Abstract
How the integrin head transitions to the high-affinity conformation is debated. Although experiments link activation with the opening of the hinge angle between the βA and hybrid domains in the ligand-binding headpiece, this hinge is closed in the liganded αvβ3 integrin crystal structure. We replaced the RGD peptide ligand of this structure with the 10th type III fibronectin module (FnIII10) and discovered through molecular dynamics (MD) equilibrations that when the conformational constraints of the leg domains are lifted, the βA/hybrid hinge opens spontaneously. Together with additional equilibrations on the same nanosecond timescale in which small structural variations impeded hinge-angle opening, these simulations allowed us to identify the allosteric pathway along which ligand-induced strain propagates via elastic distortions of the α1 helix to the βA/hybrid domain hinge. Finally, we show with steered MD how force accelerates hinge-angle opening along the same allosteric pathway. Together with available experimental data, these predictions provide a novel framework for understanding integrin activation.
Introduction
The interplay of mechanical forces between cells and their surroundings regulates crucial processes such as cell shape, differentiation, and protein expression (Bershadsky et al., 2003; Ingber, 2003; Katsumi et al., 2004; Vogel and Sheetz, 2006). Integrins are large transmembrane adhesion proteins that provide the physical link between the extracellular matrix and the contractile cytoskeleton. Integrins are heterodimers, composed of α and β subunits that associate noncovalently to form an extracellular, ligand-binding headpiece, followed by two multidomain “legs,” two transmembrane helices, and two cytoplasmic tails (Fig. 1 A).
Figure 1.
Set up. (A) Diagram of the FnIII10 crystal structure (yellow; Leahy et al., 1996) docked to the liganded αvβ3 integrin crystal structure (Xiong et al., 2002). α and β subunits are indicated in blue and gray, respectively. Transmembrane domains and extracellular β-subunit domains not resolved in the original crystal structure were added in pink. (B) The FnIII10–αvβ3 integrin headpiece complex (908 residues) was simulated in a box of explicit water molecules.
Integrins are not constitutively active. Rather, ligand-binding affinity is regulated via conformational change that can originate from cytoplasmic or extracellular interactions (Hynes, 2002). For example, ligand binding has been shown to induce the opening of the hinge angle between the βA (also called the I-like or βI domain) and hybrid domains in the integrin headpiece (Takagi et al., 2002, 2003; Mould et al., 2003b). Opening of this hinge has been linked to the switch to high binding affinity (Luo et al., 2003, 2004b; Mould et al., 2003b). The x-ray crystallographic structures of the unliganded αvβ3 and the liganded αIIbβ3 integrins display the integrin headpiece conformations before and after the ligand-induced transition from the closed to the open βA/hybrid domain hinge, respectively (Xiong et al., 2001; Xiao et al., 2004). The α1 and α7 helices of the βA domain are known to be involved in this transition (Mould et al., 2002, 2003b; Yang et al., 2004).
Because of a lack of dynamic, nonequilibrium information, the sequential details by which structural change in the ligand-binding pocket is allosterically coupled to the remote hinge conformation at the βA/hybrid interface have remained unknown, as have the details of force-induced integrin conformational change, which would enable insights into how integrin function is kinetically regulated during cellular mechanosensing. With no techniques currently available to gain these details experimentally, we used MD and steered MD (SMD) to derive high-resolution, dynamic structural models computationally.
Our simulations are based on the liganded crystal structure of the αvβ3 integrin, which was obtained by soaking RGD peptide into unliganded integrin crystals that were preformed with bent legs and a closed βA/hybrid hinge (Xiong et al., 2002). Although comparison of this crystal structure with the unliganded αvβ3 integrin structure reveals ligand-induced conformational changes local to the binding site (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1), both structures share the same long-range conformation of bent legs and a closed βA/hybrid hinge. Thus, although unequivocally recognized as a milestone in integrin research, the liganded αvβ3 integrin crystal structure has sparked much debate; does it represent the final, high-affinity conformation (Calzada et al., 2002; Xiong et al., 2003; Adair et al., 2005), or were the long-range, ligand-induced structural changes that underlie integrin activation arrested by conformational constraints (Takagi et al., 2002; Mould et al., 2003b, 2005; Xiao et al., 2004; Iwasaki et al., 2005)?
RGD ligands bind to the integrin β subunit via a divalent metal ion located at the top of the βA domain, termed the “metal ion–dependent adhesion site” (MIDAS; Xiong et al., 2002). Two additional ion-binding sites border the βA domain MIDAS on either side, which are termed the “ligand-induced metal-binding site” (LIMBS) and the “adjacent to the MIDAS” (ADMIDAS; Xiong et al., 2002). Although all three sites are occupied by ions when the ligand is bound (Xiong et al., 2002; Xiao et al., 2004), only the ADMIDAS is occupied in unliganded αvβ3 crystal structures (Xiong et al., 2001, 2002). Moreover, the ADMIDAS of unliganded αvβ3 crystal structures directly contacts the β6-α7 loop (Met335), whereas this contact is lost and the ADMIDAS is shifted inward by ∼4 Å in both liganded β3 integrin crystal structures (Xiong et al., 2001, 2002; Xiao et al., 2004).
Because integrins in a cellular context bind to extracellular matrix proteins, we replaced the RGD peptide in the liganded αvβ3 integrin crystal structure (Xiong et al., 2002) with the RGD-containing 10th type III fibronectin (FnIII10) module (Leahy et al., 1996) and equilibrated this complex in a box of explicit water molecules and in the absence of the integrin legs. Domains included were the propeller domain from the α subunit and the βA and hybrid domains from the β subunit (Fig. 1, A and B). Based on these simulations, together with equilibrations of the same domains from the unliganded αvβ3 integrin crystal structure (Xiong et al., 2001), we propose that the energy barrier to ligand-induced hinge opening in the αvβ3 headpiece is lowered by (a) the MIDAS conformation found in liganded crystal structures (Xiong et al., 2002; Xiao et al., 2004), (b) Mg2+ in place of Ca2+ at the LIMBS and ADMIDAS, or (c) a stabilizing contact outside of the RGD-binding pocket between the top of the α1 helix and FnIII10, which is reported for the first time in this study. Finally, we show how ligand-mediated mechanical force accelerates hinge opening along the same allosteric pathway. Through comparison with current experimental data, we propose a structural model that provides new insight into existing notions of integrin activation.
Results
Docking was performed without any major steric clashes by fitting the RGD loop of FnIII10 (Leahy et al., 1996), which is known to be flexible (Copie et al., 1998), to the conformation of the RGD sequence in the liganded αvβ3 crystal structure (Fig. 2 C; Xiong et al., 2002) The solvated FnIII10–αvβ3 integrin complex (Fig. 1 B) was investigated in 16 separate simulations of at least 6 ns each, consisting of 7 equilibrations (regular MD) and 9 simulations in which mechanical force was applied to an equilibrated complex (SMD). In the latter case, tensile stress was applied with a spring attached to the C or N terminus of FnIII10 and pulled at a constant velocity. Because Mg2+ ions are known to regulate integrin activation (Mould et al., 1995) and are present in the integrin under physiological conditions, all integrin metal ion–binding sites were occupied with Mg2+, unless specified otherwise. In all figures, FnIII10 is yellow and the β subunit domains are colored according to the time point of the particular structural snapshot, with gray and black representing the starting conformations of the liganded and unliganded αvβ3 crystal structures, respectively.
Figure 2.
Integrin headpiece hinge opening and receptor–ligand contact stability during MD equilibration. (A) After 6 ns of MD, the equilibrated integrin complex (blue, purple, and yellow) is aligned with the starting crystal structure (gray; Xiong et al., 2002) via βA domain β-strands. For clarity, the βA domain is shown in transparent surface representation with the secondary structure of select regions visible in cartoon representation. (B) A close-up of the βA/hybrid domain interface shows the location inside the hinge of the hydrogen bond between Asn303 and Lys417. This equilibrated complex, in which the Asn303–Lys417 bond broke when the hinge spontaneously increased by ∼20°, is referred to in the text as the “reference complex.” (C, middle) In all of the equilibrated FnIII10–integrin complexes we investigated, independent of structural variations, the RGD loop of FnIII10 (yellow) displayed close alignment with the conformation of the RGD sequence found in the original crystal structure (gray), and the two principle RGD–integrin binding contacts (AspRGD-MIDAS ion and ArgRGD–α-Asp218) remained stable. (C, top and bottom). In the same, but separate, views of the equilibrated complex (top) and the liganded crystal structure (bottom), two additional AspRGD-βA domain–binding contacts that have been shown by pharmacophore refinement to be vital to the RGD-αvβ3 ligand–integrin recognition process (Marinelli et al., 2003) displayed stability. These are hydrogen bonds between AspRGD–β-Arg216 and AspRGD–β-Tyr122. Bonds present in the original crystal structure complex are shown in green. Also visible is a hydrogen bond between Fn-Glu62 and β-Tyr129 (orange) that formed in the equilibrated complex and was found to promote hinge angle opening on the nanosecond time frame.
The βA/hybrid domain hinge opens spontaneously in the isolated FnIII10–αvβ3 integrin complex
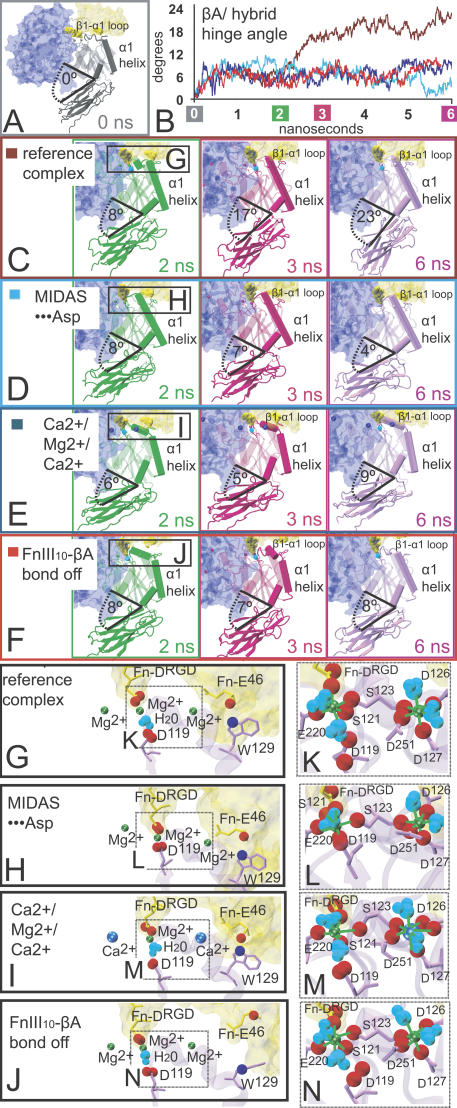
A hydrogen bond between Asn303 on the βA domain and Lys417 on the hybrid domain, located where the closed hinge is most acute (Fig. 2 B), was permanently disrupted after 2.5 ns. Sequence analysis reveals that this Asn is completely conserved across the eight known integrin β subunits, whereas the Lys is conserved in four integrin β subunits (β3, β4, β6, and β8) and is replaced by the similarly basic Arg in the other four β subunits (β1, β2, β5, and β7). Once this bond was lost, the βA/hybrid domain hinge angle increased to an average of ∼20° greater than its value in the starting crystal structure for the remainder of the 7-ns equilibration (Fig. 3 C and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1). We shall refer to this FnIII10–αvβ3 integrin complex as the “reference” complex, to distinguish it from additional complexes described in the article, in which the effect of various structural perturbations on spontaneous hinge opening was investigated.
Figure 3.
Minor structural changes perturb the α1 helix and deter hinge opening. (A) The starting complex. (B) Hinge angle values traced over time for four separate equilibrations of the integrin complex. (C–F) Snapshots corresponding to each of these four equilibrations are shown with the hinge-angle values indicated. Only in the reference complex (C) did the hinge angle increase by ∼20°. When, instead, the α1 helix was shifted slightly outward as a result of a 2-Å MIDAS shift (D), or was interrupted as a result of Ca2+ at the ADMIDAS (E) or instability of the E46–W129 bond (F), the hinge did not significantly increase in the same 6-ns time window. β-Subunit domains, shown in cartoon representation, are colored by time point. Within the βA domain, the β1-α1 loop, α1, and α7 helix are emphasized by opaque shading. In each snapshot, the α-subunit domain and FnIII10 are shown in transparent surface blue and yellow representations, respectively. (G) Close-up of the βA domain–binding pocket in the reference complex reveals the MIDAS···H20···Asp conformation, which is visible by the water molecule (cyan) between the MIDAS ion and Asp119; Mg2+ (green) at the LIMBS, MIDAS, and ADMIDAS; and the FnIII10-βA domain–binding contact between E46 and W129. (H–J) Close-ups from the three additional equilibrations, corresponding to the snapshots in D–F, respectively, indicate the specific structural alterations that made each complex unique, which were the switch to the MIDAS···Asp conformation (H), Ca2+ (blue) in place of Mg2+ at the LIMBS and ADMIDAS (I), and (J) no E46–W129 bond. (K–N) MIDAS- and ADMIDAS-coordinating residues are shown for each complex. These residues are identical in every case except in L, where Asp119 joined the MIDAS coordination sphere in the switch to the MIDAS···Asp conformation, and in M, where the second oxygen from Asp126 joined the ADMIDAS coordination in the switch from Ca2+ to Mg2+ at this position.
In all complexes we investigated, ligand–receptor contacts that have been identified as vital to RGD-αvβ3 integrin–binding remained stable during equilibration (Fig. 2 C). In addition, a hydrogen bond consistently formed outside of the RGD-binding pocket, at a location between FnIII10 and Trp129 at the top of the βA domain α1 helix (residues 128–145). This contact is shown in Fig. 2 C (top and middle) and described in more detail in the section entitled An FnIII10-α1 helix contact promotes spontaneous hinge opening.
The helical structure of the β1−α1 loop is restored
Between the unliganded αvβ3 and the liganded αIIbβ3 integrin crystal structures, a single turn of the 310 helix in the βA domain β1−α1 loop (residues 121 to 127) is moved inward en bloc, and a bend between this region and the α1 helix is straightened, such that the β1−α1 loop and α1 helix are joined into one continuous helix (Xiong et al., 2001; Xiao et al., 2004). In both of these structures, the ADMIDAS ion is directly coordinated by Asp127, which is located where the β1−α1 loop joins the α1 helix. In contrast, the 310 helix in the β1−α1 loop is disrupted, and Asp127 does not coordinate the ADMIDAS ion in the liganded αvβ3 integrin crystal structure (Xiong et al., 2002). We found that when the coordinates of this structure are minimized for 2,000 steps before equilibration with MD, direct contact between Asp127 and the ADMIDAS ion formed (six out of six times) and the helical structure of the β1−α1 loop was restored (Fig. 3, A vs. C–F), regardless of a change in the ADMIDAS ion from Mg2+ to Ca2+ (Fig. 3, K–N). At the beginning of MD equilibrations in Mg2+-occupied complexes, the α1 helix joined together with the β1−α1 loop to form one uninterrupted helical structure (Fig. 3, C and D).
The MIDAS conformation found in liganded crystal structures promotes hinge opening
In both available crystal structures that contain the ligand-bound βA domain, the distance between the MIDAS ion and the carboxyl oxygens of Asp119 is ∼4 Å. It was suggested that a water molecule likely bridged this distance in the αvβ3 structure and, indeed, in the αIIbβ3 structure that was resolved at a higher resolution, a water molecule was found to be present between Asp119 and the MIDAS ion. In two out of three separate equilibrations that we initially performed on the αvβ3–FnIII10 complex, ranging from 1 to 8 ns, a water molecule diffused into this location between the MIDAS ion and Asp119 early on and remained there. In subsequent equilibrations, we placed a water molecule there manually. In 17 separate equilibrations (1 ns each; unpublished data), we found that this water molecule either remained between the ion and Asp119 (nine times) or diffused away (eight times).
When the water diffused away, the MIDAS ion shifted ∼2 Å into direct contact with Asp119. Because the two different MIDAS conformations obtained in this study are thus distinguished by their Asp119 contact, we refer hereafter to the original MIDAS conformation resolved in crystal structures and maintained in the reference complex that was previously described as “MIDAS⋅⋅⋅H2O⋅⋅⋅Asp” (Fig. 3, K, M, and N), and to the shifted MIDAS conformation as “MIDAS⋅⋅⋅Asp” (Fig. 3 L). For a comparison between these two βA domain MIDAS conformations and the two αA domain MIDAS conformations that were previously established (Lee et al., 1995), see Fig. S2 (available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1).
Aside from differences in Asp119 contact, all ion-coordinating residues are identical between MIDAS⋅⋅⋅H2O⋅⋅⋅Asp and MIDAS⋅⋅⋅Asp complexes. In other words, all residues other than Asp119 that directly coordinate the MIDAS, LIMBS, or ADMIDAS ions remained the same in the presence of both MIDAS conformations (Fig. 3, K and N vs. L).
Over the first 2.5 ns of every equilibration that we performed, both unliganded and liganded and independent of the 2-Å MIDAS shift or other structural perturbations, the hinge angle between the βA and hybrid domains increased by ∼6°. At this value, the conserved βA/hybrid domain hydrogen bond between Asn303 and Lys417 was able to remain formed. When an equilibration containing the MIDAS⋅⋅⋅Asp conformation was extended to 6 ns, the βA/hybrid hinge increase of this FnIII10 integrin complex remained at 6 ± 2°. Thus, we asked how the 2-Å MIDAS shift deters βA/hybrid domain hinge opening. We found that over the first 2.5 ns, the α1 helix of the MIDAS⋅⋅⋅Asp complex moved outward relative to its position in the starting crystal structure (Fig. 4 C). In contrast, during the same timeframe of the reference complex equilibration, before the substantial hinge increase that this complex later displayed, the α1 helix moved inward (Fig. 4, A and B and Video 1).
Figure 4.
Hinge opening follows the inward lever-arm movement of the α1 helix. In all images, gray, green, red, and purple indicate the conformation at 0, 1, 2.5, and 6 ns, respectively. The MIDAS···H20···Asp conformation is visible by the water molecule (cyan) located between the MIDAS ion and Asp119. (A) 10 snapshots over the first 2.5 ns of the reference equilibration are aligned with the starting crystal structure (PDB code 1L5G) via the βA domain β-strands. The α1 and α7 helices are shown and colored according to 250-ps intervals. From gray to red, the path of the α1 helix is inward and upward. (B) The inward movement of the α1 helix occurred before a significant jump in the hinge value, as indicated in the trace of the hinge over time and (far right) depicted by alignment with the starting crystal structure of the α1 helix at 2.5 ns (red) and the α7 helix and β/hybrid domain at 6 ns (purple). (C–H) The same pictorial scheme is used to depict the three additional equilibrations in which various structural perturbations (Fig. 3) occurred and hinge opening was hindered. Notably, as evident by the presence of gray inside the α1-helix close-ups shown in C, E, and G, the path of the α1 helix was outward over the first 2.5-ns of these equilibrations. For dynamic, 3D views, see Videos 1–4, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1.
At the top of the α1 helix, ADMIDAS ion-coordinating residues Asp127 and Asp126 are linked along the β1−α1 loop to MIDAS residues Ser123, Ser121, and Asp119. Thus, when the β1−α1 loop was repositioned in association with the shift to the MIDAS⋅⋅⋅Asp conformation, the α1 helix was repositioned as well (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1). Notably, the α1 helix also contacts the C-terminal α7 helix and the outside of the βA/hybrid hinge through noncovalent interactions at its middle and bottom, respectively. In this fashion, the MIDAS position of the reference complex promotes an inward “lever-arm” motion of the joined β1–α1 loop/α1 helix; the key to how the MIDAS shift deterred hinge opening was by disruption of this lever-arm mechanism. This finding was reproduced in a second 6-ns MIDAS⋅⋅⋅Asp equilibration. For further depiction, see Fig. S3.
Ca2+ at the ADMIDAS impedes hinge opening
Ca2+ at the ADMIDAS has been shown to inhibit integrin activation (Mould et al., 2002) and mutation of the ADMIDAS Asp residues has been shown to override this effect (Chen et al., 2003). Mutation of the ADMIDAS has also been shown to disrupt the transduction of conformational change between the MIDAS and the βA/hybrid hinge (Mould et al., 2003a). To investigate the impact of Ca2+ on the conformation of the integrin headpiece complex, we replaced Mg2+ with Ca2+ in all ion-binding sites, except for the MIDAS, because occupation of the MIDAS by Mg2+ is known to be important for ligand binding (Mould et al., 1995, 2002). Hereafter, we shall refer to this ion arrangement as “Ca2+–Mg2+–Ca2+,” in reference to the occupancies of the LIMBS, MIDAS, and ADMIDAS, respectively. With Ca2+ in place of Mg2+, the second carboxylate oxygen of the Asp126 side chain joined the ADMIDAS coordination sphere (Fig. 3 M). This is consistent with crystallographic data of the Ca2+-occupied ADMIDAS, in both liganded and unliganded integrin structures (Xiong et al., 2001, 2002; Xiao et al., 2004).
Although the water in the MIDAS⋅⋅⋅H2O⋅⋅⋅Asp conformation remained consistently in place (Fig. 3 M) in three out of three separate Ca2+–Mg2+–Ca2+ equilibrations (1 ns each), the βA/hybrid hinge did not increase beyond 6 ± 2° when one of these equilibrations was extended to 8 ns.
In every Ca2+–Mg2+–Ca2+ equilibration we observed (ranging from 1 to 8 ns), a break between the β1−α1 loop and α1-helix structure occurred early on, in the vicinity of Asp126 (Fig. 3, E vs. C and D). In the first 2.5 ns, the β1−α1 loop displayed little change from its position in the starting crystal structure, whereas the α1 helix moved outward (Fig. 4 E and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1). Thus, although the movement of the β1−α1 loop resembled that of the reference complex in which the hinge increased, the movement of the α1 helix resembled that of the MIDAS⋅⋅Asp complex, and just as in that complex, the hinge did not substantially increase (Video 4).
An FnIII10–α1 helix contact promotes hinge opening
In all equilibrations of the integrin complex described thus far, a stable hydrogen bond consistently formed between Glu46, which is a conserved acidic residue on the same face of FnIII10 as the RGD sequence, and Trp129, which is located at the top of the α1−helix (Fig. 2 C, orange line). In two repeat reference complex equilibrations, this Glu46–Trp129 bond broke at either 1 or 3 ns, and the hinge increase of ∼20° either did not occur in the 6-ns time window or occurred but was delayed by ∼2 ns relative to the original reference complex, respectively (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1). In a third reference complex repeat equilibration, we deliberately disrupted the Glu46–Trp129 bond at 1 ns by turning off the charge of the nitrogen in the Trp129 side chain. When this computationally “mutated” complex was equilibrated for 5 ns, the α1 helix became distorted (Fig. 3 F) and moved outward (Fig. 4 G), and the hinge angle did not substantially increase (Fig. 4 H). From these studies, we linked the stability of the Glu46–Trp129 contact with an inward movement of the α1 helix early on in the equilibration and with an ∼20° hinge-angle increase on a 6-ns timeframe. In other words, we found that, like MIDAS–Asp and Ca2+–Mg2+–Ca2+ complexes, the absence of substantial hinge opening in Mg2+-occupied complexes was also linked with an outward α1-helix shift (Fig. 3, E and F; and Fig. 4, E and G). Sequence analysis indicates that the Glu46–Trp129–binding contact is unique to β3 integrins (Fig. S4).
Traces of the βA/hybrid domain hinge-angle value over time for the aforementioned four distinct integrin complexes are overlaid in Fig. 3 B and shown separately in Fig. 4.
Comparison to the unliganded αvβ3 integrin headpiece
To gain a sense of how these structural snapshots fit along the allosteric pathway of ligand-induced integrin activation, we equilibrated the same headpiece domains from the unliganded αvβ3 crystal structure occupied with Mg2+ (Xiong et al., 2001). The α1 helix of this structure is split in half and this split remained throughout the 6-ns equilibration (Fig. 5 A). Coordination of the ADMIDAS ion by Ser123 was lost early on, and the β1-α1 loop exhibited a substantially wider range of movement relative to the dynamics of this loop in liganded integrin complexes (Fig. 5 B vs. Fig. 4; and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1), illustrating the stabilizing effect that ligand-binding together with LIMBS and MIDAS occupancy bring to this structural region. Aside from Ser123, the starting ADMIDAS coordination remained unchanged, and the βA/hybrid domain hinge varied by 8 ± 3° relative to its starting value. Like the aforementioned integrin complexes, in which the hinge also did not substantially increase, the conserved Asn303–Lys417 hydrogen bond located where the closed hinge is most acute (Fig. 2 B) was still able to form after 6 ns (Video 6).
Figure 5.
Significant fluctuations of the α1 helix and β1-α1 loop in the unliganded integrin equilibration. (A) Snapshots from the equilibration of the unliganded αvβ3 integrin headpiece domains (Xiong et al., 2001) are shown in the same pictorial scheme as Fig. 3 (C–F), for comparison. (B) 10 snapshots over the first 2.5 ns of this equilibration are aligned with the starting crystal structure (PDB code 1JV2) via the βA domain β-strands and colored according to 250-ps intervals, as described in Fig. 4. From black to red, significant outward and downward movement of the β1-α1 loop and α1 helix is evident. (C) The hinge angle is traced over time. The equilibrated complex is aligned with the starting crystal structure. Black, red, and purple indicate time points at 0, 2.5, and 6 ns, respectively. (D) The increased solvent-accessible surface area at the bottom of the α1 helix during equilibration of the unliganded integrin domains (black) is compared with that of the four FnIII10-bound integrin complexes. For dynamic 3D views, see Videos 5 and 6, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1.
A comparison across all equilibrations described thus far reveals a higher degree of floppiness at the bottom of the α1 helix in structures in which hinge opening was not observed during the nanosecond time window, as indicated by comparison over time of the average solvent-accessible surface area in this location (residues 139–147; Fig. 5 D). In other words, the bottom of the α1 helix displayed the least fluctuations in the solvent-accessible surface area (indicating structural stability), as well as the least accessibility to water (in agreement with the inward helical shift) in the reference complex in which the hinge opened spontaneously (Fig. 5 D, red line), whereas the same structural region in the unliganded integrin displayed considerable fluctuations (Fig. 5 D, black line).
Mechanical force opens the hinge
To see if the closed hinge could be opened by mechanical force, we applied force between the C-terminal ends of FnIII10 and the β-hybrid domain after 1 ns of equilibration, at which time the hinge angle was only ∼6° greater than its starting value. In a total of seven separate SMD simulations (three of the reference complex, two of the MIDAS⋅⋅⋅Asp complex, and two of the Ca2+–Mg2+–Ca2+ complex), we found that pulling with a constant velocity of 10 Å/ns caused the βA/hybrid domain hinge to increase by ∼70° over 5 ns (Fig. 6, A and B; and Videos 7 and 8, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1). At this value, the βA and hybrid domains were aligned along the vector of force and continued strain did not increase the hinge further.
Figure 6.
Mechanical force accelerates hinge opening. (A) Traces of the change in the hinge angle over time from both MD- and SMD-simulations are overlaid. SMD simulations were begun after complexes were equilibrated for 1 ns. (B) Snapshots from the SMD simulation in which the hinge was opened by ∼70° by pulling for 5 ns on the C terminus of FnIII10 at 10 Å/ns. β-Subunit domains are colored according to time point. For dynamic 3D views, see Videos 7 and 8. (C) Corresponding time points from the SMD simulation in which the hinge angle was opened by ∼50° by pulling for 5 ns on the N terminus of FnIII10 at 10 Å/ns. In the last frame of both C and D, the SMD-derived structures, after 6 ns for each pulling vector (shown with β-subunit domains in purple), are aligned with the starting crystal structure (gray) via the βA domain β-strands. Videos 7 and 8 are available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1.
In two additional SMD simulations, the hinge was increased by ∼50° over 5 ns when force was applied to the N-terminal end of FnIII10 instead (Fig. 6, A and C). In every SMD simulation (nine in total), the hinge increased in the same characteristic fashion, independent of the FnIII10 terminus to which force was applied and whether or not FnIII10 started to unravel under the strain (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1). Also, in every case, the principle force-bearing binding contact between AspRGD of FnIII10 and the MIDAS ion was well maintained. When the force was applied between FnIII10 and the C terminus of the α-subunit propeller domain instead, the hinge could not be pulled open (unpublished data).
As a control, we deleted FnIII10 and applied force directly to the MIDAS ion in the reference, MIDAS···Asp, and Ca2+–Mg2+–Ca2+ integrin structures. Each time (three out of three), force applied between the MIDAS and the β-hybrid domain increased the hinge in the same characteristic fashion of ∼70° over 3 ns (unpublished data).
Thus, the key finding of these investigations is that the βA/hybrid hinge opens when force is applied between the MIDAS ion and the hybrid domain.
Force-induced opening of the hinge reverses α1-helix distortion
We have shown that a characteristic feature of spontaneous βA/hybrid hinge opening is the inward shift of the α1 helix (Fig. 4, A and B). This finding is further illustrated in Fig. 7 B by alignment of the reference and MIDAS⋅⋅⋅Asp integrin complexes after 6 ns of equilibration each. Because the hinge angle of the MIDAS⋅⋅⋅Asp complex did not increase beyond 6 ± 2° under equilibration conditions on the nanosecond timescale, we asked if opening the hinge by force would induce the α1 helix of this complex to resemble that of the reference complex in which the hinge angle had increased spontaneously.
Figure 7.
Force-induced opening of the hinge reverses outward α1-helix distortion. (A) Traces of the hinge angle over time show that although the hinge of the MIDAS···Asp complex did not open under equilibration conditions (cyan), mechanical force induced the hinge angle of this complex to open (pink) by the same amount that was found to occur spontaneously in the MIDAS··H20···Asp complex under equilibration conditions (dark red). (B) The MD-derived MIDAS···Asp and MIDAS··H20···Asp complexes, aligned after 6 ns of equilibration each and shown in cyan and dark red, respectively. The hinge angle difference between these complexes is 21° at this time point. (top) Close-up of the aligned βA domains reveal a difference between the α1-helix conformations. The β1-α1 loop, α7 helix, ions, and Asp119 of each structure are also shown as opaque, for orientation. (C) The SMD-derived MIDAS···Asp complex (pink) and the MD-derived MIDAS··H20···Asp complex (dark red) are aligned after 6 ns of simulation time each. The hinge angle of each complex is now 21°. (Top) Close-up of the aligned βA domains reveal a new similarity between the α1-helix conformations.
After 1 ns of equilibration, force was applied to the MIDAS⋅⋅⋅Asp complex by pulling on the FnIII10 C terminus with a constant velocity of 5 Å/ns. After 5 ns of pulling, the hinge of this complex was increased by 21°, which is the same hinge value that the reference complex displayed at 6 ns without force (Fig. 7 A). Comparison of the α1 helices of these two structures at this time point reveals a striking similarity (Fig. 7 C). Thus, we found that force-induced opening of the βA/hybrid hinge reverses the elastic distortion in the α1 helix that occurred when the hinge remained closed.
Discussion
Integrin activation is known to have a structural origin (Takagi and Springer, 2002; Luo et al., 2005), yet the key sequential details of activating conformational change have been controversial. For example, experiments have shown that ligand binding to the integrin head induces the opening of the hinge between the βA and hybrid headpiece domains (Takagi et al., 2002, 2003; Mould et al., 2003c; Xiao et al., 2004; Iwasaki et al., 2005), but this hinge is closed in the liganded αvβ3 crystal structure (Xiong et al., 2002). We replaced the peptide ligand of this structure with FnIII10 and revealed in MD simulations of the isolated integrin headpiece complex how ligand- induced structural change propagates across the βA domain via the α1 helix, to then be amplified by an increase in the remote βA/hybrid hinge. In separate equilibrations of the same complex, three different structural perturbations at the top of the βA domain induced small, elastic distortions in the α1 helix, and the hinge did not increase substantially in the nanosecond time window. The structural variations that we found to impede hinge opening on the nanosecond timescale are a 2-Å MIDAS shift, Ca2+ in place of Mg2+ at the LIMBS and ADMIDAS, and disruption of an FnIII10–α1 helix–binding contact. In equilibration of the unliganded integrin headpiece domains (Xiong et al., 2001), maximum fluctuations of the β1−α1 loop and α1 helix were found to occur. Together, these investigations enabled us to deduce the allosteric pathway whereby a small inward (vs. outward) movement of the α1 helix promotes (or impedes) βA/hybrid domain hinge opening. In addition to predicting a role for a new FnIII10–β3 integrin contact located outside of the RGD-binding pocket, these findings offer a structural basis for mAb experimental data that also link an outward α1-helix position with Ca2+ in place of Mg2+ at the ADMIDAS and an inward α1-helix position with hinge opening and integrin activation (Mould et al., 2002, 2003b).
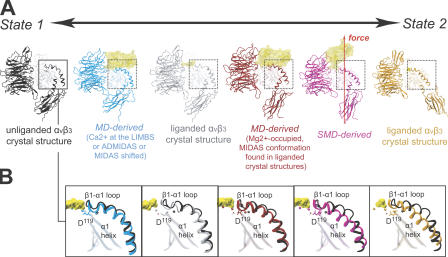
The crystal structures of the unliganded αvβ3- and the liganded αIIbβ3 integrins provide stationary endpoints before and after the transition of the integrin headpiece to the open conformation of the βA/hybrid hinge, respectively (Xiong et al., 2001; Xiao et al., 2004). Together with experiments that have linked integrin activation with βA/hybrid hinge opening (Mould et al., 2003b; Luo et al., 2004b; Iwasaki et al., 2005), we propose that the unliganded αvβ3 structure represents a low-affinity state of the integrin headpiece (which we shall call “state 1”), and the liganded-αIIbβ3 structure represents a high-affinity state of the integrin headpiece (hereafter referred to as “state 2”). In this framework, the MD-derived conformations presented in this study are snapshots along the ligand-induced transition from state 1 to state 2 (Fig. 8).
Figure 8.
Transitional snapshots along the hinge-opening pathway. (A) The unliganded αvβ3-, liganded αvβ3-, and liganded αIIbβ3 integrin crystal structures are shown in black, gray, and orange, respectively (Xiong et al., 2001, 2002; Xiao et al., 2004). A representative MD-derived snapshot, in which a structural perturbation hindered hinge angle opening, is indicated in blue. The MD-derived snapshot in which the hinge angle increased spontaneously is indicated in dark red. The SMD-derived snapshot in which the hinge angle was increased by mechanical force is shown in pink. (B). Close-ups of βA domain α1 helices for each structural snapshot are shown overlaid with the α1 helix of the unliganded αvβ3 integrin crystal structure (which is proposed to represent state 1). MIDAS and ADMIDAS ions are visible in each close-up.
Quite unusual for domain–domain contacts, βA connects to the hybrid domain via two peptide linkers, at both the C and N terminus. In this fashion, floppiness of the βA/hybrid hinge itself is greatly restricted. A key finding of this study is the allosteric link between floppiness and distortion in the βA domain α1 helix and the opening of the βA/hybrid hinge. Although we found that Ca2+ in place of Mg2+ at the LIMBS and ADMIDAS resulted in the outward shift of the α1 helix and no substantial hinge-angle increase on the nanosecond timeframe, experiments have shown that ligand-induced opening of the hinge does ultimately result in Ca2+ in the LIMBS and ADMIDAS (Xiao et al., 2004). Importantly, although experimental results were obtained under equilibrium conditions in which the strain built-up by ligand binding had sufficient time to be released by the opening of the βA/hybrid hinge, the nonequilibrium snapshots described in this study probe structural dynamics on the nanosecond timescale and, thus, reveal how the ligand-induced strain either propagates to the βA/hybrid interface and is released by hinge-angle opening or, as a result of conformational variations near the binding site, produces elastic α1-helix distortions. Notably, the liganded αIIbβ3 integrin crystal structure, which presents the open-hinge conformation with Ca2+ at the LIMBS and ADMIDAS, also displays the inward-shifted α1 helix joined together with the β1−α1 loop, which is not found in integrin crystal structures with closed βA/hybrid hinges (Xiong et al., 2001, 2002). Also unique to the αIIbβ3 crystal structure is a molecule of glycerol bound directly to the ADMIDAS ion at the top of the α1 helix (Xiao et al., 2004), which may lend additional stability to this key regulatory region.
These findings address a long-standing debate regarding a feature of the liganded αvβ3−integrin crystal structure that surprised the integrin community; a severe bend in the legs (Fig. 1 A). Because the structure is liganded, it has been considered to be in the active conformation (Xiong et al., 2002, 2003). However, because the structure was obtained by soaking RGD peptides into unliganded integrin crystals that were preformed with bent legs and closed hinges (Xiong et al., 2002), it has been suggested that the conformation of this structure is the result of constraint imposed by the preexisting crystal lattice (Liddington, 2002; Takagi et al., 2002; Mould et al., 2003b). In support of the latter notion, the legless, liganded αIIbβ3 crystal structure, which presents the same MIDAS⋅⋅⋅H2O⋅⋅⋅Asp conformation as the liganded αvβ3−integrin crystal structure, but was formed by cocrystallization with a ligand, displays the open hinge (Xiao et al., 2004). We have shown that once the conformational constraints of the leg domains are lifted, the α1 helix joins together with the β1−α1 loop and moves inward, and the hinge opens spontaneously on the 6-ns timeframe when the liganded αvβ3 integrin crystal structure is occupied with Mg2+ and equilibrated in complex with FnIII10. Thus, the liganded αvβ3 crystal structure may depict a nonequilibrium headpiece conformation that was constrained along the transition from state 1 to state 2 at a point after RGD peptide–binding had induced a change in the ADMIDAS and β1−α1 loop and before the strain built-up by this structural perturbation could be relieved by the opening of the βA/hybrid hinge (Fig. S1).
Although the bend in the integrin legs is known to be physiologically relevant (Takagi et al., 2002; Adair et al., 2005), the function of the bend during activation is debated. Currently, two models are widespread; in the “deadbolt” model, activation results from the loss of a constraining contact between the β-tail and βA domains (Xiong et al., 2003), whereas in the “switchblade” model, activation results from extension and separation of the legs and the opening of the βA/hybrid hinge (Takagi and Springer, 2002). Because the legs remain bent in the former and become extended in the latter, these models are often presented as incongruent. However, in both paradigms, the conformation with the bent legs and the closed headpiece hinge angle is low affinity (state 1), and activation necessitates disruption of headpiece–tailpiece contacts. Mutational and mAb experiments have shown that the low-affinity state is stabilized by a close association between the α and β cytoplasmic tails (Hughes et al., 1995; Kim et al., 2003), α and β transmembrane helical packing (Luo et al., 2004a, 2005; Partridge et al., 2005), and a close association in multiple points along the α and β legs (Beglova et al., 2002; Xie et al., 2004; Clark et al., 2005; Mould et al., 2005). According to the switchblade model, α- and β-subunit contacts stabilize the low-affinity state by constraining the integrin legs in the bent conformation. Recently, electron microscopy studies of the intact, Fn-bound αvβ3 integrin ectodomain were found to display the bent-leg conformation together with a βA/hybrid hinge angle increase of ∼11 ± 4°, relative to the αvβ3 crystal structures (Adair et al., 2005). We have linked an inward shift of the α1 helix with a hinge increase of ∼20° (Fig. 7), indicating that hinge opening by this amount might already be sufficient to induce affinity-regulating conformational change. Aside from the findings by Adair et al. (2005), studies correlating movement in the headpiece hinge with changes in affinity have been done on integrin ectodomains with modified, truncated, or entirely absent legs (Takagi et al., 2002, 2003; Mould et al., 2003c; Xiao et al., 2004). In consideration of these experimental findings, together with our computational snapshots, we propose that the major physiological role of the of the bent-leg conformation is to tune, via domain–domain contacts, the height of the energy barrier that has to be overcome for the βA/hybrid hinge to increase and thereby transition the integrin head to the high-affinity state.
Integrin activation is bidirectional and can originate from either the cytoplasmic or the extracellular end of the molecule (Hynes, 2002). The transition of integrins from the low- (state 1) to the high-affinity state (state 2) before ligand binding has been referred to as “priming” (Humphries et al., 2003). When induced from inside the cell, the structural mechanisms underlying priming have been shown to be generated by conformational change in the cytoplasmic tails, transmembrane helices, and leg domains (Takagi et al., 2002; Kim et al., 2003; Partridge et al., 2005). However, conformational changes in these structural regions distal to the ligand-binding site are not required when integrin priming is induced extracellularly (Luo et al., 2004a). In other words, extracellular factors that prime integrins, such as Mn2+ ions, conformation-dependent mAbs that map to the integrin headpiece (Mould et al., 2002, 2003b; Clark et al., 2005), and mutations that induce the shortening of the α7 helix (Yang et al., 2004) or the opening of the headpiece hinge (Luo et al., 2003) do so by inducing the high-affinity conformation directly. As expected for an allosterically regulated protein, we thus propose that any mechanism which causes the headpiece hinge angle to open can thereby transition the RGD-binding pocket from state 1 to state 2, even in the absence of bound ligand. Hinge opening and the ensuing inward movement of the α1 helix would then constitute the final step in integrin priming.
Finally, we asked how mechanical force might impact the transition between state 1 and state 2, and we found in SMD simulations that force greatly accelerated hinge opening. The force-accelerated transition to the open hinge conformation was accompanied by the same inward movement of the α1 helix that we found to be characteristic of hinge opening under equilibrium conditions (Fig. 7). This finding implies that force accelerates hinge opening along the same allosteric pathway. Moreover, we have now shown that a 2-Å MIDAS shift or Ca2+ in place of Mg2+ at the ADMIDAS can prevent spontaneous hinge opening, but does not inhibit force-induced hinge opening on the same nanosecond timescale. This finding implies that these structural perturbations raise the energy barrier and that force acting along the β-subunit domains lowers the energy barrier to transition the ligand-bound integrin headpiece from state 1 to state 2. Considering the high sensitivity to minor structural perturbations that these findings reveal, the way in which point mutations to this region enhance (Chen et al., 2003) or disrupt (Mould et al., 2003a) the allosteric activation pathway remains enigmatic.
Because ample experimental data, together with the MD simulations presented in this work, demonstrate that ligand binding can induce the transition from the closed to the open hinge conformation, what would be the advantage of accelerating this transition with force? Kinetic measurements have shown that the ligand-induced conversion of αIIbβ3 integrin extracellular domains to the high-affinity conformation occurs on a timescale of ∼10 s (Müller et al., 1993; Bednar et al., 1997). Yet turnover rates at the timescale of seconds have been reported for constituents in newly formed adhesion sites, and a recent study shows that cells lose their ability to sense the rigidity of their surrounding Fn matrix when the binding of αvβ3 integrins to Fn is blocked (Jiang et al., 2006). Thus, considering the shortness of the time window during which cells have to form adhesions that can sustain cell-generated tension, and assuming that integrins are in state 1 (i.e., not primed) when they first bind to matrix-exposed ligands, mechanical force could then play a physiologically important role in up-regulating the kinetics by which ligand-bound integrins transition from the low- to the high-affinity state. Do integrins then form catch bonds with their RGD ligands, bonds that are switched from a short-lived, low-affinity state to a long-lived, high-affinity state under mechanical force? A preliminary comparison between the αvβ3 integrin complex investigated in this study and the catch bond characteristics of the bacterial adhesin FimH when bound to mannose reveals striking similarities (Tchesnokova et al., 2006) that will be considered in detail elsewhere (unpublished data).
Materials and methods
System setup
The crystal structures of the αvβ3 integrin in complex with an RGD-containing mimetic ligand (Protein Data Bank [PDB] code 1L5G; 3.2 Å resolution [Xiong et al., 2002]) and of FNIII10 from the FNIII7-10 tetramer (PDB code 1FNF; 2.0 Å resolution [Leahy et al., 1996]) were adopted to build the FNIII10–αvβ3 starting structure with the program VMD (Humphrey et al., 1996). First, the RGD tripeptides of the two separate structures were aligned, and then the mimetic ligand from the αvβ3 structure was removed. The resulting protein–protein complex contained slight overlap between the C and D strands of FnIII10 and the β6-α7 loop of the I-like domain in the integrin β3 subunit. The steric conflict was readily resolved by fixing the RGD tripeptide and rotating the remainder of the FnIII10 module around the Cγ atom of the ArgRGD, away from the integrin. The rotation caused an ∼10° decrease in the angle formed by the Cα carbons at the N-terminal end, the AspRGD, and the C-terminal end. The two backbone bonds that connect the RGD sequence to the rest of the FnIII10 module were lengthened after the rotation, but a short minimization of 600 steps restored these bonds to their normal length. The key RGD–integrin–binding contacts, the bond between the AspRGD and the MIDAS cation and the bidentate salt bridge between the ArgRGD and αv-Asp218, were well maintained. However, the salt bridge observed in the crystal structure between the ligand and αv Asp150 was broken when the peptide was replaced by the FnIII10 module. To reduce the integrin to a size that can be feasibly simulated, we used only the βA and hybrid domains of the β3 integrin subunit, the β-propeller domain of the αv integrin subunit. The starting structure was solvated in a 116 × 115 × 122 Å TIP3 (Jorgensen et al., 1983) water box, resulting in 153,570 atoms. The water molecule manually added to the MIDAS conformation, between Asp119 and the MIDAS cation, was placed according to the location of the water molecule in the αIIbβ3 integrin crystal structure (Xiao et al., 2004). Seven cations resolved in the headpiece, three resolved in the MIDAS-, LIMBS-, and ADMIDAS- binding pocket motifs, and four resolved in the solvent-exposed β hairpin loops at the bottom of the α-propeller, were occupied by Mn2+ in the original αvβ3 structure and by Mg2+ in these simulations. For comparison, a solvated complex was also built, in which Mg2+ occupied the MIDAS and Ca2+ occupied the other six ion binding sites. This complex is called Ca2+–Mg2+–Ca2+ in the text, in reference to the occupancies of the LIMBS, MIDAS, and ADMIDAS, respectively.
Simulation procedure and parameters
All MD simulations were performed with the program NAMD (Phillips et al., 2005) using the CHARMM27 force fields (MacKerell et al., 1998). The system was first minimized for three consecutive 2,000-conjugate gradient steps, during which the protein was initially held fixed and water molecules were allowed to move; next, only the protein backbone was held fixed, and finally, all atoms were allowed to move. After the minimizations, the system was heated from 0 to 300 K in 10 ps and subsequently equilibrated under constant pressure and temperature conditions (NPT). The bidentate salt bridge between ArgRGD and Asp218 of the integrin αv domain was held with harmonic constraints until 10 ps into the equilibration and remained stable in all equilibrated structures (Fig. 2 C). A more detailed explanation of the simulation protocol is described elsewhere (Craig et al., 2004). During equilibrations the complex remained stable, exhibiting an overall Cα RMSD in the head domains (the β-propeller from the αv subunit and βA domain from the β3 subunit) of <2.0 Å.
To open the closed hinge under force external forces were applied by SMD protocols at a constant pulling velocity of 5 and 10 Å/ns (Isralewitz et al., 2001). The spring constant was set to 6 kcal/mol/Å2. To examine the mechanical response of the complex in various pulling geometries, we fixed either one or both of the Cα atoms of at the C-terminal residues of the integrin headpiece (αv-Arg438 and/or β3-Asp434) and attached the spring to a Cα atom at either terminal residue of FnIII10 (Val1 or Thr93). Generally, the direction of stretching force was chosen along the vector pointing from the fixed atom to the pulling atom. In the special case when both C-terminal ends of the α and β subunit were fixed, the direction of force was pointed from the midpoint of the two fixed atoms to the pulling atom.
Simulations presented in this work lasted 107 ns altogether. Simulations were completed at the National Center for Supercomputing Applications at the University of Illinois and on the Gonzales cluster at the Swiss Federal Institute of Technology. 1-ns simulation required ∼12 h on 128 nodes (IBM )with 1.5-GHz processors (Itanium; Intel), or 24 h on 64 Dlco nodes with 2.4-GHz processors (Opteron; AMD), respectively. All structure alignments were done in VMD (Humphrey et al., 1996) via the backbone atoms of the 6 βA domain β-strands (unless otherwise noted). Hinge angles were measured using Hingefind (Wriggers and Schulten, 1997). Figures were rendered using VMD.
Online supplemental material
Fig. S1 shows the headpiece domains of the liganded αvβ3 integrin structure (1L5G.pdb) aligned with those of the unliganded αvβ3 integrin structure via the backbone atoms of the 6 βA domain β-strands. Fig. S2 compares the two previously established conformations of the αA domain MIDAS with the two βA domain MIDAS conformations presented in this study. Fig. S3 depicts the “lever-arm” scenario, in which a 2-Å MIDAS shift causes a characteristic repositioning of the ADMIDAS, β1-α1 loop, and α1 helix that we found to impede hinge angle opening. Fig. S4 considers the FnIII10-βA domain contact between Glu46 and Tyr129, together with additional data and sequence analysis. Within the additional data is a repeat equilibration in which Mg2+ occupied all binding sites, the MIDAS⋅⋅⋅H2O⋅⋅⋅Asp conformation was maintained, and the hinge angle again spontaneously increased by ∼20°. Fig. S5 describes FnIII10 domain deformation under force. Videos 1, 3, and 5 show trajectories from the equilibrations of the reference complex, the Ca2+–Mg2+–Ca2+ complex, and the unliganded integrin, respectively. In each case, the MD complex is overlaid with the starting crystal structure. Videos 2, 4, and 6 are 180° rotations around the y axis of the final frames from Videos 1, 3, and 5, respectively. Videos 7 and 8 are the trajectory and the 180° rotation, respectively, of the SMD simulation in which the hinge was opened 70° under force. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200602071/DC1.
Supplementary Material
Acknowledgments
We acknowledge Drs. V. Hytönen and M.L. Smith for helpful discussions and C. Faucher for help with figure preparation.
This work was supported by funding from the Swiss Federal Institute of Technology, the Swiss National Computing Centre, the National Institutes of Health (NIH P41RR05969), the National Science Foundation (MCAP3S028), and the Large Resource Allocation Committee (LRAC MC935028).
M. Gao's present address is the Center for the Study of Systems Biology, School of Biology, Georgia Institute of Technology, Atlanta, GA.
Abbreviations used in this paper: ADMIDAS, adjacent to the MIDAS; LIMBS, ligand-induced metal-binding site; MIDAS, metal ion–dependent adhesion site; MD, molecular dynamics; SMD, steered molecular dynamics.
References
- Adair, B., J.-P. Xiong, C. Maddock, S. Goodman, M.A. Arnaout, and M. Yeager. 2005. Three-dimensional EM structure of the ectodomain of integrin αVβ3 in a complex with fibronectin. J. Cell Biol. 168:1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar, B., M.E. Cunningham, P.A. McQueney, M.S. Egbertson, B.C. Askew, R.A. Bednar, G.D. Hartman, and R.J. Gould. 1997. Flow cytometric measurement of kinetic and equilibrium binding parameters of arginine-glycine-aspartic acid ligands in binding to glycoprotein IIb/IIIa on platelets. Cytometry. 28:58–65. [DOI] [PubMed] [Google Scholar]
- Beglova, N., S.C. Blacklow, J. Takagi, and T.A. Springer. 2002. Cysteine-rich module structure reveals a fulcrum for integrin rearrangement upon activation. Nat. Struct. Biol. 9:282–287. [DOI] [PubMed] [Google Scholar]
- Bershadsky, A.D., N.Q. Balaban, and B. Geiger. 2003. Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 19:677–695. [DOI] [PubMed] [Google Scholar]
- Calzada, M.J., M.V. Alvarez, and J. Gonzalez-Rodriguez. 2002. Agonist-specific structural rearrangements of integrin alpha IIb beta 3. Confirmation of the bent conformation in platelets at rest and after activation. J. Biol. Chem. 277:39899–39908. [DOI] [PubMed] [Google Scholar]
- Chen, J., A. Salas, and T.A. Springer. 2003. Bistable regulation of integrin adhesiveness by a bipolar metal ion cluster. Nat. Struct. Biol. 10:995–1001. [DOI] [PubMed] [Google Scholar]
- Clark, K., R. Pankov, M.A. Travis, J.A. Askari, A.P. Mould, S.E. Craig, P. Newham, K.M. Yamada, and M.J. Humphries. 2005. A specific alpha5beta1-integrin conformation promotes directional integrin translocation and fibronectin matrix formation. J. Cell Sci. 118:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copie, V., Y. Tomita, S.K. Akiyama, S. Aota, K.M. Yamada, R.M. Venable, R.W. Pastor, S. Krueger, and D.A. Torchia. 1998. Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure. J. Mol. Biol. 277:663–682. [DOI] [PubMed] [Google Scholar]
- Craig, D., M. Gao, K. Schulten, and V. Vogel. 2004. Structural insights into how divalent ions stabilize integrin binding of an RGD peptide under force. Structure. 12:2049–2058. [DOI] [PubMed] [Google Scholar]
- Hughes, P.E., T.E. O'Toole, J. Ylanne, S.J. Shattil, and M.H. Ginsberg. 1995. The conserved membrane-proximal region of an integrin cytoplasmic domain specifies ligand binding affinity. J. Biol. Chem. 270:12411–12417. [DOI] [PubMed] [Google Scholar]
- Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD - Visual Molecular Dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
- Humphries, M.J., P.A. McEwan, S.J. Barton, P.A. Buckley, J. Bella, and A.P. Mould. 2003. Integrin structure: heady advances in ligand binding, but activation still makes the knees wobble. Trends Biochem. Sci. 28:313–320. [DOI] [PubMed] [Google Scholar]
- Hynes, R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687. [DOI] [PubMed] [Google Scholar]
- Ingber, D.E. 2003. Mechanobiology and diseases of mechanotransduction. Ann. Med. 35:564–577. [DOI] [PubMed] [Google Scholar]
- Isralewitz, B., M. Gao, and K. Schulten. 2001. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 11:224–230. [DOI] [PubMed] [Google Scholar]
- Iwasaki, K., K. Mitsuoka, Y. Fujiyoshi, Y. Fujisawa, M. Kikuchi, K. Sekiguchi, and T. Yamada. 2005. Electron tomography reveals diverse conformations of integrin alphaIIbbeta3 in the active state. J. Struct. Biol. 150:259–267. [DOI] [PubMed] [Google Scholar]
- Jiang, G., A.H. Huang, Y. Cai, M. Tanase, and M.P. Sheetz. 2006. Rigidity sensing at the leading edge through alphavbeta3 integrins and RPTPalpha. Biophys. J. 90:1804–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, W.L., J. Chandrasekhar, J.D. Madura, R.W. Impey, and M.L. Klein. 1983. Comparison of simple potential functions for simulating water. J. Chem. Phys. 79:926–935. [Google Scholar]
- Katsumi, A., A.W. Orr, E. Tzima, and M.A. Schwartz. 2004. Integrins in mechanotransduction. J. Biol. Chem. 279:12001–12004. [DOI] [PubMed] [Google Scholar]
- Kim, M., C.V. Carman, and T.A. Springer. 2003. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 301:1720–1725. [DOI] [PubMed] [Google Scholar]
- Leahy, D.J., I. Aukhil, and H.P. Erickson. 1996. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 84:155–164. [DOI] [PubMed] [Google Scholar]
- Lee, J.O., L.A. Bankston, M.A. Arnaout, and R.C. Liddington. 1995. Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure. 3:1333–1340. [DOI] [PubMed] [Google Scholar]
- Liddington, R.C. 2002. Will the real integrin please stand up? Structure. 10:605–607. [DOI] [PubMed] [Google Scholar]
- Luo, B.-H., T.A. Springer, and J. Takagi. 2003. Stabilizing the open conformation of the integrin headpiece with a glycan wedge increases affinity for ligand. Proc. Natl. Acad. Sci. USA. 100:2403–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, B.H., T.A. Springer, and J. Takagi. 2004. a. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2:e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, B.H., K. Strokovich, T. Walz, T.A. Springer, and J. Takagi. 2004. b. Allosteric beta1 integrin antibodies that stabilize the low affinity state by preventing the swing-out of the hybrid domain. J. Biol. Chem. 279:27466–27471. [DOI] [PubMed] [Google Scholar]
- Luo, B.H., C.V. Carman, J. Takagi, and T.A. Springer. 2005. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc. Natl. Acad. Sci. USA. 102:3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell, A.D., D. Bashford, M. Bellott, R.L. Dunbrack, J.D. Evanseck, M.J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Physiol. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- Marinelli, L., A. Lavecchia, K.-E. Gottschalk, E. Novellino, and H. Kessler. 2003. Docking studies on alphavbeta3 integrin ligands: pharmacophore refinement and implications for drug design. J. Med. Chem. 46:4393–4404. [DOI] [PubMed] [Google Scholar]
- Mould, A.P., S.K. Akiyama, and M.J. Humphries. 1995. Regulation of integrin alpha 5 beta 1-fibronectin interactions by divalent cations. Evidence for distinct classes of binding sites for Mn2+, Mg2+, and Ca2+. J. Biol. Chem. 270:26270–26277. [DOI] [PubMed] [Google Scholar]
- Mould, A.P., J. Askari, S.J. Barton, A.D. Kline, P.A. McEwan, S.E. Craig, and M.J. Humphries. 2002. Integrin activation involves a conformational change in the alpha-1 helix of the beta-subunit A-domain. J. Biol. Chem. 277:19800–19805. [DOI] [PubMed] [Google Scholar]
- Mould, A.P., S. Barton, J. Askari, S. Craig, and M.J. Humphries. 2003. a. Role of ADMIDAS Cation-binding site in ligand recognition by integrin alpha 5 beta 1. J. Biol. Chem. 278:51622–51629. [DOI] [PubMed] [Google Scholar]
- Mould, A.P., S.J. Barton, J.A. Askari, P.A. McEwan, P.A. Buckley, S.E. Craig, and M.J. Humphries. 2003. b. Conformational changes in the integrin betaA domain provide a mechanism for signal transduction via hybrid domain movement. J. Biol. Chem. 278:17028–17035. [DOI] [PubMed] [Google Scholar]
- Mould, A.P., E.J. Symonds, P.A. Buckley, J.G. Grossmann, P.A. McEwan, S.J. Barton, J.A. Askari, S.E. Craig, J. Bella, and M.J. Humphries. 2003. c. Structure of an integrin-ligand complex deduced from solution x-ray scattering and site-directed mutagenesis. J. Biol. Chem. 278:39993–39999. [DOI] [PubMed] [Google Scholar]
- Mould, A.P., M.A. Travis, S.J. Barton, J.A. Hamilton, J.A. Askari, S.E. Craig, P.R. Macdonald, R.A. Kammerer, P.A. Buckley, and M.J. Humphries. 2005. Evidence that monoclonal antibodies directed against the integrin beta subunit plexin/semaphorin/integrin domain stimulate function by inducing receptor extension. J. Biol. Chem. 280:4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, B., H.-G. Zerwes, K. Tangemann, J. Peter, and J. Engel. 1993. Two step binding mechanism of fibrinogen to alpha IIb beta3 integrin reconstituted into planar lipid bilayers. J. Biol. Chem. 268:6800–6808. [PubMed] [Google Scholar]
- Partridge, A.W., S. Liu, S. Kim, J.U. Bowie, and M.H. Ginsberg. 2005. Transmembrane domain helix packing stabilizes integrin alphaIIbbeta3 in the low affinity state. J. Biol. Chem. 280:7294–7300. [DOI] [PubMed] [Google Scholar]
- Phillips, J.C., R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R.D. Skeel, L. Kale, K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, J., and T.A. Springer. 2002. Integrin activation and structural rearrangement. Immunol. Rev. 186:141–163. [DOI] [PubMed] [Google Scholar]
- Takagi, J., B.M. Petre, T. Walz, and T.A. Springer. 2002. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 110:599–611. [DOI] [PubMed] [Google Scholar]
- Takagi, J., K. Strokovich, T.A. Springer, and T. Walz. 2003. Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 22:4607–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokova, V., P. Aprikian, O. Yakovenko, B. Kidd, V. Vogel, W.E. Thomas, and E. Sokurenko. 2006. Catch Bond-Mediating FimH Adhesin of E. coli is a One-Ligand Allosteric. Protein. In press. [DOI] [PubMed]
- Vogel, V., and M. Sheetz. 2006. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7:265–275. [DOI] [PubMed] [Google Scholar]
- Wriggers, W., and K. Schulten. 1997. Protein domain movements: detection of rigid domains and visualization of hinges in comparisons of atomic coordinates. Proteins. 29:1–14. [PubMed] [Google Scholar]
- Xiao, T., J. Takagi, B.S. Coller, J.H. Wang, and T.A. Springer. 2004. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 432:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, C., M. Shimaoka, T. Xiao, P. Schwab, L.B. Klickstein, and T.A. Springer. 2004. The integrin alpha-subunit leg extends at a Ca2+-dependent epitope in the thigh/genu interface upon activation. Proc. Natl. Acad. Sci. USA. 101:15422–15427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, J.-P., T. Stehle, R. Zhang, A. Joachimiak, M. Frech, S.L. Goodman, and M.A. Arnaout. 2001. Crystal structure of the extracellular segment of integrin alpha Vbeta 3. Science. 294:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, J.-P., T. Stehle, R. Zhang, A. Joachimiak, M. Frech, S.L. Goodman, and M.A. Arnaout. 2002. Crystal structure of the extracellular segment of integrin alpha Vbeta 3 in complex with an Arg-Gly-Asp ligand. Science. 296:151–155. [DOI] [PubMed] [Google Scholar]
- Xiong, J.-P., T. Stehle, S. Goodman, and A.M. Arnaout. 2003. New insights into the structural basis of integrin activation. Blood. 102:1155–1159. [DOI] [PubMed] [Google Scholar]
- Yang, W., M. Shimaoka, J.-F. Chen, and T.A. Springer. 2004. Activation of integrin beta-subunit I-like domains by one-turn C-terminal alpha-helix deletions. Proc. Natl. Acad. Sci. USA. 101:2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.