10 μM anisomycin, 5 ng/ml Fas-L, 1 μg/ml TRAIL, and 50 ng/ml TNFα; or cultured in glucose-free medium for 2 d, as indicated. They were then stained with Hoechst to determine the fraction of apoptotic cells, as described in Materials and methods. The mean + SEM from at least three experiments is shown. *, P < 0.001. (B) Primary FIP200 KO and WT MEFs were either left untreated or treated for 1 d with 50 ng/ml TNFα. The fraction of apoptotic cells, necrotic cells, or dead cells (including both apoptotic and necrotic cells) was determined by acridine orange and ethidium bromide costaining. The mean + SEM from at least three experiments is shown. (C) FIP200 KO MEFs were transfected with plasmid encoding HA-tagged FIP200 (fifth and sixth lanes) or empty vector as a control (third and fourth lanes). 1 d after transfection, these and WT MEFs (first and second lanes) were treated with or without 50 ng/ml TNFα for another day, as indicated. Cell lysates were prepared and analyzed by Western blotting using antibodies against cleaved caspase-3, FIP200, and vinculin, as indicated. (D) FIP200 KO MEFs were transfected with empty vector or vector encoding HA-FIP200, along with pEGFP (3:1 ratio). The cells were either left untreated or treated for 1 d with 50 ng/ml TNFα, as indicated. They were then stained with Hoechst to determine the fraction of apoptotic cells in EGFP-positive cells. The mean + SEM from at least three experiments is shown.
Defective JNK signaling is responsible for increased TNFα-stimulated apoptosis in FIP200 KO cells
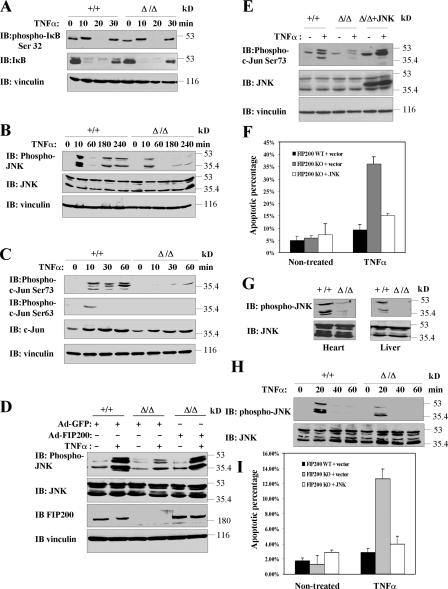
To investigate the mechanisms by which FIP200 deletion leads to increased susceptibility to TNFα-induced apoptosis, we first examined potential changes in the NF-κB pathway, which is one of the major cell survival pathways activated by TNFα (Chen and Goeddel, 2002). We tested whether TNFα induced IκBα phosphorylation and degradation is affected in FIP200 KO MEFs, which is the biochemical indicator for NF-κB activation by TNFα stimulation. Fig. 6 A shows rapid induction of phosphorylation (very significant given the dramatically reduced levels of IκBα protein upon stimulation) and degradation of IκBα at 10 min after stimulation in both FIP200 KO and WT MEFs. At 30 min after TNFα treatment, IκBα protein reappeared as expected because of translocation of freed NF-κB to the nucleus and its activation of the transcription of IκBα itself in both cells. Furthermore, TNFα stimulation increased NF-κB transcription activity to similar levels in both FIP200 KO and WT MEFs (unpublished data). These results suggest that deletion of FIP200 did not affect TNFα- induced NF-κB activation.
Figure 6.
Reduced JNK activation in response to TNFα stimulation in FIP200 KO cells and embryos. (A–C) FIP200 KO and WT MEFs were serum starved overnight. They were then left untreated or treated with 50 ng/ml TNFα for the different periods of time, as indicated. Cell lysates were then analyzed by Western blotting with various antibodies as indicated. (D) FIP200 KO and WT MEFs were infected with recombinant adenoviruses encoding FIP200 (Ad-FIP200) or GFP (Ad-GFP, as control), as indicated. 1 d after infection, cells were serum starved overnight and were left untreated or treated with 50 ng/ml TNFα for 10 min. Cell lysates were analyzed by Western blotting with various antibodies as indicated. (E and F) FIP200 KO MEFs were transfected with empty vector or vector encoding Myc-JNK1, along with pEGFP (3:1 ratio). The cells were either left untreated or treated for 1 d with 50 ng/ml TNFα, as indicated. Cell lysates were prepared and analyzed by Western blotting with various antibodies, as indicated (E). Alternatively, the cells were stained with Hoechst to determine the fraction of apoptotic cells in EGFP-positive populations. The mean + SEM from at least three experiments is shown (F). (G) Western blotting analysis of heart and liver protein extracts from FIP200 WT (+/+) and KO (Δ/Δ) embryos using antibodies against phospho-JNK or JNK, as indicated. (H) Hepatocytes isolated from FIP200 WT (+/+) and KO (Δ/Δ) embryos were serum starved overnight. They were then left untreated or treated with 50 ng/ml TNFα for the different periods of time, as indicated. Cell lysates were then analyzed by Western blotting with phospho-JNK or JNK antibodies, as indicated. (I) FIP200 KO hepatocytes were transfected with empty vector or vector encoding Myc-JNK1, along with pEGFP (3:1 ratio). The cells were either left untreated or treated for 1 d with 50 ng/ml TNFα, as indicated. They were then stained with Hoechst to determine the fraction of apoptotic cells in EGFP-positive populations. The mean + SEM from at least three experiments is shown.
TNFα stimulation also leads to JNK activation, which can promote either cell survival or death, depending on the cellular context (Davis, 2000). Interestingly, JNK1 and -2 double-KO MEFs also exhibit more sensitivity to TNFα-induced apoptosis (Lamb et al., 2003). We therefore examined whether JNK signaling is affected in FIP200 KO MEFs. Fig. 6 B shows that induction of JNK phosphorylation (both the transient and robust JNK activation within the first 30 min, and the later sustained and weaker JNK activation) in response to TNFα stimulation was significantly reduced in FIP200 KO MEFs compared with WT MEFs. Consistent with this, phosphorylation of c-Jun on both serine 63 and 73 upon TNFα stimulation was also lower in FIP200 KO MEFs than that in WT MEFs (Fig. 6 C). We also observed that TNFα stimulation increased c-Jun protein level, which is consistent with the previous observations that JNK-mediated c-Jun phosphorylation protected c-Jun from ubiquitin- dependent degradation, resulting in increased c-Jun protein levels (Fig. 6 C; Musti et al., 1997). The increase of c-Jun protein level in response to TNFα stimulation was reduced in FIP200 KO MEFs, consistent with the reduced JNK phosphorylation in response to TNFα stimulation in FIP200 KO MEFs (Fig. 6 C). Furthermore, we found that reexpression of FIP200 in the FIP200 KO MEFs restored JNK activation in response to TNFα stimulation (Fig. 6 D), suggesting the JNK phosphorylation defect observed in FIP200 KO MEFs is a direct consequence of FIP200 deletion.
To determine whether the reduced JNK activation in FIP200 MEFs is responsible for the increased TNFα-induced apoptosis, we examined the effect of restoration of JNK activity on TNFα-induced apoptosis in these cells. Transient transfection of FIP200 KO MEFs with an expression vector encoding JNK restored JNK activity as measured by the increased c-Jun phosphorylation in these cells (Fig. 6 E). This increased JNK activity significantly suppressed TNFα-induced apoptosis in FIP200 KO MEFs (Fig. 6 F), suggesting that the increased sensitivity of FIP200 KO MEFs to TNFα-induced apoptosis is caused by the defective JNK activation in these cells.
The aforementioned results from analyses in MEFs raised the possibility that the increased apoptosis in the liver and heart of FIP200 KO embryos is also due to reduced JNK activity in these embryos. To test this possibility directly, we first examined JNK activation status in embryonic liver and heart of FIP200 KO. Consistent with results in MEFs, Western blotting analysis of liver and heart protein extracts showed that JNK phosphorylation was decreased in FIP200 KO embryos compared with WT littermate embryos (Fig. 6 G). Immunohistochemical analysis of liver sections by anti–phospho-JNK also showed reduced JNK activity in FIP200 KO embryos (unpublished data). We then isolated primary liver cells from FIP200 KO and control WT embryos and examined their JNK activation and apoptosis in response to TNFα stimulation. Similar to the results in MEFs, we found that TNFα-induced JNK activation was reduced in liver cells from FIP200 KO embryos compared with that from control WT embryos (Fig. 6 H). Also consistent with results in MEFs, liver cells from FIP200 KO embryos exhibited increased sensitivity to TNFα-induced apoptosis and overexpression of JNK suppressed the increased apoptosis in these cells (Fig. 6 I). Collectively, these results suggest that FIP200 is required for TNFα-induced JNK activation, and reduced JNK activation and its prosurvival function upon FIP200 deletion may be responsible for the increased susceptibility to TNFα-induced apoptosis in FIP200 KO MEFs and hepatocytes, which may contribute to the increased apoptosis and embryonic lethality observed in FIP200 KO embryos.
Interaction of FIP200 with ASK1 and TRAF2 and its role in TNFα signaling to JNK
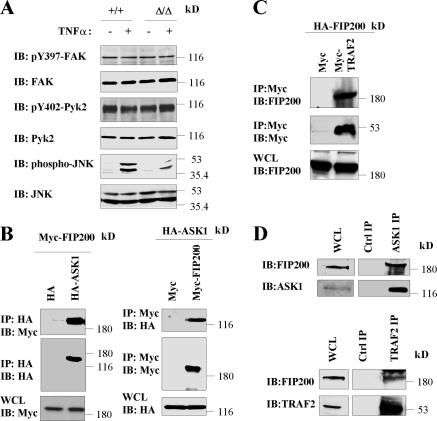
To explore potential mechanisms by which FIP200 participates in the stimulation of JNK signaling by TNFα, we first examined whether FIP200 could affect JNK signaling through its effects on FAK and/or Pyk2. We found that TNFα stimulation did not affect FAK and Pyk2 activation, whereas under the same condition, JNK was potently activated (Fig. 7 A). Furthermore, similar phosphorylation levels of FAK and Pyk2 were observed in FIP200 KO and WT MEFs, although previous studies showed inhibition of FAK and Pyk2 by overexpression of FIP200 in other cells (Ueda et al., 2000; Abbi et al., 2002). These results suggested that FAK and Pyk2 were not involved in FIP200 regulation of TNFα-induced JNK activation.
Figure 7.
Association of FIP200 with ASK1 and TRAF2. (A) FIP200 KO and WT MEFs were serum starved overnight. They were then left untreated or treated with 50 ng/ml TNFα for 10 min, as indicated. Cell lysates were then analyzed by Western blotting with various antibodies, as indicated. (B) 293T cells were cotransfected with vector encoding HA-ASK1 or empty vector and plasmid encoding Myc-FIP200 or empty vector, as indicated. Cell lysates were immunoprecipitated with anti-HA or anti-Myc and analyzed by Western blotting with anti-Myc and anti-HA, as indicated. Aliquots of whole cell lysates (WCL) were also analyzed directly by Western blotting with anti-Myc and anti-HA, as indicated. (C) 293T cells were cotransfected with vector encoding Myc-TRAF2 or empty vector and plasmid encoding HA-FIP200. Cell lysates were immunoprecipitated with anti-Myc and analyzed by Western blotting with anti-FIP200 or anti-Myc, as indicated. Aliquots of the lysates were also analyzed directly by Western blotting with anti-FIP200. (D) Lysates from MEFs were immunoprecipitated by anti-ASK1 or anti-TRAF2 or an irrelevant antibody as control, as indicated. They were then analyzed by Western blotting with anti-FIP200, anti-ASK1, or anti-TRAF2, as indicated. Aliquots of the lysates were also analyzed directly.
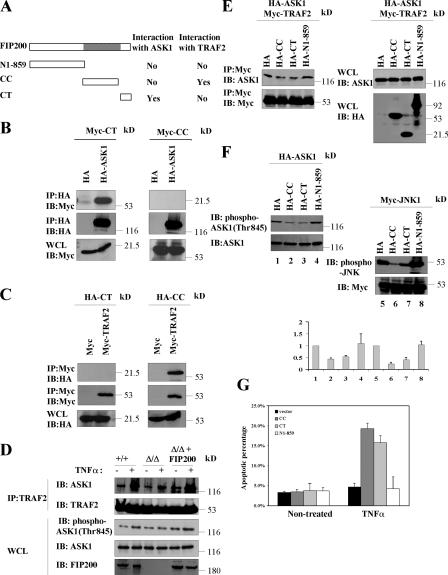
Previous studies documented that TNFα activates JNK through TNFR-TRAF2-ASK1-MKK4/MKK7-JNK signaling cascade (Davis, 2000). Thus, we examined whether FIP200 was capable of interacting with any of the components in the cascade. Interestingly, we observed that FIP200 could interact with both ASK1 and TRAF2. Fig. 7 B shows coimmunoprecipitation of Myc-FIP200 with HA-ASK1 by anti-HA precipitation (left) and reciprocal coimmunoprecipitation of HA-ASK1 with Myc-FIP200 by anti-Myc precipitation (right), when both were transfected into 293T cells. Similar coimmunoprecipitation studies showed association of HA-FIP200 with Myc-TRAF2 in 293T cells (Fig. 7 C). Consistent with these transfection studies, we also detected the interaction of endogenous FIP200 with the endogenous ASK1 and TRAF2 in MEFs (Fig. 7 D). We next determined the regions of FIP200 responsible for its association with ASK1 and TRAF2. We found that the FIP200 C-terminal region (CT), but not the N-terminal 859 residues (N1-859) or the CC (Fig. 8 A), could associate with ASK1 (Fig. 8, A and B). In contrast, CC, but not CT or N1-859, coprecipitated with TRAF2 (Fig. 8, A and C). These results demonstrated that FIP200 could interact with TRAF2 and ASK1 through two separate regions of the molecule.
Figure 8.
Role of FIP200 interaction with TRAF2 and ASK1 in its regulation of TNFα signaling to JNK. (A) Schematic diagram showing the different regions of FIP200 and a summary of their ability to interact with ASK1 or TRAF2. (B and C) 293T cells were cotransfected with vector encoding HA-ASK1 or empty vector and plasmid encoding Myc-CT or -CC (B) or vector encoding Myc-TRAF2 or empty vector and plasmid encoding HA-CT or HA-CC (C), as indicated. Cell lysates were immunoprecipitated with anti-HA or anti-Myc and analyzed by Western blotting with anti-Myc and anti-HA, as indicated. Aliquots of whole cell lysates (WCL) were also analyzed directly by Western blotting with anti-Myc or anti-HA, as indicated. (D) FIP200 KO MEFs were transfected with plasmid encoding HA-tagged FIP200 (fifth and sixth lanes) or empty vector as a control (third and fourth lanes), and WT MEFs were transfected with empty vector. HA-ASK1 was also cotransfected in each transfection. 1 d after transfection, cells were serum starved overnight. They were then left untreated or treated with 50 ng/ml TNFα for 10 min, as indicated. Cell lysates were then immunoprecipitated with anti-TRAF2 and analyzed by Western blotting with anti-ASK1 and anti-TRAF2, as indicated. Aliquots of the lysates were also analyzed by Western blotting with phospho-ASK1 (Thr845), ASK1, or FIP200 antibodies, as indicated. (E) Lysates from cells transfected with HA-CC, HA-CT, HA-N1-859, or empty vector were mixed with lysates from cells cotransfected with HA-ASK1 and Myc-TRAF2. Cell lysates were then immunoprecipitated with anti-Myc and analyzed by Western blotting with anti-Myc and anti-ASK1, as indicated. Aliquots of the lysates were also analyzed directly by Western blotting, as indicated. (F) 293T cells were cotransfected with vectors encoding HA-CC, HA-CT, HA-N1-859, or empty vector and plasmid encoding HA-ASK1 (left) or Myc-JNK1 (right). Cell lysates were then analyzed by Western blotting with various antibodies, as indicated. The phosphorylation levels of ASK1 and JNK are normalized to the expression levels of HA-ASK1 and Myc-JNK, and mean + SEM from three independent experiments are shown on the bottom. (G) MEFs were transfected with empty vector or vector encoding HA-CC, HA-CT, or HA-N1-859, along with pEGFP (3:1 ratio). The cells were either left untreated or treated for 1 d with 50 ng/ml TNFα, as indicated. They were then stained with Hoechst to determine the fraction of apoptotic cells in EGFP-positive cells. The mean + SEM from at least three experiments is shown.
Previous studies have shown that TRAF2 stimulates TNFα-induced JNK activation through TRAF2 interaction with ASK1 and TRAF2-mediated ASK1 activation (Nishitoh et al., 1998). Thus, our identification of FIP200 interaction with both TRAF2 and ASK1 through different regions raised the interesting possibility that FIP200 may play a role in TNFα-induced JNK activation by serving as a scaffold to facilitate TRAF2-ASK1 signaling to JNK activation. To test this possibility directly, we first evaluated whether TNFα-induced TRAF2–ASK1 interaction and ASK1 activation is reduced in FIP200 KO MEFs compared with WT control MEFs. Consistent with previous reports (Nishitoh et al., 1998; Noguchi et al., 2005), we found that TNFα treatment increased TRAF2 interaction with ASK1 and stimulated activation of ASK1 as measured by phosphorylation of ASK1 at threonine 845 in WT MEFs (Fig. 8 D). Deletion of FIP200 significantly reduced TNFα-stimulated TRAF2–ASK1 interaction and ASK1 phosphorylation in FIP200 KO MEFs compared with that in WT MEFs, which could be rescued by reexpression of FIP200 in FIP200 KO MEFs. We then determined whether overexpression of FIP200 CC or CT segment, which is expected to compete with endogenous FIP200 to reduce its interaction with TRAF2 or ASK1, respectively, will affect TRAF2–ASK1 interaction and ASK1 activation. Fig. 8 E shows that overexpression of either of these two segments, but not N1-859 segment, functioned in a dominant-negative manner to inhibit TRAF2 interaction with ASK1. Furthermore, overexpression of FIP200 CC or CT, but not N1-859 segment, could reduce ASK1 phosphorylation (Fig. 8 F, left). Consistent with the reduced ASK1 activation, they also reduced JNK activation in these cells (Fig. 8 F, right). Finally, we found that overexpression of FIP200 CC or CT segment increased TNFα stimulation–induced apoptosis in MEFs (Fig. 8 G) and hepatocytes (not depicted). Together, these results identified novel interactions of FIP200 with ASK1 and TRAF2, which might mediate FIP200 regulation of ASK1 and JNK activation in response to TNFα stimulation.
Discussion
Our analysis of the FIP200 KO embryos showed their defective cardiac and liver development and mid/late embryonic lethality caused by heart failure and liver degeneration. A role for FIP200 in cardiac formation and development is indicated by the severely affected myocardium with sparsely cellular myocardium forming thinner ventricular walls, as well as ventricular dilation and generalized edema, suggesting cardiac failure in the FIP200 KO embryos. Interestingly, on the other hand, TSC1 and -2 KO embryos also exhibited severe heart defects with thickened ventricular walls (Kobayashi et al., 1999, 2001). The opposite defective cardiac phenotypes suggested that FIP200 might function to antagonize TSC in the regulation of the thickness of the ventricular wall during heart development. This is consistent with our recent findings showing FIP200 interactions with TSC1 that inhibit TSC1–TSC2 complex function in vitro (Gan et al., 2005). They also suggest that a balance between positive and negative regulation of ventricular wall thickness by FIP200 and TSC, respectively, is critical for the normal cardiac development in embryogenesis.
Consistent with the opposite cardiac phenotype of the FIP200 and TSC KO embryos and our previous in vitro findings of inhibition of TSC functions by FIP200 (Gan et al., 2005), we found increased TSC activity in the FIP200 KO embryos as measured by a decreased activation of S6K in cardiomyocytes and hepatocytes as well as when the whole FIP200 KO embryo extracts were analyzed. Furthermore, we observed reduced cell size in the heart and liver of FIP200 KO embryos (Fig. 3, D and E), as well as isolated FIP200 KO MEFs (Fig. 3 C). We noted that the reduction in the size of FIP200 KO MEFs (∼5%) is not as pronounced as liver and heart cells in the FIP200 KO embryos. The relatively modest changes in the size of MEFs may account for their apparently normal functions in vivo. It is also possible that the functions of fibroblasts in vivo are less dependent on the changes in these signaling pathways (thus apparently lack of any defective phenotypes in earlier embryogenesis) than cells in the heart and liver, which are organs characterized by high metabolic activities in mid/late gestation that may be more tightly regulated by protein synthesis and cell growth. Consistent with this notion, both TSC1 and -2 KO embryos also showed major defects in heart and liver development (Kobayashi et al., 1999, 2001; Onda et al., 1999; Kwiatkowski et al., 2002). Therefore, a role of FIP200 in the regulation of cell size/cell growth may contribute to its potential function in heart and liver development as revealed in the current study.
It has been shown that cell size increase is a prerequisite for cell proliferation during normal organ growth (Conlon and Raff, 1999). Thus, it would be expected that the cell size decrease observed in FIP200 KO embryos would also lead to a cell proliferation defect. However, we did not detect any defects in cell cycle progression in the heart or liver in the FIP200 KO embryos (unpublished data). Although cell size and proliferation are coordinately regulated in many cases, these two cellular processes have also been shown to be uncoupled under some conditions where change in cell size does not necessarily affect cell proliferation. For example, deletion of S6K1 reduces the size of myoblasts without affecting their proliferation (Ohanna et al., 2005). In other cases, cell size and proliferation are regulated in an opposite manner. For example, it was shown that Rb triple-KO MEFs (lacking all three Rb family proteins pRb, p107, and p130) showed a significantly reduced cell size but an increased cell proliferation, when compared with control MEFs (Sage et al., 2000). Thus, it is likely that FIP200 function in these two cellular processes might be uncoupled during embryonic development.
Large truncation deletion of the FIP200 gene has been observed in ∼20% of primary breast cancers screened in a recent study, implicating a potential role of FIP200 as a tumor suppressor (Chano et al., 2002b). However, heterozygous deletion of FIP200 has not led to development of mammary or any other tumors within 1 yr of age so far (unpublished data). It is possible that one WT allele remaining in FIP200 heterozygous mice is sufficient to maintain its potential tumor suppressor function. It has been shown that, for many tumor suppressor genes, the germline single-allele loss in combination with stochastic somatic loss would lead to an increased tumor incidence in certain organs. For example, total deletion of tumor suppressor gene Brca1 leads to embryonic lethality phenotype, and Brac1 heterozygous mice don't develop tumors. Notably, introduction of a p53-null allele significantly enhances mammary gland tumor formation in Brca1 mammary gland conditional KO mice (Deng and Scott, 2000). Therefore, it is possible that combination with another allele that enhances breast cancer development might be synergistic in breast cancer development in FIP200 heterozygous mice. Future studies using FIP200 conditional KO mice combined with crossing with other mice tumor models will be necessary to overcome the embryonic lethality of homozygous deletion of FIP200 and clarify its potential role as a tumor suppressor in vivo.
Our results suggested that increased apoptosis in FIP200 KO embryos may be caused by an increased susceptibility to TNFα-induced apoptosis. Furthermore, defective JNK signaling was shown to be responsible for increased TNFα-stimulated apoptosis in FIP200 KO cells, which may contribute to the increased apoptosis and embryonic lethality observed in FIP200 KO embryos. The exact role of JNK in TNFα-regulated cell survival/death is complex and remains somewhat controversial. Some studies suggest an essential role for JNK in TNFα-induced cell death (De Smaele et al., 2001; Deng et al., 2003). Other studies indicate that JNK is not essential for TNFα-induced cell death (Liu et al., 1996) and may suppress TNFα-stimulated apoptosis (Lee et al., 1997; Roulston et al., 1998; Reuther-Madrid et al., 2002; Lamb et al., 2003). These studies together suggest a model in which JNK regulates TNFα-induced cell death in a temporal fashion such that early transient JNK activation upon TNFα stimulation suppresses TNFα-stimulated apoptosis, whereas late sustained JNK activation promotes TNFα-induced cell death (Ventura et al., 2006). Consistent with this model, our data showed that FIP200 KO cells exhibited decreased transient JNK activation (Fig. 6, B and H) and increased apoptosis upon TNFα stimulation (Fig. 6, F and I). Our study thus suggests that the increased apoptosis in FIP200 KO cells is mainly mediated by the reduction of transient JNK activation and that overexpression of JNK in FIP200 KO cells can suppress TNFα-induced apoptosis possibly by restoration of transient JNK activation.
Materials and methods
Construction of the targeting vector, and generation of FIP200 KO mice
Based on the available mouse genome sequence in the database, a DNA fragment in the intron between exons 4 and 5 was obtained by PCR. This fragment was then used to isolate mouse FIP200 genomic clones from BAC library derived from isogenic 129SvJ mice (Genome Systems, Inc.) by PCR. A targeting vector was then constructed that contains a 1.7-kb left arm, with exons 4 and 5 flanked by two loxP sites followed by a neo cassette gene and a third loxP site, a 5-kb right arm, and a HSV-TK gene. The linearized targeting vector was transfected into the E14.1 embryonic stem (ES) cells, and homologous recombinant clones were identified by Southern blotting analysis using a fragment outside of the left arm as probe (Fig. 1 A). The recombinant clone was used to generate chimeric mice by injection into C57BL/6 blastocytes. Male chimeras (identified by the presence of agouti coat color) were then mated with C57BL/6 females to determine germline transmission. The targeted allele was successfully transmitted to the germline cells, as identified by Southern blotting analysis using the same conditions for the detection of the homologous recombination event in the ES cells, which generated the heterozygote targeted mice designated as FIP200 floxNeo/+ mice. All mice used in the study were bred and maintained at Cornell University Transgenic Animal Core Facility (Ithaca, NY) under specific pathogen-free conditions in accordance with institutional guidelines.
Genotype analysis by PCR
The following primers were used in PCR genotyping: P1, 5′-GGAACCACGCTGACATTTGACACTG-3′; P2, 5′-CAAAGAACAACGAGTGGCAGTAG-3′; and P3, 5′-CATCAGATACACTAGAGCTGG-3′. The combination of primers P1 and P3 amplifies an ∼800-bp fragment from FIP200Δ allele. The combination of primers P2 and P3 amplifies 262- and 225-bp fragments from WT and FIP200flox alleles, respectively. The PCR condition is as follows: 3 cycles at 94°C for 3 min, 60°C for 1 min, and 72°C for 2 min, followed by 33 cycles at 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min, and 1 cycle at 94°C for 1 min, 60°C for 1 min, and 72°C for 10 min.
Antibodies and reagents
The rabbit antiserum against FIP200 and Pyk2 have been described previously (Zheng et al., 1998; Ueda et al., 2000). The mouse monoclonal α-vinculin and Tyrosine397 phospho-FAK antibodies were obtained from Upstate Biotechnology. The rabbit polyclonal α-HA (Y11) antibody, the mouse monoclonal α-c-Myc-tag (9E10) antibody, rabbit polyclonal α-FAK (C20) antibody, rabbit polyclonal and mouse monoclonal ASK1 antibodies, rabbit polyclonal and mouse monoclonal TRAF2 antibodies, and rabbit polyclonal S6K antibody were obtained from Santa Cruz Biotechnology, Inc. JNK1/2 antibody was obtained from BD Biosciences. Cleaved caspase-3, Tyr402 phospho-Pyk2, phospho-JNK, c-Jun, Ser63 phospho-c-Jun, Thr845 phospho-ASK1, Ser73 phospho-c-Jun, IκBα, Ser32-phospho-IκBα, Ser473 phospho-Akt, Akt, Thr1462 phospho-TSC2, Ser240/244 phospho-S6, S6, Thr389 phospho-S6K, Thr37/46 phospho-4EBP1, and 4EBP1 antibodies were purchased from Cell Signaling Technology, Inc. Sorbitol, acridine orange, and anisomycin were obtained from Sigma-Aldrich. TNFα was obtained from Calbiochem, TRAIL was provided by J. Yuan (Harvard Medical School, Boston, MA), and Fas-L was provided by Z. Liu (National Cancer Institute, Bethesda, MD). d-glucose–free DME and ethidium bromide were obtained from Invitrogen.
Cell culture
The FIP200 KO and WT MEFs were isolated from E12.5 embryos and cultured in DME supplemented with 10% FBS. 293T cells were cultured in DME supplemented with 10% FBS.
Plasmids and recombinant adenoviruses
Vectors encoding HA-FIP200, HA-FIP200 N1-859, and HA-FIP200 CT were described previously (Abbi et al., 2002; Gan et al., 2005). The CC of FIP200 (860–1401 aa) was amplified by PCR and then subcloned into HA tag–containing vector pKH3 to generate plasmid HA-FIP200 CC. Full-length FIP200, FIP200 N1-859, CC, and CT fragments were then subcloned from corresponding pKH3 vectors into Myc tag–containing pHAN vector (Han et al., 2002), resulting in Myc-FIP200, Myc-FIP200 N1-859, Myc-FIP200 CC, and Myc-FIP200 CT. The HA-ASK1 construct was provided by H. Fu (Emory University, Atlanta, GA), and the NF-κB reporter construct was provided by A. Lin (University of Chicago, Chicago, IL). Full-length mouse JNK1 and TRAF2 cDNA were amplified by RT-PCR and then subcloned into Myc tag–containing pHAN vector to generate the plasmids Myc-JNK1 and -TRAF2, respectively. The recombinant adenovirus Ad-FIP200 was generated as previously described (Melkoumian et al., 2005).
Immunoprecipitation and Western blotting
Tissue samples and embryos were homogenized, and extracts were used for Western blotting analysis as described previously (Peng et al., 2004). Preparation of whole cell lysates, immunoprecipitation, and Western blotting were performed as previously described (Abbi et al., 2002).
Histological and immunohistochemical analysis
Histological and immunohistochemical analysis were performed as previously described (Peng et al., 2004). The histological and immunohistochemical slides were examined under a microscope (model BX41; Olympus) with UplanF1 10×/0.3 objective lens at RT, and the images were captured using a camera (model DP70; Olympus) with DP Controller version 1.2.1.108.
Determination of cell size by WGA-TRITC staining and FACS analysis
To determine cell size by WGA staining, after deparaffinization and rehydration, tissue sections were stained with 150 μg/ml WGA tetramethylrhodamine conjugate (Invitrogen) at 37°C for 30 min and washed with PBS three times, each for 4 min. The samples were mounted for analyses by immunofluorescent microscopy. The area of cross section of cells was quantified by NIH Image program. To determine the cell size of cultured cells, FACS analysis with Cell Quest software (BD Biosciences) was performed as previously described (Inoki et al., 2003).
Apoptosis assays
Apoptosis assays were done as previously described (Ueda et al., 2000). The numbers of apoptotic and necrotic cells were determined by fluorescent dye costaining with acridine orange and ethidium bromide as described previously (Xu et al., 1996).
Acknowledgments
We are grateful to Ke-Yu Deng of Cornell Core Transgenic Mouse Facility for ES cell injection. We thank Huei Jin Ho for technical support and Zara Melkoumian for assistance in luciferase assays. We thank our colleagues Z. Melkoumian, H. Wei, Y. Yoo, X. Wu, D. Rhoads, and H. Ho for their critical reading of the manuscript and helpful comments.
This research was supported by National Institutes of Health grants GM52890 and HL73394 to J.-L. Guan.
J.-L. Guan's present address is University of Michigan Medical School, Ann Arbor, MI 48109.
Abbreviations used in this paper: 4EBP1, 4E binding protein 1; ASK1, apoptosis signal–regulating kinase 1; CC, coiled-coil region; CT, C-terminal region; E, embryonic day; ES, embryonic stem; FIP200, FAK family interacting protein of 200 kD; KO, knockout; MKK, MAPK kinase; MEF, mouse embryo fibroblast; mTOR, mammalian target of rapamycin; RB1CC1, RB1-inducible coiled-coil 1; S6K, S6 kinase; TNFR, TNF receptor; TRAF2, TNFR-associated factor 2; TRITC, tetramethyl rhodamine isothiocyanate; TSC, tuberous sclerosis complex; WGA, wheat germ agglutinin; WT, wild-type.
References
- Abbi, S., H. Ueda, C. Zheng, L.A. Cooper, J. Zhao, R. Christopher, and J.L. Guan. 2002. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol. Biol. Cell. 13:3178–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamba, N., T. Chano, T. Taga, S. Ohta, Y. Takeuchi, and H. Okabe. 2004. Expression and regulation of RB1CC1 in developing murine and human tissues. Int. J. Mol. Med. 14:583–587. [PubMed] [Google Scholar]
- Beg, A.A., W.C. Sha, R.T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 376:167–170. [DOI] [PubMed] [Google Scholar]
- Bueno, O.F., L.J. De Windt, K.M. Tymitz, S.A. Witt, T.R. Kimball, R. Klevitsky, T.E. Hewett, S.P. Jones, D.J. Lefer, C.F. Peng, et al. 2000. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 19:6341–6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chano, T., S. Ikegawa, K. Kontani, H. Okabe, N. Baldini, and Y. Saeki. 2002. a. Identification of RB1CC1, a novel human gene that can induce RB1 in various human cells. Oncogene. 21:1295–1298. [DOI] [PubMed] [Google Scholar]
- Chano, T., K. Kontani, K. Teramoto, H. Okabe, and S. Ikegawa. 2002. b. Truncating mutations of RB1CC1 in human breast cancer. Nat. Genet. 31:285–288. [DOI] [PubMed] [Google Scholar]
- Chen, G., and D.V. Goeddel. 2002. TNF-R1 signaling: a beautiful pathway. Science. 296:1634–1635. [DOI] [PubMed] [Google Scholar]
- Conlon, I., and M. Raff. 1999. Size control in animal development. Cell. 96:235–244. [DOI] [PubMed] [Google Scholar]
- Davis, R.J. 2000. Signal transduction by the JNK group of MAP kinases. Cell. 103:239–252. [DOI] [PubMed] [Google Scholar]
- De Smaele, E., F. Zazzeroni, S. Papa, D.U. Nguyen, R. Jin, J. Jones, R. Cong, and G. Franzoso. 2001. Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature. 414:308–313. [DOI] [PubMed] [Google Scholar]
- Deng, C.X., and F. Scott. 2000. Role of the tumor suppressor gene Brca1 in genetic stability and mammary gland tumor formation. Oncogene. 19:1059–1064. [DOI] [PubMed] [Google Scholar]
- Deng, Y., X. Ren, L. Yang, Y. Lin, and X. Wu. 2003. A JNK-dependent pathway is required for TNFalpha-induced apoptosis. Cell. 115:61–70. [DOI] [PubMed] [Google Scholar]
- Fingar, D.C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 23:3151–3171. [DOI] [PubMed] [Google Scholar]
- Gan, B., Z.K. Melkoumian, X. Wu, K.L. Guan, and J.L. Guan. 2005. Identification of FIP200 interaction with the TSC1-TSC2 complex and its role in regulation of cell size control. J. Cell Biol. 170:379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell. 109(Suppl.):S81–S96. [DOI] [PubMed] [Google Scholar]
- Han, D.C., T.L. Shen, H. Miao, B. Wang, and J.L. Guan. 2002. EphB1 associates with Grb7 and regulates cell migration. J. Biol. Chem. 277:45655–45661. [DOI] [PubMed] [Google Scholar]
- Hay, N., and N. Sonenberg. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945. [DOI] [PubMed] [Google Scholar]
- Hoeflich, K.P., J. Luo, E.A. Rubie, M.S. Tsao, O. Jin, and J.R. Woodgett. 2000. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 406:86–90. [DOI] [PubMed] [Google Scholar]
- Inoki, K., T. Zhu, and K.L. Guan. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 115:577–590. [DOI] [PubMed] [Google Scholar]
- Inoki, K., H. Ouyang, Y. Li, and K.L. Guan. 2005. Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 69:79–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., O. Minowa, J. Kuno, H. Mitani, O. Hino, and T. Noda. 1999. Renal carcinogenesis, hepatic hemangiomatosis, and embryonic lethality caused by a germ-line Tsc2 mutation in mice. Cancer Res. 59:1206–1211. [PubMed] [Google Scholar]
- Kobayashi, T., O. Minowa, Y. Sugitani, S. Takai, H. Mitani, E. Kobayashi, T. Noda, and O. Hino. 2001. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc. Natl. Acad. Sci. USA. 98:8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski, D.J. 2003. Tuberous sclerosis: from tubers to mTOR. Ann. Hum. Genet. 67:87–96. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski, D.J., H. Zhang, J.L. Bandura, K.M. Heiberger, M. Glogauer, N. el-Hashemite, and H. Onda. 2002. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11:525–534. [DOI] [PubMed] [Google Scholar]
- Lakso, M., J.G. Pichel, J.R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F.W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA. 93:5860–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, J.A., J.J. Ventura, P. Hess, R.A. Flavell, and R.J. Davis. 2003. JunD mediates survival signaling by the JNK signal transduction pathway. Mol. Cell. 11:1479–1489. [DOI] [PubMed] [Google Scholar]
- Lee, S.Y., A. Reichlin, A. Santana, K.A. Sokol, M.C. Nussenzweig, and Y. Choi. 1997. TRAF2 is essential for JNK but not NF-kappaB activation and regulates lymphocyte proliferation and survival. Immunity. 7:703–713. [DOI] [PubMed] [Google Scholar]
- Li, Q., D. Van Antwerp, F. Mercurio, K.F. Lee, and I.M. Verma. 1999. Severe liver degeneration in mice lacking the IkappaB kinase 2 gene. Science. 284:321–325. [DOI] [PubMed] [Google Scholar]
- Liu, Z.G., H. Hsu, D.V. Goeddel, and M. Karin. 1996. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell. 87:565–576. [DOI] [PubMed] [Google Scholar]
- Manning, B.D., and L.C. Cantley. 2003. United at last: the tuberous sclerosis complex gene products connect the phosphoinositide 3-kinase/Akt pathway to mammalian target of rapamycin (mTOR) signalling. Biochem. Soc. Trans. 31:573–578. [DOI] [PubMed] [Google Scholar]
- Melkoumian, Z.K., X. Peng, B. Gan, X. Wu, and J.L. Guan. 2005. Mechanism of cell cycle regulation by FIP200 in human breast cancer cells. Cancer Res. 65:6676–6684. [DOI] [PubMed] [Google Scholar]
- Musti, A.M., M. Treier, and D. Bohmann. 1997. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 275:400–402. [DOI] [PubMed] [Google Scholar]
- Nishitoh, H., M. Saitoh, Y. Mochida, K. Takeda, H. Nakano, M. Rothe, K. Miyazono, and H. Ichijo. 1998. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol. Cell. 2:389–395. [DOI] [PubMed] [Google Scholar]
- Noguchi, T., K. Takeda, A. Matsuzawa, K. Saegusa, H. Nakano, J. Gohda, J. Inoue, and H. Ichijo. 2005. Recruitment of tumor necrosis factor receptor-associated factor family proteins to apoptosis signal-regulating kinase 1 signalosome is essential for oxidative stress-induced cell death. J. Biol. Chem. 280:37033–37040. [DOI] [PubMed] [Google Scholar]
- Ohanna, M., A.K. Sobering, T. Lapointe, L. Lorenzo, C. Praud, E. Petroulakis, N. Sonenberg, P.A. Kelly, A. Sotiropoulos, and M. Pende. 2005. Atrophy of S6K1(−/−) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat. Cell Biol. 7:286–294. [DOI] [PubMed] [Google Scholar]
- Onda, H., A. Lueck, P.W. Marks, H.B. Warren, and D.J. Kwiatkowski. 1999. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J. Clin. Invest. 104:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, X., H. Ueda, H. Zhou, T. Stokol, T.L. Shen, A. Alcaraz, T. Nagy, J.D. Vassalli, and J.L. Guan. 2004. Overexpression of focal adhesion kinase in vascular endothelial cells promotes angiogenesis in transgenic mice. Cardiovasc. Res. 64:421–430. [DOI] [PubMed] [Google Scholar]
- Reuther-Madrid, J.Y., D. Kashatus, S. Chen, X. Li, J. Westwick, R.J. Davis, H.S. Earp, C.Y. Wang, and A.S. Baldwin Jr. 2002. The p65/RelA subunit of NF-kappaB suppresses the sustained, antiapoptotic activity of Jun kinase induced by tumor necrosis factor. Mol. Cell. Biol. 22:8175–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulston, A., C. Reinhard, P. Amiri, and L.T. Williams. 1998. Early activation of c-Jun N-terminal kinase and p38 kinase regulate cell survival in response to tumor necrosis factor alpha. J. Biol. Chem. 273:10232–10239. [DOI] [PubMed] [Google Scholar]
- Rudolph, D., W.C. Yeh, A. Wakeham, B. Rudolph, D. Nallainathan, J. Potter, A.J. Elia, and T.W. Mak. 2000. Severe liver degeneration and lack of NF-kappaB activation in NEMO/IKKgamma-deficient mice. Genes Dev. 14:854–862. [PMC free article] [PubMed] [Google Scholar]
- Sage, J., G.J. Mulligan, L.D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14:3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H., S. Abbi, C. Zheng, and J.L. Guan. 2000. Suppression of Pyk2 kinase and cellular activities by FIP200. J. Cell Biol. 149:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, J.J., A. Hubner, C. Zhang, R.A. Flavell, K.M. Shokat, and R.J. Davis. 2006. Chemical genetic analysis of the time course of signal transduction by JNK. Mol. Cell. 21:701–710. [DOI] [PubMed] [Google Scholar]
- Xu, J., Y. Xu, Q. Nguyen, P.M. Novikoff, and M.J. Czaja. 1996. Induction of hepatoma cell apoptosis by c-myc requires zinc and occurs in the absence of DNA fragmentation. Am. J. Physiol. 270:G60–G70. [DOI] [PubMed] [Google Scholar]
- Zheng, C., Z. Xing, Z.C. Bian, C. Guo, A. Akbay, L. Warner, and J.L. Guan. 1998. Differential regulation of Pyk2 and focal adhesion kinase (FAK). The C-terminal domain of FAK confers response to cell adhesion. J. Biol. Chem. 273:2384–2389. [DOI] [PubMed] [Google Scholar]