Abstract
In eukaryotic nuclei, DNA is wrapped around a protein octamer composed of the core histones H2A, H2B, H3, and H4, forming nucleosomes as the fundamental units of chromatin. The modification and deposition of specific histone variants play key roles in chromatin function. In this study, we established an in vitro system based on permeabilized cells that allows the assembly and exchange of histones in situ. H2A and H2B, each tagged with green fluorescent protein (GFP), are incorporated into euchromatin by exchange independently of DNA replication, and H3.1-GFP is assembled into replicated chromatin, as found in living cells. By purifying the cellular factors that assist in the incorporation of H2A–H2B, we identified protein phosphatase (PP) 2C γ subtype (PP2Cγ/PPM1G) as a histone chaperone that binds to and dephosphorylates H2A–H2B. The disruption of PP2Cγ in chicken DT40 cells increased the sensitivity to caffeine, a reagent that disturbs DNA replication and damage checkpoints, suggesting the involvement of PP2Cγ-mediated histone dephosphorylation and exchange in damage response or checkpoint recovery in higher eukaryotes.
Introduction
In eukaryotic nuclei, DNA is wrapped around a protein octamer containing two copies of each of the core histones H2A, H2B, H3, and H4, forming nucleosomes, the fundamental units of chromatin (Luger et al., 1997). Nucleosomes are assembled with the assistance of chaperones or assembly complexes. During de novo nucleosome assembly, DNA is first wrapped around the H3–H4 tetramer before the addition of two H2A–H2B dimers (Loyola and Almouzni, 2004). Once assembled, these core histones are tightly bound to DNA, and the interactions must be loosened or remodeled to allow the access of molecular machineries (e.g., polymerases) to DNA. In living cells, the histone–DNA interaction and chromatin structure are expected to be continually altered during transcription, genome duplication, and damage recovery, and the remodeling of chromatin is often associated with histone exchange (Belotserkovskaya and Reinberg, 2004; Flaus and Owen-Hughes, 2004; Loyola and Almouzni, 2004; Henikoff and Ahmad, 2005; Kimura, 2005). On the other hand, nucleosome contexts on specific loci must be preserved to maintain epigenetic marks on histone tails (Turner, 2002). The modification of histones, including acetylation, methylation, and phosphorylation, plays essential roles in chromatin functions such as gene expression and chromosome segregation. Thus, fluidity and stability seem to be well balanced in nucleosomes in living cells.
Early studies of histone deposition and exchange in living cells used radiochemical labeling. The stable association of [3H]arginine-labeled H3–H4 with chromatin was demonstrated by cell fusion (Manser et al., 1980). In a series of studies analyzing the deposition of the newly synthesized radio-labeled histones into nucleosomes, it was found that linker histone H1 and H2A–H2B undergo exchanges independently of DNA replication and transcription (Louters and Chalkley, 1985; Jackson, 1990). Such different behaviors of different histone species are also seen in living cells using GFP-tagged proteins (Kimura, 2005). The linker histone H1 is rapidly exchanged within a few minutes, and core histones are more stably bound. Long-term observation and cell fusion experiments further revealed that a substantial fraction of H2B-GFP exchanges slowly in euchromatin, whereas most H3-GFP (which is the H3.1 variant and is referred to below as H3.1-GFP) and H4-GFP remain bound to chromatin. In addition to the slowly exchanging fraction of H2B-GFP, which exchanges independently of DNA replication and transcription, another rapidly exchanging fraction, probably coupled to transcription, has been observed (Kimura and Cook, 2001), which is in agreement with the dimer eviction observed during transcription (Kireeva et al., 2002; Belotserkovskaya and Reinberg, 2004).
The exchange and assembly of histones are regulated in a development- and differentiation-specific manner (Meshorer et al., 2006) by chaperones or assembly factors that are distinct for each histone variant. Histone H3 has several variants, which are deposited into specialized chromatin loci mediated differentially through the action of deposition complexes (Loyola and Almouzni, 2004; Tagami et al., 2004; Henikoff and Ahmad, 2005; Thiriet and Hayes, 2005). The modification pattern in the conserved tail is also distinctive in each variant (Hake et al., 2006). H3.3 has modifications associated with transcriptionally active chromatin, which is consistent with its localization on active genes; in contrast, replication-coupled H3.2 has mostly silencing modifications. Although the modification pattern of each histone is established after its assembly into nucleosomes (influenced by the surrounding chromatin state), some specific modifications are associated with nucleosome-free deposition forms. Such modifications are typically found in H4, whose deposition form is diacetylated throughout eukaryotes, and some acetylation is associated with newly synthesized H3 in human cells (Benson et al., 2006).
In addition to H3, variant-specific deposition and modification are found in H2A. Nucleosome assembly protein 1 (Nap1) and the related proteins are known as somatic H2A–H2B chaperones after Nap1's purification from HeLa cells on the basis of nucleosome assembly activity in vitro (Ishimi et al., 1984; Loyola and Almouzni, 2004; Henikoff and Ahmad, 2005). Although Nap1 is not essential for yeast viability, its disruption affects the expression level of ∼10% of genes in clusters, suggesting a nucleosome maintenance function of Nap1 by depositing H2A–H2B (Ohkuni et al., 2003). Although Nap1 assists nucleosome assembly without ATP in vitro, complexes containing ATP-dependent chromatin remodeling activity have recently been shown to mediate the exchange of H2A–H2B dimers (Bruno et al., 2003; Kobor et al., 2004; Krogan et al., 2004; Mizuguchi et al., 2004). A complex containing yeast SWR1 (Swi2/Snf2-related ATPase 1) exchanges canonical H2A with H2AZ in nucleosome arrays, and SWR1 and H2AZ regulate an overlapping subset of genes. Another complex containing Tip60 (Tat-interacting protein 60) is involved in the exchange of phosphorylated H2Av (a Drosophila melanogaster histone H2A variant homologous to H2AX) with the unphosphorylated form at DNA lesions in Drosophila (Kusch et al., 2004). Thus, multiple mechanisms appear to exist to control the exchange of H2A variants at appropriate chromatin loci and in response to various stimuli, including DNA damages.
To understand the molecular mechanisms that regulate the assembly and exchange of histones in higher eukaryotes, we set out to establish an in vitro system that mimics in vivo histone dynamics using permeabilized cells. When cells are treated with nonionic detergents such as Triton X-100 or saponin, cellular membranes are permeabilized and some proteins are extracted, but many nuclear functions remain active (Jackson and Cook, 1985; Pombo et al., 1999), and some nuclear structures can be manipulated by adding exogenous factors (Misteli and Spector, 1996; Maison et al., 2002). Therefore, we expected that exogenously added histones might be incorporated into chromatin in permeabilized cells by exchange or replication-coupled assembly. As expected, GFP-tagged histones were indeed incorporated into chromatin in permeabilized cells with the assistance of cellular factors. By purifying the factors assisting GFP-H2A–H2B incorporation, we identified the type 2C protein phosphatase (PP) 2Cγ/PPM1G (Travis and Welsh, 1997; Murray et al., 1999) in addition to the Nap1 family members. PP2Cγ directly bound to and dephosphorylated H2A–H2B, and its disruption in chicken DT40 cells caused hypersensitivity to checkpoint abrogation. Although PP2Cγ did not appear to be the major phosphatase for H2AX and H2B, dephosphorylation and exchange via PP2Cγ may function to allow full recovery from DNA damage.
Results
Histone assembly and exchange in permeabilized cells
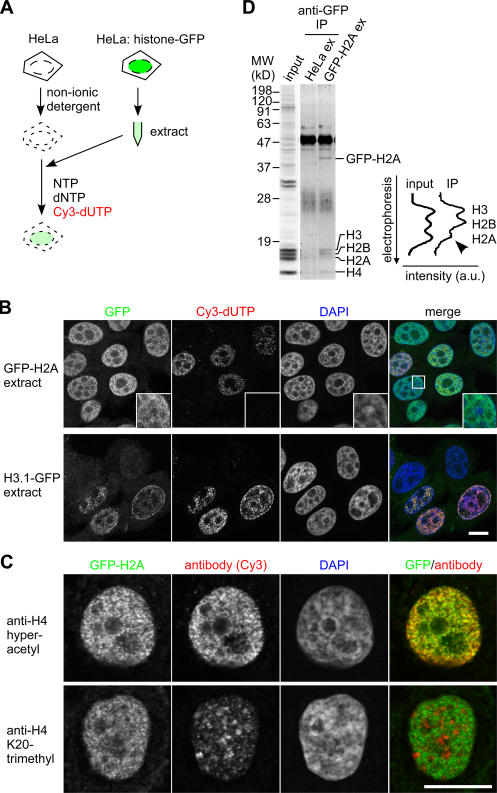
As illustrated in Fig. 1 A, HeLa cells were permeabilized and incubated in cell extracts prepared from cells stably expressing GFP-tagged core histones whose expression levels were <10% of their endogenous counterparts (Kimura and Cook, 2001). After washing out the unincorporated materials, GFP-H2A localized to euchromatin, which was devoid of DAPI-dense heterochromatin (Fig. 1 B, inset), in most permeabilized cells (Fig. 1 B). In contrast, H3.1-GFP highlighted replicated chromatin, which was labeled with Cy3-dUTP. The euchromatic localization of GFP-H2A was confirmed by the overlapping signals with specific antibodies recognizing acetylated histone H3 (not depicted) and H4 (Fig. 1 C), which are associated with transcriptionally active chromatin (Turner, 2002). In contrast, GFP-H2A was excluded from inactive chromatin rich in K20-trimethylated H4 (Fig. 1 C; Turner, 2002). To confirm that GFP-H2A replaced the endogenous H2A in chromatin in permeabilized cells, GFP-containing mononucleosomes were prepared by immunoprecipitation using antibody directed against GFP, and the ratio of core histones was analyzed by SDS-PAGE and Coomassie staining (Fig. 1 D). The amount of H2A was roughly halved in GFP-H2A nucleosomes (Fig. 1 D), suggesting the incorporation of a dimer of GFP-H2A and H2B (GFP-H2A–H2B) into chromatin by exchange. This stoichiometry is unlikely to be created by the nonspecific aggregation of a GFP-H2A–containing histone octamer from the extract onto chromatin, as H2A–H2B and H3–H4 are present in different complexes in the chromatin-free fraction (see Fig. 5 B; Chang et al., 1997; Tagami et al., 2004).
Figure 1.
Visualization of histone exchange and assembly by permeabilized cells incubated in cell extract containing GFP-tagged histones. (A) Strategy. HeLa cells are permeabilized and incubated in extract prepared from cells expressing histone-GFP. Cy3-dUTP is incorporated into replicated chromatin. (B) Localization of histone-GFP in permeabilized cells incubated in cell extracts. Permeabilized cells were incubated in S100 extract prepared from cells expressing GFP-H2A or H3.1-GFP. After washing and fixation, DNA was counterstained with DAPI. Four views of single confocal sections are shown. Insets show magnified views of the boxed areas. (C) Localization of GFP-H2A in euchromatin. After incubating permeabilized cells with GFP-H2A–containing extract and Cy5-dUTP, cells were immunolabeled with rabbit polyclonal antibody specific to hyperacetylated H4 (top) or K20-trimethylated H4 (bottom) and Cy3-conjugated donkey anti–rabbit IgG. Typical examples of cells outside the S phase (without Cy5-dUTP incorporation) are shown. (B and C) Bars, 10 μm. (D) GFP-H2A replaces H2A in chromatin. Permeabilized cells were incubated in S100 extract from HeLa cells or cells expressing GFP-H2A. Mononucleosomes (input) and GFP-containing nucleosomes precipitated using anti-GFP (IP) were separated by SDS-PAGE. The line intensity profiles of histones stained with Coomassie are shown.
Figure 5.
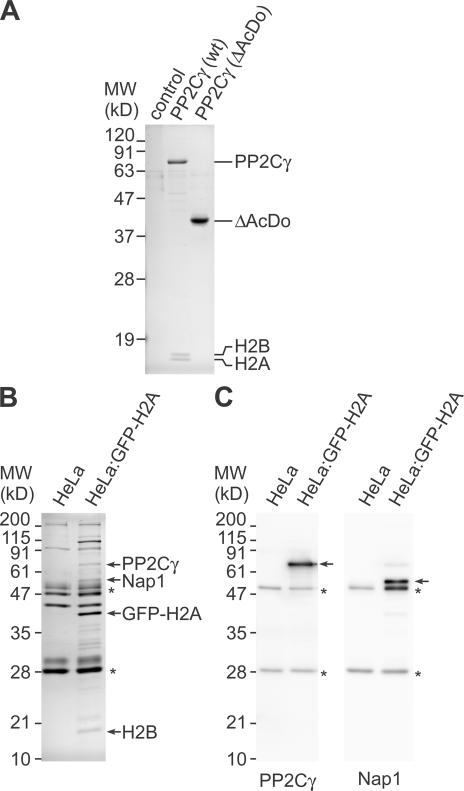
Binding of histones with PP2Cγ. (A) SDS-PAGE analysis of anti-FLAG immunoprecipitation. Immunoprecipitates from 293T cells transfected with GFP (control), FLAG-PP2Cγ (wild type [wt]), and FLAG-PP2Cγ (ΔAcDo) were separated by SDS-PAGE and stained with Coomassie. The two bands smaller than 19 kD that coprecipitated with FLAG-PP2Cγ were identified as histone H2B and H2A by mass spectrometry and immunoblotting. (B and C) Immunoprecipitation of GFP-H2A and its binding proteins. GFP-H2A was immunoprecipitated from S100 extract using anti-GFP agarose beads. The immunoprecipitated materials from control (HeLa) and extract prepared from cells stably expressing GFP-H2A (HeLa:GFP-H2A) were separated by SDS-PAGE and stained with Coomassie (B) or analyzed by immunoblotting (C). (B) Coomassie staining. The positions of size standards are indicated on the left. Proteins identified by mass spectrometry are indicated on the right. Asterisks indicate IgG heavy and light chains released from beads. (C) Immunoblotting. The membranes were probed with anti-PP2Cγ (left) and anti-Nap1 (right). The target proteins of the antibodies and IgG from beads are indicated by arrows and asterisks, respectively.
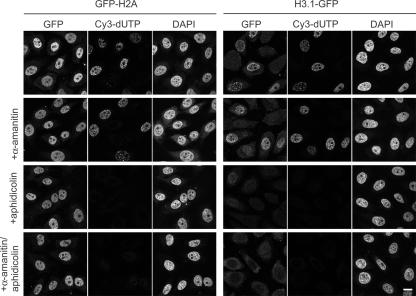
We next used specific inhibitors to examine whether the incorporation of histones depends on transcription and/or DNA replication in permeabilized cells. Most H2A–H2B appeared to be exchanged independently of ongoing RNA polymerase II transcription and DNA replication, as the incorporation of GFP-H2A and H2B-GFP was still observed in the presence of α-amanitin and aphidicolin, respectively (Fig. 2 and not depicted). The incorporation of H3.1-GFP into chromatin was coupled to DNA replication, as the signal almost disappeared in the presence of aphidicolin (Fig. 2). These results are reminiscent of the different behaviors of H2A–H2B and H3.1–H4 observed in living mammalian cells (Louters and Chalkley, 1985; Jackson, 1990, Kimura and Cook, 2001; Benson et al., 2006).
Figure 2.
Effects of polymerase inhibitors on the incorporation of GFP histones. Permeabilized HeLa cells were incubated in extracts prepared from cells expressing GFP-H2A and H3.1-GFP as in Fig. 1. In some cases, inhibitors of RNA polymerase II (2 μg/ml α-amanitin) and/or DNA polymerase (5 μg/ml aphidicolin) were added. Three views of single confocal sections are shown. Bar, 10 μm.
Purification of the activity required for histone H2A–H2B incorporation from HeLa cell extract
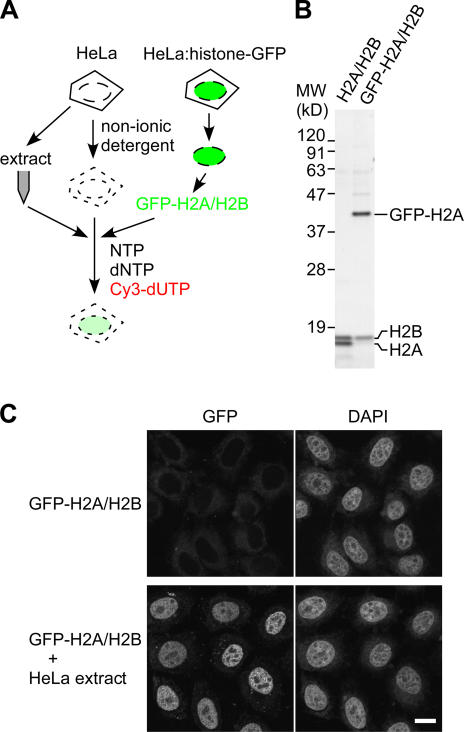
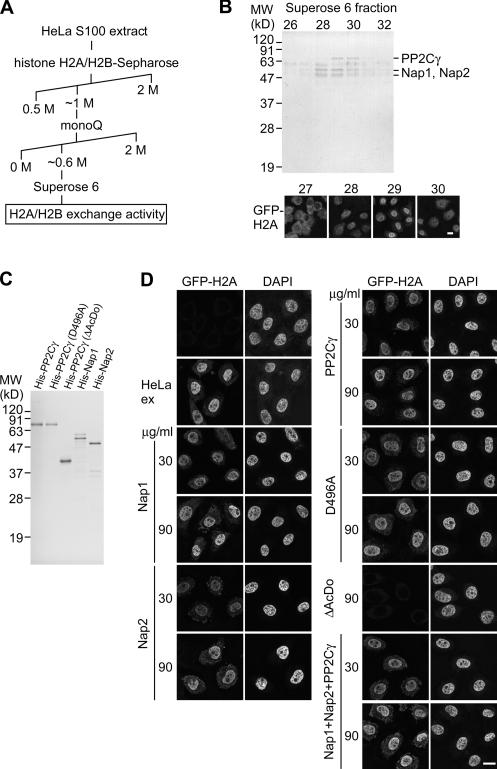
To analyze whether GFP-H2A–H2B dimer alone can be incorporated into chromatin, permeabilized cells were incubated with GFP-H2A–H2B purified from HeLa cells expressing GFP-H2A (Fig. 3). Although GFP-H2A–H2B alone failed to be incorporated, its incorporation was restored when supplemented with HeLa cell extract (Fig. 3 C), suggesting the presence of soluble factors required for H2A–H2B exchange in the extract. By following the incorporation of GFP-H2A under a fluorescent microscope, we purified the activity required for H2A–H2B exchange using column chromatography (Fig. 4 A). The purest active fraction consisted of three major bands by SDS-PAGE (Fig. 4 B). Mass spectrometry analysis identified these polypeptides as PP2Cγ/PPM1G (Travis and Welsh, 1997; Murray et al., 1999), Nap1/Nap1L1 (Ishimi et al., 1984), and Nap2/Nap1L4 (Rodriguez et al., 1997). It was not surprising to find Nap1 and Nap2 in the active fractions, as they have been described as histone chaperones that assist nucleosome assembly in vitro (Ishimi et al., 1984; Rodriguez et al., 1997). In contrast, no link between PP2Cγ and histones was previously established, which prompted us to focus on the function of PP2Cγ in histone H2A–H2B exchange. The phosphatase might regulate the chaperone activity of Nap1/2 by altering their phosphorylation state. On the other hand, PP2Cγ might also mediate the H2A–H2B exchange as such because it has a unique acidic domain (Travis and Welsh, 1997) that could potentially interact with histone H2A–H2B.
Figure 3.
The incorporation of purified GFP-H2A–H2B into permeabilized cell chromatin requires HeLa cell extract. (A) Strategy. Permeabilized HeLa cells are incubated in the nucleotide mixture containing Cy3- dUTP with GFP-H2A–H2B dimer in the presence or absence of extract from nonfluorescent HeLa cells. (B) SDS-PAGE. H2A–H2B and GFP-H2A–H2B dimers purified from chromatin of HeLa cells and cells stably expressing GFP-H2A, respectively, were analyzed by SDS-PAGE and Coomassie staining. (C) GFP-H2A incorporation. Permeabilized cells were incubated with 10 μg/ml of purified GFP-H2A–H2B ± HeLa S100 extract. Two views (GFP and DAPI) of single confocal sections are shown. Bar, 10 μm.
Figure 4.
Identification of histone exchange activity. (A) Chromatography procedure to purify the activity assisting H2A–H2B exchange. (B) Identification of proteins associated with H2A–H2B exchange activity. The Superose 6 fractions were separated by SDS-PAGE and stained with Coomassie (top), and the activity supporting GFP-H2A incorporation was followed under a microscope (bottom). The 76- and 54/50-kD bands were identified as PP2Cγ and Nap1/Nap2 by mass spectrometry as indicated. (C) Recombinant proteins. His-tagged PP2Cγ, the mutants, Nap1, and Nap2 expressed in and purified from E. coli were analyzed by SDS-PAGE and Coomassie staining. (D) GFP-H2A–H2B incorporation supported by Nap1, Nap2, and PP2Cγ. Permeabilized cells were incubated in 10 μg/ml GFP-H2A–H2B ± HeLa extract or indicated recombinant proteins (30 or 90 μg/ml). In the case of mixing three proteins, an equal amount of each protein (10 or 30 μg/ml) was added to give the final concentration (30 or 90 μg/ml, respectively). Two views of single confocal sections are shown. (B and D) Bars, 10 μm.
Recombinant Nap1, Nap2, and PP2Cγ individually support H2A–H2B incorporation
To examine the relationship between H2A–H2B exchange activity and the individual proteins, we incubated permeabilized cells with the purified GFP-H2A–H2B and each recombinant protein fused to a histidine hexamer (His) tag expressed in and purified from Escherichia coli (Fig. 4, C and D). GFP-H2A was incorporated into chromatin in the presence of either His-Nap1, -Nap2, or -PP2Cγ (Fig. 4 D), and similar results were obtained when ATP was omitted from the system (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200608001/DC1), suggesting that PP2Cγ itself possesses ATP-independent chaperone activity, as do Nap1 and Nap2. Although the cofractionation of these three proteins by gel filtration chromatography (Fig. 4 B) suggests their presence in a complex, only additive effects on the incorporation of GFP-H2A–H2B were observed when these three recombinant proteins were mixed (Fig. 4 D).
As the acidic domain is unique to PP2Cγ among PP2C family members (Travis and Welsh, 1997; Murray et al., 1999), this domain might be essential for the chaperone function. Indeed, a phosphatase mutant lacking the acidic domain (ΔAcDo) did not support GFP-H2A incorporation (Fig. 4 D). Furthermore, coimmunoprecipitation analysis confirmed that the physical interaction of PP2Cγ with H2A–H2B requires this domain (Fig. 5 A). When FLAG-tagged phosphatase was transiently expressed in human 293T cells and recovered using anti-FLAG agarose beads, substantial amounts of endogenous H2A and H2B were coprecipitated with FLAG-PP2Cγ but not with FLAG-ΔAcDo (Fig. 5 A). The interaction between basic proteins like histones and the acidic domain could occur through nonspecific binding as a result of the positive and negative charges. However, the immunoprecipitation experiments show specific binding of the phosphatase to H2A–H2B because only these two, but not the other histones (i.e., H1, H3, and H4), were coprecipitated even though all histone subtypes are positively charged. The complex formation between GFP-H2A–H2B and PP2Cγ in the cell extract (used in Fig.1) was observed by immunoprecipitation using anti-GFP agarose beads (Fig. 5, B and C). The presence of PP2Cγ as well as Nap1 in the immunoprecipitates was confirmed by mass spectrometry (Fig. 5 B) and immunoblotting (Fig. 5 C). A two-hybrid cDNA library screen also yielded histone H2B as an interactor with PP2Cγ, and the interaction required the phosphatase's acidic domain (unpublished data).
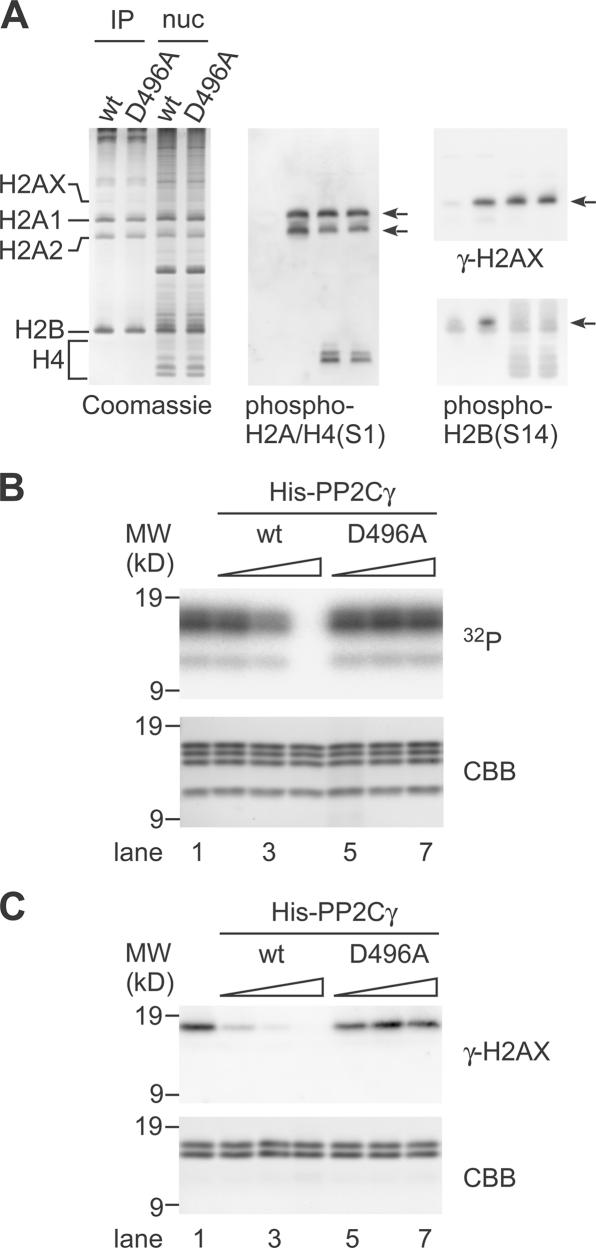
PP2Cγ dephosphorylates nucleosome-free histone H2A–H2B
The aforementioned results showing the physical interaction between PP2Cγ and H2A–H2B suggest that these histones could be substrates for the phosphatase. Therefore, we analyzed the phosphorylation state of FLAG-PP2Cγ–bound histones using acid-urea-Triton (AUT) gel electrophoresis and immunoblotting with specific antibodies directed against phosphorylated histones (Fig. 6 A). As expected, histones coprecipitated with the wild-type phosphatase were poorly recognized by antiphosphohistone antibodies. In contrast, histones bound to a phosphatase-inactive mutant (D496A) comprised detectable levels of phosphorylated molecules, including those related to DNA damage response and apoptosis such as Ser139-phosphorylated H2AX (called γ-H2AX; Rogakou et al., 1999) and Ser14-phosphorylated H2B (Cheung et al., 2003; Fernandez-Capetillo et al., 2004), although the overall migration pattern was similar to those bound to the wild-type phosphatase. As bulk nucleosomal histones were still phosphorylated in cells overexpressing the wild-type phosphatase (Fig. 6 A and by immunofluorescence; not depicted), only nucleosome-free H2A–H2B may be dephosphorylated by the phosphatase. Consistently, the purified His-PP2Cγ efficiently dephosphorylated nucleosome-free histones, including γ-H2AX in vitro (Fig. 6, C and D). As the D496A mutant still supported histone exchange in permeabilized cells (Fig. 4 D), the histone exchange and dephosphorylation do not appear to be coupled. These results suggest that a nucleosome-free H2A–H2B that binds to PP2Cγ may be dephosphorylated before its deposition into a nucleosome. Although we did not obtain positive data indicating the dephosphorylation of nucleosomal histones by PP2Cγ in overexpression and in vitro assays, it is also possible that additional cellular factors, which may be limited in the assays, stimulate the phosphatase activity or targeting toward the nucleosomal histones.
Figure 6.
Dephosphorylation of histones by PP2Cγ. (A) PP2Cγ-bound histones analyzed by AUT gel electrophoresis. After transfecting FLAG-PP2Cγ (wild type [wt]) or FLAG-PP2Cγ (D496A) into 293T cells, PP2Cγ-bound and nucleosomal histones were separated in an AUT gel. Coomassie staining (left) and immunoblots with the indicated phospho-specific antibodies are shown. The positions of histone subtypes and their phosphorylated forms (arrows) are indicated. (B) Dephosphorylation of histones by PP2Cγ. 2 μg of 32P-labeled histones, which were phosphorylated by MSK1, were incubated with different amounts (lanes 2 and 5, 12.5 ng; lanes 3 and 6, 50 ng; lanes 4 and 7, 250 ng) of His-PP2Cγ (wt; lanes 2–4) or mutant (D496A; lanes 5–7) and separated by SDS-PAGE. The radioactivity and Coomassie-stained histones are shown. The 32P signals from all of the histones disappear by dephosphorylation with increasing amounts of His-PP2Cγ but not with His-PP2Cγ (D496A). (C) Dephosphorylation of H2AX by PP2Cγ. 1 μg histone H2A–H2B fraction prepared from 12 Gy–irradiated HeLa cells was incubated alone (lane 1) or with different amounts (lanes 2 and 5, 1 ng; lanes 3 and 6, 5 ng; lanes 4 and 7, 25 ng) of His-PP2Cγ (wt; lanes 2–4) or mutant (D496A; lanes 5–7) and separated by SDS-PAGE. The phosphorylation level of H2AX was analyzed by immunoblotting with anti–γ-H2AX. The Coomassie-stained gel (CBB) is shown as a loading control.
We next tested whether PP2Cγ has in vitro nucleosome assembly activity using a supercoiling assay (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200608001/DC1) in which the assembly of nucleosomes can be assessed by the formation of supercoils from relaxed circular DNA (Ishimi et al., 1984; Rodriguez et al., 1997). Most of the plasmid DNA became supercoiled in the presence of Nap1, but only some supercoiled molecules accumulated even in the presence of high levels of PP2Cγ (Fig. S2), indicating that PP2Cγ has only weak de novo nucleosome assembly activity.
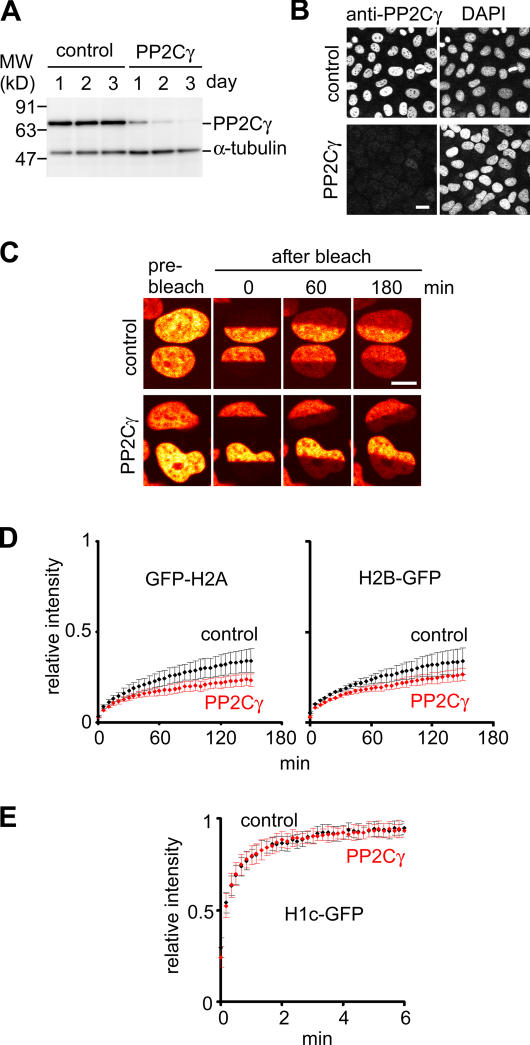
Effect of PP2Cγ knockdown on H2A–H2B mobility in living HeLa cells
To investigate whether PP2Cγ is involved in the regulation of H2A–H2B kinetics in living cells, we knocked down PP2Cγ in HeLa cells expressing histone-GFP using RNAi; 3 d after the transfection of a specific siRNA, the level of PP2Cγ decreased substantially to <5% of the normal level (Fig. 7, A and B). The mobility of H2A–H2B was analyzed by fluorescence recovery after photobleaching (Kimura and Cook, 2001). The recovery kinetics of both GFP-H2A and H2B-GFP decreased in cells transfected with PP2Cγ-specific siRNA compared with those with the control siRNA (Fig. 7, C and D), whereas the mobility of the linker histone H1c-GFP was unaffected (Fig. 7 E). These observations in living cells reflect the results from in vitro assays, suggesting that at least a part of H2A–H2B exchange is mediated by PP2Cγ as a histone chaperone in HeLa cells.
Figure 7.
Effects of PP2Cγ knockdown on kinetics of GFP-tagged H2A and H2B in living cells. (A and B) Knockdown of PP2Cγ by siRNA. The amount of PP2Cγ in HeLa cells transfected with control or PP2Cγ-specific siRNA was evaluated by immunoblotting (A) and immunofluorescence (B; 3 d after transfection) with anti-PP2Cγ. Anti–α-tubulin was used as a loading control. (C–E) FRAP. 3 d after RNA transfection, the mobility of histone-GFP was analyzed by bleaching a half of a nucleus (H2A and H2B) or a 2-μm spot (H1c) after the fluorescence recovery. Examples (GFP-H2A) and the recovery curves of GFP-H2A and H2B-GFP (D) or H1c-GFP (E) are shown. The means of the relative intensity in the bleached area are indicated with the SD (n ≥ 9). (B and C) Bars, 10 μm.
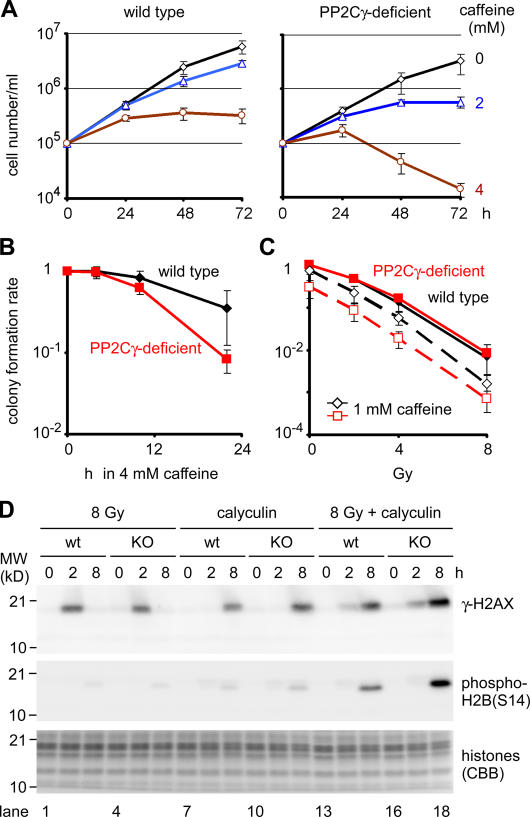
PP2Cγ-deficient DT40 cells show hypersensitivity to caffeine
To gain further insights into the biological function of PP2Cγ in vertebrate cells, we established PP2Cγ-deficient chicken DT40 cells by gene targeting (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200608001/DC1). As the deficient cells were generated by a simple knockout strategy to disrupt both alleles, PP2Cγ does not appear to be essential for cell growth. However, substantial growth defects were observed when DNA replication and damage checkpoints were abrogated by caffeine, which preferentially inhibits ataxia telangiectasia mutated– and ataxia telangiectasia and RAD3 related–dependent pathways, although its exact interfering points remain elusive (Kaufmann et al., 2003; Abraham, 2004). As shown in Fig. 8, PP2Cγ-deficient cells were more sensitive to caffeine compared with the wild type in a growth rate assay (Fig. 8 A) and in a colony formation assay (Fig. 8 B). In 2 mM caffeine, the wild-type cells continued to grow for 3 d, but PP2Cγ-deficient cells stopped growing at day 2. At a higher concentration (4 mM), the number of live cells (judged by the exclusion of trypan blue) became considerably lower after day 2 in PP2Cγ-deficient cells (Fig. 8 A). The colony formation assay revealed that the survival rate after 22 h of incubation in 4 mM caffeine was 35 ± 8 and 8.2 ± 0.3% in the wild-type and PP2Cγ-deficient cells, respectively (Fig. 8 B). As caffeine is known to sensitize cells to DNA double-strand breaks induced by ionizing radiation (Kaufmann et al., 2003; Abraham, 2004), we compared the sensitivity of these cells with γ-ray irradiation in the presence or absence of caffeine. Although PP2Cγ-deficient cells showed a similar radiation sensitivity to the wild type without caffeine, they became more sensitive when 1 mM caffeine was present in the colony-forming medium (Fig. 8 C).
Figure 8.
Phenotypes of PP2Cγ-deficient cells. (A) Growth curve. The wild-type (Cl18; left) and PP2Cγ-deficient cells (clone KO30; right) were plated (105 cells/ml), grown in caffeine (0, 2, or 4 mM), and the number of cells excluding trypan blue was counted every 24 h until 72 h. n = 4. (B and C) Colony formation assay. The number of colonies 10–12 d after plating was expressed as the relative value to that in controls without treatments. n = 3. (A–C) The mean and SD (error bars) are shown. (B) Caffeine sensitivity. Cells were treated with caffeine for 4, 10, or 22 h, diluted, and plated. (C) Radiation sensitivity. Cells were plated in methylcellulose medium ± 1 mM caffeine, irradiated (0, 2, 4, or 8 Gy), and incubated. (D) Effect of calyculin A on the phosphorylation of H2AX and H2B. The wild-type (wt; lanes 1–3, 7–9, and 13–15) or PP2Cγ-deficient (lanes 4–6, 10–12, and 16–18) cells were irradiated (8 Gy; lanes 1–6 and 13–18) or not irradiated (lanes 7–12), and calyculin A was added (lanes 7–18). Cells were collected either immediately (0 h), 2, or 4 h after irradiation, and the levels of γ-H2AX and phosphorylated H2B (S14) were analyzed by immunoblotting. The Coomassie-stained gel (CBB) is shown as a loading control. KO, knockout.
These results indicate that PP2Cγ is not essential for DNA double-strand break repair but suggest its involvement in recovery from damage. As H2AX is known to be phosphorylated around damaged chromatin, its dephosphorylation is required for full recovery from the damage response (Chowdhury et al., 2005; Keogh et al., 2005). Even though PP2A is likely to be the major γ-H2AX phosphatase in higher eukaryotes (Chowdhury et al., 2005), PP2Cγ could be involved in a backup dephosphorylation and deposition pathway. To assess the role of PP2Cγ in γ-H2AX dephosphorylation, the phosphorylation level of H2AX (i.e., the signal detected with antibody directed against γ-H2AX) was compared between the wild-type and PP2Cγ-deficient cells in response to DNA damage combined with treatment with calyculin A, an inhibitor of PP1 and PP2A (Nazarov et al., 2003; Chowdhury et al., 2005). In both cells, γ-H2AX appeared at a similar level 2 h after irradiation (8 Gy) and disappeared by 8 h (Fig. 8 D, lanes 1–6); faint signals of apoptosis-associated H2B (S14) phosphorylation appeared by 8 h. When cells were incubated with calyculin A, γ-H2AX was accumulated by 8 h even in the wild-type cells, probably as a result of spontaneous or replication-associated damages, which is consistent with the involvement of PP2A in γ-H2AX dephosphorylation (Chowdhury et al., 2005). The levels of γ-H2AX and phospho-H2B (S14) were higher in PP2Cγ-deficient cells in the presence of calyculin A (Fig. 8 D, lanes 9 and 12), suggesting that PP2Cγ is also one of the phosphatases that regulate γ-H2AX and phosphorylated H2B. The difference between wild-type and mutant cells became more evident when the cells were irradiated and incubated in calyculin A, as more γ-H2AX and phospho-H2B (S14) were accumulated in PP2Cγ-deficient cells (Fig. 8 D, lanes 13–18). These results suggest that PP2Cγ dephosphorylates γ-H2AX and phosphorylated H2B in wild-type DT40 cells.
Discussion
Identification of histone chaperones required for H2A–H2B incorporation into chromatin in permeabilized cells
To understand the biological function and molecular mechanisms of histone dynamics, we established a permeabilized cell-based assay for histone assembly and exchange. GFP-H2A and H2B-GFP were incorporated into euchromatin in permeabilized cells. This is consistent with the exchange of H2A–H2B in living cells, which can occur independently of DNA replication and transcription (Jackson, 1990; Kimura and Cook, 2001), preferentially in chromatin-containing acetylated H4 (Benson et al., 2006). H3.1-GFP assembled into replicated chromatin but contrasted to H2A–H2B, which is also reminiscent of the behavior in living cells (Kimura and Cook, 2001). By purifying the activity that assists GFP-H2A–H2B incorporation into chromatin in permeabilized cells, we identified three proteins—Nap1, Nap2, and PP2Cγ—in the purest fraction. Finding these Nap1-related proteins in our active fractions reassures us that the permeabilized cell-based assay has physiological relevance. The third protein we found was PP2Cγ, which harbors a unique acidic domain (Travis and Welsh, 1997) and was purified as a factor that stimulates spliceosome assembly in vitro (Murray et al., 1999).
Our analyses indicated that the phosphatase as such can assist the incorporation of H2A–H2B into chromatin in permeabilized cells and that it binds to and dephosphorylates histone H2A and H2B subtypes. Although the acidic domain of PP2Cγ could potentially mediate nonspecific electrostatic binding to basic proteins such as the histones, the fact that H2A–H2B was exclusively coimmunoprecipitated among all of the histones using FLAG-tagged phosphatase suggests that the interaction between PP2Cγ and H2A–H2B is specific. These histone chaperones do not require ATP for assisting H2A–H2B incorporation into chromatin in permeabilized cells as well as for in vitro nucleosome assembly with naked DNA. Because we followed the most active fractions that support GFP-H2A incorporation globally in euchromatin, other H2A–H2B exchange factors that are probably less abundant and act on more specific loci, including facilitating chromatin transcription (FACT; Belotserkovskaya and Reinberg, 2004) and ATP-dependent remodeling factors (Flaus and Owen-Hughes, 2004), were not found in the final preparation. Although Nap1/2 and PP2Cγ may mediate global H2A–H2B exchange independently of transcription and DNA replication, FACT may participate in transcription-coupled exchange. Future studies may reveal whether FACT supports H2A–H2B incorporation in a transcription-dependent manner in permeabilized cells.
A recent study revealed that ATP-dependent chromatin remodeling complexes can mediate histone exchange in addition to their remodeling function without the displacement of histone octamers (Flaus and Owen-Hughes, 2004). Therefore, it is also possible that the function of ATP-independent chaperones like Nap1/2 and PP2Cγ is solely to escort H2A–H2B and transfer the dimer to the ATP-dependent machineries, such as the yeast SWR1 complex that catalyzes the exchange between a canonical dimer and an H2AZ–H2B dimer (Mizuguchi et al., 2004). However, several lines of evidence suggest that the chaperones might also mediate H2A–H2B incorporation by themselves in addition to their escorting function. First, yeast Nap1 has the ability to exchange H2A–H2B in mononucleosomes in vitro (Park et al., 2005). Second, additional ATP is not required for H2A–H2B incorporation supported by Nap1/2 and PP2Cγ in permeabilized cells. Third, a substantial H2B-GFP recovery was still observed in living cells by FRAP even when the cellular ATP pool was depleted by treatment with sodium azide (unpublished data). Thus, although ATP-dependent factors might be required for the exchange of a dimer containing H2AZ at specific loci or during gene activation, ATP-independent chaperones may participate in the basal level of exchange of the major H2A and other variants. Alternatively, the major role of ATP-independent chaperones may be to deposit an H2A–H2B dimer into an incomplete nucleosome lacking a dimer, which can result from positive torsional stress (Jackson et al., 1994) or through ATP-driven eviction. This may account for the slow exchange rate of H2A–H2B in living cells despite the presence of a large pool of PP2Cγ (∼106 molecules/HeLa cell) diffusing almost freely in the nucleus (unpublished data).
Involvement of PP2Cγ in DNA damage response
To understand the biological function of PP2Cγ at the cellular level, we used chicken DT40 cells to create knockout cells by gene targeting. Although the deficient cells are viable, they show subtle growth retardation and a remarkable hypersensitivity to caffeine, which abrogates DNA replication and damage checkpoints. One possible mechanism to explain these phenomena is that the chaperone function together with the phosphatase activity plays a role in completing chromatin formation after DNA repair and/or replication by depositing dephosphorylated H2A–H2B molecules (Fig. 9). H2AX is phosphorylated around damaged chromatin (Rogakou et al., 1999), and its dephosphorylation is required for full recovery from damage responses. Also, H2AX molecules outside the damaged area are kept from undergoing phosphorylation for several hours. Although PP2A seems to play a major role in γ-H2AX dephosphorylation on chromatin (Chowdhury et al., 2005), we showed that PP2Cγ likewise mediates γ-H2AX and H2B dephosphorylation, as PP2Cγ-deficient cells showed a greater accumulation of γ-H2AX and phosphorylated H2B (S14) compared with wild-type cells when PP1 and PP2A were inhibited by calyculin A.
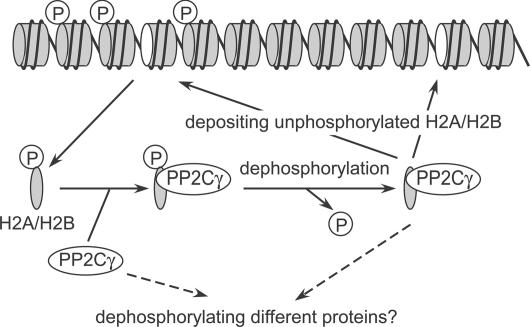
Figure 9.
A model for PP2Cγ function. PP2Cγ binds to nucleosome-free H2A–H2B (or H2AX–H2B) and removes phosphate groups (indicated by circled P) before the next deposition. Alternatively, the phosphatase activity and/or substrate specificity might be controlled by binding with histones. See Discussion for details.
Although the eviction of γ-H2AX or phosphorylated H2B may be mediated by other proteins such as the Drosophila Tip60-containing complex (Kusch et al., 2004), PP2Cγ may passively deposit dephosphorylated H2A–H2B or H2AX–H2B to incomplete nucleosomes lacking one dimer. This view is consistent with the observed uncoupling of the chaperone function and phosphatase activity of PP2Cγ; histone dephosphorylation can occur at any time after the binding of PP2Cγ until deposition (Fig. 9). Most H2A–H2B that bound to PP2Cγ but away from nucleosomes was indeed dephosphorylated. The lack of PP2Cγ in the DT40 knockout cells may thus delay the recovery from damage. When checkpoints are functional, such a subtle repair defect would not be critical and might only cause a subtle delay in cell growth. However, when checkpoints are abrogated, more cells with damaged chromatin would enter into mitosis for catastrophe.
An alternative possibility is that the substrate specificity or phosphatase activity of PP2Cγ is regulated by binding to H2A–H2B (Fig. 9); the level of nucleosome-free H2A–H2B could be altered by damage or replication fork arrest. The type 2C phosphatase family members are indeed involved in checkpoint responses (Leroy et al., 2003; Lu et al., 2005), and the γ subtype in particular might take part in inactivating checkpoints by sensing the free H2A–H2B level in the nucleus. Finally, a link between chromatin-remodeling factors and alternative pre-mRNA splicing was recently reported (Batsche et al., 2006). Consistent with this observation, PP2Cγ was previously identified as a factor that stimulates pre-mRNA splicing in vitro (Murray et al., 1999), raising the interesting possibility that PP2Cγ coordinately regulates stress responses in mammalian cells at the level of chromatin and RNA splicing.
Concluding remarks
It is now widely acknowledged that histone modification is key for the regulation of chromatin functions. Recent studies further indicate that the deposition and exchange of appropriate histone variants to specific chromosome loci are also important for gene expression and genome integrity (Loyola and Almouzni, 2004; Henikoff and Ahmad, 2005). A connection between the histone modification and deposition has been shown typically in the case of histone H4; before replication-coupled assembly, the newly synthesized molecules are diacetylated by HAT1 histone acetylase in the H3.1–H4 deposition complex (Chang et al., 1997; Tagami et al., 2004). Although diacetylation is not a prerequisite for assembly (Ma et al., 1998), this modification contributes to the recovery from replication block-mediated DNA damage (Barman et al., 2006). Similarly, in the case of H2A–H2B and H2AX–H2B, the deposition of unphosphorylated forms mediated by PP2Cγ appears to play a role in DNA damage responses. Thus, controlling the incorporation of appropriately modified histones seems to be important for maintaining genome integrity. Future studies should reveal how individual ATP-independent chaperones and ATP-dependent remodeling complexes function in distinct exchange processes in different chromatin contexts. Although differences in histone exchange kinetics in vivo were shown decades ago (Manser et al., 1980; Louters and Chalkley, 1985), the biological significance of the exchange and the underlying molecular mechanisms are just emerging. The approach presented in this study may contribute to bridging the gap between live cell observations and biochemical analyses.
Materials and methods
Histone exchange and assembly in permeabilized cells
In typical experiments, HeLa cells were plated in a 12-well plate containing 15-mm coverslips and were grown up to subconfluence. Cells were chilled on ice, washed twice in ice-cold physiological buffer (PB; 100 mM CH3COOK, 30 mM KCl, 10 mM Na2HPO4, 1 mM DTT, 1 mM MgCl2, and 1 mM ATP; Jackson and Cook, 1985) containing 5% Ficoll (PBF; pH 7.4; 1 ml per well; Nacalai Tesque), permeabilized in PBF containing 0.1% Triton X-100 (1 ml; for 5 min on ice), and washed twice in 1 ml PBF on ice. Cells were incubated for 1 h at 30°C in a reaction mixture containing cell extract (40%) or purified proteins supplemented with 100 μM each of NTP and dNTP (GE Healthcare), 0.4 μM Cy3-dUTP (PerkinElmer), and 800 μM MgCl2 in PBF. For incubation, a coverslip was overlaid (cell side down) on a 100-μl drop of the reaction mixture on Parafilm covering a flat aluminum block in a water bath at 30°C. After washing twice in 1 ml PBF for 5 min on ice in a 12-well plate, cells were fixed in 4% PFA (Electron Microscopy Sciences) in 250 mM Hepes-NaOH, pH 7.4 (Wako), for 20 min at room temperature, washed three times in 1 ml PBS, and DNA was counterstained with DAPI (12.5 ng/ml in PBS; 1 ml for 15 min; Nacalai Tesque). After washing twice in 1 ml PBS, coverslips were mounted using Prolong Gold (Invitrogen). In some cases, ATP and the other nucleotides were omitted from PBF and the reaction mixture.
For immunolabeling (Fig. 1 C), permeabilized cells were incubated in the reaction mixture containing 40% GFP-H2A extract and 2 μM Cy5-dUTP instead of Cy3-dUTP for 30 min at 30°C. After fixation, cells were treated with 1% Triton X-100 in PBS for 20 min, washed five times in PBS, and incubated in blocking buffer (0.2% gelatin, 1% BSA, and 0.05% Tween 20 in PBS, pH 8.0) for 30 min and then with rabbit polyclonal antibodies directed against hyperacetylated H4 (1:1,000; Upstate Biotechnology) or H4-trimethylated K20 (1:500; Abcam) in the same buffer for 3 h. Cells were washed in PBS containing 0.05% Tween 20 (PBST) three times for 10 min, incubated in Cy3-conjugated donkey anti–mouse IgG (1:500; Jackson ImmunoResearch Laboratories) overnight at 4°C, and washed with PBST three times for 10 min before DAPI staining.
Fluorescence images were sequentially collected using a confocal microscope featuring 405-, 488-, 543-, and 633-nm laser lines with the optimized pinhole setting operated by the built-in software: either a microscope (LSM510 META; Carl Zeiss MicroImaging, Inc.) with a C-Apo 40× NA 1.2 objective lens (for Figs. 1 B and 7 B) or a microscope (FV-1000; Olympus) with a UPlanSApo 60× NA 1.35 lens (for Figs. 1 C and 2–4 ). Image files were converted to tiff format using the operating software, merged, linearly contrast stretched (with the same setting in each set of experiments) using Photoshop version 7.01 (Adobe), and imported into Canvas 8 (Deneva) for assembly.
For chromatin immunoprecipitation, cells were centrifuged at 1,300 g for 10 min at 4°C after each step for buffer replacement. After the incubation and washing, nucleosomes were prepared, and GFP-containing nucleosomes were precipitated as described previously (Kanda et al., 1998; Kimura and Cook, 2001).
Preparation of cell extracts and protein purification
HeLa cells and derivatives expressing H2B-GFP (Kanda et al., 1998) and H3.1-GFP were grown as described previously (Kimura and Cook, 2001), and lines expressing GFP-H2A and H1c-GFP were established by transfecting the expression vectors (Misteli et al., 2000; Perche et al., 2000). Cell extracts were prepared based on the study by Dignam et al. (1983) with modifications. The S100 extract was prepared using a 1.5 cell-packed volume of 10 mM CH3COOK, 3 mM KCl, 1 mM Na2HPO4, 1 mM MgCl2, 1 mM ATP, 1 mM DTT, 10 mM Hepes-KOH, pH 7.4, and Complete protease inhibitor cocktail (EDTA-free; Roche) and dialyzed against PB plus inhibitors (1.5 μg/ml leupeptin, 2.5 μg/ml aprotinin, and 1 μg/ml pepstatin A; Wako). The nuclear pellet was extracted using an equal volume of 20 mM Hepes-KOH, 0.6 M KCl, 0.2 mM EDTA, 25% glycerol, 1 mM DTT, 1.5 mM MgCl2, and protease inhibitor cocktail (Roche) to yield the nuclear extract, which was also dialyzed against PB. Histone H2A–H2B and H3–H4 were separately purified from the nuclear pellet essentially according to Simon and Felsenfeld (1979), and the GFP-H2A–H2B fraction was separated from untagged H2A–H2B using gel filtration column chromatography (HiLoad Superdex 75; GE Healthcare).
To purify the activity assisting histone H2A–H2B incorporation in permeabilized cells, S100 extract was first fractionated through a histone H2A–H2B column, which was prepared by coupling 3 mg of the purified H2A–H2B to 1 ml N-hydroxysuccinimide ester–activated Sepharose (GE Healthcare) according to the manufacturer's instructions. Approximately 6 mg/ml HeLa S100 extract was mixed with 5 M NaCl to yield a final salt concentration of 0.5 M before applying to the column (2 ml per run). After washing with five column volumes of PB containing 0.5 M NaCl, bound proteins were eluted with a linear gradient of NaCl (0.5–2 M in PB; 20 column volumes). Each fraction was concentrated, and the buffer was substituted to PB using Ultrafree 0.5 (Millipore) before use in the permeabilized cell assay with 10–20 μg/ml of purified GFP-H2A–H2B. The activity supporting the nuclear localization of GFP-H2A except in nucleoli was followed under a fluorescence microscope (Axiovert2; Carl Zeiss MicroImaging, Inc.). The active fractions eluted in ∼1 M NaCl were further separated using a MonoQ 5/50 GL column (GE Healthcare) with a linear gradient of NaCl (0–2 M in PB; 20 column volumes). The most active fraction eluted in ∼0.6 M NaCl was concentrated and separated (0.2 ml in each fraction) on a Superose 6 gel filtration column (GE Healthcare). Proteins were identified by mass spectrometric analysis using a mass spectrometer (Ultraflex TOF/TOF; Bruker Daltonics) and by comparison between the determined molecular weights and theoretical peptide masses from the proteins registered in the National Center for Biotechnology Information.
Recombinant proteins and phosphatase assay
The cDNAs encoding human Nap1 (Nap1L1) and Nap2 (Nap1L4) were amplified by reverse transcription (Revertra Ace and oligo-dT18; TOYOBO) of HeLa RNA and PCR (high fidelity PCR master; Roche) using the following primers designed from GenBank/EMBL/DDBJ accession no. BT007023 (Nap1) and BC022090 (Nap2): Nap1 forward, TTACCATATGGCAGACATTGACAACAAAGAACAGTC; Nap1 reverse, CATCAAGCTTCACTGCTGCTTGCACTCTGCTGGGTT; Nap2 forward, CCTTCATATGGCAGATCACAGTTTTTCAGATGGGGT; and Nap2 reverse, GACAAAGCTTACACCTTGGGGTTAATTTCCGCATCA.
The amplified products were purified (QIAGEN), digested with NdeI and HindIII, and ligated into pHIT51 (containing T7 promoter, D box, His sequence, and multicloning site; provided by H. Tabara, Kyoto University, Kyoto, Japan) digested with the same enzymes. The resulting plasmids were verified by nucleotide sequencing. The expression plasmids for PP2Cγ and the mutants were also constructed by inserting the PP2Cγ sequence (Murray et al., 1999) into pHIT51. Each plasmid was introduced into BL21-Gold (Stratagene), and expression was induced with 1 mM IPTG at 30°C. The His-tagged proteins were purified using Ni-agarose beads (Sigma-Aldrich) according to the standard protocol by the manufacturer followed by MonoQ chromatography (GE Healthcare) and elution with a linear NaCl gradient (0.1–1 M NaCl in 50 mM Tris-Cl, pH 8.0).
For the phosphatase assay of bulk histones, histones purified from 20 μg HeLa cells were phosphorylated using 1 μg MSK1 (Upstate Biotechnology) in 12 mM MOPS, pH 7.0, 15 mM MgCl2, 0.2 mM EDTA, 1 mM EGTA, 0.2 mM DTT, 0.1 mM ATP, and 7.4 MBq/ml γ-[32P]ATP for 10 min at 30°C. The unincorporated ATP was removed, and the buffer was substituted with TMD (10 mM Tris-Cl, pH 8.0, 10 mM MgCl2, and 1 mM DTT) using Ultrafree filters 0.5 (Millipore). The phosphorylated histones (2 μg in 10 μl) were mixed with a serial dilution of His-tagged proteins (2.5 μl) and incubated for 1 h at 37°C. After stopping the reactions by adding 12.5 μl of 2× SDS gel loading buffer (Sambrook et al., 1989) and boiling, the samples were separated by SDS-PAGE and stained with Coomassie. The radioactivity was detected using an imaging analyzer (BAS2000; Fujifilm). For the γ-H2AX dephosphorylation assay, the γ-H2AX–containing H2A–H2B fraction was prepared from HeLa cells irradiated (12 Gy) using a 137Cs source at a dose rate of 1.13 Gy/min (Gammacell 40 Exactor; MDS Nordion). The H2A–H2B sample (1 μg in 10 μl) was mixed with 2.5 μl phosphatases in TMD buffer, incubated for 30 min at 37°C, and the level of γ-H2AX was analyzed by immunoblotting using antiphospho-H2AX Ser139 antibody (1:1,000; Upstate Biotechnology).
Immunoprecipitation and AUT gel electrophoresis
To immunoprecipitate GFP-H2A and its binding proteins, 1 ml S100 extract from HeLa cells (control) or cells expressing GFP-H2A was mixed with 50 μl anti-GFP agarose beads (Nacalai Tesque). After incubation for 1.5 h at 4°C with rotation, the beads were collected by centrifugation at 1,600 g for 5 min at 4°C. After washing four times for 10 min at 4°C in PB containing 0.05% Tween 20, 0.2 M NaCl, and protease inhibitor cocktail (Nacalai Tesque), the immunoprecipitates were eluted from the beads by boiling for 10 min in 60 μl of 2× SDS gel loading buffer (Sambrook et al., 1989).
For Fig. 5 (A and B), the FLAG-PP2Cγ, -ΔAcDo, and -D496A plasmids were generated by inserting the corresponding cDNAs into a modified version of pcDNA3.1/Hygro (Invitrogen) that contains N-terminal FLAG and V5 tags. 293T cells (4 × 90-cm2 dishes; 20% confluent) were transfected with these constructs using a calcium phosphate precipitation method (Sambrook et al., 1989). 3 d later, cells were washed with ice-cold PB, lysed in 2 ml PB containing 0.1% Triton X-100 and protease inhibitor cocktail, incubated for 5 min on ice, and cleared by centrifugation at 1,600 g for 10 min at 4°C. The supernatant was collected and mixed with 100 μl anti-FLAG agarose M2 beads (Sigma-Aldrich). After incubation and washing in the same buffer four times for 10 min at 4°C, the immunoprecipitated material was eluted with 100 μg/ml 3× FLAG peptide in PB (three times at 150 μl). The elution was pooled and either mixed with 2× SDS gel loading buffer for SDS-PAGE or with 20 mg/ml Casamino acids (final concentration of 100 μg/ml; Difco) and 100% trichloroacetic acid (final concentration of 20%) for AUT gel electrophoresis. After incubation for 1 h on ice and centrifugation at 20,000 g for 30 min at 4°C, the pellet was washed with acetone chilled at −20°C and dissolved in AUT sample buffer (Pilch et al., 2004). Immunoblotting was performed as described previously (Kimura and Cook, 2001) using the following primary antibodies: rabbit antiphospho-H2A/H4 Ser1 (1:1,000; Upstate Biotechnology), mouse antiphospho-H2AX Ser139 (1:1,000; Upstate Biotechnology), and mouse antiphospho-H2B Ser14 (clone 6C9; 1:20 hybridoma supernatant).
To produce antiphospho-H2B Ser14, mice were immunized with a synthetic peptide KSAPAPKKG(phospho-S)KKAVTKAQKC (Sigma-Genosys) coupled to keyhole limpet hemocyanin (Kimura et al., 1994), and a hybridoma clone 6C9 was obtained by ELISA screening using the phosphorylated and unphosphorylated peptides. As H2B Ser14 is phosphorylated during apoptosis (Cheung et al., 2003), the specificity was then checked by the specific appearance of positive signals in apoptosis-induced (etoposide treated) HeLa cells by immunoblotting and immunofluorescence.
siRNA transfection and photobleaching
PP2Cγ-specific Stealth RNA (Invitrogen; nucleotide number 351-376 or 642-667 of GenBank EMBL/DDBJ accession no. NM_177983) and the control RNA (Invitrogen; number 12935-300) were transfected using LipofectAMINE2000 (Invitrogen). Total cellular proteins were prepared 1–3 d after transfection, separated on an 8% SDS-polyacrylamide gel, and immunoblotted (Kimura and Cook, 2001) with mouse monoclonal antibody directed against PP2Cγ (1:10,000; Murray et al., 1999) or α-tubulin (1:1,000; Oncogene Research Products) as a control. Cells grown on coverslips were transfected with Stealth RNA and fixed for immunofluorescence using the mouse anti-PP2Cγ (1:30,000) and Cy3-conjugated anti–mouse IgG (1:500; Jackson ImmunoResearch Laboratories).
For photobleaching studies, HeLa cells expressing GFP-H2A, H2B-GFP, or H1c-GFP (Misteli et al., 2000) grown on glass-bottom dishes (Mat-Tek) were transfected with Stealth RNA. 3 d later, the dish was set on an inverted microscope (LSM510 META; Carl Zeiss MicroImaging, Inc.) in an air chamber at 37°C, and the mobility was analyzed by photobleaching using the inverted microscope with a plan-Neofluar 40× NA 1.3 objective. For H2A and H2B, five confocal images were collected (512 × 512 pixels, zoom 3, maximum scan speed, pinhole 3.7 airy unit, LP505 emission filter, and 0.3% transmission of 458-nm Ar laser with 75% output power), one half of a nucleus was bleached using 100% transmission of 458 and 488 nm (eight iterations), and images were collected using the original setting every 5 min. For H1c, five images were collected (256 × 256 pixels, zoom 8, and scan speed 12), a 2-μm spot was bleached using 100% transmission of 458 and 488 nm (eight iterations), and images were collected every 5 s (the graph in Fig. 7 E shows the points of every 10 s for ease of comparison). The fluorescence intensity of the bleached area was measured using MetaMorph software (Molecular Devices). After subtracting the background, the intensity was normalized to the initial intensity before bleaching.
DT40 cells
PP2Cγ-deficient DT40 cells were established using standard methods (Fig. S3; Fukagawa et al., 2004) and grown at 37°C. To measure the cell density, cells were mixed with trypan blue solution (Invitrogen), and the number of live cells excluding the dye was counted. To determine the sensitivity to caffeine and irradiation, serially diluted cells were plated in methylcellulose plates with or without 1 mM caffeine (Sigma-Aldrich) and irradiated using a Gammacell 40 Exactor (Nordion). Colonies were counted 10–12 d after plating. For immunoblotting (Fig. 8 D), 4 × 105 cells/ml were irradiated, and calyculin A (Sigma-Aldrich) was immediately added (final concentration of 10 ng/ml). A 1-ml aliquot was taken at each time point, and cells were collected (600 g for 2 min) and lysed in 100 μl of 2× SDS gel loading buffer.
Online supplemental material
Fig. S1 shows that ATP is not required for GFP-H2A incorporation into chromatin in permeabilized cells assisted by PP2Cγ or Nap1. Fig. S2 shows that PP2Cγ has weak de novo nucleosome assembly activity. Fig. S3 shows evidence for the generation of PP2Cγ knockout DT40 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200608001/DC1.
Supplementary Material
Acknowledgments
We thank D.T. Brown, Y. Ishimi, T. Kanda, P.Y. Perche, H. Tabara, C. Vourc'h, and G. Wahl for materials, and Y. Agata, T. Ikura, H. Kurumizaka, T. Misteli, and S. Tashiro for valuable discussion and comments on the manuscript.
This work was prepared, in part, at the Radiation Biology Center and the Radioisotope Research Center (Kyoto University). This work was supported by Grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Special Coordination Funds for Promoting Science and Technology from the MEXT of Japan. E. Allemand and A.R. Krainer acknowledge support from the National Institutes of Health grant GM42699.
Abbreviations used in this paper: AUT, acid-urea-Triton; FACT, facilitating chromatin transcription; Nap, nucleosome assembly protein; PB, physiological buffer; PP, protein phosphatase.
References
- Abraham, R.T. 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst.). 3:883–887. [DOI] [PubMed] [Google Scholar]
- Batsche, E., M. Yaniv, and C. Muchardt. 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 13:22–29. [DOI] [PubMed] [Google Scholar]
- Barman, H.K., Y. Takami, T. Ono, H. Nishijima, F. Sanematsu, K. Shibahara, and T. Nakayama. 2006. Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem. Biophys. Res. Commun. 345:1547–1557. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya, R., and D. Reinberg. 2004. Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev. 14:139–146. [DOI] [PubMed] [Google Scholar]
- Benson, L.J., Y. Gu, T. Yakovleva, K. Tong, C. Barrows, C.L. Strack, R.G. Cook, C.A. Mizzen, and A.T. Annunziato. 2006. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J. Biol. Chem. 281:9287–9296. [DOI] [PubMed] [Google Scholar]
- Bruno, M., A. Flaus, C. Stockdale, C. Rencurel, H. Ferreira, and T. Owen-Hughes. 2003. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell. 12:1599–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, L., S.S. Loranger, C. Mizzen, S.G. Ernst, C.D. Allis, and A.T. Annunziato. 1997. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry. 36:469–480. [DOI] [PubMed] [Google Scholar]
- Cheung, W.L., K. Ajiro, K. Samejima, M. Kloc, P. Cheung, C.A. Mizzen, A. Beeser, L.D. Etkin, J. Chernoff, W.C. Earnshaw, and C.D. Allis. 2003. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 113:507–517. [DOI] [PubMed] [Google Scholar]
- Chowdhury, D., M.C. Keogh, H. Ishii, C.L. Peterson, S. Buratowski, and J. Lieberman. 2005. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell. 20:801–809. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., R.M. Lebovits, and R.G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo, O., C.D. Allis, and A. Nussenzweig. 2004. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 199:1671–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14:165–173. [DOI] [PubMed] [Google Scholar]
- Fukagawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki, Y. Takami, T. Nakayama, and M. Oshimura. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6:784–791. [DOI] [PubMed] [Google Scholar]
- Hake, S.B., B.A. Garcia, E.M. Duncan, M. Kauer, G. Dellaire, J. Shabanowitz, D.P. Bazett-Jones, C.D. Allis, and D.F. Hunt. 2006. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 281:559–568. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and K. Ahmad. 2005. Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 21:133–153. [DOI] [PubMed] [Google Scholar]
- Ishimi, Y., J. Hirosumi, W. Sato, K. Sugasawa, S. Yokota, F. Hanaoka, and M. Yamada. 1984. Purification and initial characterization of a protein which facilitates assembly of nucleosome-like structure from mammalian cells. Eur. J. Biochem. 142:431–439. [DOI] [PubMed] [Google Scholar]
- Jackson, V. 1990. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 29:719–731. [DOI] [PubMed] [Google Scholar]
- Jackson, D.A., and P.R. Cook. 1985. A general method for preparing chromatin containing intact DNA. EMBO J. 4:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S., W. Brooks, and V. Jackson. 1994. Dynamics of the interactions of histones H2A,H2B and H3,H4 with torsionally stressed DNA. Biochemistry. 33:5392–5403. [DOI] [PubMed] [Google Scholar]
- Kanda, T., K.F. Sullivan, and G.M. Wahl. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385. [DOI] [PubMed] [Google Scholar]
- Kaufmann, W.K., T.P. Heffernan, L.M. Beaulieu, S. Doherty, A.R. Frank, Y. Zhou, M.F. Bryant, T. Zhou, D.D. Luche, N. Nikolaishvili-Feinberg, et al. 2003. Caffeine and human DNA metabolism: the magic and the mystery. Mutat. Res. 532:85–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh, M.C., J.A. Kim, M. Downey, J. Fillingham, D. Chowdhury, J.C. Harrison, M. Onishi, N. Datta, S. Galicia, A. Emili, et al. 2005. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 439:497–501. [DOI] [PubMed] [Google Scholar]
- Kimura, H. 2005. Histone dynamics in living cells revealed by photobleaching. DNA Repair (Amst.). 4:939–950. [DOI] [PubMed] [Google Scholar]
- Kimura, H., and P.R. Cook. 2001. Kinetics of core histones in living cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, K., N. Nozaki, M. Saijo, A. Kikuchi, M. Ui, and T. Enomoto. 1994. Identification of the nature of modification that causes the shift of DNA topoisomerase II beta to apparent higher molecular weight forms in the M phase. J. Biol. Chem. 269:24523–24526. [PubMed] [Google Scholar]
- Kireeva, M.L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V.M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell. 9:541–552. [DOI] [PubMed] [Google Scholar]
- Kobor, M.S., S. Venkatasubrahmanyam, M.D. Meneghini, J.W. Gin, J.L. Jennings, A.J. Link, H.D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., K. Baetz, M.C. Keogh, N. Datta, C. Sawa, T.C. Kwok, N.J. Thompson, M.G. Davey, J. Pootoolal, T.R. Hughes, et al. 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA. 101:13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, T., L. Florens, W.H. Macdonald, S.K. Swanson, R.L. Glaser, J.R. Yates III, S.M. Abmayr, M.P. Washburn, and J.L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 306:2084–2087. [DOI] [PubMed] [Google Scholar]
- Leroy, C., S.E. Lee, M.B. Vaze, F. Ochsenbein, R. Guerois, J.E. Haber, and M.C. Marsolier-Kergoat. 2003. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell. 11:827–835. [DOI] [PubMed] [Google Scholar]
- Louters, L., and R. Chalkley. 1985. Exchange of histones H1, H2A, and H2B in vivo. Biochemistry. 24:3080–3085. [DOI] [PubMed] [Google Scholar]
- Loyola, A., and G. Almouzni. 2004. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta. 1677:3–11. [DOI] [PubMed] [Google Scholar]
- Lu, X., B. Nannenga, and L.A. Donehower. 2005. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 19:1162–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger, K., A.W. Mader, R.K. Richmond, D.F. Sargent, and T.J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 389:251–260. [DOI] [PubMed] [Google Scholar]
- Ma, X.J., J. Wu, B.A. Altheim, M.C. Schultz, and M. Grunstein. 1998. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. USA. 95:6693–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison, C., D. Bailly, A.H.F.M. Peters, J.P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329–334. [DOI] [PubMed] [Google Scholar]
- Manser, T., T. Thacher, and M. Rechsteiner. 1980. Arginine-rich histones do not exchange between human and mouse chromosomes in hybrid-cells. Cell. 19:993–1003. [DOI] [PubMed] [Google Scholar]
- Meshorer, E., D. Yellajoshula, E. George, P.J. Scambler, D.T. Brown, and T. Misteli. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 10:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., and D.L. Spector. 1996. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol. Cell. 7:1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D.T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W.H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 303:343–348. [DOI] [PubMed] [Google Scholar]
- Murray, M.V., R. Kobayashi, and A.R. Krainer. 1999. The type 2C Ser/Thr phosphatase PP2Cγ is a pre-mRNA splicing factor. Genes Dev. 13:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov, I.B., A.N. Smirnova, R.I. Krutilina, M.P. Svetlova, L.V. Solovjeva, A.A. Nikiforov, S.L. Oei, I.A. Zalenskaya, P.M. Yau, E.M. Bradbury, and N.V. Tomilin. 2003. Dephosphorylation of histone gamma-H2AX during repair of DNA double-strand breaks in mammalian cells and its inhibition by calyculin A. Radiat. Res. 160:309–317. [DOI] [PubMed] [Google Scholar]
- Ohkuni, K., K. Shirahige, and A. Kikuchi. 2003. Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 306:5–9. [DOI] [PubMed] [Google Scholar]
- Park, Y.J., J.V. Chodaparambil, Y. Bao, S.J. McBryant, and K. Luger. 2005. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280:1817–1825. [DOI] [PubMed] [Google Scholar]
- Perche, P.Y., C. Vourc'h, L. Konecny, C. Souchier, M. Robert-Nicoud, S. Dimitrov, and S. Khochbin. 2000. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10:1531–1534. [DOI] [PubMed] [Google Scholar]
- Pilch, D.R., C. Redon, O.A. Sdelnikova, and W.M. Bonner. 2004. Two-dimensional gel analysis of histones and other H2AX-related methods. Methods Enzymol. 375:76–88. [DOI] [PubMed] [Google Scholar]
- Pombo, A., D.A. Jackson, M. Hollinshead, Z. Wang, R.G. Roeder, and P.R. Cook. 1999. Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 18:2241–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, P., D. Munroe, D. Prawitt, L.L. Chu, E. Bric, J. Kim, L.H. Reid, C. Davies, H. Nakagama, R. Loebbert, et al. 1997. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics. 44:253–265. [DOI] [PubMed] [Google Scholar]
- Rogakou, E.P., C. Boon, C. Redon, and W.M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., E.F. Fritsch, and J. Sambrook. 1989. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 545 pp.
- Simon, R.H., and G. Felsenfeld. 1979. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 6:689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 116:51–61. [DOI] [PubMed] [Google Scholar]
- Thiriet, C., and J.J. Hayes. 2005. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 19:677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis, S.M., and M.J. Welsh. 1997. PP2Cγ: a human protein phosphatase with a unique acidic domain. FEBS Lett. 412:415–419. [DOI] [PubMed] [Google Scholar]
- Turner, B.M. 2002. Cellular memory and the histone code. Cell. 111:285–291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.