Abstract
Paracoccidioides brasiliensis is a human pathogenic fungus that switches from a saprobic mycelium to a pathogenic yeast. Consistent with the morphological transition being regulated by the cAMP-signalling pathway, there is an increase in cellular cAMP levels both transiently at the onset (< 24 h) and progressively in the later stages (> 120 h) of the transition to the yeast form, and this transition can be modulated by exogenous cAMP. We have cloned the cyr1 gene encoding adenylate cyclase (AC) and established that its transcript levels correlate with cAMP levels. In addition, we have cloned the genes encoding three Gα (Gpa1–3), Gβ (Gpb1) and Gγ (Gpg1) G proteins. Gpa1 and Gpb1 interact with one another and the N-terminus of AC, but neither Gpa2 nor Gpa3 interacted with Gpb1 or AC. The interaction of Gpa1 with Gpb1 was blocked by GTP, but its interaction with AC was independent of bound nucleotide. The transcript levels for gpa1, gpb1 and gpg1 were similar in mycelium, but there was a transient excess of gpb1 during the transition, and an excess of gpa1 in yeast. We have interpreted our findings in terms of a novel signalling mechanism in which the activity of AC is differentially modulated by Gpa1 and Gpb1 to maintain the signal over the 10 days needed for the morphological switch.
Introduction
The phylogenetically related ascomycete fungi Paracoccidioides brasiliensis, Histoplasma capsulatum, Blastomyces dermatitidis, Coccidioides immitis, Penicillium marneffei and Sporothrix schenkii from more than hundred thousand different species of environmental fungi are able to adapt for survival in mammalian hosts (Borges-Walmsley et al., 2002; Morris-Jones, 2002; San-Blas et al., 2002; Bradsher et al., 2003; DiCaudo, 2006; Vanittanakom et al., 2006; Kauffman, 2007). These are known as dimorphic fungi because they undergo extensive changes that allow them to switch from mycelium, a non-pathogenic filamentous form, to pathogenic single-cellular yeast that causes infections in millions of people across the globe every year. Infection is the result of the release from mycelium, frequently found in soil, of fragments or spores, which are inhaled by the host, exposing them to an elevated temperature that triggers the morphological switch. The pathogenicity of these fungi is intimately linked to the morphological change because strains that are unable to transform from mycelium to yeast are often avirulent (Nemecek et al., 2006). However, our knowledge of how these fungi sense and respond to the temperature change is still rudimentary.
In eukaryotes, many cell-signalling processes are mediated by guanine-nucleotide binding proteins known as G proteins (for a review, see Sprang, 1997). Generally these are activated when they interact with a G protein-coupled receptor (GPCR) and transmit signals to downstream effectors such as adenylate cyclase (AC) and protein kinases. Typically these G proteins function as heterotrimeric complexes, composed of Gα, Gβ and Gγ subunits, which are activated when GTP binds to, and replaces bound GDP on, the Gα subunit to cause its dissociation from the Gβγ dimer. The signal can be mediated by Gα-GTP, Gβγ or both, depending on the pathway, and is attenuated as the GTP is hydrolysed to allow the re-association of the trimeric complex (Sprang, 1997). Although this is a generally held view of the function of heterotrimeric G proteins, there is recent evidence that complex dissociation is not needed for signalling by all G proteins (Frank et al., 2005).
Fungi possess between two and four, but most have three, Gα proteins. However, they only appear to have a single Gβ and Gγ protein, suggesting that either some Gα proteins can act independently or multiple Gα proteins can interact with the Gβγ dimer (Lafon et al., 2006; Yu, 2006). Saccharomyces cerevisiae has two Gα proteins, Gpa1 and Gpa2, which have been shown to regulate mating, in response to pheromones, and filamentous growth, in response to glucose, via mitogen-activated protein kinase (MAPK) and cAMP-signalling pathways respectively (for a recent review, see Hoffman, 2005). Gpa1 interacts with the Gβγ dimer, composed of Ste4 and Ste18, preventing activation of the MAPK-signalling pathway by the Ste4–Ste18 dimer, which binds to the scaffold protein Ste5 (Whiteway et al., 1995; Pryciak and Huntress, 1998) and the p21-activated kinase Ste20 (Leeuw et al., 1998). In contrast, the cAMP-signalling pathway is activated by Gpa2 and Ras (Lorenz and Heitman, 1997; Colombo et al., 1998; 2004; Xue et al., 1998; Kraakman et al., 1999; Lorenz et al., 2000), both of which are presumed to bind directly to AC. Although Gpa2 does not appear to have a Gβ partner, it can bind the kelch-repeat proteins Gpb1 and Gpb2 that may mimic Gβ subunits to control the level of the free protein (Harashim and Heitman, 2002). Schizosaccharomyces pombe also possesses two Gα proteins that regulate pheromone-activated MAPK and glucose-activated cAMP-signalling pathways. However, the pheromone pathway is activated by the Gα protein Gpa1, rather than by a Gβγ dimer, but its target is unknown (Obara et al., 1991; Ladds et al., 2005); while Gpa2, which binds to the Gβγ dimer, composed of Git5 and Git11 (Landry and Hoffman, 2001), binds to and activates AC (Ogihara et al., 2004; Ivey and Hoffman, 2005). Ustilago maydis has four Gα proteins, but it is not known whether they work independently of a Gβ or whether they all interact with the same Gβ, or even if they have kelch-repeat protein partners (Regenfelder et al., 1997; Muller et al., 2004). Aspergillus nidulans possesses three Gα proteins, FadA (Yu et al., 1996), GanA and GanB (Chang et al., 2004), and apparently a single Gβ, sfaD (Rosen et al., 1999), and Gγ, GpgA (Seo et al., 2005), protein, but does not appear to possess kelch-repeat proteins that are related to Gpb1 or Gpb2. There is evidence from the phenotypes of disruption mutants that a sfaD–GpgA dimer can interact with both FadA and GanB (Lafon et al., 2005), but a direct interaction has not been demonstrated.
Gβ proteins have been found to be involved in developmental pathways of several filamentous fungi: for example, in controlling the development and/or virulence of the plant pathogens Cryphonectria parasitica (Kasahara and Nuss, 1997), Magnaporthe grisea (Nishimura et al., 2003), Fusarium oxysporum (Jain et al., 2003; Delgado-Jarana et al., 2005), U. maydis (Muller et al., 2004) and Cochliobolus heterostrophus (Ganem et al., 2004), and in the development of Neurospora crassa (Krystofova and Borkovich, 2005), A. nidulans (Rosen et al., 1999) and Cryptococcus neoformans (Wang et al., 2000). Although the Gβ proteins MBP1 and GPB1, from M. grisea and C. neoformans, appear to function through MAPK-signalling pathways in analogy with Ste4, the other Gβ proteins function, at least partially, through cAMP-signalling pathways. In several cases, exogenous cAMP can suppress some, if not all, of the defects caused by deleting these genes. For example, the filamentous growth of a bpp1 deletion in U. maydis can be suppressed by exogenous cAMP (Muller et al., 2004). A constitutively active allele of gpa3 also suppresses this phenotype, suggesting that Bpp1 and Gpa3 are components of the same heterotrimeric G protein that acts on AC. However, in contrast to Δbbp1 strains, Δgpa3 strains are impaired in pathogenicity, suggesting that Gpa3 operates independently of Bpp1. In N. crassa, deletion of the Gβ protein Gnb-1 causes a reduction in cAMP levels, but this was attributed to a reduction in Gα proteins that activate AC (Krystofova and Borkovich, 2005). In S. pombe, deletion of the Gβ protein Git5 does not affect basal cAMP levels but inhibits the glucose-induced elevation of cAMP levels (Landry et al., 2000), and one proposal is that Git5 interacts directly with AC. Thus, there is evidence that Gα and Gβ proteins can independently activate different signalling processes.
If Gα and Gβ, and the Gβγ and Gαβγ complexes can all elicit signalling, then there is no reason why these subunits should be expressed equivalently. Signalling could be brought about by the increased expression of either the Gα or Gβ subunit, above that of its cognate subunit, so that there would be a pool of uncomplexed G protein that would be free to interact with other proteins in the signalling pathway. In the case of S. cerevisiae, it has been reported that there is an excess of Gpa1 over Ste4 (Ghaemmaghami et al., 2003) that might be needed to prevent pheromone-independent signalling by the Ste4–Ste18 dimer (Hoffman, 2005). This balance in the subunits is important because a twofold increase in Ste4 is sufficient to activate the pathway (Hao et al., 2003). A similar imbalance has been noted in S. pombe, where the Gβ Git5 is transcribed at much lower levels than the Gα Gpa2 and Gγ Git11 (Hoffman, 2005). It has been hypothesized that the uncomplexed Gpa2 is associated with AC to form an inactive complex: glucose activation of the signalling pathway would lead to Gpa2-GTP, released from the Git5–Git11 dimer, being swapped for Gpa2-GDP bound to AC that would then become activated (Hoffman, 2005).
The cAMP-signalling pathway has been shown to be important in controlling morphological changes and the pathogenicity of several fungi (Borges-Walmsley and Walmsley, 2000). For example, signalling through AC controls the virulence of Candida albicans (Rocha et al., 2001), C. neoformans (Alspaugh et al., 2002) and A. fumigatus (Liebmann et al., 2003). In contrast to the ACs of C. neoformans (Vallim et al., 2005) and A. fumigatus (Liebmann et al., 2003) that only appear to be regulated via Gα proteins, C. albicans is regulated by both Ras (Rocha et al., 2001) and Gpa2 (Miwa et al., 2004; Maidan et al., 2005), which can also interact with the MAPK pathway (Sanchez-Martinez and Perez-Martin, 2002; Bennett and Johnson, 2006). In the plant pathogen M. grisea, the morphological changes that are involved in pathogenicity are dependent upon G protein-mediated cAMP signalling, and exogenous cAMP induces formation of the infective appressorium (Nishimura et al., 2003).
Paracoccidioides brasiliensis, the aetiological agent of paracoccidioidomycosis (for a review, see San-Blas et al., 2002; Borges-Walmsley et al., 2002), the most prevalent systemic mycosis in Latin America, where it is estimated that throughout the endemic region as many as 10 million individuals, out of a population of about 90 million, may be infected (Restrepo et al., 2001). The fungus is dimorphic undergoing a complex transformation in vivo, in which mycelia and conidia transform to the pathogenic yeast form (Medoff et al., 1987; Borges-Walmsley et al., 2002; San-Blas et al., 2002), while strains that are unable to undergo the mycelium-to-yeast transformation are avirulent (San-Blas and Niño-Veja, 2001). Herein we establish that this process is regulated by the cAMP-signalling pathway, and we have cloned the gene encoding AC and genes that encode a set of G proteins, which potentially function upstream of AC. In the absence of molecular tools for forward and reverse genetic approaches for studying gene function in P. brasiliensis, we have investigated the interaction of these proteins using yeast two-hybrid and pull-down assays. Our data indicate that the Gβ and Gγ proteins Gpb1 and Gpg1 interact specifically with the Gα Gpa1, but not with Gpa2 or Gpa3, to form a trimeric complex. This trimer can dissociate to release Gpa1 and Gpb1 that can independently interact with the N-terminus of AC. We propose that the ability of AC to bind both Gpa1 and Gpb1 enables maintenance of a long-term signal that is required to direct the morphological transition from the saprobic mycelium to pathogenic yeast, which occurs over a period of about 10 days.
Results
The P. brasiliensis morphological transition is controlled by cAMP
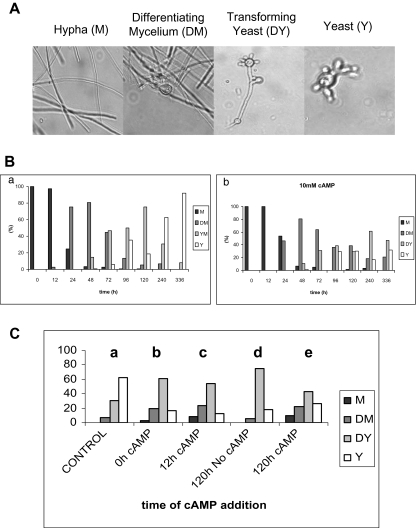
The cAMP-signalling pathway has been implicated in controlling morphological changes and the virulence of a number of fungi. We sought to determine whether cAMP would affect the morphological transition, which underlies the virulence, of P. brasiliensis. As a prerequisite for such an analysis, we monitored the morphology of P. brasiliensis cells, growing in liquid culture, which had been induced to undergo the mycelium-to-yeast transition by increasing the temperature from 26°C to 37°C, and established parameters to quantify the different morphotypes that are produced during this process. We classified the transition into four different morphological states (Fig. 1A): (i) hyphae; (ii) differentiating hyphae, characterized by the development of chlamydospore-like cells, produced by intercalary or lateral swellings in the fertile hyphae; (iii) transforming yeast, characterized by the production of multiple buds by the chlamydospore; and (iv) mature, multibudding yeast. This classification helped us establish quantitative parameters to assess the morphological transition at successive time point: after 336 h, 92% of the cells were yeast, indicating that the transition had gone to near completion (Fig. 1B, left graphic). In contrast, when the mycelial cells were treated with 10 mM dibutyryl-cAMP and the morphological switch induced, only 32% of the cells were yeast after 336 h, indicating that cAMP retarded the morphological transition (Fig. 1B, right graphic). We did not find any appreciable effect of dibutyryl-cAMP, at concentrations up to 20 mM, upon the yeast-to-mycelium transition induced by decreasing the incubation temperature from 37°C to 26°C (data not shown). This latter finding contrasts with an earlier study that indicated that cAMP retarded the yeast-to-mycelium transition (Paris and Duran, 1985), but we cannot identify any clear reason for this difference.
Fig. 1. The cAMP-signalling pathway regulates the transition from mycelium to yeast in P. brasiliensis.
A. The morphology of P. brasiliensis cells, growing in liquid culture, which had been induced to undergo the mycelium-to-yeast transition by increasing the temperature from 26°C to 37°C and was monitored to quantify the different morphotypes that are produced during this process. Cellular forms were classified into four different morphological states: (i) hyphae; (ii) differentiating hyphae, characterized by the development of chlamydospore-like cells, produced by intercalary or lateral swellings in the fertile hyphae; (iii) transforming yeast, characterized by the production of multiple buds by the chlamydospore; and (iv) mature, multibudding yeast. At the indicated times during the morphological switch, 300 morphological units were picked and the number of individual forms quantified.
B. The bar charts show the percentage of each morphological form at increasing times during the morphological transition from mycelium to yeast in the absence (a) and presence (b) of 10 mM dibutyryl-cAMP. The data indicate that exogenous dibutyryl-cAMP retards the mycelium-to-yeast morphological transition.
C. The bar charts show the percentage of each morphological form at 240 h after initiating the transition in the absence of cAMP (chart a) and for cells to which 10 mM dibutyryl-cAMP was added at the start of the transition (chart b), at 12 h (chart c) and 120 h (chart e) after initiating the transition; for comparison, the percentage of each morphological forms after 120 h is also shown (chart d). The data indicate that the addition of exogenous dibutyryl-cAMP late in the transition reverses the mycelium-to-yeast morphological transition. M-mycelium; DM-Differentiating mycelium; DY-Differentiating yeast; Y-Yeast.
Identification of the components of a cAMP-signalling pathway in P. brasiliensis
In an attempt to identify genes from the cAMP-signalling pathway, which our studies clearly implicated in the control of the morphological switch from the mycelium to pathogenic yeast form of P. brasiliensis, we used homology-based strategies to clone the genes that encode AC (e.g. CYR1) and several G proteins, including three Gα (e.g. GPA1, GPA2 and GPA3), Gβ (e.g. GPB1) and Gγ (e.g. GPG1) subunits, which might be expected to be involved in its regulation (see Supplementary material).
A phylogenetic analysis of all known fungal Gα proteins identified to date indicates that they fall into three major families (groups 1–3), each of which is represented by one of the three Gα proteins, Gpa1–3, from P. brasiliensis (Fig. S1). In contrast, only a single Gβ and Gγ have been identified in these fungi, raising the question as to whether they interact with more than one Gα. Consequently, we sought to test whether the Gβ protein Gpb1 interacts with Gpa1–3 from P. brasiliensis by two-hybrid screening in S. cerevisiae. Initially, no interactions between any of the Gα proteins and Gpb1 were detected (Fig. 2A). Although the Gβ protein Ste4 can interact independently, of its cognate Gγ protein Ste18, with the Gα protein Gpa1 in two-hybrid assays in S. cerevisiae (Ongay-Larios et al., 2000), it is possible that the failure to detect any interaction between the P. brasiliensis Gβ and Gα proteins was attributable to the requirement for a Gγ protein to stabilize Gpb1 for interaction with the Gα proteins. However, we failed to identify an interaction between Gpa1–3 and a Gpb1-linker–Gpg1 fusion protein, nor was there an interaction between Gpb1 and Gpg1 (data not shown). Consequently, we decided to test whether any of the Gpa proteins would interact with discrete domains of Gpb1, which might only be available for interaction in the Gpb1–Gpg1 complex. These experiments revealed an interaction of Gpa1, but not Gpa2 or Gpa3, with C-terminal-truncated Gpb1, with a deletion analysis indicated that Gpa1 interacted with the first two WD domains at the N-terminus (Fig. 2A). Gpa1 also interacted with a fusion protein in which the first WD domain was fused to the third, but not the seventh, WD domain (Fig. 2A). In analogy, S. cerevisiae Gpa2 has been reported to interact with an N-terminal-truncated, consisting of residues 531–740 of the, but not with the full-size, kelch-repeat protein Gpb1 (Batlle et al., 2003). We complemented this approach by constructing random mutagenesis libraries for Gpa1–3 and Gpb1 in the yeast two-hybrid vector pGADT7: screening the Gpa1–3 libraries with Gpb1 and the Gpb1 library with Gpa1–3. No point or frameshift mutations were detected that led to an interaction (data not shown). As a positive control we used the GPR1 and GPA2 genes that, respectively, encode a GPCR and its cognate Gα protein in S. cerevisiae: two-hybrid screens indicated that Gpa2 could interact with the C-terminal domain, encompassing residues 679–961, of Gpr1 (data not shown). When a random mutagenesis library of GPR1679−7961 was screened with GPA2, at least 50 positive colonies were detected. Screening the same library with P. brasiliensis GPA1–3 did not yield any positive colonies, nor did a screen of the P. brasiliensis GPA1–3 random mutagenesis libraries with S. cerevisiae GPR1 (data not shown). These results establish that there is a high degree of stringency in the Gα–GPCR interaction because none of the Gα proteins from P. brasiliensis could interact with S. cerevisiae Gpr1. We conclude that of the three Gα proteins found in P. brasiliensis, only Gpa1 interacts with Gpb1, and that it binds to the same N-terminal region of Gpb1 to which Gpg1 binds. These results suggest that Gpa1, Gpb1 and Gpg1 form a Gαβγ trimer.
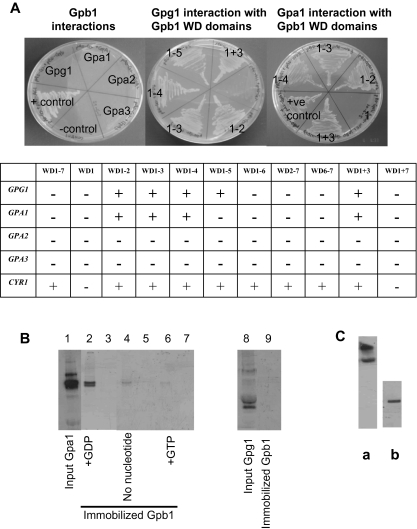
Fig. 2.
A. GPA1, but not GPA2 nor GPA3, and GPG1 interact with GPB1. Each of the P. brasiliensis proteins Gpa1, Gpa2, Gpa3, Gpg1 and Cyr1 was tested by two-hybrid screening in S. cerevisiae for interactions with Gpb1 (e.g. WD1–7) and a series of truncates in which successive WD domains were deleted from the C-terminus (e.g. WD1–6 to WD1); a construct that lacked the N-terminal WD domain (e.g. WD2–7); a construct that comprised the two C-terminal WD domains (e.g. WD6–7); and a fusion of WD domains 1 and 3 (e.g. WD1+3), and 1 and 7 (e.g. WD1+7). S. cerevisiae strain AH109, harbouring pGADT7 plasmids bearing genes that encoded proteins to test for interactions with Gpb1 and truncates of this protein, expressed from pGBKT7, were identified by auxotrophic selection on SD/–Leu/–Trp/–His/–ADE plates and Xgal assays. As illustrated by the left-hand plate, full-length Gpb1 did not interact with Gpa1, Gpa2, Gpa3 nor Gpg1. However, full-length Gpb1 did interact with the positive control Cyr1−678, consequently, establishing that the full-length protein is expressed. The middle- and right-hand plates show the interaction of Gpg1 and Gpa1 respectively with the WD domains of Gpb1. As a negative control, each pGBK protein vector was cotransformed, with pGADT7, into AH109 and screen for growth on SD/–Leu/–Trp/–His/–ADE plates. None of these control cells grew (data not shown). In the accompanying table, the (+) and (–) symbols are indicative of the presence and absence of protein–protein interactions respectively.
B. Pull-down assays to demonstrate that Gpa1 interacts with Gpb1. GST and GST-Gpb1 were purified from bacteria, loaded onto glutathione sepharose beads before incubation with in vitro translated 35S-Gpa1and 10 mM nucleotide. After washing the beads, the proteins were eluted by the addition of 4× NuPAGE LDS sample buffer, followed by boiling at 90°C for 5 min, and separated on a 4–12% NuPAGE gel under denaturing conditions. Bound Gpa1 was detected as a gel band by autoradiography. Lanes 2, 4 and 6 establish that Gpa1 binds to immobilized Gpb1, but the apparent affinity decreases in order of incubation with GDP (lane 2), no nucleotide (lane 4) and GTP (lane 6). Negative controls, using immobilized GST, are shown in lanes 3, 5, 7 and 10. Using in vitro translated 35S-Gpg1 (lane 8), there was no detectable interaction with Gpb1 (lane 9).
C. Gpb1 and Gpa1 used in pull-down assays cross-react with specific antibodies. (a) Gpb1 produced as a fusion protein with GST in E. coli and (b) Gpa1 synthesized using an in vitro transcription/translation system ran at the expected Mr on SDS-PAGE gels and cross-reacted with antibodies generated to specific sequences within these proteins.
We used pull-down assays to confirm the interaction of Gpa1, produced by in vitro translation, with Gpb1, expressed and purified as a glutathione S-transferase (GST) fusion protein from bacteria. We found that Gpa1 could interact directly with Gpb1, but this interaction appeared to be strengthened by GDP and blocked by GTP (Fig. 2B). The apparent capability of Gpa1 to bind GTP and dissociate from Gpb1 provides strong evidence that both proteins are correctly folded and functional. Furthermore, we found that both Gpa1 and Gpb1 cross-reacted with specific antibodies to these proteins (Fig. 2C). Consistent with our yeast two-hybrid assays, no interaction between Gpb1 and Gpg1, produced by in vitro translation, was detected. This is perhaps not surprising because recent studies have revealed a role for phosducins as molecular chaperones required for Gβγ dimer assembly (Lukov et al., 2005).
Adenylate cyclase interacts with Gpa1 and Gpb1
Although it has been known for some time that G proteins modulate the activity of mammalian AC by binding to the catalytic domain (Tesmer et al., 1997), only recently has it been established that S. pombe Gpa2 and S. cerevisiae Gpa2 bind to the N-terminus of AC to cause its activation in S. pombe (Ogihara et al., 2004; Ivey and Hoffman, 2005) and S. cerevisiae (Peeters et al., 2006) respectively.
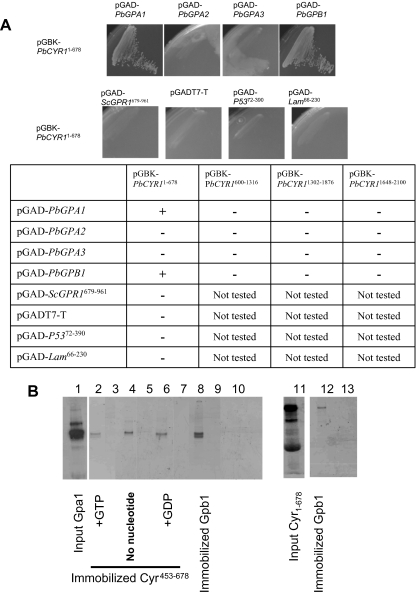
We used two-hybrid analyses to test whether any of the G proteins interact with AC from P. brasiliensis. The AC, when analysed with SMART (EMBL), contains four domains: a Ras association and Gα binding domain (RA, positions 1-678) domain (RA, positions); 14 leucine-rich repeats (LRR_TYR domains, positions 752–1244); a serine/threonine phosphatase family 2C catalytic domain (PP2Cc, positions 1341–1627); and an adenylyl/guanylyl cyclase catalytic domain (CYCc domain, positions 1574–1856). The AC cDNA was segmented into four parts with each containing an individual domain and cloned into yeast two-hybrid vectors to make constructions PbCYR11−678, PbCYR1600−1316, PbCYR11301−1876 and PbCYR11347−2100 that were used to test for interactions with Gpa1–3 and Gpb1 from P. brasiliensis. We found that the N-terminus of AC, encoded by the pGBK-PbCYR11−678 construct, interacted with Gpa1 and Gpb1, but not with Gpa2 nor Gpa3 (Fig. 3A). Furthermore, a deletion analysis indicated that AC could interact with a pair of WD domains from the extremes of either the N- or C-terminus of Gpb1 (Fig. 2A). We also tested for interactions between the PbCYR11−678, PbCYR1600−1316, PbCYR11301−1876 and PbCYR11347−2100 constructs, but none were found (data not shown).
Fig. 3.
A. Yeast two-hybrid assays indicate that full-length GPA1 and GPB1, but not GPA2 nor GPA3, directly interact with CYR11−678. Bait and prey vectors were simultaneously transformed into yeast strain AH109 and plated out on SD/–Leu/–Trp for 3 days. Yeast colonies that grew on SD/–Leu/Trp were restreaked on SD/–Ade/–His/–Leu/–Trp and incubated for a further 3 days. The results growths indicate that pGAD-PbGPA1 and pGAD-PbGPB1 could, but pGAD-PbGPA2 and pGAD-PbGPA3 could not, directly interact with pGBK-PbCYR11−678. In a series of negative controls, pGBK-PbCYR11−678 could not interact with pGAD-ScGPR1679−961, pGADT7-T, pGAD-P5372−390 and pGAD-Lam66−230. B. Pull-down assays to demonstrate that Gpa1 and Gpb1 both interact with Gpb1. GST and GST-Cyr453−678 were purified from bacteria, loaded onto glutathione sepharose beads before incubation with in vitro translated 35S-Gpa1 and 10 mM nucleotide. After washing the beads, the proteins were eluted by the addition of 4× NuPAGE LDS sample buffer, followed by boiling at 90°C for 5 min, and separated on a 4–12% NuPAGE gel under denaturing conditions. Bound Gpa1 was detected as a gel band by autoradiography. Lanes 2, 4 and 6 establish that Gpa1 binds to immobilized Cyr1, but there was little difference in apparent affinity after incubation with GTP (lane 2), no nucleotide (lane 4) or GDP (lane 6). Negative controls, using immobilized GST, are shown in lanes 3, 5, 7, 10 and 13. A control using immobilized GST-Gpb1 to pull-down 35S-Gpa1, in the presence of 10 mM GDP, shows a more intense band, suggesting that Gpa1-GDP is bound with higher affinity to Gpb1 than to Cyr. Using in vitro translated 35S-Cyr1−678 (lane 11), an interaction with immobilized GST-Gpb1 (lane 12) was detected.
To confirm these interactions, we used an N-terminal fragment of Cyr1, comprising residues 453–678, fused to GST, Cyr1(453−678)-GST, produced and purified from bacteria, for pull-down assays with in vitro translated Gpa1; and an in vitro translated N-terminal fragment of Cyr1, comprising residues 1–678, for pull-down assays with the Gpb1–GST fusion protein (Fig. 3B). These assays establishing that both Gpa1 and Gpb1 could interact with the N-terminus of AC and specifically with a region that incorporates the putative Gα and Ras binding domains, between residues 453 and 678 (Fig. 3B). Gpa1 was able to bind Cyr1 in the presence of GTP or GDP or in the absence of nucleotides but, surprisingly, the relative intensities of the bands suggested a preference for Gpa1 in the absence of nucleotides. A similar comparison suggested that Gpa1-GDP had a stronger affinity for Gpb1 than for Cyr1 (Fig. 3B, lane 8). Indeed, in a pull-down assay using immobilized Gpb1-GST with in vitro translated, 35S-labelled, Cyr11−678 and Gpa1, in the presence of excess Gpa1 and GDP, we did not detect a Cyr1 band, suggesting that Gpa1-GDP binds preferentially to Gpb1 (Fig. 3B, lane 9). However, the strength of these interactions will need to be confirmed by direct measurement when, and if, the proteins can be obtained in sufficient quantities for biophysical studies.
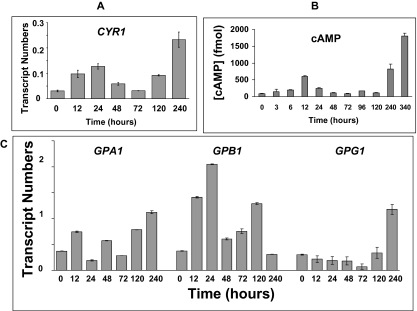
A transient increase in CYR1 transcript and cellular cAMP levels correlates with the onset of the morphological switch
Real-time reverse transcription polymerase chain reaction (RT-PCR) was used to evaluate the CYR1 transcript levels, which revealed that it is differentially expressed at higher levels in yeast than in mycelium (Fig. 4A). However, monitoring the transcript levels during the morphological transition revealed a significant transient peak in CYR1 transcripts after 24 h of the onset of the morphological transition, correlating with the peak in mycelial differentiation, and a further progressive increase in CYR1 transcripts from about 72 h, as the fungus adopted the yeast form. Considering this behaviour, we sought to determine whether the increase in CYR1 transcripts correlated with an increase in cellular cAMP levels. We found that the level of cellular cAMP peaked at about 12 h, and then progressively increased from a minimum at 72 h, strongly suggesting that increasing cAMP levels regulate the morphological transition (Fig. 4B). We noted that the CYR1 transcript levels increased at 24 and 240 h by about 3.2- and 7.5-fold respectively, while the cAMP levels were about 7.5- and 17-fold higher than in mycelium. This behaviour suggests that not only is the expression of AC upregulated, but it is also activated, presumably upon G protein binding.
Fig. 4.
The changes in intracellular cAMP levels correlate with the CYR1, GPA1, GPB1 and GPG1 transcript levels during the mycelium-to-yeast transition. The measured quantity of each P. brasiliensis gene mRNA in each of the treated samples was normalized by using the CT values obtained for the α-tubulin RNA amplifications run on the same plate. The relative quantification of each P. brasiliensis gene and α-tubulin gene expression was determined by a standard curve (i.e. Ct values plotted against the logarithm of the DNA copy number). The values represent the number of copies of the cDNAs of each P. brasiliensis gene divided by the number of copies of the cDNAs of the α-tubulin gene.
A. A bar chart showing the CYR1 transcript levels at the indicated times following an increase in temperature from 26°C to 37°C to induce the mycelium-to-yeast transformation. The data represent the average of three independent measurements.
B. The corresponding changes in the cellular cAMP levels during the morphological transition from mycelium to yeast are shown in chart B. Intracellular cAMP measurements were made using a non-acetylated EIA procedure (see Experimental procedures) and are the average of six assays.
C. A bar chart showing the GPA1, GPB1 and GPG1 transcript levels at the indicated times following an increase in temperature from 26°C to 37°C to induce the mycelium-to-yeast transformation. The data represent the average of three independent measurements.
Our results are intriguing because we previously found that the addition of exogenous dibutyryl-cAMP retarded the morphological transition. Perhaps the cells detect and respond to transient changes in cAMP levels rather than the absolute concentration of cAMP? Consistent with this proposal, we found that adding exogenous dibutyryl-cAMP 12 h after the onset of the morphological transition, when cellular cAMP levels would be maximal, had less effect in retarding the transition compared with its addition at the onset of the transition (i.e. compare charts b and c in Fig. 1C). However, the addition of dibutyryl-cAMP after 120 h, when the cAMP levels had dropped to a minimal level, similar to that in mycelium, induced a partial reversal of the transition (i.e. compare charts d and e in Fig. 1C): at this time, 0.5%, 5.5%, 75.5% and 18.5% of the morphology units were hyphae, differentiating hyphae, transforming yeast and yeast respectively (Fig. 1C, chart d); but after a further 120 h after the addition of the dibutyryl-cAMP, the proportion of these states was 9.3%, 22%, 43% and 25.7% respectively (Fig. 1C, chart e). Presumably because the cells are not synchronized in their development, some are committed to the transition and, accordingly, transform into yeast, but a large proportion of the cells convert back to hyphae. Furthermore, the addition of dibutyryl-cAMP, at concentrations up to 20 mM, did not induce the transformation of yeast at 37°C, indicating that once the cells had passed a certain point in their development, increasing dibutyryl-cAMP was insufficient to reverse this process (data not shown). Indeed, this is consistent with our hypothesis because the cellular cAMP levels are relatively high in yeast, suggesting that the yeast-to-mycelium transition is triggered by decreasing cAMP levels. Consistent with this hypothesis, we could not prevent the temperature-induced yeast-to-mycelium interconversion (e.g. upon decreasing the temperature from 37°C to 26°C) with exogenous dibutyryl-cAMP (data not shown).
Evidence for a switch from G subunit-signalling during the morphological transition
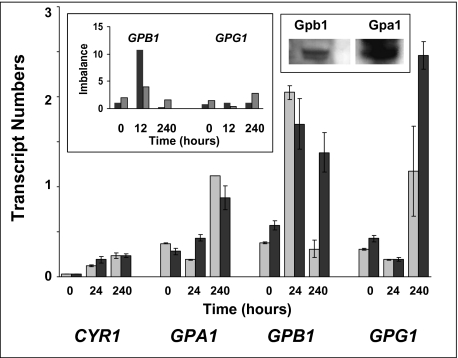
As our studies indicated that both Gpa1 and Gpb1 could interact with AC, we sought to determine whether there was an imbalance in the concentrations of the G protein subunits during the morphological switch from mycelium to yeast in P. brasiliensis. RT-PCR experiments indicated that the GPA1, GPB1 and GPG1 transcript levels were equivalent in mycelium; but there was a 5.4-fold increase in GPB1 transcript levels 24 h from the onset of the transition, while those for GPA1 and GPG1 declined 2-fold from that in mycelium, so that the GPB1 transcript levels were more than 10-fold higher than those for GPA1 and GPG1, which were still nearly equivalent (Fig. 5, left inset). Conversely, as the transition approached its end-point, after 240 h, when most cells had adopted the yeast form, the GPB1 transcript levels dropped to a level 3.7-fold lower than those for GPA1 and GPG1. This behaviour contrasts with that for RAS transcript levels that were about 50-fold higher than those for the G protein subunits and fluctuated little during the transition, suggestive of a role in controlling basal cAMP levels (see Fig. S2). We sought to confirm the imbalance in Gpa1 and Gpb1 subunits by Western blotting: comparing the intensities of the bands for the Gpa1 and Gpb1 blots indicated that there was, as predicted, greater expression of Gpa1 than Gpb1 in yeast (Fig. 5, right inset). We did not extend suchanalyses though, because they were complicated by the fact that the Gpa1 antibodies also cross-reacted with higher- and lower-molecular-weight proteins, which could be oligomers and degradation products respectively (data not shown). The presence of the latter would not be surprising if the concentration of Gpa1 needs to be finely controlled in order to modulate cAMP production. Indeed, it would be difficult to comprehend the functional significance of our finding that nucleotide-free Gpa1 and Gpb1 can bind to AC if these were always at equivalent concentrations and preferentially in complex with each other in the absence of GTP. However, we must be cautious in the interpretation of our transcript data because the imbalance in transcript levels may not reflect the difference in protein levels, especially if they are degraded at different rates, and there is a future need to ascertain whether, and how, Gpa1 and/or Gpb1 affect the catalytic activity of AC.
Fig. 5.
The hindrance of the mycelium-to-yeast transition by dibutyryl-cAMP correlates with an imbalance in Gpa1 and Gpb1 expression. A set of bar charts for the transcript levels of the CYR1, GPA1, GPB1 and GPG1 genes at the indicated times following an increase in temperature from 26°C to 37°C to induce the mycelium-to-yeast transformation in the absence (black bars) and presence (grey bars) of 10 mM dibutyryl-cAMP. The right inset shows the ratio of the GPB1 and GPG1 transcripts respectively relative to the number of GPA1 transcripts in mycelium, and at 12 and 240 h after the onset of the transition to the yeast form. The transcript numbers were determined in the absence (black bars) and presence (grey bars) of 10 mM dibutyryl-cAMP. Data are the average of three independent measurements. The right inset is a Western blot showing that Gpa1 is expressed at a higher level than Gpb1 in yeast.
As our morphology studies indicated that the addition of exogenous dibutyryl-cAMP retarded the switch from mycelium to yeast, we sought to determine whether this reflected a change in transcription of AC and/or G proteins. Accordingly, we redetermined the transcript levels in mycelium, 24 h after the onset of the transition, and in yeast, in the presence of 10 mM dibutyryl-cAMP (Fig. 5). There was little change in the CYR1 transcript levels at any stage in the morphological switch, which could have caused a reduction in cAMP levels to retard the transition. However, after 24 h, there was a notable reduction, from about 10- to 4-fold, in the imbalance in GPB1 to GPA1 transcripts. If Gpb1 has a lower affinity than Gpa1 for Cyr1, then this excess of Gpb1 might be insufficient to efficiently curtail the Gpa1 signal by displacing it from Cyr1, potentially retarding the transition. Furthermore, after 240 h there was still a 1.6-fold imbalance in GPB1 to GPA1 transcripts; whereas in the absence of exogenous dibutyryl-cAMP, the GPB1 transcript levels were 3.7-fold lower than GPA1. All of the Gpa1 should be complexed in the trimer, and none would be freely available to interact with AC as normal.
Discussion
Our data are consistent with the morphological transition in P. brasiliensis being controlled by changing cAMP levels, with the onset of the transition correlating with a transient increase in cAMP, suggesting that the cAMP-signalling pathway is activated. Furthermore, there is a clear correlation with the changes in cAMP levels and the expression of AC during the transition. However, the fold-increase in CYR1 transcripts (e.g. 3.2 and 7.5 at 24 and 240 h) was less than that in cAMP levels (e.g. 7.5 and 17 at 24 and 240 h), suggesting that increased cAMP was not simply due to more AC but because the protein is activated. We sought to identify the G proteins most likely responsible for the activation of AC. Most, if not all, filamentous fungi possess three or more Gα proteins, but, to date, only single Gβ and Gγ proteins have been identified. It has remained a mystery as to whether the Gβ and Gγ proteins can form trimeric complexes with the different Gα proteins. In this study, we have established for the first time that the Gβ and Gγ proteins, Gpb1 and Gpg1, interact with only a single Gα protein, Gpa1, in P. brasiliensis, presumably to form a Gpa1/Gpb1/Gpg1 trimeric complex. We did not find an interaction between Gpb1 and the other Gα proteins, Gpa2 and Gpa3, which work either independently or perhaps in association with other Gβ mimics (Harashima and Heitman, 2002; Palmer et al., 2006; Slessareva et al., 2006; reviewed by Hoffman, 2007). We then established that both the Gα and Gβ proteins, Gpa1 and Gpb1, could interact with the N-terminal domain of AC. Previous studies established that, in S. Pombe, Gpa2 interacts with the N-terminus of AC to cause its activation (Ogihara et al., 2004; Ivey and Hoffman, 2005), while more recently, an interaction between Gpa2 and AC from S. cerevisiae was confirmed (Peeters et al., 2006). Although there is genetic evidence for the Gβ protein Git5 interacting with and activating AC in S. pombe (Landry et al., 2000), we have shown a direct interaction between Gβ and a fungal AC that has not previously been demonstrated.
Our studies demonstrate that both Gα and Gβ proteins Gpa1 and Gpb1 bind to a site that lies between residues 453 and 678 of P. brasiliensis AC. Similarly, the activity of mammalian ACs is regulated by the binding of Gα and Gβ proteins (Dessauer et al., 2002; Diel et al., 2006). However, mammalian ACs are integral membrane proteins that share little sequence homology with fungal ACs, which are only associated with the periphery of the membrane. Mammalian ACs have a common topology consisting of two transmembrane domains, M1 and M2, each followed by a cytosolic catalytic domain, C1 and C2 (Krupinski et al., 1989). The catalytic activity of mammalian ACs is regulated by the binding of Gαs and Gαi proteins to the C2 and C1 cytoplasmic domains respectively, to increase or decrease the interactions of these domains (Dessauer et al., 2002). The effect of binding the Gβγ subunits, apparently to C1, depends upon the AC subtype, for example, activating ACII but inhibiting ACIII (Diel et al., 2006). Our data indicate that fungal ACs employ a different mechanism of modulation that involves the binding of G proteins to the N-terminus, but it is not clear how they modulate the activity of the catalytic domain, which is about 1000 residues away. We did not detect any interaction between the RA, LRR_TYR, PP2Cc and CYCc domains of AC, but these might only be induced by the binding of effectors, such as Gpa1 and Gpb1, to the N-terminal domain. The activity of AC in S. cerevisiae has been shown to be controlled by its interaction with Sgt1 that binds directly to the LRR domain (Dubacq et al., 2002). Similarly, Git7, a homologue of Sgt1, has been shown to control cAMP levels and play a role in glucose-triggered cAMP signalling in S. pombe (Schadick et al., 2002). Sequence analyses indicated that Sgt1 has features of a co-chaperone, and it has been proposed to act as a co-chaperone or factor in the assembly or the conformational activation of specific multiprotein complexes (Dubacq et al., 2002). Consistent with such a role, recent studies have shown that Sgt1 interacts with Hsp90 (Bansal et al., 2004;Lingelbach and Kaplan, 2004). Failure to detect interactions between the different domains of AC in the present investigation might be attributable to the involvement of Sgt1–Hsp90 in stabilizing inter-domain interactions and complex assembly. Considering the growing number of proteins identified as interacting with fungal ACs, this is an attractive hypothesis, as the interactions must occur in a controlled manner.
Our studies indicate that there is an imbalance in Gpa1, Gpb1 and Gpg1 G protein subunits as P. brasiliensis undergoes the morphological transition from mycelium to yeast. In mycelium the transcript levels for GPA1, GPB1 and GPG1 are nearly equivalent, but GPB1 predominates during the transition, while GPA1 predominates in yeast. It is notable that the CYR1, GPA1, GPB1 and GPG1 transcripts have extensive leader sequences that incorporate motifs likely to be targeted by RNA binding proteins that can be used to regulate the life times of these transcripts (see Supplementary material data). It seems plausible that signal progression is effected by regulating the longevity of these transcripts and of the translated proteins. Presumably, activation of the cAMP-signalling pathway in mycelium will depend upon GTP-induced dissociation of the Gpa1/Gpb1/Gpg1 trimer to release ‘free’ Gpa1 and Gpb1 that can interact with AC. However, remodelling of the cell, as it changes from mycelium to yeast, is a relatively lengthy process, requiring about 10 days for completion. A requirement for GTP to maintain the signal would constitute a metabolic waste over such a time period. An alternative strategy would be to increase the concentration of the protein that activates the signalling process, so that GTP-induced dissociation of the trimeric complex was not subsequently required. Early on in the transition Gpb1 is produced at higher levels than Gpa1 and, as we have established that it only interacts with the Gpa1, and not with Gpa2 or Gpa3, this excess will be ‘free’ to interact directly with AC. However, as we have found that Gpa1 can interact with AC in the absence of GTP, why does not the cell simply produce an excess of Gpa1 as it does in yeast? We note that the excess of Gpa1 in yeast correlates with an increase in the basal cAMP level, suggesting that nucleotide-free Gpa1 can activate AC. Consequently, if Gpa1 was produced in excess during the early stages of the transformation, it would be expected to maintain a high cAMP level, which we have shown, via the addition of dibutyryl-cAMP, actually retards the morphological transition. It seems plausible that Gpb1 serves a role in attenuating the cAMP signal by inactivating AC. Although we have not mapped the precise binding sites for Gpa1 and Gpb1, we have found that both G proteins bind to a domain of AC that incorporates residues 453–678, raising the possibility that they bind to the same site in a competitive manner. Considering the fact that the cAMP levels undergo cyclical changes during the morphological switch, and the effect of exogenous dibutyryl-cAMP in retarding the transition, particularly when cAMP levels are low, it seems likely that these changes in cAMP levels are needed to co-ordinate the activation of sequential steps in the morphological change.
Interestingly, while it is generally believed that it is the Gβγ dimer that is the functional unit, we have found that Gpb1 alone can interact with AC. This behaviour is, however, consistent with the expression of the Gpb1 exceeding that of Gpa1 and Gpg1 during the morphological switch. Gpg1 has a CCAAX box at its C-terminus (e.g. CCMIM) that is the site for prenylation, which is important in targeting the Gβγ dimer to the membrane (Whiteway and Thomas, 1994; Hirschman and Jenness, 1999; Manahan et al., 2000). A recent study indicated that there is considerable heterogeneity in the prenylation process, suggesting that this can affect the targeting of the Gβγ dimer, possibly as a means to switch between different signalling pathways (Cook et al., 2006). Accordingly, it is worth considering whether the increased expression of Gpb1, above that of Gpg1 and Gpa1, during the morphological transition is not only used to differentially modulate the activity of AC, but could be used to alter the targeting of Gpb1. Recent studies have shown that Gα proteins are segregated into distinct pools that allow specific signalling pathways to be activated at the plasmamembrane and at intracellular membranes (Slessareva et al., 2006; Slessareva and Dohlman, 2006).
Experimental procedures
Strain and culture
Paracoccidioides brasiliensis strain ATCC 90659 was grown as a mycelium form at 26°C and as a yeast form at 37°C in a modified liquid YPD medium (1% yeast extract, 2% neo-peptone and 2% dextrose, pH 6.5) with shaking at 110 r.p.m. for up to 15 days. The primers and plasmids used in this investigation are described in Tables 1 and 2 respectively, and the primers used for gene cloning are shown in Table S1. Exogenous cAMP was added to cultures of P. brasiliensis as the non-metabolite cAMP analogue dibutyryl-adenosine 3′-5′-cyclic monophosphoric acid (Sigma).
Table 1.
Primers used for constructing yeast two-hybrid vectors, quantitative RT-PCR and expression constructs for pull-down assays.
| Primer | Sequence (5′→3′; restriction sites underlined) | Description |
|---|---|---|
| PbAC-F26(KpnI) | GGTACCAAAATGTCTAGGAGACAGCGGGAGAAAGATAGG | Plasmid construction |
| PbAC-R34(NotI) | GCGGCCGCGCCGTGCTCGAAGAACTAGAACCAC | Plasmid construction |
| PbAC-GADF1(NcoI) | GGTACCACCATGGCAAGGAGACAGCGGGAGAAAG | Plasmid construction |
| PbAC-GADF2(NcoI) | TCGTCCATGGAAGATGAGCTGAATAACTAC | Plasmid construction |
| PbAC-GADR1(BamHI) | TGATTTGGATCCCGTTGATTCCCAAGTCAGC | Plasmid construction |
| PbAC-GADR2(BamHI) | CCCCAGGATCCTAAGATGTTTCAAAGAGTTG | Plasmid construction |
| PbAC-EF1(NdeI) | GGGATTCATATGGTTAATAGCACAGATCTG | Plasmid construction |
| PbAC-EF2(NdeI) | AGTATCCATATGGGACTGTCTCCATTAACTG | Plasmid construction |
| PbAC-ER1(XhoI) | CCATGGCTCGAGCGCCGTGCTCGAAGAACT | Plasmid construction |
| PbAC-ER2(XhoI) | CTCGCGCTCGAGATTGTGATTAAGATAGTC | Plasmid construction |
| PbGPA1-ExGPAF1 | CATATGGGTTGTGGAATGAGC | Plasmid construction |
| PbGPA1-pYER3 | GCCTAGGTCATATCAGTCCACAGAGGCGAAG | Plasmid construction |
| PbGPA2-F6 | CATGGGTTGCGCAAGTTCTCAACCAGTGGA | Plasmid construction |
| PbGPA2-F7(EcoRI) | CTAGGAATTCATGGGTTGCGCAAGTTCTCA | Plasmid construction |
| PbGPA2-R6 | CTGTTGGCACCTACAGAATCAGGTTGTTGA | Plasmid construction |
| PbGPA3-F5 | TGGTATCAGATAGGATGGGTGGGTGTTGCA | Plasmid construction |
| PbGPA3-R5 | CCCTTTCACAAAATACCAGAATCTTTCAGG | Plasmid construction |
| PbRAS-F1(NdeI) | CCGCGCTTCATATGCAGCTTTGTCTAAACC | Plasmid construction |
| PbRAS-R1(EcoRI) | GTGATACTGTAGAATTCCACGAAACCCTC | Plasmid construction |
| PbGPB-NdeI-F | ACGCTCATATGGCGGCCGATTTGAGCGG | Plasmid construction |
| PbGPB-BanHI-R | CGATGGATCCCTACCATGCCCAGACCTTGAG | Plasmid construction |
| PbGPG-NdeI-F | CATATGGCCCCTGCCTACGAGCTTCGAC | Plasmid construction |
| PbGPG-BamHI-R | GGATCCTTACATGATCATACAGCAGCCACCTGAT | Plasmid construction |
| pbgpb-NdeI-F | ACGCTCATATGGCGGCCGATTTGAGCGG | pAD-WD1 |
| gpbWD-1R | GCTAGGATCCTAATCGGAGATGATTAGTTTCC | pAD-WD1 |
| pbgpb-NdeI-F | ACGCTCATATGGCGGCCGATTTGAGCGG | pAD-WD2 |
| gpbWD-2R | GCTAGGATCCTAATTATAGATGGAACAGATG | pAD-WD2 |
| pbgpb-NdeI-F | ACGCTCATATGGCGGCCGATTTGAGCGG | pAD-WD3 |
| gpbWD-3R | GCTAGGATCCTAATCCCATAGCATACAGGTC | pAD-WD3 |
| pbgpb-NdeI-F | ACGCTCATATGGCGGCCGATTTGAGCGG | pAD-WD4 |
| gpbWD-4R | GCTAGGATCCTAATCCCAGAGTTTAGCAAAGG | pAD-WD4 |
| pbgpb-NdeI-F | ACGCTCATATGGCGGCCGATTTGAGCGG | pAD-WD5 |
| gpbWD-5R | CGTAGGATCCTAATCGAATAGACGGCAGGTGG | pAD-WD5 |
| pbgpb-NdeI-F | ACGCTCATATGGCGGCCGATTTGAGCGG | pAD-WD6 |
| gpbWD-6R | GCTAGGATCCTAGTCCCAGACCTTGCACTCAT | pAD-WD6 |
| pbgpb-NdeI-F | ACGCTCATATGGCGGCCGATTTGAGCGG | pAD-WD13 |
| gpbWD-1R | GCTAGGATCCTAATCGGAGATGATTAGTTTCC | pAD-WD13 |
| Gpb-WD3-F | GGATCCCTTTCCTCTCGAGAAGGTCC | pAD-WD13 |
| gpbWD-3R | GCTAGGATCCTAATCCCATAGCATACAGGTC | pAD-WD13 |
| Gpb-WD27-R | CATATGGCATACACAACAAACAAAGTGCAC | pAD-WD27 |
| pbgpb-BanHI-R | CGATGGATCCCTACCATGCCCAGACCTTGAG | pAD-WD27 |
| gpbWD-67-F | CATATG ATCCGCGCGGATAGAGAACTTAATAC | pAD-WD67 |
| pbgpb-BanHI-R | CGATGGATCCCTACCATGCCCAGACCTTGAG | pAD-WD67 |
| ScGPR1-F1 | CGGGATCCGAAGTGTGACGAATAAAGC | Plasmid construction |
| ScGPR1-F3 | CGGGATCCATATGATAACTGAGGGATTTCCCCCG | Plasmid construction |
| ScGPR1-F4 | CCGGATCCGTGAAAGTAAAAGAATTAAAGCGC | Plasmid construction |
| ScGPR1-F5 | CCGGATCCAGGAAAAACCTTGGAACTATTCATG | Plasmid construction |
| ScGPR1-R1 | CGGGATCCATTTTCAAACATCGCGATAC | Plasmid construction |
| ScGPR1-R3 | CCGGATCCTTAGATTCTTTTGAATTTGTGCC | Plasmid construction |
| ScGPA2-F1 | CCGGATCCTGGGTCTCTGCGCATCTTCA | Plasmid construction |
| ScGPA2-R1 | CCGGATCCGCTGTGCATTCATTGTAACAC | Plasmid construction |
| 3′BD screening | TGGCTGCAAGCGCGCAAAAAACCCCTCAAGAC | Plasmid construction |
| 5′BD screening | TCATCGGAAGAGAGTAGTAACAAAGGTCAAAGA | Plasmid construction |
| pGADT-linker-gamma-F1 | CCGCTCCGGCCCTGACCCCGGCGGATGTGGCCCGCAGCAT GGCCCCTGCCTACGAGCTTCGACCCG | For construct of PbGpb-linker-PbGpgamma fusion gene |
| pGADT-linker-gamma-F2 | GGATCCCGTATTAAAAACGGCAGCGGTGCGGCAGCCCCGAAA GCCGCTCCGGCCCTGACCCCGGCGGATG | For construct of PbGpb-linker-PbGpgamma fusion gene |
| pGADT-Gpb-R2(TAG) | GGATCCCCATGCCCAGACCTTGAGCAGAGAATC | For construct of PbGpb-linker-PbGpgamma fusion gene |
| Real-time PCR | ||
| Alpha P-alpha | CCAGAACCAGGCAGTCCAAA FAM-CACCTGCCTAACAAGATTGGACCAGG5G | Real-time PCR of α-tubulin |
| P-gpa1_Pb p-gpa1_Pb | GTACCGCCACCTACGTCGAACATACGG5AC-FAM ATGTCCTTCGCTCCCGTGTTA | Real-time PCR of gpa1 |
| P-Gpb_Pb p-Gpb_Pb | CACGGCGAGAAGGTCCAACCCG5G-FAM TCGCCGGAAGAAGTGATGATT | Real-time PCR of gpb1 |
| P-Gpg_Pb p-Gpg_Pb | CGGTTGTCAATCTGTCCCCAAAC5G-FAM CGGCCATGTCACTCATCAACT | Real-time PCR of gpg1 |
| P-PbAden_cyc p-PbAden_cyc | CACATTCAAAGTTTCGTGGGAAGGAATG5G-FAM AGAGGCCGATTCTCATGCAA | Real-time PCR of cyr1 |
| Protein overexpression | ||
| pGEX-6P-3 Cyr1453–678_Pb (BamHI and NotI) | AAGGATGGATCCGATAAAACCCATCAGGATAACTTTG CATATCGCGGCCGCTTAGTGGCTAAACTTTTGGTTCTCGTG | Plasmid construction |
| pGEX-6P-3 Gpb1 (BamHI and NotI) | ATACATGGATCCATGGCGGCCGATTTGAGCGGCG ATATCTGCGGCCGCCTACCATGCCCAGACCTTG | Plasmid construction |
In most cases, the same primers were used to make constructs in both pGADT7 and pGBKT7 for yeast two-hybrid screens.
Table 2.
Plasmid used in this study.
| Plasmid name | Vector | Insert and cloning description |
|---|---|---|
| pGEMTE-cPbAC-F26R34-1 | pGEM T Easy (Promega) | The insert is the full-length PbCYR1 cDNA amplified with PbAC-F26 and PbAC-R34 |
| pGBK-PbCYR1(1−678) | pGBKT7 (Clontech) | PbCYR1 cDNA fragment (NcoI/BamHI) obtained by PCR with PbAC-GADF1 and PbAC-GADR1 cloned into pGBKT7 (NcoI/BamHI) |
| pGBK-PbCYR1(600−1316) | pGBKT7 (Clontech) | PbCYR1 cDNA fragment (NcoI/BamHI) obtained by PCR with PbAC-GADF2 and PbAC-GADR2 cloned into pGBKT7 (NcoI/BamHI) |
| pGBK-PbCYR1(1302−1876) | pGBKT7 (Clontech) | PbCYR1 cDNA fragment (NdeI/XhoI) obtained by PCR with PbAC-EF1 and PbAC-ER2 cloned into pGBKT7 (NdeI/SalI) |
| pGBK-PbCYR1(1648−2100) | pGBKT7 (Clontech) | PbCYR1 cDNA fragment (NdeI/XhoI) obtained by PCR with PbAC-EF2 and PbAC-ER1 cloned into pGBKT7 (NdeI/SalI) |
| pGAD-PbCYR1(1−678) | pGADT7 (Clontech) | PbCYR11−678 fragment from pGBK-PbCYR1(1−678) (NdeI/BamHI) cloned into pGADT7 (NdeI/BamHI) |
| pGAD-PbCYR1(600−1316) | pGADT7 (Clontech) | PbCYR1600−1316 fragment from pGBK-PbCYR1(600−1316) (NdeI/BamHI) cloned into pGADT7 (NdeI/BamHI) |
| pGAD-PbCYR1(1302−1876) | pGADT7 (Clontech) | PbCYR1 cDNA fragment (NdeI/XhoI) obtained by PCR with PbAC-EF1 and PbAC-ER2 cloned into pGADT7 (NdeI/XhoI) |
| pGAD-PbCYR1(1648−2100) | pGADT7 (Clontech) | PbCYR1 cDNA fragment (NdeI/XhoI) obtained by PCR with PbAC-EF2 and PbAC-ER1 cloned into pGBKT7 (NdeI/XhoI) |
| pGEMTE-cPbGPA1-ExGPAF1pYER3-9(T7>SP6) | pGEM T Easy (Promega) | RT-PCR product with PbGPA1-ExGPAF1 and PbGPA1-pYER3 from Pb mRNA cloned into pGEM T Easy. The insert is the full-length PbGPA1 cDNA |
| pGEMTE-cPbGPA2-F6R6-2(SP6>T7) | pGEM T Easy (Promega) | RT-PCR product with PbGPA2-F6 and PbGPA2-R6 from Pb mRNA cloned into pGEM T Easy. The insert is the full-length PbGPA2 cDNA |
| pGEMTE-cPbGPA3-F5R5-2(T7>SP6) | pGEM T Easy (Promega) | RT-PCR product with PbGPA3-F5 and PbGPA3-R5 from Pb mRNA cloned into pGEM T Easy. The insert is the full-length PbGPA3 cDNA |
| pGBK-PbGPA1 | pGBKT7 (Clontech) | Insert (NdeI) from pGEMTE-cPbGPA1-ExGPAF1pYER3-9 cloned into pGBKT7 (NdeI) |
| pGAD-PbGPA1 | pGADT7 (Clontech) | PbGPA1 insert (NdeI digested) from pGBK-PbGPA1 cloned into pGADT7 (NdeI) |
| pGBK-PbGPA2 | pGBKT7 (Clontech) | PCR product (EcoRI digested) with PbGPA2-F7 and T7 as primers and pGEMTE-cPbGPA2-F6R6-2 as template, cloned into pGBKT7 (EcoRI digested) |
| pGAD-PbGPA2 | pGADT7 (Clontech) | PbGPA2 insert (NdeI/SalI digested) from pGBK-PbGPA2 cloned into pGADT7 (NdeI/XhoI) |
| pGBK-PbGPA3 | pGBKT7 (Clontech) | PCR product (EcoRI digested) with PbGPA3-F8 and SP6 as primers and pGEMTE-cPbGPA3-F5R5-2 as template, cloned into pGBKT7 (EcoRI digested) |
| pGAD-PbGPA3 | pGADT7 (Clontech) | PbGPA3 insert (NdeI/BamHI digested) from pGBK-PbGPA3 cloned into pGADT7 (NdeI/BamHI) |
| pGBK-PbGPB1 | pGBKT7 (Clontech) | PbGPB1 cDNA (NdeI/BamHI) cloned into pGBKT7 (NdeI/BamHI) |
| pGAD-PbGPB1 | pGADT7 (Clontech) | PbGPB1 cDNA (NdeI/BamHI) cloned into pGADT7 (NdeI/BamHI) |
| pGBK-PbRAS | pGBKT7 (Clontech) | PbRAS cDNA (NdeI/EcoRI) cloned into pGBKT7 (NdeI/EcoRII) |
| pGAD-PbRAS | pGADT7 (Clontech) | PbRAS insert (NdeI/EcoRI digested) from pGBK-PbRAS cloned into pGADT7 (NdeI/EcoRI) |
| pGBKT7-53 | pGBKT7 (Clontech) | From Clontech |
| pGAD-P53(72−390) | pGADT7 (Clontech) | P53 insert (NdeI/BamHI digested) from pGBKT7-P53, cloned into pGADT7 (NdeI/BamHI) |
| pGBKT7-Lam | pGBKT7 (Clontech) | From Clontech |
| pGAD-Lam(66−230) | pGADT7 (Clontech) | Lam insert (NdeI/BamHI) from pGBKT7-Lam cloned into pGADT7 (NdeI/BamHI) |
| pGADT7-T | pGADT7 (Clontech) | From Clontech |
| pGEMTE-ScGPR1-F1R1-17 | pGEM T Easy (Promega) | The insert is the full-length ScGPR1 amplified with Sc-GPR1-F1 and ScGPR1-R1 |
| pGAD-ScGPR1(679−961)-F5R1 | pGADT7 (Clontech) | PCR product (BamHI digested) with ScGPR1-F5 and ScGPR1-R1 as primers, and pGEMTE-ScGPR1-F1R1-17 as template, cloned into pGADT7 (BamHI digested); corresponding to C-terminal cytoplasmic domain |
| pGAD-ScGPR1(274−621)-F4R3 | pGADT7 (Clontech) | PCR product (BamHI digested) with ScGPR1-F4 and ScGPR1-R3 as primers, and pGEMTE-ScGPR1-F1R1-17 as template, cloned into pGADT7 (BamHI digested); corresponding to the third cytoplasmic loop |
| pGAD-ScGPR1(1−961)-F3R1 | pGADT7 (Clontech) | PCR product (BamHI digested) with ScGPR1-F3 and ScGPR1-R1 as primers, and pGEMTE-ScGPR1-F1R1-17 as template, cloned into pGADT7 (BamHI digested); corresponding to full-length ScGPR1 |
| pGBK-ScGPR1(1−961)-F3R1 | pGBKT7 (Clontech) | Insert (BamHI digested) from pGAD-ScGPR1(1−961)-F3R1 cloned into pGBKT7 |
| pGBK-ScGPA2-F1R1 | pGBKT7 (Clontech) | PCR product (BamHI digested) with ScGPA2-F1 and ScGPA2-R1 as primers, and Sc DNA as template, cloned into pGBKT7 (BamHI digested); corresponding to full-length ScGPA2 |
| pAD-PbGPB1 | pGADT7 (Clontech) | 1062 bp PbGPB1 gene linked into pGADT-7 at the sites of NdeI and BamHI |
| pBD-PbGPB1 | pGBKT7 (Clontech) | 1062 bp PbGPB1 gene linked into pGBKT-7 at the sites of NdeI and BamHI |
| pAD-PbGPB1-TAG | pGADT7 (Clontech) | 1059 bp PbGPB1 gene without stop code was linked into pGADT-7 at the site of NdeI and BamHI |
| pBD-PbGPB1-TAG | pGBKT7 (Clontech) | 1059 bp PbGPB1 gene without stop code was linked into pGBDT-7 at the site of NdeI and BamHI |
| pAD-PbGPG1 | pGADT7 (Clontech) | 276 bp PbGPG1 gene linked into pGADT-7 at the sites of NdeI and BamHI |
| pBD-PbGPG1 | pGBKT7 (Clontech) | 276 bp PbGpg gene linked into pGBKT-7 at the sites of NdeI and BamHI |
| pAD-PbGPB1-link-PbGPG1 | pGADT7 (Clontech) | PbGpb-flexible linker-PbGpg was ligated into pGADT-7 at the site of NdeI and BamHI |
| pBD-PbGPB1-link-PbGPG1 | pGBKT7 (Clontech) | PbGpb-flexible linker-PbGpb was ligated into pGBKT-7 at the site of NdeI and BamHI |
| pAD-WD1 | pGBKT7 (Clontech) | The first WD domain of PbGpb1 with a stop codon linked into pGADT-7 at the sites of NdeI and BamHI |
| pAD-WD2 | pGADT7 (Clontech) | The region encoding from the N-termini to the end of the second WD domain of PbGpb1 with a stop codon linked into pGADT-7 vector at the sites of NdeI and BamHI |
| pAD-WD3 | pGADT7 (Clontech) | The region encoding from the N-termini to the end of the third WD domain of PbGpb1 with a stop codon linked into pGADT-7 vector at the sites of NdeI and BamHI |
| pAD-WD4 | pGADT7 (Clontech) | The region encoding from the N-termini to the end of the fourth WD domain of PbGpb1 with a stop codon linked into pGADT-7 vector at the sites of NdeI and BamHI |
| pAD-WD5 | pGADT7 (Clontech) | The region encoding from the N-termini to the end of the fifth WD domain of PbGpb1 with a stop codon linked into pGADT-7 vector at the sites of NdeI and BamHI |
| pAD-WD6 | pGADT7 (Clontech) | The region encoding from the N-termini to the end of the sixth WD domain of PbGpb1 with a stop codon linked into pGADT-7 vector at the sites of NdeI and BamHI |
| pAD-WD13 | pGADT7 (Clontech) | The region encoding a fragment containing the first and third WD domains of PbGpb1 with a start codon linked into pGADT-7 vector at the sites of NdeI and BamHI |
| pAD-WD27 | pGADT7 (Clontech) | The region encoding a fragment from the second WD domain to the end of PgGpb1 with a start codon linked into pGADT-7 vector at the sites of NdeI and BamHI |
| pAD-WD67 | pGADT7 (Clontech) | The region encoding a fragment from the sixth WD domain to the end of PgGpb1 with a start codon linked into pGADT-7 vector at the sites of NdeI and BamHI |
| pAD-WD17 | pGADT7 (Clontech) | Deletion of a SalI fragment from the region encoding the fragment between the first and seventh WD domains of PbGpb1 in pAD-PbGPB1 |
| pGEX-6P-3 Cyr1(453−678) | pGEX-6P-3 (GE Healthcare) | The region encoding the cDNA fragment of CYR1, 225 bp (Gα binding domain and Ras association domain) linked to pGEX-6P-3 vector (GST fusion) at the sites of BamHI and NotI |
| pGEX-6P-3 Gpb1 | pGEX-6P-3 (GE Healthcare) | 1062 bp PbGPB1 gene linked into pGEX-6P-3 (GST fusion) vector at the sites of BamHI and NotI |
Microscopy
For the microscopic assays, the different morphotypes were transferred to fixative solution (3.7% formaldehyde, 50 mM sodium phosphate buffer pH 7.0, 0.2% Triton X-100) for 120 min at room temperature. Then, they were briefly rinsed with PBS buffer (140 mM NaCl, 2 mM KCl, 10 mM NaHPO4, 1.8 mM KH2PO4, pH 7.4) and mounted on the slides. The material was photographed using a Zeiss epifluorescence microscope.
RNA extraction
For the real-time RT-PCR experiments, yeast cells and mycelium were disrupted with glass beads and grinding in liquid nitrogen respectively, and immediately mixed with Trizol (Gibco-BRL) for RNA extraction following the supplier's recommendations. To verify the RNA integrity, 20 μg of RNA from each treatment was fractionated in 2.2 M formaldehyde, 1.2% agarose gel, stained with ethidium bromide, and visualized with UV light. The presence of intact 28S and 18S ribosomal RNA bands was used as a criterion to verify whether the RNA was intact. RNase-free DNase treatment was performed in a final volume of 100 μl containing 40 mM Tris-HCl pH 7.5 and 6 mM MgCl2, 1 μl of RNasin (40 U μl−1, Promega, USA), 10 μl of RNase-free DNase (1 U μl−1, Promega or Life Technologies, USA), 2.5 μl of 200 mM DTT, and 10 μg of total RNA. The reaction was incubated at 37°C for 60 min and stopped by incubating at 70°C for 30 min. The absence of DNA contamination after the RNase-free DNase treatment was verified by PCR amplification of the GP43 gene.
Construction of cDNA libraries
A P. brasiliensis yeast cDNA library was constructed in the vector pDNR-LIB using the Creator SMART cDNA Library (Clontech) according to the manufacturer's instructions. The CYR1, GPA1, GPA2, GPA3, GPB1, GPG1 and RAS genes were cloned from the Creator pDNR Library as described in Supplementary methods.
Real-time PCR and RT-PCR reactions
All the real-time PCR and RT-PCR reactions were performed using an ABI Prism 7700 Sequence Detection System (Perkin-Elmer Applied Biosystems, USA). Taq-ManR EZ RT-PCR kits (Applied Biosystems, USA) were used for RT-PCR reactions. The thermocycling conditions comprised an initial step at 50°C for 2 min, followed by 30 min at 60°C for reverse transcription, 95°C for 5 min, and 40 cycles at 94°C for 20 s and 60°C for 1 min. Taq ManR PCR reagent kits were used for PCR reactions. As there is no ideal control for gene expression, we first compared several genes as normalizers for the expression experiments, such as those encoding α-tubulin, hexokinase and a translation factor. We have seen no difference by using any of these normalizers. Accordingly, the calibrator gene used for the expression experiments was the α-tubulin gene (data not shown). The reactions and calculations were performed according to Semighini et al. (2002). Primer and probe sequences are described in Table 1.
Subcloning of genes for yeast two-hybrid analysis
The GPA1, GPA2, GPA3, GPB1 and GPG1 genes, and fragments of the CYR1 and GPB1 gene, were subcloned into the vectors pGBKT7 and pGADT7 for use in yeast two-hybrid screens (Table 2).
Construction of random mutagenesis libraries for yeast two-hybrid screening
Random mutagenesis libraries for GPA1, GPA2, GPA3 and GPB1 from P. brasiliensis, GPR1 from S. cerevisiae, and mammalian P53 and LAM were constructed using the GeneMorph II Random Mutagenesis kit (Stratagene) and cloned into the prey vector pGADT7 to create the libraries pGAD-PbGPA1-RM-Lib, pGAD-PbGPA2-RM-Lib, pGAD-PbGPA3-RM-Lib, pGAD-PbGPB-RM-Lib, pGAD-ScGPR1-F5R1-RM-Lib, pGAD-P53-RM-Lib and pGAD-Lam-RM-Lib respectively for yeast two-hybrid screening.
Yeast two-hybrid analysis and screening
The Matchmaker Two-Hybrid System 3 (Clontech) was used to test for protein–protein interactions and to screen librariesfor potential interacting protein partners. For the former purpose, transformants were generated by introducing both bait and prey vectors into yeast strain AH109 simultaneously. For the latter, bait vectors were introduced into AH109 first, followed by sequential transformation with 20 μg of prey library plasmids. Experimental procedures were conducted in accord with the Matchmaker GAL4 Two-Hybrid System 3 manual and the Yeast Protocol Handbook (Clontech). Protein interactions were identified by observing the growth of transformants on SD-Ade/–His/–Leu/–Trp plates. After screening random mutagenesis libraries, candidate transformants were twice restreaked on SD-Ade/–His/–Leu/–Trp plates to allow loss of non-interacting prey vectors. Interacting prey vectors were purified using a Yeast Plasmid Isolation kit (Clontech) and subjected to DNA sequence analysis to confirm the identities of the interacting gene products.
Assays for cAMP production
Paracoccidioides brasiliensis mycelium growing in modified liquid YPD media at 26°C was subjected to an increase in temperature to 37°C to induce the morphological transition to the yeast form. Cells were harvested at different times during the transition and immediately stored at −80°C. To thawed cells, collected by centrifugation, 4% formic acid was added and agitated for 5 h to disrupt the cells. The cell debris was removed by centrifugation and the supernatant was lyophilized. Subsequently, the lyophilized pellet was made up in assay buffer, containing 2.5% dodecyl-trimethyl-ammonium-bromide, and was assayed using a Biotrak Enzyme Immuno Assay (EIA) kit from Amersham, according to the manufacturer's protocol 3. Measurements were normalized by using an equivalent wet weight of cells during the disruption procedure.
Expression of GST-Gpb1 and GST-Cyr1453−678
GPB1 and CYR1453−5678 were cloned into pGEX6p-3, to enable expression of GST fusion proteins, and transformed into Escherichia coli codon plus cells, which were grown in 2YT, at 25°C with shaking at 200 r.p.m., before induction with 0.1 mM IPTG. Cells were harvested by centrifugation, resuspended and disrupted by passage through a Constant Systems cell disrupter; 0.1% Triton X-100 was added to the disrupted cells and the debris collected by ultracentrifugation. The supernatant was mixed with GST beads and incubated on a rotator for 30 min at 4°C, loaded into a glass column, and washed with PBS and finally with GST elution buffer (50 mM Tris/HCl pH 8.0, 10 mM glutothione). The elution fractions were run on 4–12% SDS-PAGE (NuPAGE precast) polyacrylamide gels. The protein concentrations were measured using a BCATM protein assay kit (Pierce). These procedures typically yielded 7 mg ml−1 GST-Cyr1453−678, 5 mg ml−1 GST-Gpb and 3 mg ml−1 GST.
In vitro translation of Gpa1, Gpg1 and Cyr11−678
Gpa1, Gpg1 and Cyr11−678 were synthesized by an in vitro coupled transcription and translation system (Promega), labelling the proteins with RedivueTMl-35S-methionine (Amersham), using rabbit reticulocyte lysate. This was necessary because our attempts to overexpress these proteins, as well as full-length Cyr1, either as His-tagged or as GST-tagged proteins, always resulted in the production of inclusion bodies. The yeast two-hybrid pGBKT7 vectors, into which each gene or gene fragment had been cloned, were used as the template for the in vitro translation. The reaction was incubated at 30°C for 2 h, and 2.5 μl of the translated samples was loaded onto a gel to verify the translation. The translated proteins were stored at 4°C.
Pull-down assays
GST pull-down assays were performed with GST fusion proteins as bait and proteins labelled with 35S, produced by TNT-coupled transcription/translation (Promega), as prey. The GST proteins were immobilized on 40 μl glutathione sepharose 4B beads (GE Healthcare), which had been preblocked with 200 μl of binding buffer (20 mM HEPES pH 7.9, 600 mM NaCl, 0.1% Tween 20, 5% glycerol, 1 mM DTT, 5% milk and 1% BSA) for 10–15 min at room temperature with 10 μl of EDTA-free protease inhibitor (1/4 tablet in 0.2 mlPBS) (Roche); to which was added 10 μl of in vitro translated protein and, in some assays, 10 mM GTP or GDP, and incubated on an end-over rotator at room temperature for 2 h. To facilitate comparisons between different pull-down experiments, we always utilized the same batch of in vitro translated product at an equivalent concentration. The beads were washed seven times with buffer (20 mM HEPES pH 7.9, 600 mM NaCl, 0.1% Tween 20, 5% glycerol, 1 mM DTT), and then proteins were eluted by the addition of 4× NuPAGE LDS sample buffer (Invitrogen), followed by boiling at 90°C for 5 min. The proteins were separated on 4–12% NuPAGE (precast Bis-Tris) gels. The gels were fixed with 20% ethanol and 10% acetic acid for 30 min, and then soaked in 5–10 ml of fluorographic reagent NAMP 100 (Amersham Biosciences) to amplify the signal. The gels were dried at 80°C for 35 min under vacuum and autoradiographed (2–3 days exposed at −80°C). Each assay was repeated three times with a different batch of in vitro translated product to confirm the results.
Western blots
Antibodies were to the GST tag (Novagen) or commercially produced polyclonal antibodies (Invitrogen) raised in rabbits to specific oligopeptides: Gpa1 – CFR RSR EYQ LND SAR and Gpb1 – CDI RAD REL NTY QSD.
Proteins were separated by SDS-PAGE (on 4–12% polyacrylamide gels) and electrotransferred to PVDF membranes. Blots were incubated with the respective antibodies (e.g. anti-GST at 1:12 500 dilution and specific antibodies at 1:2500 dilution). Alkaline-phosphatase-conjugated anti-mouse IgG (1:2500 dilution) and horseradish peroxidase-conjugated anti-rabbit IgG (1:2500 dilution) were used as secondary antibodies for GST and specific protein blots respectively.
Nucleotide sequence accession number
The GenBank accession numbers for the P. brasiliensis genes used in this study are: CYR1 (AAS01025), GPA1 (AAT40562), GPA2 (AAT40564), GPA3 (AAT40563), GPB1 (AAT40565), GPG1 (EF687895) and RAS (AY547438).
Acknowledgments
This work was supported by grants from the Wellcome Trust to M.I.B.W. and A.R.W., and from Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), both from Brazil, to G.H.G. D.C. and G.C. were recipients of travelling fellowships from the Wellcome Trust. This collaboration was also supported by an international project grant from the Royal Society (UK).
Supplementary material
The following supplementary material is available for this article:
The phylogenetic relationship between Paracoccidioides brasiliensis and other fungal cAMP-signalling proteins – a neighbour-joining bootstrap tree is derived from the amino-acid sequence alignments of fungal (A) adenylate cyclase (AC) (B) Gb, (C) Gg and (D) Ga proteins, using the VectorNTi 6.0 align program (Informax).
The RAS transcript levels during the mycelium-toyeast transition.
Primers used for gene cloning.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05824.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, et al. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal PK, Abdulle R, Kitagawa K. Sgt1 associates with Hsp90: an initial step of assembly of the core kinetochore complex. Mol Cell Biol. 2004;24:8069–8079. doi: 10.1128/MCB.24.18.8069-8079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle M, Lu A, Green DA, Xue Y, Hirsch JP. Krh1p and Krh2p act downstream of the Gpa2p G (alpha) subunit to negatively regulate haploid invasive growth. J Cell Sci. 2003;116:701–710. doi: 10.1242/jcs.00266. [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Johnson AD. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol Microbiol. 2006;62:100–119. doi: 10.1111/j.1365-2958.2006.05367.x. [DOI] [PubMed] [Google Scholar]
- Borges-Walmsley MI, Walmsley AR. cAMP signaling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 2000;8:133–141. doi: 10.1016/s0966-842x(00)01698-x. [DOI] [PubMed] [Google Scholar]
- Borges-Walmsley MI, Chen D, Shu X, Walmsley AR. The pathobiology of Paracoccidioides brasiliensis. Trends Microbiol. 2002;10:80–87. doi: 10.1016/s0966-842x(01)02292-2. [DOI] [PubMed] [Google Scholar]
- Bradsher RW, Chapman SW, Pappas PG. Blastomycosis. Infect Dis Clin North Am. 2003;17:21–40. doi: 10.1016/s0891-5520(02)00038-7. vii. [DOI] [PubMed] [Google Scholar]
- Chang MH, Chae KS, Han DM, Jahng KY. The GanB Galpha-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetics. 2004;167:1305–1315. doi: 10.1534/genetics.103.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, Ma P, Cauwenberg L, Winderickx J, Crauwels M, Teunissen A, et al. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose- and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:3326–3341. doi: 10.1093/emboj/17.12.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S, Ronchetti D, Thevelein JM, Winderickx J, Martegani E. Activation state of the Ras2 protein and glucose-induced signaling in Saccharomyces cerevisiae. J Biol Chem. 2004;279:46715–46722. doi: 10.1074/jbc.M405136200. [DOI] [PubMed] [Google Scholar]
- Cook LA, Schey KL, Wilcox MD, Dingus J, Ettling R, Nelson T, et al. Proteomic analysis of bovine brain G protein γ subunit processing heterogeneity. Mol Cell Proteomics. 2006;5:671–685. doi: 10.1074/mcp.M500223-MCP200. [DOI] [PubMed] [Google Scholar]
- Delgado-Jarana J, Martinez-Rocha AL, Roldan-Rodriguez R, Roncero MI, Di Pietro A. Fusarium oxysporum G-protein beta subunit Fgb1 regulates hyphal growth, development, and virulence through multiple signalling pathways. Fungal Genet Biol. 2005;42:61–72. doi: 10.1016/j.fgb.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Dessauer CW, Chen-Goodspeed M, Chen J. Mechanism of Galpha i-mediated inhibition of type V adenylyl cyclase. J Biol Chem. 2002;277:28823–28829. doi: 10.1074/jbc.M203962200. [DOI] [PubMed] [Google Scholar]
- DiCaudo DJ. Coccidioidomycosis: a review and update. J Am Acad Dermatol. 2006;55:929–942. doi: 10.1016/j.jaad.2006.04.039. [DOI] [PubMed] [Google Scholar]
- Diel S, Klass K, Wittig B, Kleuss C. Gβγ activation site in adenylyl cyclase type II. Adenylyl cyclase type III is inhibited by Gβγ. J Biol Chem. 2006;281:288–294. doi: 10.1074/jbc.M511045200. [DOI] [PubMed] [Google Scholar]
- Dubacq C, Guerois R, Courbeyrette R, Kitagawa K, Mann C. Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot Cell. 2002;1:568–582. doi: 10.1128/EC.1.4.568-582.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M, Thumer L, Lohse MJ, Bunemann M. G-protein-activation without subunit dissociation depends on a Galpha i-specific region. J Biol Chem. 2005;280:24584–24590. doi: 10.1074/jbc.M414630200. [DOI] [PubMed] [Google Scholar]
- Ganem S, Lu SW, Lee BN, Chou DY, Hadar R, Turgeon BG, Horwitz BA. G-protein beta subunit of Cochliobolus heterostrophus involved in virulence, asexual and sexual reproductive ability, and morphogenesis. Eukaryot Cell. 2004;3:1653–1663. doi: 10.1128/EC.3.6.1653-1663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hao N, Yildirim N, Wang Y, Elston TC, Dohlman HG. Regulators of G protein signaling and transient activation of signaling: experimental and computational analysis reveals negative and positive feedback controls on G protein activity. J Biol Chem. 2003;278:46506–46515. doi: 10.1074/jbc.M308432200. [DOI] [PubMed] [Google Scholar]
- Harashima T, Heitman J. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol Cell. 2002;10:163–173. doi: 10.1016/s1097-2765(02)00569-5. [DOI] [PubMed] [Google Scholar]
- Hirschman JE, Jenness DD. Dual lipid modification of the yeast Gγ subunit Ste18p determines membrane localization of Gβγ. Mol Cell Biol. 1999;19:7705–7711. doi: 10.1128/mcb.19.11.7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS. Except in every detail: comparing and contrasting G-protein signaling in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Eukaryot Cell. 2005;4:495–503. doi: 10.1128/EC.4.3.495-503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS. Propping up our knowledge of G protein signaling pathways: diverse functions of putative noncanonical Gβ subunits in fungi. Sci STKE. 2007;370:pe3. doi: 10.1126/stke.3702007pe3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey FD, Hoffman CS. Direct activation of fission yeast adenylate cyclase by the Gpa2 Galpha of the glucose signaling pathway. Proc Natl Acad Sci USA. 2005;102:6108–6113. doi: 10.1073/pnas.0502270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Akiyama K, Kan T, Ohguchi T, Takata R. The G protein beta subunit FGB1 regulates development and pathogenicity in Fusarium oxysporum. Curr Genet. 2003;43:79–86. doi: 10.1007/s00294-003-0372-9. [DOI] [PubMed] [Google Scholar]
- Kasahara S, Nuss DL. Targeted disruption of a fungal G-protein beta subunit gene results in increased vegetative growth but reduced virulence. Mol Plant Microbe Interact. 1997;10:984–993. doi: 10.1094/MPMI.1997.10.8.984. [DOI] [PubMed] [Google Scholar]
- Kauffman CA. Histoplasmosis: a clinical and laboratory update. Clin Microbiol Rev. 2007;20:115–132. doi: 10.1128/CMR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, Van Dijck P, et al. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- Krupinski J, Coussen F, Bakalyar HA, Tang WJ, Feinstein PG, Orth K, et al. Adenylyl cyclase amino acid sequence: possible channel- or transporter-like structure. Science. 1989;244:1558–1564. doi: 10.1126/science.2472670. [DOI] [PubMed] [Google Scholar]
- Krystofova S, Borkovich KA. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gbetagamma dimer required for normal female fertility, asexual development, and galpha protein levels in Neurospora crassa. Eukaryot Cell. 2005;4:365–378. doi: 10.1128/EC.4.2.365-378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladds G, Davis K, Das A, Davey J. A constitutively active GPCR retains its G protein specificity and the ability to form dimers. Mol Microbiol. 2005;55:482–497. doi: 10.1111/j.1365-2958.2004.04394.x. [DOI] [PubMed] [Google Scholar]
- Lafon A, Seo JA, Han KH, Yu JH, d'Enfert C. The heterotrimeric G-protein GanB ({alpha})-SfaD ({beta})-GpgA ({gamma}) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics. 2005;171:71–80. doi: 10.1534/genetics.105.040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A, Han KH, Seo JA, Yu JH, d'Enfert C. G-protein and cAMP-mediated signaling in aspergilli: a genomic perspective. Fungal Genet Biol. 2006;43:490–502. doi: 10.1016/j.fgb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Landry S, Hoffman CS. The git5 Gbeta and git11 Ggamma form an atypical Gbetagamma dimer acting in the fission yeast glucose/cAMP pathway. Genetics. 2001;157:1159–1168. doi: 10.1093/genetics/157.3.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S, Pettit MT, Apolinario E, Hoffman CS. The fission yeast git5 gene encodes a Gbeta subunit required for glucose-triggered adenylate cyclase activation. Genetics. 2000;154:1463–1471. doi: 10.1093/genetics/154.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T, Wu C, Schrag JD, Whiteway M, Thomas DY, Leberer E. Interaction of a G-protein beta-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- Liebmann B, Gattung S, Jahn B, Brakhage AA. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol Genet Genomics. 2003;269:420–435. doi: 10.1007/s00438-003-0852-0. [DOI] [PubMed] [Google Scholar]
- Lingelbach LB, Kaplan KB. The interaction between Sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol Cell Biol. 2004;24:8938–8950. doi: 10.1128/MCB.24.20.8938-8950.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Pan X, Harashima T, Cardenas ME, Xue Y, Hirsch JP, Heitman J. The G protein-coupled receptor gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukov GL, Hu T, McLaughlin JN, Hamm HE, Willardson BM. Phosducin-like protein acts as a molecular chaperone for G protein betagamma dimer assembly. EMBO J. 2005;24:1965–1975. doi: 10.1038/sj.emboj.7600673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidan MM, De Rop L, Serneels J, Exler S, Rupp S, Tournu H, et al. The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol Biol Cell. 2005;16:1971–1986. doi: 10.1091/mbc.E04-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manahan CL, Patnana M, Blumer KJ, Linder ME. Dual lipid modification motifs in G(alpha) and G(gamma) subunits are required for full activity of the pheromone response pathway in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:957–968. doi: 10.1091/mbc.11.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medoff G, Painter A, Kobayashi GS. Mycelial-to-yeast-phase transitions of the dimorphic fungi Blastomyces dermatitidis and Paracoccidioides brasiliensis. J Bacteriol. 1987;169:4055–4060. doi: 10.1128/jb.169.9.4055-4060.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa T, Takagi Y, Shinozaki M, Yun CW, Schell WA, Perfect JR, et al. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot Cell. 2004;3:919–931. doi: 10.1128/EC.3.4.919-931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Jones R. Sporotrichosis. Clin Exp Dermatol. 2002;27:427–431. doi: 10.1046/j.1365-2230.2002.01087.x. [DOI] [PubMed] [Google Scholar]
- Muller P, Leibbrandt A, Teunissen H, Cubasch S, Aichinger C, Kahmann R. The Gbeta-subunit-encoding gene bpp1 controls cyclic-AMP signaling in Ustilago maydis. Eukaryot Cell. 2004;3:806–814. doi: 10.1128/EC.3.3.806-814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek JC, Wuthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Park G, Xu JR. The G-beta subunit MGB1 is involved in regulating multiple steps of infection-related morphogenesis in Magnaporthe grisea. Mol Microbiol. 2003;50:231–243. doi: 10.1046/j.1365-2958.2003.03676.x. [DOI] [PubMed] [Google Scholar]
- Obara T, Nakafuku M, Yamamoto M, Kaziro Y. Isolation and characterization of a gene encoding a G-protein alpha subunit from Schizosaccharomyces pombe: involvement in mating and sporulation pathways. Proc Natl Acad Sci USA. 1991;88:5877–5881. doi: 10.1073/pnas.88.13.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara H, Shima F, Naito K, Asato T, Kariya K, Kataoka T. Direct activation of fission yeast adenylyl cyclase by heterotrimeric G protein gpa2. Kobe J Med Sci. 2004;50:111–121. [PubMed] [Google Scholar]
- Ongay-Larios L, Savinon-Tejeda AL, Williamson MJ, Jr, Duran-Avelar M, Coria R. The Leu-132 of the Ste4(Gbeta) subunit is essential for proper coupling of the G protein with the Ste2 alpha factor receptor during the mating pheromone response in yeast. FEBS Lett. 2000;467:22–26. doi: 10.1016/s0014-5793(00)01106-6. [DOI] [PubMed] [Google Scholar]
- Palmer DA, Thompson JK, Li L, Prat A, Wang P. Gib2, a novel Gβ-like/RACK1 homolog, functions as a Gβ subunit in cAMP signaling and is essential in Cryptococcus neoformans. J Biol Chem. 2006;281:32596–32605. doi: 10.1074/jbc.M602768200. [DOI] [PubMed] [Google Scholar]
- Paris S, Duran S. Cyclic adenosine 3′,5′ monophosphate (cAMP) and dimorphism in the pathogenic fungus Paracoccidioides brasiliensis. Mycopathologia. 1985;92:115–120. doi: 10.1007/BF00444093. [DOI] [PubMed] [Google Scholar]
- Peeters T, Louwet W, Gelade R, Nauwelaers D, Thevelein JM, Versele M. Kelch-repeat proteins interacting with the Gα protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc Natl Acad Sci USA. 2006;103:13034–11309. doi: 10.1073/pnas.0509644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak PM, Huntress FA. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gbetagamma complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenfelder E, Spellig T, Hartmann A, Lauenstein S, Bolker M, Kahmann R. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 1997;16:1934–1942. doi: 10.1093/emboj/16.8.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo A, McEwen JG, Castañeda E. The habitat of Paracoccidioides brasiliensis: how far from solving the riddle? Med Mycol. 2001;39:233–241. doi: 10.1080/mmy.39.3.233.241. [DOI] [PubMed] [Google Scholar]
- Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, et al. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen S, Yu JH, Adams TH. The Aspergillus nidulans sfaD gene encodes a G protein beta subunit that is required for normal growth and repression of sporulation. EMBO J. 1999;18:5592–5600. doi: 10.1093/emboj/18.20.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Blas G, Niño-Veja G. Paracoccidioides brasiliensis: virulence and host response. In: Cihlar RL, Calderone RA, editors. Fungal Pathogenesis: Principles and Clinical Applications. New York, N.Y.: Marcel Dekker Inc.; 2001. pp. 205–226. [Google Scholar]
- San-Blas G, Niño-Veja G, Iturriaga T. Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med Mycol. 2002;40:225–242. doi: 10.1080/mmy.40.3.225.242. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martinez C, Perez-Martin J. Gpa2, a G-protein alpha subunit required for hyphal development in Candida albicans. Eukaryot Cell. 2002;1:865–874. doi: 10.1128/EC.1.6.865-874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadick K, Fourcade HM, Boumenot P, Seitz JJ, Morrell JL, Chang L, et al. Schizosaccharomyces pombe Git7p, a member of the Saccharomyces cerevisiae Sgtlp family, is required for glucose and cyclic AMP signaling, cell wall integrity, and septation. Eukaryot Cell. 2002;1:558–567. doi: 10.1128/EC.1.4.558-567.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semighini CP, Marins M, Goldman MH, Goldman GH. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl Environ Microbiol. 2002;68:1351–1357. doi: 10.1128/AEM.68.3.1351-1357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JA, Han KH, Yu JH. Multiple roles of a heterotrimeric G protein {gamma} subunit in governing growth and development of Aspergillus nidulans. Genetics. 2005;171:81–89. doi: 10.1534/genetics.105.042796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slessareva JE, Dohlman HG. G protein signaling in yeast: new components, new connections, new compartments. Science. 2006;314:1412–1413. doi: 10.1126/science.1134041. [DOI] [PubMed] [Google Scholar]
- Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein α subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: insights from structural analyses. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha. GTPgammaS. Science. 1997;278:1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- Vallim MA, Nichols CB, Fernandes L, Gramer KL, Alspaugh JA. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high temperature growth in the pathogenic fungus cryptococcus neoformans. Eukaryot Cell. 2005;4:1066–1078. doi: 10.1128/EC.4.6.1066-1078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanittanakom N, Cooper CR, Fisher MC, Sirisanthana T. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin Microbiol Rev. 2006;19:95–110. doi: 10.1128/CMR.19.1.95-110.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Perfect JR, Heitman J. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol Cell Biol. 2000;20:352–362. doi: 10.1128/mcb.20.1.352-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway MS, Thomas DY. Site-directed mutations altering the CAAX box of Ste18, the yeast pheromone-response pathway Gγ subunit. Genetics. 1994;137:967–976. doi: 10.1093/genetics/137.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway MS, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas DY, Leberer E. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Xue Y, Batlle M, Hirsch JP. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Galpha subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans. J Microbiol. 2006;44:145–154. [PubMed] [Google Scholar]
- Yu JH, Wieser J, Adams TH. The Aspergillus FIbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 1996;15:5184–5190. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The phylogenetic relationship between Paracoccidioides brasiliensis and other fungal cAMP-signalling proteins – a neighbour-joining bootstrap tree is derived from the amino-acid sequence alignments of fungal (A) adenylate cyclase (AC) (B) Gb, (C) Gg and (D) Ga proteins, using the VectorNTi 6.0 align program (Informax).
The RAS transcript levels during the mycelium-toyeast transition.
Primers used for gene cloning.