Abstract
This study demonstrates that the eukaryotic translation initiation factor eIF4E is a critical node in an RNA regulon that impacts nearly every stage of cell cycle progression. Specifically, eIF4E coordinately promotes the messenger RNA (mRNA) export of several genes involved in the cell cycle. A common feature of these mRNAs is a structurally conserved, ∼50-nucleotide element in the 3′ untranslated region denoted as an eIF4E sensitivity element. This element is sufficient for localization of capped mRNAs to eIF4E nuclear bodies, formation of eIF4E-specific ribonucleoproteins in the nucleus, and eIF4E-dependent mRNA export. The roles of eIF4E in translation and mRNA export are distinct, as they rely on different mRNA elements. Furthermore, eIF4E-dependent mRNA export is independent of ongoing RNA or protein synthesis. Unlike the NXF1-mediated export of bulk mRNAs, eIF4E-dependent mRNA export is CRM1 dependent. Finally, the growth-suppressive promyelocytic leukemia protein (PML) inhibits this RNA regulon. These data provide novel perspectives into the proliferative and oncogenic properties of eIF4E.
Introduction
RNA regulons have been proposed as a means by which eukaryotic cells coordinate gene expression (Keene and Tenenbaum, 2002; Keene and Lager, 2005). In contrast to prokaryotes, in which the coordinated regulation of genes is achieved by genomic organization, eukaryotes coordinate the regulation of subsets of mRNAs involved in the same biological processes at the posttranscriptional level by manipulating compositions and activities of discrete subsets of RNPs. It has been postulated that related RNA sequences termed untranslated sequence elements for regulation (USER) codes, which are similar to zip codes for RNA localization, are used for specific association with a variety of regulatory proteins involved in different levels of posttranscriptional regulation (Keene and Lager, 2005). mRNA nuclear export is one level of control that could be coordinated in this way. Initially, mRNA export was thought to be a general process by which all mRNAs were transported from the nucleus to the cytoplasm irregardless of sequence-specific features. More recent findings indicate that mRNA export can be coordinated with other events in RNA metabolism, particularly transcription and splicing, and, thus, that nuclear history of transcripts can modulate the cytoplasmic fate of targeted mRNAs (Hieronymus and Silver, 2003, 2004; Hieronymus et al., 2004; Keene and Lager, 2005). This way, nuclear export can be coordinated through compartmentalization via mRNP organization, coupling the coordinated export of functional classes of mRNAs with their functions in biological processes such as proliferation, differentiation, and development.
Studies with eukaryotic translation initiation factor eIF4E provide an example of a factor that differentially affects the expression of a subset of mRNAs. Although it associates with all transcripts through the common 5′ methyl-7-guanosine (m7G) cap structure (Sonenberg and Gingras, 1998), many groups showed that eIF4E overexpression does not lead to global increases in protein expression (Rousseau et al., 1996; Clemens and Bommer, 1999). In the cytoplasm, mRNAs deemed eIF4E sensitive have their protein levels modulated by eIF4E more so than other mRNAs. This sensitivity is attributed to the complexity of the 5′ untranslated regions (UTRs) in these transcripts (Sonenberg and Gingras, 1998). Up to 68% of eIF4E is found in the nucleus in a broad variety of species ranging from yeast to humans (Cohen et al., 2001; Iborra et al., 2001). Here, eIF4E overexpression leads to the increased export of cyclin D1 but not glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (Rousseau et al., 1996; Topisirovic et al., 2002; Culjkovic et al., 2005). Specific association of eIF4E with cyclin D1 mRNA in the nucleus requires the m7G cap and a small element in its 3′ UTR referred to as an eIF4E sensitivity element (4E-SE; Culjkovic et al., 2005).
Overexpression of eIF4E is correlated with oncogenic transformation in tissue culture, cancers in animal models, and poor prognosis in several human cancers (Sonenberg and Gingras, 1998; Topisirovic et al., 2003b). Several lines of evidence suggest that the mRNA export function of eIF4E contributes to its oncogenic potential. For instance, cyclin D1 mRNA export is up-regulated in specific subtypes of human leukemia (Topisirovic et al., 2003b). These specimens contain unusually high levels of eIF4E, the vast majority of which is located in the nucleus (Topisirovic et al., 2003b). Also, inhibitors of eIF4E-dependent mRNA export, the promyelocytic leukemia protein (PML), and proline-rich homeodomain (PRH) bind eIF4E in the nucleus, inhibit eIF4E-dependent mRNA export, and inhibit eIF4E-mediated oncogenic transformation (Cohen et al., 2001; Topisirovic et al., 2003a; Culjkovic et al., 2005). Furthermore, mutagenesis studies link the activity of eIF4E in mRNA export to its ability to oncogenically transform cells (Cohen et al., 2001; Culjkovic et al., 2005).
Although cyclin D1 plays a key role in the cell cycle that links eIF4E's proliferative properties and its mRNA export function, it is possible that eIF4E coordinately alters the expression of some other growth-promoting mRNAs as well to drive its proliferative potential. This study shows that several mRNAs involved in cell cycle progression are also targets of eIF4E-dependent mRNA export and that the subsets of mRNAs regulated at the level of eIF4E-dependent mRNA export are distinct from those that are preferentially translated in the cytoplasm. We identified an underlying USER code for the export of eIF4E-sensitive transcripts. This code is required for the subnuclear distribution of these RNAs as well as for the formation of relevant eIF4E RNPs. Interestingly, the 4E-SE USER code is a structurally conserved element rather than a sequence-based one. eIF4E-dependent mRNA export can be decoupled from translation. Finally, eIF4E-dependent mRNA export occurs via an alternative mRNA export pathway than bulk mRNA. These results provide the basis for a novel paradigm of eIF4E-mediated tumorigenesis.
Results
eIF4E alters the mRNA transport of a wide variety of transcripts
eIF4E-dependent mRNA export is potentially a broadly based mechanism by which eIF4E controls gene expression and, thereby, modulates growth and proliferation. We sought to determine whether mRNAs other than cyclin D1 might be regulated in an eIF4E-dependent manner. Using nuclear lysates, we isolated mRNAs associated with endogenous eIF4E via immunoprecipitation (IP) or with recombinant eIF4E using a GST pull down–based method (the SNAAP method of Trifillis et al. [1999]) and identified these by differential display. Given that many of the identified genes are involved in cell cycle progression, eIF4E-immunoprecipitated fractions were also tested for other genes known to be involved in these processes as well as for known growth-inhibitory mRNAs (Table I). All target identification was confirmed by eIF4E IP and quantitative or semiquantitative RT-PCR analysis (Fig. 1 A and Fig. S1 A, available at http://www.jcb.org/cgi/conent/full/jcb.200607020/DC1). Importantly, the list provided in Table I is not intended to be totally inclusive but rather to represent a sampling of the target mRNA population because results from differential display data suggest that hundreds of mRNAs are likely regulated in this manner; in this study, we identified only a subset (Fig. S2).
Table I.
List of mRNAs that associate (or do not) with nuclear eIF4E
| Target RNA | Function/growth-promoting properties | Translationally sensitive to eIF4E | Data for interaction | |||
|---|---|---|---|---|---|---|
| RNAs associated with the nuclear fraction of eIF4E | ||||||
| Cyclin D1 (gi: 77628152) |
G1/S cell cycle progression (Liang and Slingerland, 2003) |
No (Rousseau et al., 1996) |
Fig. 1 A | |||
| Cyclin E1 (gi: 17318558) |
G1/S cell cycle progression (Liang and Slingerland, 2003) |
ND | Fig. 1 A | |||
| Cyclin A2 (gi: 16950653) |
S/G2/M cell cycle progression (Liang and Slingerland, 2003) |
ND | Fig. S1 | |||
| Cyclin B1 (gi: 34304372) |
G2/M cell cycle progression (Liang and Slingerland, 2003) |
Yes (Cao and Richter, 2002) |
Fig. S1 | |||
| ODC (gi: 4505488) |
Polyamine synthesis/tumor promoting (Pegg, 2006) |
Yes (Clemens and Bommer, 1999) |
Fig. 1 A | |||
| Pim-1 (gi: 31543400) |
S/T kinase (Bachmann and Moroy, 2005) |
Yes (Clemens and Bommer, 1999) |
Fig. 1 A | |||
| Mdm2 (gi: 46488903) |
Survival/apoptotic rescue (Liang and Slingerland, 2003) |
ND | Fig. S1 | |||
| c-Myc (gi: 71774082) |
Facilitates G1/S progression and transcriptionally up-regulates eIF4E (Liang and Slingerland, 2003; Schmidt, 2004) |
Yes (Clemens and Bommer, 1999) |
Fig. 1 A | |||
| Nibrin/NBS1 (gi: 67189763) |
DNA repair/Akt activation/promotes growth (Chen et al., 2005) |
ND | Fig. S1 | |||
| Fbox1 (gi: 16306583) |
Promotes cell cycle progression (Liang and Slingerland, 2003) |
ND | Fig. S1 | |||
| CGGbp1 (gi: 56550052) |
Influences FMR1 expression (Naumann et al., 2004) |
ND | Fig. S1 | |||
| P54nrb/NONO.1 (gi: 34932413) |
RNA-binding protein/promotes survival (Stier et al., 2005) |
ND | Fig. S1 | |||
| Selenoprotein S (gi: 45439347) |
Glucose-regulated ER protein (Gao et al., 2004) |
ND | Fig. S1 | |||
| RNAs not associated with the nuclear fraction of eIF4E | ||||||
| GAPDH (gi: 83641890) |
Housekeeping/apoptotic | No (Rousseau et al., 1996; Clemens and Bommer, 1999) |
Fig. 1 A | |||
| VEGF (gi: 71051577) |
Mitogen/angiogenesis/tumor invasion (Roy et al., 2006) |
Yes (Clemens and Bommer, 1999) |
Fig. S1 | |||
| p53 (gi: 8400737) |
Proapoptotic/reduces eIF4E transcription (Zhu et al., 2005) |
No (Clemens and Bommer, 1999) |
Fig. 1 A | |||
| β-Actin (gi: 5016088) |
Cytoskeletal | No (Rousseau et al., 1996) |
Fig. S1 | |||
| α-Tubulin (gi: 57013275) |
Cytoskeletal | No | Fig. S1 | |||
| eIF4E (gi: 54873625) |
Translation and mRNA export promotes growth and survival (Strudwick and Borden, 2002) |
No (Clemens and Bommer, 1999; Strudwick and Borden, 2002) |
Fig. 1 A | |||
| PML (gi: 67089161) |
Proapoptotic/G1 arrest (Borden, 2002) |
No (Strudwick and Borden, 2002) |
Fig. 1 A | |||
| α-Globin (gi: 14456711) |
Housekeeping | No | Fig. S1 | |||
| c-ebpa (gi: 28872793) |
Arrests proliferation (Wang et al., 2001) |
ND | Fig. S1 | |||
Figure 1.
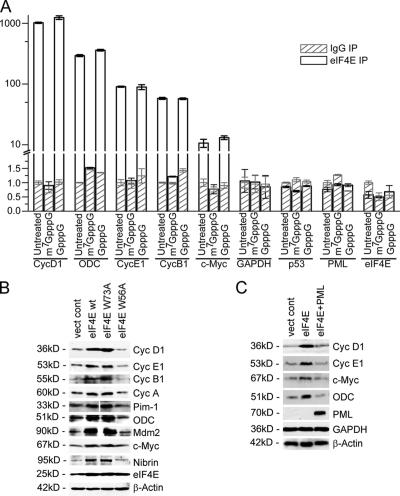
Enhanced mRNA export corresponds to elevated protein levels of eIF4E-sensitive targets. (A) Relative fold difference of mRNAs bound to nuclear eIF4E. mRNAs were immunoprecipitated from untreated nuclear lysates or those treated with 50 μM m7GpppG or GpppG. Values represent relative fold ± SD (error bars; normalized against untreated immunoprecipitated IgG, which was set to 1). Calculations of fold were performed using the relative standard curve method (User Bulletin #2 ABI Prism 7700; Applied Biosystems). Relative amounts of the target mRNA = 10[C (t)-b]/a were determined for each PCR reaction. Average values ± SD were calculated for each set of triplicates. Average values obtained for the IPs (i.e., average relative amount of immunoprecipitated target mRNA) were divided by values obtained for 5% nuclear input (i.e., average relative amount of target mRNA present in the 5% of the amount of nuclear extract used for IP). Obtained values ± SD (i.e., average IP/average 5% nuclear) were normalized by setting the untreated IgG IP value to 1. (B and C) eIF4E-enhanced mRNA transport leads to up-regulated protein levels of corresponding mRNAs. Total cell lysates from U937 (B) or NIH3T3 (C) cells transfected as indicated were analyzed for protein content by Western methods. Note that in C, where human PML was overexpressed, the 5E10 mAb PML antibody only recognizes the human PML, not the endogenous mouse PML.
Many of the mRNAs that physically associate with the nuclear fraction of eIF4E code for gene products that act in cell cycle progression and survival, which is consistent with the physiological functions associated with eIF4E (Table I). Importantly, eIF4E does not bind all mRNAs tested (Table I). For instance, eIF4E does not associate with the mRNAs corresponding to negative regulators of growth such as PML or p53 or housekeeping genes such as GAPDH, β-actin, or α-tubulin. Also, this specificity is not a simple reflection of the sensitivity of mRNAs for regulation at the translational level, as mRNAs sensitive only at the translation level (such as VEGF; Clemens and Bommer, 1999) are not associated with the nuclear fraction of eIF4E (Table I). It is important to note that mRNAs that were not found in the eIF4E-immunoprecipitated fractions were readily detected in our nuclear lysates (Table II and Fig. S1 A). Note that that the estimated efficiency of IP with anti-eIF4E mAb is up to 80%.
Table II.
Cytoplasmic/nuclear ratio of different mRNAs in U937 cells overexpressing eIF4E wild type or mutants
| mRNA | MSCV vector control | 4E wild type | W73A | W56A |
|---|---|---|---|---|
| Cyclin D1 | 1 ± 0.035 | 11.995 ± 0.860 | 11.450 ± 860 | 1.110 ± 0.036 |
| Cyclin E1 | 1 ± 0.022 | 3.442 ± 0.05 | 3.172 ± 0.208 | 1.200 ± 0.015 |
| Cyclin A2 | 1 ± 0.044 | 5.472 ± 0.580 | 7.736 ± 0.540 | 1.002 ± 0.058 |
| Cyclin B1 | 1 ± 0.108 | 4.720 ± 0.750 | 4.073 ± 0.434 | 1.475 ± 0.122 |
| ODC | 1 ± 0.010 | 6.847 ± 0.373 | 7.138 ± 0.852 | 1.272 ± 0.018 |
| Pim-1 | 1 ± 0.051 | 3.435 ± 0.194 | 3.391 ± 0.251 | 1.029 ± 0.029 |
| Mdm2 | 1 ± 0.325 | 15.698 ± 0.160 | 15.097 ± 0.793 | 1.379 ± 0.014 |
| c-Myc | 1 ± 0.033 | 2.980 ± 0.233 | 2.857 ± 0.226 | 0.925 ± 0.042 |
| Nibrin | 1 ± 0.030 | 4.728 ± 0.145 | 4.958 ± 0.230 | 1.226 ± 0.024 |
| F-box 1 | 1 ± 0.069 | 11.202 ± 0.866 | 10.713 ± 0.633 | 1.363 ± 0.062 |
| Selenoprotein S | 1 ± 0.072 | 14.520 ± 1.164 | 11.839 ± 0.257 | 1.193 ± 0.234 |
| VEGF | 1 ± 0.111 | 0.835 ± 0.063 | 0.980 ± 0.261 | 1.387 ± 0.022 |
| β-Actin | 1 ± 0.173 | 1.020 ± 0.238 | 1.220 ± 0.203 | 1.313 ± 0.180 |
| p53 | 1 ± 0.016 | 0.892 ± 0.006 | 1.392 ± 0.230 | 0.994 ± 0.008 |
| α-Globin | 1 ± 0.379 | 0.861 ± 0.237 | 1.265 ± 0.232 | 1.275 ± 0.346 |
Cytoplasmic/nuclear values represent relative fold ± SD normalized to vector control (MSCV), which was set to 1. Average values ± SD were calculated for each set of triplicates. Average values of all analyzed mRNAs obtained for each fraction of each sample were divided by GAPDH mRNA values obtained for the same fraction/sample. After dividing cytoplasmic with nuclear values of each sample, obtained cytoplasmic/nuclear values ± SD were normalized by setting the MSCV vector control cytoplasmic/nuclear value to 1.
Because eIF4E associates with the m7G cap of mRNAs, we examined whether this was required for the association of eIF4E with mRNAs in the nuclear fraction (Fig. 1 A). eIF4E was immunoprecipitated from the nuclear fraction, and mRNAs were treated with excess m7GpppG or an analogue that does not bind eIF4E (GpppG). All mRNAs tested associate with eIF4E in a cap-dependent manner (i.e., m7GpppG competes for binding, whereas GpppG does not). These data indicate that the association of eIF4E with mRNAs in the nucleus is m7G cap dependent.
Physical association of eIF4E with mRNAs is correlated with enhanced mRNA export
To test whether there is a correlation between the ability of eIF4E to associate with mRNAs in the nuclear fraction and the ability of eIF4E overexpression to enhance eIF4E-dependent mRNA export, the subcellular distribution of identified mRNAs as a function of eIF4E overexpression was analyzed (Table II and Fig. S1 B). U937 and NIH3T3 cells overexpressing eIF4E or appropriate mutants were fractionated, and mRNAs levels were monitored by real-time PCR or Northern analysis. eIF4E overexpression increases the amount of eIF4E-sensitive mRNAs in the cytoplasmic fraction versus vector controls (Table II and Fig. S1 B). Conversely, transcripts that did not associate with eIF4E in the nuclear fraction did not have their export altered by eIF4E overexpression (Table II and Fig. S1 B). As expected, the subcellular distribution of β-actin, GAPDH, U6snRNA, and tRNALys were unaffected (Table II and Fig. S1 B). There is no alteration in total mRNA levels (Fig. S1 C). Consistently, when eIF4E could not bind these mRNAs because of a mutation in its cap-binding site (W56A), the subcellular distribution of these mRNAs was not altered (Table II and Fig. S1 B). Furthermore, the dorsal surface mutant W73A, which does not act in translation but promotes cyclin D1 mRNA export (Sonenberg and Gingras, 1998; Topisirovic et al., 2002), also promotes the export of other eIF4E-sensitive mRNAs (Table II and Fig. S1 B). Thus, it is likely that all sensitive mRNAs will require the m7G cap-binding activity of eIF4E but not W73 on the dorsal surface for their interaction with eIF4E in the nucleus. Importantly, a circular dichroism study indicates that both W73A and W56A mutants have structures indistinguishable from wild-type eIF4E (Kentsis et al., 2001).
One of the consequences of the eIF4E-dependent promotion of mRNA export is increased availability of these mRNAs to the translation machinery, leading to increased protein levels. Thus, we examined whether protein levels for a subset of identified genes are elevated by eIF4E. Consistent with enhanced mRNA export, the overexpression of wild-type eIF4E or the W73A mutant leads to increased protein levels of a subset of examined genes (Fig. 1 B and Fig. S1 D), whereas there is no increase in protein levels when the cap-binding mutant (W56A) is overexpressed. Importantly, wild-type eIF4E and the W73A and W56A mutants were expressed to similar levels (about threefold overexpression) for all experiments (Fig. 1 B and Fig. S1 D).
To determine whether these mRNAs are regulated through the same mechanism, it was important to examine the effect of PML. Our previous studies showed that PML colocalizes and coimmunoprecipitates with nuclear eIF4E and that this interaction is important for the ability of PML to inhibit eIF4E-dependent cyclin D1 mRNA export and eIF4E-mediated transformation (Cohen et al., 2001; Topisirovic et al., 2002). Thus, we examined whether PML inhibits the export of a range of target mRNAs. In this way, we could determine whether PML was a general inhibitor of eIF4E-dependent export or whether these activities were limited to cyclin D1 mRNA. We observed decreased mRNA export (Fig. S1 B) and reduced protein levels of ornithine decarboxylase (ODC), c-myc, cyclin D1, and cyclin E1 (Fig. 1 C and Fig. S1 D) in cells overexpressing PML. Also, PML did not reduce levels of eIF4E, β-actin, or GAPDH proteins (Fig. 1 C and Fig. S1 D), and there was no alteration in total mRNA levels for any of these transcripts when PML was overexpressed (Fig. S1 C). Thus, PML acts as an inhibitor of eIF4E-dependent mRNA export, not just as an inhibitor of cyclin D1 mRNA export.
In summary, the physical association of mRNAs with the nuclear fraction of eIF4E is strongly correlated with their enhanced nuclear export. In the cytoplasm, these mRNAs may (i.e., ODC) or may not (i.e., cyclin D1) be a subject of modulation by eIF4E at the level of translation. Thus, eIF4E-mediated modulation at the nuclear level neither precludes nor necessitates such modulation at the cytoplasmic level.
The RNA USER code for eIF4E-dependent mRNA export
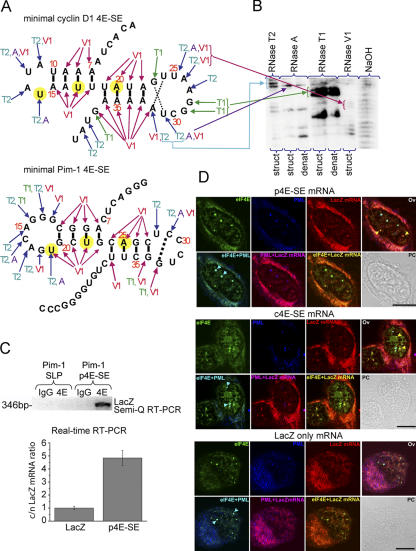
Because we previously identified a 100-nucleotide 4E-SE in the 3′ UTR of cyclin D1, which sensitizes cyclin D1 and corresponding chimeric lacZ constructs to regulation by eIF4E at the mRNA export level (Culjkovic et al., 2005), we performed an extensive bioinformatics analysis to identify 4E-SE–like elements in the other target RNAs identified in Table I. Sequence analysis indicated that the 4E-SE was well conserved in cyclin D1 transcript (from birds to humans; Culjkovic et al., 2005), but a comparison of cyclin D1 with the other eIF4E-sensitive transcripts identified here failed to reveal any shared sequence homology. Therefore, we examined the possibility that the 4E-SE is a structurally conserved element. Using the PatSearch algorithm (Grillo et al., 2003), we found that the cyclin D1 4E-SE had a predicted pattern of stem loop pairs (SLPs). Using this same strategy, we found that Pim-1 also contained a similar putative element in its 3′ UTR. As for the cyclin D1 4E-SE mapping (Culjkovic et al., 2005), we made the corresponding constructs of Pim-1–lacZ fusions and showed that one of these elements (Pim-1–p4E-SE; Fig. 2 C) was a functional 4E-SE (see next paragraph). Note that a construct derived from a similar element in Pim-1's 3′ UTR (SLP) did not associate with eIF4E (Fig. 2 C).
Figure 2.
A common secondary structure for the 4E-SE that acts as a zip code for eIF4E nuclear bodies. (A) Secondary structure for cyclin D1 4E-SE (c4E-SE) and Pim-1 4E-SE (p4E-SE) as determined by RNase mapping experiments. Conserved set of A and U nucleotides (UX2UX2A) are highlighted in yellow. (B) A sample gel. (C, top) Only one of the two predicted SLPs from Pim-1 3′ UTR (p4E-SE) immunoprecipitates with eIF4E. (bottom) eIF4E promotes the export of lacZ mRNA that contains minimal p4E-SE. Cytoplasmic/nuclear (c/n) values represent relative fold ± SD (error bars) normalized to the lacZ control, which was set to 1. Calculations were performed as described in Table II. (D) Colocalization of lacZ–p4E-SE, lacZ–c4E-SE, or lacZ transcripts with PML and eIF4E protein was examined in U2OS cells transfected with lacZ/lacZ–4E-SE. LacZ mRNA was detected using in situ hybridization with a biotin-labeled nick-translated probe to lacZ (red). Cells were then immunostained using an eIF4E mAb conjugated directly to FITC (green) and PML mAb 5E10 (blue). Importantly, lacZ mRNAs containing the 4E-SE from either cyclin D1 or Pim-1 colocalize to eIF4E nuclear bodies (arrowheads). As we have shown previously for endogenous cyclin D1 mRNA, there are two populations of eIF4E nuclear bodies: those that colocalize with lacZ mRNA and those that colocalize with PML. Magnification was 100× and 3× (for lacZ and c4ESE) or 4× (p4ESE) digital zoom. Bars, 10 μM.
To best identify the common structural elements in the target mRNAs, we decided to compare the cyclin D1 4E-SE with the 4E-SE from one of the newly identified target mRNAs, Pim-1. We mapped the 4E-SE from cyclin D1 and the 4E-SE from Pim-1 to a minimal ∼50-nucleotide region (Fig. 2 A). These minimal domains, when fused to heterologous lacZ mRNA, immunoprecipitate with eIF4E and have their mRNA export promoted by eIF4E (Fig. 2 C; Culjkovic et al., 2005). Thus, these are functional 4E-SEs. In summary, although there was no sequence homology observed, both elements contain putative SLPs as predicted by PatSearch.
We used nuclease digestion methods to determine whether these two functional 4E-SEs had conserved secondary structural features such as the predicted stem loops. Importantly, these studies revealed that both elements fold into similar secondary structures. We refer to this element as an adjacent SLP (Fig. 2, A and B; and Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200607020/DC1). Consistently, biophysical analysis indicates that Pim-1 and cyclin D1 4E-SEs have similar biophysical properties. For instance, circular dichroism analysis of thermal melting curves using purified RNA oligomers for cyclin D1 and Pim-1 4E-SEs revealed multiphase behavior consistent with the presence of multiple structural elements with different melting temperatures (unpublished data). Thus, both Pim-1 and cyclin D1 4E-SEs have similar secondary structures, which is consist with two adjacent stem loop elements.
An initial problem we encountered in these studies is that the presence of stem loop elements is common in the 3′ UTRs of cyclin D1 and Pim-1. In cyclin D1 alone, the PatSearch program (Grillo et al., 2003) predicts 10 potential SLPs, but our previous findings indicate that the only part of the cyclin D1 3′ UTR that can impart eIF4E sensitivity is the aforementioned 4E-SE (Culjkovic et al., 2005). Similarly, the Pim-1 3′ UTR contains two predicted adjacent SLPs, whereas only one is a functional 4E-SE (Fig. 2 C). Thus, we compared the secondary structures of Pim-1 and cyclin D1 4E-SEs to determine features that would enable us to distinguish functional 4E-SEs from other SLPs. Visual inspection of the secondary structures reveal the conservation of a set of A and U nucleotides (UX2UX2A, highlighted in Fig. 2 A). Importantly, these patterns of nucleotides were not found in any of the other SLPs found in cyclin D1 or Pim-1 3′ UTRs. Thus, these are features that can be used to distinguish functional 4E-SEs from other elements that have the potential to fold into similar secondary structures.
Further bioinformatics analyses showed that the predicted SLP structure with the conserved pattern of nucleotides is also present in other eIF4E-sensitive targets identified here (Table I, top). Importantly, none of the mRNAs that are not eIF4E sensitive contain SLPs with the conserved pattern of nucleotides found in the functional 4E-SEs. In summary, we have identified a structural motif consisting of two adjacent SLPs, which impart eIF4E sensitivity. Importantly, there exist in this motif sequence features of 4E-SEs that can be used to distinguish functional 4E-SEs from other paired stem loop structures.
The 4E-SE is sufficient for localization with eIF4E nuclear bodies
To assess whether the 4E-SE acted as an RNA zip code for eIF4E nuclear bodies, lacZ chimeric constructs with either Pim-1 or cyclin D1 4E-SE were expressed in U2OS cells. Both chimeric mRNAs colocalize with eIF4E nuclear bodies (Fig. 2 D). In the absence of the 4E-SE, no localization of lacZ transcripts to eIF4E nuclear bodies is observed (Fig. 2 D). Importantly, lacZ–4E-SE does not associate with eIF4E bodies that contain the negative regulator PML. This is consistent with our previous studies showing that there are two classes of eIF4E nuclear bodies: those that colocalize with PML and those that colocalize with endogenous cyclin D1 mRNA (Cohen et al., 2001; Topisirovic et al., 2002; Culjkovic et al., 2005). Thus, endogenous cyclin D1 mRNAs colocalize with eIF4E nuclear bodies that do not contain PML (Culjkovic et al., 2005). In this way, lacZ–4E-SE transcripts and endogenous mRNAs behave similarly.
These experiments demonstrate that the 4E-SE is sufficient to localize capped mRNAs into eIF4E nuclear bodies irrespective of the rest of the mRNA sequence. Moreover, the 4E-SE from Pim-1 and cyclin D1 are functionally equivalent in terms of localization activity. Thus, the 4E-SE provides an RNA zip code for localization to eIF4E nuclear bodies.
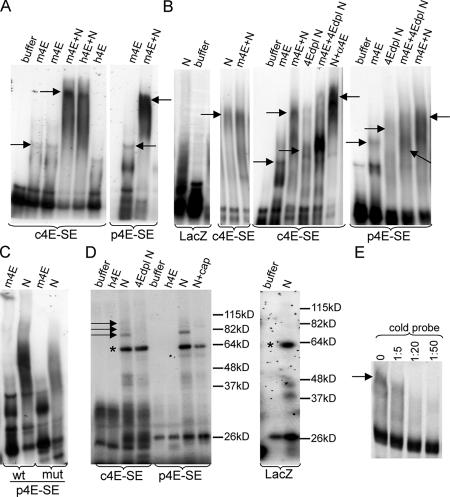
The 4E-SE makes eIF4E-dependent complexes
To establish whether the 4E-SE functions simply as a localization signal or whether it acts in the formation of eIF4E-dependent mRNPs, we performed electromobility shift assays (EMSAs). Studies were performed with both the lacZ–cyclin D1–4E-SE (c4E-SE) and the lacZ–Pim-1–4E-SE (p4E-SE) to ensure that the assembly of these complexes is dependent on the 4E-SE itself and not on features specific to either 4E-SE. RNA probes were 32P 3′ end labeled and m7G capped. The addition of either mouse eIF4E with a 6-kD solubility tag (m4E) or untagged human eIF4E (h4E) led to the formation of slower migrating species for both lacZ–4E-SE constructs (Fig. 3, A and B). Importantly, the addition of nuclear lysates led to the formation of substantially higher molecular weight complexes, indicating that proteins other than eIF4E are likely to be present. Complex sizes were approximately the same for both 4E-SE constructs. The addition of cold competitor 4E-SE RNAs led to a reduction in signal, which is consistent with the 4E-SE element competing for the labeled 4E-SE–containing transcripts (Fig. 3 E). The addition of nuclear lysates to lacZ transcripts lacking the 4E-SE did not lead to the formation of these complexes (Fig. 3 B and Fig. S3 B). Although the addition of purified eIF4E to the nuclear lysate supplemented with 4E-SE–containing transcripts led to a considerable increase in the amount of RNA undergoing the shift, it did not alter the position of the shift (Fig. 3 B).
Figure 3.
The 4E-SE is required for the formation of eIF4E-dependent complexes. (A and B) EMSA analysis indicates that lacZ transcripts, which contain either the cyclin D1 4E-SE (c4E-SE) or the Pim-1 4E-SE (p4E-SE), formed high molecular weight complexes in the presence of nuclear lysates (N). LacZ transcripts (control) without the 4E-SE did not form these complexes. The addition of purified mouse eIF4E with a 6-kD solubility tag (m4E) or untagged human eIF4E (h4E) causes partial shifts relative to shifts observed with nuclear lysate. With nuclear lysates immunodepleted of eIF4E (dpl N), gel shifts were not observed. Complexes could be supershifted by an anti-eIF4E antibody (N + α4E). Arrows indicate protein–RNA complexes. (C) Mutation of Pim-1 4E-SE (p4E-SE) reduces the efficacy of the gel shift. No molecular weight markers are shown, as A–C and E are native gels. (D) UV cross-linking experiments showed the formation of specific complexes in the 75–90-kD mass range (indicated by arrows). These complexes are specifically depleted in the presence of excess m7GpppG cap (cap) or if lysates are immunodepleted of eIF4E (dpl N). Asterisks indicate complexes that are cap and 4E-SE independent. (E) The addition of unlabeled p4E-SE ribooligonucleotide (cold probe) to the p4E-SE nuclear complexes indicates that this element can efficiently compete for complex formation. All transcripts were capped and 3′ end labeled.
To determine whether the 4E-SE complexes formed from nuclear lysates were dependent on eIF4E, EMSAs were performed with nuclear lysates depleted of eIF4E via IP. We estimated that lysates were at least 80% depleted of eIF4E (unpublished data). Lysates immunodepleted of eIF4E did not produce high molecular weight complexes (Fig. 3 B). The addition of purified tagged eIF4E to immunodepleted lysates led to a partial restoration of the complex, which could be expected because only eIF4E but not other factors that were depleted during the anti-eIF4E IP were reintroduced. Thus, eIF4E and associated factors are required for the formation of these RNPs. In addition, an antibody to eIF4E leads to a super shift of complexes formed from nuclear lysates (Fig. 3 B). Identical results are observed for lacZ–p4E-SE. Finally, a mutant that disrupts the first stem loop (G10C11G12 mutated to CAC; Fig. 2 A) in the p4E-SE is defective in complex formation (Fig. 3 C). Thus, the 4E-SE element forms complexes that are dependent on eIF4E and on the structure of the 4E-SE.
To further characterize these complexes, lacZ–4E-SE constructs were UV cross-linked followed by RNase digestion and SDS gel electrophoresis (Fig. 3 D). As for the EMSA studies, transcripts were m7G capped and 3′ end labeled, and the effects of addition of purified eIF4E or nuclear lysates to the size of cross-linked complexes was monitored. Because mRNAs were 3′ end labeled, binding of the cap only by purified eIF4E was not sufficient to protect the rest of the RNA from RNase digestion. The addition of the nuclear lysate leads to substantial shifts in molecular weight. Importantly, the lacZ–c4E-SE and the lacZ–p4E-SE form complexes similar in size. Three discrete species between 75 and 90 kD are observed (Fig. 3 D, arrows). The same complexes are absent in eIF4E-depleted nuclear lysate, indicating that these require eIF4E to form. Consistently, treatment of the nuclear lysate with the m7GpppG cap analogue (nuclear lysates + cap) also disrupts 75–90-kD range complexes. These species are absent from the lacZ controls, which lack the 4E-SE. A lower band at ∼64 kD is present in all of the experiments, likely indicating the formation of some general RNP not directly involved with eIF4E and the 4E-SE. In summary, we observe two types of complexes: those that can form in the absence of eIF4E and are cap and 4E-SE independent (Fig. 3 D, asterisk) and the second type that depends on eIF4E, the m7G cap, and a structurally intact 4E-SE. The UV cross-linking experiments together with the EMSA results indicate that the 4E-SE acts both as a zip code localizing mRNAs to bodies (Fig. 2 D) and as a USER code for the eIF4E nuclear mRNP (Fig. 3).
eIF4E-dependent mRNA export is independent of ongoing protein or RNA synthesis
We examined the importance of new protein synthesis and transcription for eIF4E-dependent mRNA export. To inhibit protein synthesis, cells were treated with 100 μg/ml cycloheximide for 1 h. Under these conditions, eIF4E-dependent mRNA export of lacZ–c4E-SE is not altered (Fig. 4 A). Also, the export of endogenous cyclin D1 mRNA was not modulated by cycloheximide treatment (unpublished data). Similarly, actinomycin D treatment (10 μg/ml) did not affect the export of these mRNAs (Fig. 4 A). Although cycloheximide treatment did not modify export, it is still possible that the 4E-SE could modulate polysomal loading in an eIF4E-dependent manner. Thus, we monitored polysomal profiles of lacZ as a function of the 4E-SE and of eIF4E overexpression. The profiles of lacZ and lacZ–c4E-SE are indistinguishable and are not altered by eIF4E overexpression (unpublished data). This is consistent with the finding that eIF4E overexpression does not change cyclin D1 mRNA polysomal loading (Rousseau et al., 1996). Given that eIF4E-dependent mRNA export is independent of ongoing protein synthesis and that the 4E-SE does not alter polysomal loading, the functions of eIF4E in mRNA export and translation appear to be decoupled.
Figure 4.
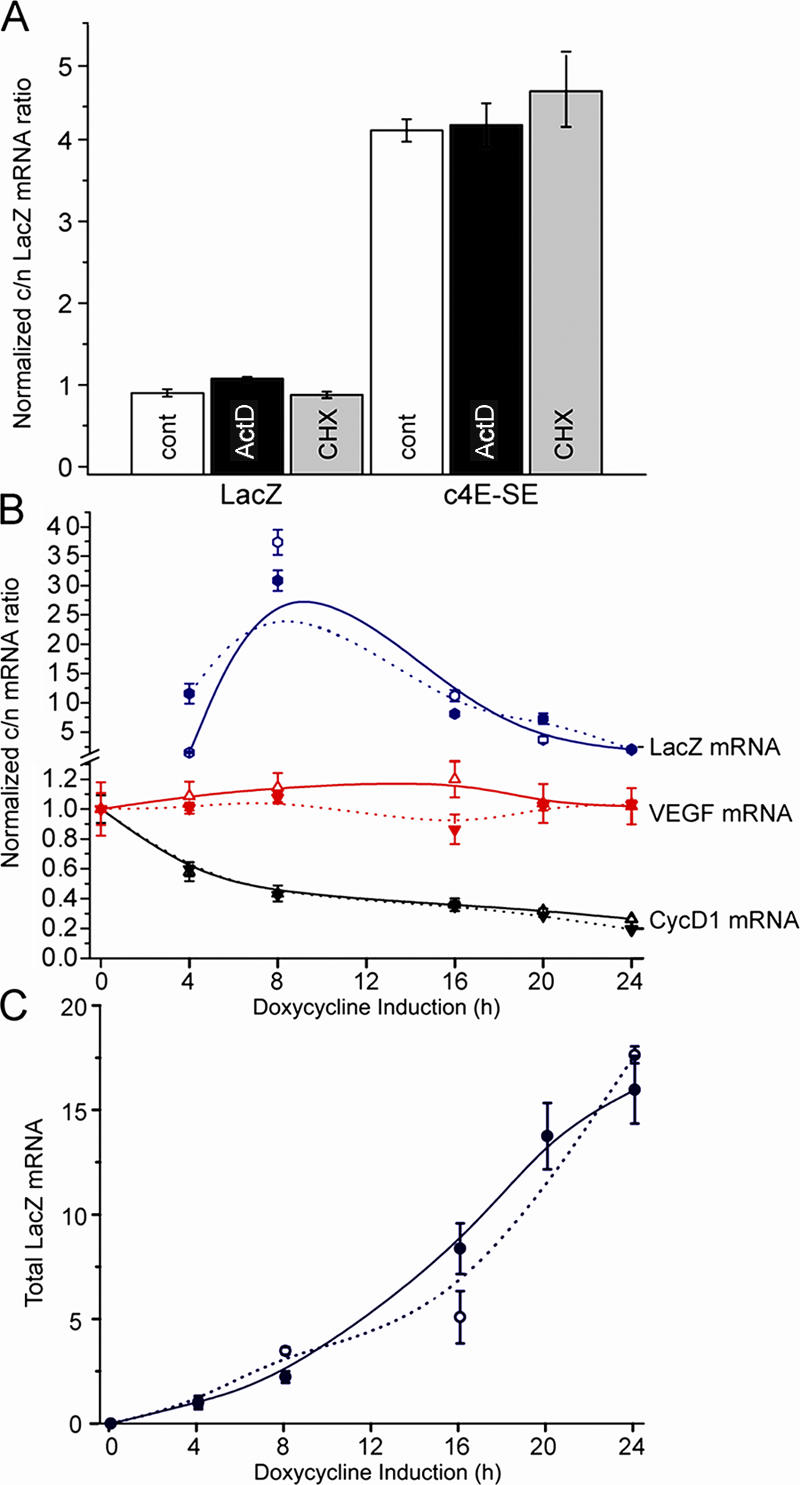
Export of 4E-SE–containing mRNAs is independent of ongoing RNA and protein synthesis, and the pathway is saturated by excess 4E-SE. (A) Quantitative real-time PCR analysis of mRNA export of lacZ–c4E-SE and lacZ in eIF4E-overexpressing cells. Cytoplasmic/nuclear (c/n) ratios represent relative fold ± SD (error bars) normalized to the lacZ untreated control, which was set to 1. Calculations were performed as described in Table II. Treatments were 10 μg/ml actinomycin D for 1 h and 100 μg/ml cycloheximide (CHX) for 1 h. (B and C) LacZ mRNA export was monitored as a function of both time and expression of lacZ transcripts ± 4E-SE induced with doxycycline. Expression as a function of time is shown. In parallel, the extent of export was monitored as the ratio of cytoplasmic/nuclear mRNA for each case. Solid lines represent trends in cells expressing lacZ–c4E-SE; dotted lines represent cells expressing lacZ–p4E-SE. Endogenous mRNAs from the same samples were also examined. Cyclin D1 mRNA export was reduced in cells expressing either lacZ–c4E-SE or lacZ–p4E-SE. Importantly, VEGF, which does not contain a 4E-SE, did not have its export affected in either case. Clearly, as the amount of 4E-SE–containing mRNAs increases in the cell (C), the ability to export these is reduced presumably because 4E-SE–dependent export becomes saturated (B). Cytoplasmic/nuclear values represent relative fold ± SD normalized to lacZ only for each time point as described in Table II. For total RNAs, values represent relative fold ± SD normalized to the first time point of induction for each transcript (4 h), which was set to 1. Average values of lacZ mRNA obtained for each time point were normalized by GAPDH mRNA values obtained for the same sample.
We previously demonstrated that lacZ–c4E-SE transcripts did not have altered stability relative to lacZ transcripts using actinomycin D over the course of several hours (Culjkovic et al., 2005). However, it is still possible that mRNA turnover could be substantially more rapid than several hours. Thus, we constructed lacZ and lacZ–4E-SE TetON-inducible cell lines and examined the stability of these mRNAs immediately upon doxycycline addition. The presence of the 4E-SE does not substantially alter the stability of the lacZ transcripts in either the short (minutes) or long term (hours; unpublished data).
eIF4E-dependent mRNA export pathway is saturated by excess 4E-SE
We reasoned that if the 4E-SE is required for export, the overexpression of lacZ–c4E-SE or lacZ–p4E-SE should specifically inhibit the export of other (endogenous) 4E-SE–containing mRNAs by competing for the 4E-SE–specific export machinery (Fig. 4, B and C). Using our TetON-inducible lacZ, lacZ–p4E-SE, or lacZ–c4E-SE constructs, we monitored the export of chimeric mRNAs as a function of total mRNA levels. The expression of lacZ–p4E-SE and lacZ–c4E-SE mRNAs as a function of time is shown in Fig. 4 C. At early time points, when levels of lacZ mRNAs are low, 4E-SE export is more efficient with higher ratios of cytoplasmic to nuclear chimeric mRNAs. As the levels of these mRNAs increase, 4E-SE export becomes saturated, and the ratio of cytoplasmic to nuclear chimeric mRNAs decreases (Fig. 4 B). At the same time, the export of endogenous cyclin D1 mRNA was impaired by the expression of 4E-SE chimeric mRNAs (Fig. 4 B). Furthermore, the export of VEGF mRNA was not affected, which is consistent with its insensitivity to eIF4E at the mRNA export level (Fig. 4 B). Thus, the overexpression of 4E-SE leads to competition for the 4E-SE–specific export machinery.
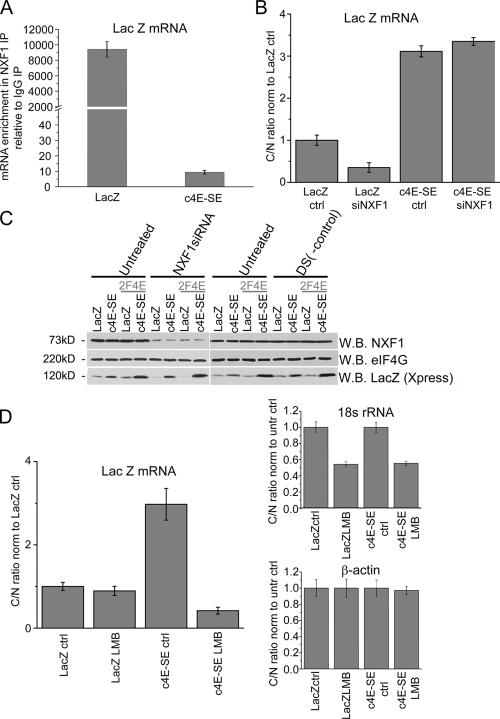
4E-SE–mediated export is NXF1 independent but CRM1 dependent
Because the best-described cellular mRNA export pathway involves the NXF1/p15 heterodimer, which appears to mediate bulk mRNA export (Cullen, 2000, 2003a), the dependence of 4E-SE mRNA export on NXF1 was examined (Fig. 5, A–C). Consistent with previous findings as well as our own, eIF4E does not immunoprecipitate with NXF1 in the nuclear fraction of cells (Lejeune et al., 2002; and unpublished data). However, this does not preclude an NXF1-dependent mechanism in which eIF4E does not need to physically associate with NXF1. To further investigate NXF1 involvement in 4E-SE export, Flag-tagged NXF1- or NXF1/p15-overexpressing cells were immunoprecipitated with anti-Flag antibodies, and the presence of lacZ or lacZ–c4E-SE mRNAs was monitored by real-time PCR (Fig. 5 A). In contrast to lacZ mRNA that is enriched in the NXF1 fractions, lacZ–c4E-SE mRNA appears to be almost completely excluded. These results are independent of the presence or absence of p15 (unpublished data).
Figure 5.
eIF4E-dependent export is NXF1 independent and CRM1 dependent. (A) Comparison of lacZ mRNA in the NXF1 IP fractions. Cells were cotransfected with FlagNXF1/Flagp15 and lacZ or lacZ–c4E-SE. IPs were performed with anti-Flag antibody. LacZ/lacZ–c4E-SE mRNA was monitored by real-time PCR and normalized to IgG controls (as described in Fig. 1 A). (B) NXF1 siRNA treatment (72 h) inhibits the export of lacZ but not lacZ–c4E-SE–containing mRNAs. The cytoplasmic/nuclear (C/N) ratios of lacZ or lacZ–c4E-SE mRNAs in cells overexpressing eIF4E as a function of siRNA treatment are shown. LacZ mRNA levels were normalized to 18S rRNA, whose cytoplasmic/nuclear ratio is unaffected by NXF1 siRNA. (C) Western blot (WB) analysis indicates that NXF1 protein levels are reduced by siRNA treatment but not by scrambled controls (DS (-control)). A Western blot for eIF4G is shown as a negative control. LacZ protein levels correspond to alterations in mRNA export shown in B. (D) Dependence of c4E-SE export on leptomycin B (LMB). The cytoplasmic/nuclear ratio of lacZ–c4E-SE mRNA in eIF4E-overexpressing U2OS cells indicated that 4E-SE export was sensitive to LMB (10 ng/ml for 4 h), whereas lacZ was not sensitive. 18S rRNA export was inhibited by LMB as expected, and β-actin mRNA export was consistently not affected. All RNAs were normalized to GAPDH mRNA (as described in Table II). (B and D) Cytoplasmic/nuclear lysate values represent relative fold ± SD (error bars) normalized to the lacZ untreated control, which was set to 1.
We extended these studies to examine the effects of knocking down NXF1 expression on lacZ–c4E-SE export (Fig. 5 B). Overexpression of eIF4E enhanced the export of lacZ–c4E-SE transcripts even when NXF1 levels were substantially reduced, indicating that the export of lacZ–c4E-SE in the presence of overexpressed eIF4E is independent of NXF1. In the absence of the 4E-SE, the lacZ mRNA cytoplasmic/nuclear ratio was substantially reduced by NXF1 depletion. Analysis of lacZ protein levels confirmed the aforementioned findings (Fig. 5 C). As expected, siRNA treatment led to a reduction in NXF1 levels, whereas treatment with scrambled controls did not (Fig. 5 C). Furthermore, levels of eIF4G were not altered, which is consistent with a study showing that longer siRNA treatments (>72 h) are needed to reduce eIF4G levels (Herold et al., 2001). Thus, the export of 4E-SE–containing transcripts is independent of the NXF1 pathway. This does not rule out the possibility that a subset of 4E-SE transcripts do transit through this pathway but simply that they do not require this pathway to be exported.
Because many RNAs can be exported through the CRM1 pathway, we examined this possibility by using leptomycin B (LMB), a specific inhibitor of CRM1 (Cullen, 2003a,b). The export of lacZ or lacZ–c4E-SE mRNAs as a function of overexpressed eIF4E and LMB treatment was monitored using real-time PCR (Fig. 5 D). Strikingly, LMB suppressed the export of lacZ–4E-SE constructs but not of lacZ or β-actin transcripts. LMB leads to the retention of 18S ribosomal RNA (rRNA; Fig. 5 D), which is consistent with previous studies showing that rRNA export requires CRM1 (Moy and Silver, 1999, 2002).
Discussion
Novel export pathway involving eIF4E and 4E-SE–containing mRNAs
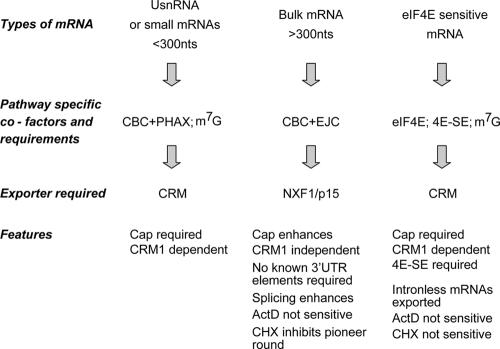
Since it was described, no underlying mechanism for eIF4E-dependent export has been determined (Rousseau et al., 1996). There are several characteristic features that differentiate eIF4E-mediated export from the pathway used for bulk mRNA (summarized in Fig. 6): (1) 4E-SE saturates export of the eIF4E pathway but does not affect the export of bulk mRNA (Fig. 4 B); (2) LMB inhibits eIF4E-dependent export (Fig. 5 D); and (3) the m7G cap is required for the eIF4E pathway (Fig. 1 B and Table II). Interestingly, there are many parallels between the eIF4E pathway and U small nuclear RNA (UsnRNA) export: both are CRM1 dependent, and both require the m7G cap. However, in contrast to the eIF4E pathway, UsnRNA export depends on RNAs being cap-binding complex bound in complex to PHAX, which acts as an adaptor for CRM1 (Izaurralde et al., 1995; Cullen, 2000, 2003a,b; Ishigaki et al., 2001).
Figure 6.
Schematic representation of mechanisms for the export of different classes of RNA. Overviews of characteristic features delineating the export of mRNAs via CRM1 or NXF1/p15 pathways are shown together with features of the eIF4E-mediated export of mRNAs.
In general, CRM1-mediated mRNA export requires cofactors that depend on the type of RNA being exported (i.e., large rRNA, small rRNA, 5S rRNA, or UsnRNA; Cullen, 2003a,b). Our previous studies indicate that eIF4E overexpression does not modulate the export of 18S or 28S rRNA, which is CRM1 dependent, or tRNA, which is exported using the exportin-t receptor (Sarkar and Hopper, 1998; Topisirovic et al., 2002, 2003a). Thus, we hypothesize that eIF4E or some subset of factors associated with the 4E-SE RNP require CRM1 adaptor proteins specific to the eIF4E-dependent pathway. Furthermore, these adaptors are found in limiting amounts and are titratable by high 4E-SE levels or by immunodepletion of eIF4E. Identifying such adaptor proteins will be an area of intense future work.
A conundrum in understanding eIF4E-dependent mRNA export results from the observation that eIF4E stimulates the export of mRNAs that can be still exported under physiological eIF4E levels. Thus, eIF4E-dependent mRNA export is a means by which the cell rapidly up-regulates gene expression by stimulating the export of mRNAs that can be exported through other pathways, albeit less efficiently. When eIF4E levels are low, or in the absence of the m7G cap or 4E-SE, transcripts are exported (presumably) through the NXF1 pathway. This idea is consistent with previous suggestions that the NXF1 pathway is a default RNA export pathway for those RNAs that do not have any special features associated with them (Cullen, 2003b). In this way, eIF4E levels can act as a cellular rheostat. As levels increase, eIF4E-sensitive mRNAs are exported much more efficiently and in a coordinated fashion through the eIF4E-dependent CRM1-sensitive pathway described in this study. A recent study indicates that CRM1-dependent mRNA export can occur during T cell activation, indicating that external cellular signals can lead to alterations in mRNA export pathways (Schutz et al., 2006).
The role of eIF4E in 4E-SE–containing mRNA export and implications for cancer
The studies reported here suggest the possibility that the proliferative and transforming properties associated with eIF4E are, at least partially, a result of the dysregulation of eIF4E-dependent mRNA export. These studies indicate a role for eIF4E in coordinating the export and expression of transcripts involved in cell cycle progression, proliferation, and survival. Importantly, eIF4E does not promote the expression of negative regulators of itself (i.e., PML). eIF4E also promotes the expression of c-myc, a factor that up-regulates the transcription of eIF4E in some cellular growth conditions (Schmidt, 2004). Thus, eIF4E modulates the expression of many genes involved in multiple points of cell cycle progression.
The 4E-SE provides a USER code for targeting these transcripts for export in an eIF4E-sensitive manner. Other transcripts may be regulated by eIF4E at the translation level using USER codes that are different from the 4E-SE. Furthermore, the 4E-SE may associate with other, as yet unidentified, RNPs. In this way, the effects of eIF4E and regulation of 4E-SE–containing transcripts are likely to be complex and combinatorial. For instance, the translation of export-sensitive mRNAs does not depend on the 4E-SE but rather on the complexity of the 5′ UTR. Transcripts such as Pim-1 and ODC (Rousseau et al., 1996; Hoover et al., 1997) serve as examples of the combinatorial use of USER codes for modulating gene expression and support the idea of the use of such a network. Consistently, our experiments indicate that the translation and export functions of eIF4E can be decoupled based on the composition of the 3′ and 5′ UTRs (i.e., eIF4E enhances the export of cyclin D1 but enhances the translation of VEGF).
Several key regulators of eIF4E-dependent mRNA export have been identified, including PML (Cohen et al., 2001) and several homeodomain proteins that contain conserved eIF4E-binding sites (Topisirovic et al., 2003a, 2005). These regulators are positioned to modulate the entire RNA regulon, potently modulating cell cycle progression and cell survival. Our studies demonstrate that PML and PRH impede the eIF4E-dependent export of cyclin D1 and other 4E-SE–containing transcripts (Topisirovic et al., 2003a; Culjkovic et al., 2005; and this study). Further in vitro, PML can also inhibit eIF4E-sensitive translation (Kentsis et al., 2001) and, thus, is positioned to control the regulon at different levels. Stimulators of this growth regulon include HOXA9, which promotes both the mRNA export and translation of genes in the regulon (Topisirovic et al., 2005). The far-reaching activities of these regulators, particularly those that regulate multiple eIF4E functions simultaneously, likely lies in their ability to modulate eIF4E, a key nexus in this regulon. The physiological importance of this regulation is clear. In primary specimens from acute myeloid leukemia patients, PRH is both down-regulated and delocalized from eIF4E nuclear bodies (Topisirovic et al., 2003b). At the same time, HOXA9 is up-regulated and becomes associated with eIF4E in both the nuclear and cytoplasmic compartments, leading to the up-regulation of both eIF4E-dependent mRNA export and translation (Topisirovic et al., 2005).
In conclusion, eIF4E-dependent modulation of mRNA export could provide an immediate response system by which the cell responds to extracellular stimuli before transcriptional reprogramming. Our results indicate that the modulation of mRNA export allows the coordinated modulation of cellular proliferation and provide one of the first examples of an RNA regulon that is positioned to directly impact human disease. The ability of eIF4E to modulate coordinated gene expression impacting proliferation and cell survival pathways ensures maximum efficiency for its growth-promoting potential. Certainly, these findings do not preclude but rather complement critical modulation of gene expression by eIF4E at other levels of mRNA metabolism, particularly translation and mRNA stability/sequestration. In summary, we define a novel mRNA export pathway that is used for the coordinate expression of genes that govern cell cycle progression and survival.
Materials and methods
Reagents and constructs
Chimeric constructs in pcDNA3.1lacZ vector (Invitrogen) were positioned 3′ of the coding region of lacZ. Cyclin D1 minimal 4E-SE (c4E-SE) was amplified using primers containing EcoRI or XbaI restriction sites at the 5′ ends and the lacZ 3′ UTR construct as a template (Culjkovic et al., 2005). The same approach was used for the cloning of Pim-1 constructs, in which pRBK–Pim-1 (a gift from N. Magnuson, Washington State University, Pullman, WA; Hoover et al., 1997) was used as a template. Primer sequences are given in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200607020/DC1). For the TetON system, chimeric lacZ constructs were cloned into pTREMyc vector (CLONTECH Laboratories, Inc.) using EcoRI and XbaI. pcDNA2Flag-eIF4E, pMV, pMV-eIF4E wild type or mutants, pLINKSV40-PML, MSCV, MSCV-eIF4E wild type or mutants, and bacterial expression constructs were previously described (Cohen et al., 2001; Topisirovic et al., 2003b; Culjkovic et al., 2005). Reagents used were all analytical grade from Sigma-Aldrich unless otherwise stated.
Antibodies
Antibodies for immunoblotting were as follows: mAb anti-PML (5E10; Stuurman et al., 1992), mAb anti-eIF4E (BD Biosciences), mAb anticyclin D1 (BD Biosciences), mAb anti-Xpress (Invitrogen), rabbit pAb anticyclin E1 (M20; Santa Cruz Biotechnology, Inc.), mAb anti-GAPDH (MAB374; Chemicon), mAb anti–c-myc (9E10; Santa Cruz Biotechnology, Inc.), rabbit pAb anticyclin A (C-19; Santa Cruz Biotechnology, Inc.), rabbit pAb antinibrin (Cell Signaling), mAb anti–Pim-1 (19F7; Santa Cruz Biotechnology, Inc.), mAb cyclin B1 (GNS1; Santa Cruz Biotechnology, Inc.), mAb anti-eIF4G (BD Biosciences), and mAb anti-NXF1/TAP (BD Biosciences).
Cell culture and transfection
Stably eIF4E- and PML-transfected NIH3T3 and U937 cells were as described previously (Topisirovic et al., 2002, 2003a). U937 cells were used to analyze endogenous Pim1, which is not expressed in NIH3T3 cells. LacZ/lacZ–4E-SE with or without 2Flag-eIF4E as well as the TetON lacZ system were stably transfected in U2OS cells. For NXF1 depletion, U2OS cells were transfected with LipofectAMINE 2000 and 10 nM siRNA duplex HSC.RNAI.N006362.1.3 (Integrated DNA Technologies) according to the manufacturer's instructions. Cells were analyzed 72 h after transfection. Actinomycin D, cycloheximide, and LMB were all cell culture grade (Sigma-Aldrich). Time and concentrations used in treatments are described in the Results and Figs. 4 and 5.
Immunopurification of eIF4E and RT-PCR
Immunopurification was performed as previously described (Culjkovic et al., 2005). Real-time PCR analyses were performed using Sybr Green PCR Master mix (Applied Biosystems) in Mx3000P thermal cycler (Stratagene), and data were analyzed with MxPro software (Stratagene). All conditions were described previously (Culjkovic et al., 2005). Primer sequences are listed in Table S1. All calculations were performed using the relative standard curve method described in User Bulletin #2 (Applied Biosystems) and are more precisely described in the corresponding figure legends. Differential display of immunopurified RNA was performed using the RNAimage kit (GeneHunter Corporation) according to the manufacturer's instructions. The SNAAP protocol used for differential display was performed as described previously (Trifillis et al., 1999), and Western blots were also performed as described previously (Topisirovic et al., 2002, 2003a).
Cellular fractionation and Northern analysis
Fractionation and RNA isolation were performed as described previously (Lai and Borden, 2000; Topisirovic et al., 2002). Probes for U6 and tRNAlys for Northern blot analysis were previously described (Topisirovic et al., 2002).
Immunofluorescence, in situ hybridization, and laser-scanning confocal microscopy
Experiments were performed as described previously (Cohen et al., 2001; Topisirovic et al., 2002). Fluorescence was observed using 100× optical magnification and 3 or 4× digital zoom as indicated on an inverted laser-scanning confocal microscope (LSM510 META; Carl Zeiss MicroImaging, Inc.) exciting at 488, 543, or 405 nm (at RT). All channels were detected separately, and no cross talk between the channels was observed. The confocal micrographs represent a single optical section through the plane of the cell. Images were obtained using LSM510 software (Carl Zeiss MicroImaging, Inc.) and were displayed using Photoshop CS 8.0 (Adobe). In situ hybridization was performed as previously described (Culjkovic et al., 2005) using nick-translated biotin-11-dUTP–labeled probes (Nick Translation kit; Roche). Probes were detected using Cy3-conjugated IgG fraction mouse mAb antibiotin (1:100; Jackson ImmunoResearch Laboratories). PML was detected using 5E10 mAb followed by Cy5-conjugated donkey anti–mouse IgG (heavy and light chains; 1:100; Jackson ImmunoResearch Laboratories). eIF4E was detected using FITC-conjugated mouse mAb anti-eIF4E (BD Transduction Laboratories). Cells were mounted in Vectashield supplemented with DAPI (Vector Laboratories).
EMSA analyses were performed as previously described (Wein et al., 2003) with the following modifications: 20–50 μg of nuclear lysate were incubated with 32P 3′ end–labeled lacZ, lacZ–c4E-SE, or lacZ–p4E-SE transcript (∼50,000 cpm) in 25 μl NET-2 buffer supplemented with 5 mg yeast tRNA (Sigma-Aldrich) and 3 mM MgCl2 for 30 min at RT with an additional 15 min after the addition of 2 mg/ml heparin. For competition studies, the unlabeled competitor RNAs were preincubated for 10 min with nuclear lysates before labeled RNAs were added. For supershift experiments, nuclear lysates were preincubated with mAb anti-eIF4E (BD Biosciences) for 15 min before the addition of labeled RNAs. Immunodepleted lysates were from IPs with rabbit pAb anti-eIF4E (Abcam). All mRNAs were in vitro transcribed using the mMessage mMachine T7 kit (Ambion) and were 3′ end labeled using [32P]pCp and T4 RNA ligase (GE Healthcare). Samples were separated by electrophoresis on 5% native (19:1) polyacrylamide gels for 2 h at 250 V using 1× Tris-borate-EDTA buffer.
UV cross-linking
50 μg of nuclear lysates were incubated with radiolabeled probes (1–2 × 105 cpm) using the same conditions as for the EMSA. After incubation with heparin, samples were placed on ice and UV irradiated for 15 min in a cross-linker (Stratalinker 1800 UV; Stratagene). Cross-linked RNA–protein complexes were treated with 10 U RNase A and 10 U RNase T1 for 15 min at 37°C. The reactions were stopped by the addition of 30 μl of 2× SDS sample buffer and heating 10 min at 95°C. Samples were loaded on 10 or 12% SDS polyacrylamide gels and separated at 50 V for 16 h at RT.
RNase mapping
Analyses were performed as described previously (Clever et al., 1995) and according to the manufacturer's instructions (Ambion). In brief, ∼0.5–105 cpm 32P 5′ end–labeled c4ESE or p4ESE RNA oligonucleotide probes (Integrated DNA Technologies) were mixed with 3 μg yeast tRNA and incubated with 1, 0.1, or 0.01 U RNase V1 (Ambion) for 15 min at RT; 1, 0.1, or 0.01 U RNase A (Ambion) for 5 min at RT; 1, 0.1, or 0.01 U RNase T1 (Sigma-Aldrich) for 15 min at RT; 1, 0.1, or 0.01 U RNase T2 (Invitrogen) for 5 min at RT; or alkaline buffer for 1, 2, or 5 min at 95°C (alkaline hydrolysis). For RNase digestions, denaturated RNA oligonucleotide probes (5 min at 75°C) were analyzed together with their native counterparts and used as an additional ladder control. Reactions were stopped by EtOH/NaAc precipitation. Samples were resolved on 6% polyacrylamide 8 M urea gels in 1× Tris-borate-EDTA buffer.
Online supplemental material
Table S1 lists the PCR primers used in this study. Fig. S1 shows mRNAs regulated by eIF4E in the nucleus. Fig. S2 shows differential display from RNAs immunoprecipitated with eIF4E mAb, mouse IgG as a negative control, and 0.5% of input nuclear RNA from U2OS nuclear lysates. Fig. S3 shows that the secondary structure of Pim-1 4E-SE confirmed by RNase mapping is necessary for nuclear 4E RNP formation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200607020/DC1.
Supplementary Material
Acknowledgments
We are grateful for the gifts of antibodies, constructs, and cell lines from Paul Freemont, Nahum Sonenberg, Nancy Magnuson, Michael Kiledjian, Ze'ev Ronai, and Gerhard Wagner. We thank Trang Hoang for use of the Mx3000P thermal cycler. We are grateful for technical assistance from Amri Abdellatif and for critical reading of the manuscript and helpful discussions with Jack Keene and Nahum Sonenberg.
K.L.B. Borden holds a Canada Research Chair. Financial support was provided by the National Institutes of Health (grants CA 98571 and 80728).
Abbreviations used in this paper: 4E-SE, eIF4E sensitivity element; EMSA, electromobility shift assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IP, immunoprecipitation; LMB, leptomycin B; ODC, ornithine decarboxylase; PML, promyelocytic leukemia protein; PRH, proline-rich homeodomain; rRNA, ribosomal RNA; SLP, stem loop pair; USER, untranslated sequence elements for regulation; UsnRNA, U small nuclear RNA; UTR, untranslated region.
References
- Bachmann, M., and T. Moroy. 2005. The serine/threonine kinase Pim-1. Int. J. Biochem. Cell Biol. 37:726–730. [DOI] [PubMed] [Google Scholar]
- Borden, K.L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Q., and J.D. Richter. 2002. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 21:3852–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y.C., Y.N. Su, P.C. Chou, W.C. Chiang, M.C. Chang, L.S. Wang, S.C. Teng, and K.J. Wu. 2005. Overexpression of NBS1 contributes to transformation through the activation of phosphatidylinositol 3-kinase/Akt. J. Biol. Chem. 280:32505–32511. [DOI] [PubMed] [Google Scholar]
- Clemens, M.J., and U.A. Bommer. 1999. Translational control: the cancer connection. Int. J. Biochem. Cell Biol. 31:1–23. [DOI] [PubMed] [Google Scholar]
- Clever, J., C. Sassetti, and T.G. Parslow. 1995. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 69:2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, N., M. Sharma, A. Kentsis, J.M. Perez, S. Strudwick, and K.L. Borden. 2001. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 20:4547–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culjkovic, B., I. Topisirovic, L. Skrabanek, M. Ruiz-Gutierrez, and K.L. Borden. 2005. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3′UTR. J. Cell Biol. 169:245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B.R. 2000. Nuclear RNA export pathways. Mol. Cell. Biol. 20:4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B.R. 2003. a. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419–424. [DOI] [PubMed] [Google Scholar]
- Cullen, B.R. 2003. b. Nuclear RNA export. J. Cell Sci. 116:587–597. [DOI] [PubMed] [Google Scholar]
- Gao, Y., H.C. Feng, K. Walder, K. Bolton, T. Sunderland, N. Bishara, M. Quick, L. Kantham, and G.R. Collier. 2004. Regulation of the selenoprotein SelS by glucose deprivation and endoplasmic reticulum stress - SelS is a novel glucose-regulated protein. FEBS Lett. 563:185–190. [DOI] [PubMed] [Google Scholar]
- Grillo, G., F. Licciulli, S. Liuni, E. Sbisa, and G. Pesole. 2003. PatSearch: a program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 31:3608–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold, A., T. Klymenko, and E. Izaurralde. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- Hieronymus, H., and P.A. Silver. 2003. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33:155–161. [DOI] [PubMed] [Google Scholar]
- Hieronymus, H., and P.A. Silver. 2004. A systems view of mRNP biology. Genes Dev. 18:2845–2860. [DOI] [PubMed] [Google Scholar]
- Hieronymus, H., M.C. Yu, and P.A. Silver. 2004. Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 18:2652–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, D.S., D.G. Wingett, J. Zhang, R. Reeves, and N.S. Magnuson. 1997. Pim-1 protein expression is regulated by its 5′-untranslated region and translation initiation factor elF-4E. Cell Growth Differ. 8:1371–1380. [PubMed] [Google Scholar]
- Iborra, F.J., D.A. Jackson, and P.R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science. 293:1139–1142. [DOI] [PubMed] [Google Scholar]
- Ishigaki, Y., X. Li, G. Serin, and L.E. Maquat. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 106:607–617. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., J. Lewis, C. Gamberi, A. Jarmolowski, C. McGuigan, and I.W. Mattaj. 1995. A cap-binding protein complex mediating U snRNA export. Nature. 376:709–712. [DOI] [PubMed] [Google Scholar]
- Keene, J.D., and S.A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 9:1161–1167. [DOI] [PubMed] [Google Scholar]
- Keene, J.D., and P.J. Lager. 2005. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 13:327–337. [DOI] [PubMed] [Google Scholar]
- Kentsis, A., E.C. Dwyer, J.M. Perez, M. Sharma, A. Chen, Z.Q. Pan, and K.L. Borden. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312:609–623. [DOI] [PubMed] [Google Scholar]
- Lai, H.K., and K.L. Borden. 2000. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene. 19:1623–1634. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Y. Ishigaki, X. Li, and L.E. Maquat. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 21:3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, J., and J.M. Slingerland. 2003. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2:339–345. [PubMed] [Google Scholar]
- Moy, T.I., and P.A. Silver. 1999. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 13:2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy, T.I., and P.A. Silver. 2002. Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci. 115:2985–2995. [DOI] [PubMed] [Google Scholar]
- Naumann, F., R. Remus, B. Schmitz, and W. Doerfler. 2004. Gene structure and expression of the 5′-(CGG)(n)-3′-binding protein (CGGBP1). Genomics. 83:106–118. [DOI] [PubMed] [Google Scholar]
- Pegg, A.E. 2006. Regulation of ornithine decarboxylase. J. Biol. Chem. 281:14529–14532. [DOI] [PubMed] [Google Scholar]
- Rousseau, D., R. Kaspar, I. Rosenwald, L. Gehrke, and N. Sonenberg. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA. 93:1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, H., S. Bhardwaj, and S. Yla-Herttuala. 2006. Biology of vascular endothelial growth factors. FEBS Lett. 580:2879–2887. [DOI] [PubMed] [Google Scholar]
- Sarkar, S., and A.K. Hopper. 1998. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol. Biol. Cell. 9:3041–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, E.V. 2004. The role of c-myc in regulation of translation initiation. Oncogene. 23:3217–3221. [DOI] [PubMed] [Google Scholar]
- Schutz, S., J. Chemnitz, C. Spillner, M. Frohme, J. Hauber, and R.H. Kehlenbach. 2006. Stimulated expression of mRNAs in activated T cells depends on a functional CRM1 nuclear export pathway. J. Mol. Biol. 358:997–1009. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N., and A.C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268–275. [DOI] [PubMed] [Google Scholar]
- Stier, S., G. Totzke, E. Gruewald, T. Neuhaus, S. Fronhoffs, S. Schoneborn, H. Vetter, and Y. Ko. 2005. Identification of p54(nrb) and the 14-3-3 Protein HS1 as TNF-alpha-inducible genes related to cell cycle control and apoptosis in human arterial endothelial cells. J. Biochem. Mol. Biol. 38:447–456. [DOI] [PubMed] [Google Scholar]
- Strudwick, S., and K.L. Borden. 2002. The emerging roles of translation factor eIF4E in the nucleus. Differentiation. 70:10–22. [DOI] [PubMed] [Google Scholar]
- Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773–784. [DOI] [PubMed] [Google Scholar]
- Topisirovic, I., A.D. Capili, and K.L. Borden. 2002. Gamma interferon and cadmium treatments modulate eukaryotic initiation factor 4E-dependent mRNA transport of cyclin D1 in a PML-dependent manner. Mol. Cell. Biol. 22:6183–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic, I., B. Culjkovic, N. Cohen, J.M. Perez, L. Skrabanek, and K.L. Borden. 2003. a. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J. 22:689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic, I., M.L. Guzman, M.J. McConnell, J.D. Licht, B. Culjkovic, S.J. Neering, C.T. Jordan, and K.L. Borden. 2003. b. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol. Cell. Biol. 23:8992–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic, I., A. Kentsis, J.M. Perez, M.L. Guzman, C.T. Jordan, and K.L. Borden. 2005. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol. Cell. Biol. 25:1100–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifillis, P., N. Day, and M. Kiledjian. 1999. Finding the right RNA: identification of cellular mRNA substrates for RNA-binding proteins. RNA. 5:1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., P. Iakova, M. Wilde, A. Welm, T. Goode, W.J. Roesler, and N.A. Timchenko. 2001. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell. 8:817–828. [DOI] [PubMed] [Google Scholar]
- Wein, G., M. Rossler, R. Klug, and T. Herget. 2003. The 3'-UTR of the mRNA coding for the major protein kinase C substrate MARCKS contains a novel CU-rich element interacting with the mRNA stabilizing factors HuD and HuR. Eur. J. Biochem. 270:350–365. [DOI] [PubMed] [Google Scholar]
- Zhu, N., L. Gu, H.W. Findley, and M. Zhou. 2005. Transcriptional repression of the eukaryotic initiation factor 4E gene by wild type p53. Biochem. Biophys. Res. Commun. 335:1272–1279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.