Abstract
Eilatin-containing ruthenium complexes bind to a broad range of different nucleic acids including: calf thymus (CT) DNA, tRNAPhe, polymeric RNAs and DNAs, and viral RNAs including the HIV-1 RRE and TAR. The nucleic acid specificity of Λ- and Δ-[Ru(bpy)2eilatin]2+ have been compared to that of the ‘free’ eilatin ligand, and to the classic intercalating agent ethidium bromide. Interestingly, all four compounds appear to bind to nucleic acids by intercalation, but the trends in nucleic acid binding specificity are highly diverse. Unlike ethidium bromide, both eilatin and the eilatin-containing coordination complexes bind to certain single-stranded RNAs with high affinity (Kd ≤ 1 µM). Eilatin itself is selective for electron-poor polymeric purines, while the eilatin-coordination complexes exhibit preference for the polypyrimidine r(U). These results show how the binding specificity of an intercalating ligand can change upon its incorporation into an octahedral metal complex.
INTRODUCTION
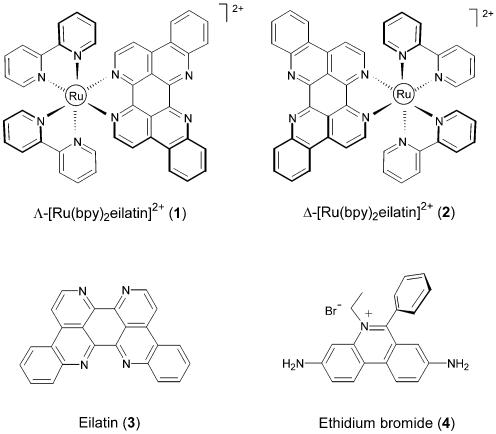
Octahedral transition metal complexes that contain one or more intercalating ligands have drawn considerable interest due to their potential uses as nucleic acid probes (1–3) and as therapeutic agents (4–6). Their unique photophysical and redox properties have facilitated the study of charge transfer reactions in DNA (7–10). The impact of incorporating an intercalating ligand into an octahedral metal complex is largely unknown. To the best of our knowledge, no side-by-side specificity comparisons of an intercalator to a metal complex containing the same intercalator have previously been made. In this paper, we explore the nucleic acid binding mode and specificity of two eilatin-containing ruthenium complexes Λ-[Ru(bpy)2eilatin]2+ (1) and Δ-[Ru(bpy)2eilatin]2+ (2). Their binding specificity is compared to those of the parent heterocyclic eilatin (3) and to ethidium bromide (4).
Eilatin (3) is a nearly planar, heptacyclic alkaloid that was isolated from the Red Sea tunicate Eudistoma sp. (11). It is known to possess cytotoxic and antiproliferation activities in a broad range of tissue cultures (12–14). Eilatin is a bi-facial metal chelator. Upon incorporation into octahedral metal complexes of the type [Ru(L)2eilatin]2+ (where L= bpy, phen, etc.), only the less hindered face of eilatin binds to the metal ion (Fig. 1) (15,16). The enantiomerically pure metal complexes Λ-[Ru(bpy)2eilatin]2+ (1) and Δ-[Ru(bpy)2eilatin]2+ (2) were synthesized from chiral precursors (16), and their expected assignments as Λ and Δ were confirmed using circular dichroism spectroscopy (4). Complexes 1 and 2 inhibit the HIV-1 Rev-RRE interaction in vitro (17) and are inhibitors of HIV replication in certain cell cultures (4). In previous studies, numerous indirect methods (including ethidium bromide displacement and thermal denaturation) were used to examine the relative DNA and RNA affinities of 1 and 2 (4,17). When compared to similar bipyridyl-containing ruthenium complexes, it was revealed that the eilatin moiety of 1 and 2 is essential to both the nucleic acid binding affinity and anti-viral activity of these compounds (4). In this paper we conduct a broad survey of the nucleic acid affinity and specificity of the eilatin-containing metal complexes 1 and 2 and compare them to eilatin (3) and to the classic intercalating agent ethidium bromide (4). These studies indicate that the nucleic acid specificity of eilatin (3) changes upon its incorporation into octahedral metal complexes, and that all compounds exhibit unique trends in nucleic acid affinity and specificity.
Figure 1.
Eilatin and eilatin-containing metal complexes. The dichoride salts of 1 and 2 were used for all experiments. Unlike eilatin (3), the chloride salts of 1, 2 and 4 are freely soluble in water.
MATERIALS AND METHODS
Nucleic acids
A sonicated solution of calf thymus (CT) DNA was purchased from Gibco BRL and quantified in 50 mM sodium phosphate (pH 7.5) using a molecular extinction coefficient of 13 100 cm–1 M–1 per base pair. All other polymeric DNAs and RNAs were purchased from Pharmacia and Sigma-Aldrich, dissolved in 1× TE, and quantified in 50 mM sodium phosphate (pH 7.5) using the following extinction coefficients (260 nm): poly (dA)–(dT) = 12 200 cm–1 M–1 per base pair, poly (dG)–(dC) = 14 800 cm–1 M–1 per base pair, poly (dAT)–(dAT) = 13 300 cm–1 M–1 per base pair, poly (dGC)–(dGC) = 16 800 cm–1 M–1 per base pair, poly (rA)–Poly (rU) = 14 280 cm–1 M–1 per base pair, poly (rG)–Poly (rC) = 14 800 cm–1 M–1 per base pair, poly (rI)–(rC) = 10 000 cm–1 M–1 per base pair, poly (rA) = 10 500 cm–1 M–1 per base, poly (rC) 6200 cm–1 M–1 per base, poly (rU) = 9500 cm–1 M–1 per base, poly (dA) = 8600 cm–1 M–1 per base, poly (dT) = 8520 cm–1 M–1 per base, poly (rI) 10 200 cm–1 M–1 per base. The oligomeric homopolymers were purchased from Gensetoligos and purified using denaturing PAGE, extraction and multiple rounds of ethanol precipitation. Stock solutions were made in 1× TE and quantified according to the extinction coefficients (260 nm) of the corresponding polymer (above). tRNAPhe was purchased from Sigma-Aldrich, dissolved in 1× TE and quantified in 50 mM sodium phosphate (pH 7.5) at 260 nm using an extinction coefficient of 733 000 cm–1 M–1. TAR31 and RREJW were transcribed from a synthetic DNA template using T7 RNA polymerase. Transcription products were purified using denaturing PAGE, extraction and multiple rounds of ethanol precipitation. The expected molecular weights of the isolated products were confirmed using mass spectrometry. The transcripts were quantified in buffer using the following extinction coefficients: TAR31 = 264 000 cm–1 M–1 and RREJW = 276 000 cm–1 M–1. All extinction coefficients are for the native (non-hydrolyzed) form of each nucleic acid.
Buffer conditions
All titrations (except for the viscosity experiments) were conducted at 22°C in a buffer containing 30 mM HEPES (pH 7.5), KCl (100 mM), sodium phosphate (10 mM), NH4OAc (20 mM), guanidinium HCl (20 mM), MgCl2 (2 mM), NaCl (20 mM), EDTA (0.5 mM) and Nonidet P-40 (0.001%).
Viscosity experiments
The viscosity of a DNA solution was determined by measuring the time needed for it to flow through a capillary viscometer. A Cannon-Manning semi-micro size 75 viscometer was submerged in a glass beaker and thermoregulated at 25.9 ± 0.1°C. Concentrated stocks (10 mM) of either ethidium (4) or rac-(1/2) were titrated into 0.5 ml of buffered solutions (50 mM sodium phosphate pH 7.5) of CT DNA (0.5 mM bases). Flow times ranged from 126.4 ± 0.3 s to 160.5 ± 0.5 s depending on the concentration of small molecule. The value η is the viscosity of the solution in the presence of a ligand; ηo is the viscosity of the DNA-only solution. Viscosity is calculated as η = t – to where t is the time for the sample to flow through the viscometer and to is the time measured for the buffer only (114.7 ± 0.3 s).
UV-vis titrations
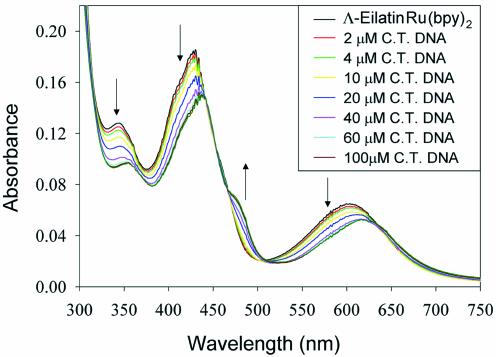
UV-vis spectra were collected using a Hewlett Packard 8452A diode array spectrophotometer. 8 µM of either 1, 2 or 4 in buffer were monitored as small volumes of concentrated nucleic acids were added. The fractional change in absorbance was averaged at multiple wavelengths to calculate each C50 value. Upon titration of TAR31, RREJW and tRNAPhe, both 1 and 2 exhibited approximately the same spectral changes as observed for CT DNA (Fig. 4). Significant diversity, however, was observed in the magnitudes of red-shifting and the changes in absorbance upon titration of different polymeric nucleic acids (see Supplementary Material for additional spectral data). Small changes in the absorbance spectrum of ethidium are observed for some nucleic acids. For these particular nucleic acids [poly r(U), poly r(A) and poly r(I)] the increase in fluorescence intensity of the ethidium bromide solution (upon addition of each nucleic acid) was used to measure binding (Ex 480 nm, Em 606 nm).
Figure 4.
UV-vis absorbance spectrum of 2 (8 µM) as a function of calf thymus DNA (concentration in base-pairs). See the Supplementary Material for the spectral changes observed upon titration of other nucleic acids.
Fluorescence titrations
In a Perkin Elmer LS-50B luminescence spectrometer, a dilute solution of eilatin (3) (0.1 µM) in buffer was excited at 417 nm using maximum slit widths. The emission of the solution was monitored at 470 nm as small volumes of highly concentrated nucleic acids were titrated. For all titrations, a small increase in the final volume (≤5%) of the sample occured.
RESULTS
Viscometry experiments
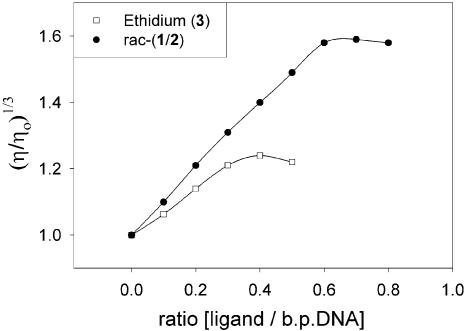
Octahedral metal complexes bind to nucleic acids through various non-covalent modes, including: intercalation, non-specific electrostatic association and groove binding (1,7,18–25). To probe the binding mode of 1 and 2, the viscosity of a concentrated DNA solution was measured in the presence of increasing concentrations of rac-1/2 (Fig. 2). In accordance with the hypothesis proposed by Lerman (26), the viscosity of a DNA solution increases upon the addition of an intercalating agent (27). Upon intercalation of a small molecule, the axial length of the DNA increases and it becomes more rigid. Both factors increase its frictional coefficient, and hence, the viscosity of the DNA in solution (27). Since the change in viscosity should, in theory, be proportional to the change in the axial length of the DNA molecule to the third power (27), viscosity results are often reported as (η/ηo)1/3 versus the molar fraction of the ligand, where η is the viscosity of the solution in the presence of a ligand and ηo is the viscosity of the DNA-only solution. Cohen and Eisenberg predicted that for an ‘ideal’ intercalating agent, this plot should be linear with a slope of 1.0 (27). Interestingly, rac-1/2 has a slope of 1.0, and the ‘classic’ intercalating agent ethidium bromide (4) displays a slope of 0.7 (Fig. 2). Other groups have also observed a slope of less than 1.0 for ethidium (21,23). At low ionic strengths, ethidium can bind to DNA by both intercalation and by surface-binding via electrostatic interactions (28). Compounds that bind to surfaces of duplexes through electrostatic interactions can decrease the viscosity of the nucleic acid solution (29). This may explain why, under these conditions, viscometric analysis of the ethidium–DNA complex yields a slope significantly lower than 1.0 (Fig. 2, right). It appears, however, that rac-1/2 fits the viscosity profile of an ‘ideal’ intercalating agent (Fig. 2) (27).
Figure 2.
Changes in the viscosity of a solution of calf thymus DNA with increasing concentrations of ligand. Since the concentration of DNA used in these experiments (0.5 mM) is much higher than the affinities of these interactions (Kd = ∼5 µM), the saturation points in the curves are sensitive only to the maximum binding stoichiometry of these interactions. A low-salt buffer (50 mM sodium phosphate pH 7.5) was used for these measurements.
Another interesting difference between the eilatin- containing metal complexes (rac-1/2) versus ethidium (4) is apparent in the viscosity titrations (Fig. 2). The ratio at which the DNA becomes saturated is significantly different for these compounds. Ethidium bromide (4) reaches saturation at ∼3 ethidium molecules per 10 base pairs of DNA. This value is consistent with ethidium’s previously reported binding stoichiometry to CT DNA (30,31). This is also the same binding stoichiometry as reported by Haq et al. (20) for the binding of [Ru(phen)2DPPZ]2+ (a metal complex similar to 1 and 2) to CT DNA. For rac-1/2, on the other hand, saturation is reached at much higher ratios (Fig. 2), indicating a maximum binding stoichiometry of ∼6 equivalents of ligand for every 10 base pairs of DNA. This is an unusually high binding stoichiometry for such a large metal complex! Only one other example of such a high binding stoichiometry by a metal complex is found in the literature (32). Barton and Lippard report that ∼6.7 equivalents of [(terpy)Pt(HET)]1+ are bound per every 10 base pairs of poly r(A)–poly r(U) (32). [(terpy)Pt(HET)]1+ is, however, a square planar metal complex, and the intercalation of this compound into duplex nucleic acids should be much less sterically demanding when compared to 1 and 2.
Eilatin (3) was previously reported to be an intercalating agent because of an apparent increase in its fluorescence emission intensity upon addition of DNA (14). It should be noted, however, that some intercalating agents, including 9-amino acridine, have a lower quantum yield when bound to DNA (33). Unfortunately, the limited water solubility of 3 is not conducive to the use of simple viscometry experiments to examine its binding mode. Intercalation of 3 can, however, be inferred, since [Ru(bpy)3]2+ does not intercalate into nucleic acids (7), and [Ru(bpy)2eilatin]2+ appears to be an intercalating agent.
Binding studies: 1 and 2 with polymeric nucleic acids
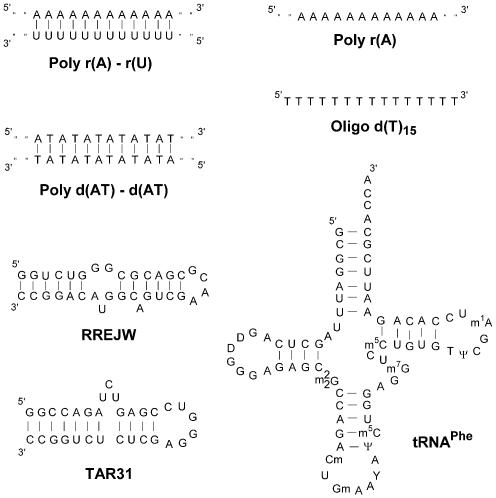
To examine the nucleic acid binding specificity of 1–4, we have measured the concentration-dependent binding of each compound to a large number of different nucleic acids. A variety of commercially available duplex and single-stranded polymeric nucleic acids, as well as three biologically relevant RNA molecules, have been evaluated (Fig. 3). Binding studies have been conducted by monitoring the UV-vis absorbance spectrum of the enantiomerically pure complexes 1 and 2 upon titration of each nucleic acid (see Fig. 4 for a representative titration). Complexes 1 and 2 exhibit multiple electronic transitions in their absorbance spectra that change upon addition of nucleic acids. The presence of isosbestic points, and saturation at high DNA concentrations, suggests a simple two-state transition between the free and bound ligand (Fig. 4). By averaging the fractional change in absorbance intensity at multiple wavelengths (as a function of nucleic acid concentration), binding isotherms are generated (Figs 5 and 6). For most nucleic acids tested, the UV-vis absorbance spectra of both 1 and 2 show changes very similar to those observed upon titration of CT DNA (see Supplementary Material for titrations using other polymeric nucleic acids). The absolute changes in intensity and the degree of red-shifting are slightly different for each nucleic acid tested, but the spectral changes for all reported titrations suggest simple two-state equilibria. The concentration of nucleic acid needed to bind half of each compound (defined as the C50 value) is determined by assuming a linear relationship between its spectral changes and the fraction of ligand bound (Figs 4–6). The C50 values for 17 different polymeric nucleic acids are summarized in Table 1. Lower C50 values indicate a higher affinity and/or higher binding stoichiometry (Table 1). Differences in binding stoichiometry can, in principle, make the C50 values in Table 1 disproportionate to the differences in binding affinities (Kd). To examine the maximum binding stoichiometries of 1 and 2 to single-stranded and duplex polymeric nucleic acids, the UV-vis absorbance spectrum of a more concentrated solution of 1 or 2 (40 µM) was monitored upon addition of either a single-stranded RNA [poly r(U)] or upon titration of the duplex DNA [poly d(AT)–poly d(AT)] (see Fig. 5 for a representative titration). Since 40 µM of 1 and 2 is a much higher concentration than the binding affinities of these binding interactions (concentration >>Kd), the association isotherms reflect the maximum binding stoichiometry of each nucleic acid (Fig. 5). Interestingly, a very high stoichiometry for single-stranded polymeric RNA is indicated, where ∼5 equivalents of 2 are bound per every 10 bases of poly r(U) (the same result is obtained for 1). Surprisingly, this is almost the same stoichiometry as observed for CT DNA (Fig. 2). Another DNA duplex, poly d(AT)–poly d(AT), also exhibits the same binding stoichiometry for both 1 and 2 (at 5 equivalents per every 10 base pairs). This suggests a very consistent binding stoichiometry of 1 and 2 to these different types of polymeric nucleic acids. We have used the C50 value for each nucleic acid (Table 1) along with a binding stoichiometry of one molecule of 1 or 2 per two bases (or base pairs) to estimate Kd values for the eilatin-containing metal complexes (Table 2). All estimates are equal to the C50 value divided by the binding stoichiometry, minus half of the total concentration. Issues related to potential sequence-dependent differences in binding stoichiometry as well as the cooperative binding of 1 and 2 to polymeric nucleic acids might, however, influence some of these estimates (Table 2). Consistent with the results from a solid-phase assay (17), thermal denaturation (4) and ethidium bromide displacement experiments (4), these ‘direct’ titrations indicate that 1 has a higher affinity for CT DNA as compared to 2 (Table 2). Interestingly, this is the opposite enantiomeric selectivity as reported for most other metal complex–DNA interactions to date (1,7,19–21).
Figure 3.
Examples of the nomenclature used for the polymeric RNA and DNA used in theses studies (top). The polymeric species (Poly) are enzymatically synthesized and typically extend, unbroken, for thousands of bases. The oligomeric species (Oligo) are chemically synthesized and are, for these studies, 15 bases in length. Secondary structures of three biologically important RNAs (bottom). RREJW and TAR31 are minimized versions of two important RNAs from HIV-1 (47,48). tRNAPhe is a well-characterized, natural RNA with diverse structural features that provide well-defined binding sites for small molecules (49).
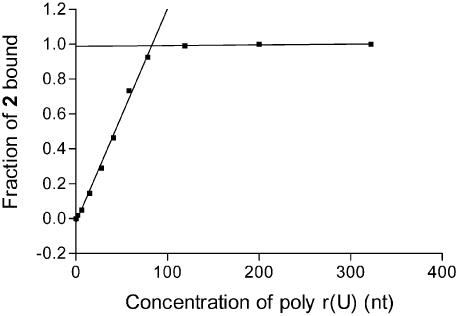
Figure 5.
At a relatively high concentration of 2 (40 µM), the binding isotherm for poly r(U) is composed of two linear elements. The intersection of these lines [at 80 µM of poly r(U)] indicates the maximum binding stoichiometry for 2 is 1 equivalent for every 2 bases of poly r(U). The other enantiomer 1 yields the same result. The same experiment was conducted with duplex DNA and also yielded a binding stoichiometry of 1 equivalent of either 1 or 2 for every 2 base pairs of poly d(AT)–d(AT).
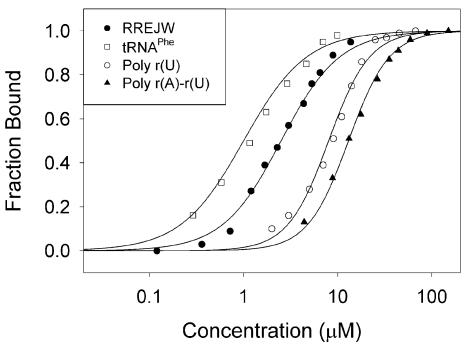
Figure 6.
Binding of 2 (8 µM) by poly r(A)-r(U), poly r(U), RREJW and tRNAPhe. For poly r[U] and poly r(A)–r(U) the data fit to a Hill coefficient of n = 2 which is also observed when the concentration of 2 is 4 µM. For each of the non-polymeric species (RREJW and tRNAPhe) a Hill coefficient of ∼1.2 is observed. Similar results are obtained for 1. The Hill coefficient is apparent as the slope of the linear region of each plot.
Table 1. C50 values (µM)a for 8 µM of Λ-[Ru(bpy)2eilatin]2+ (1), 8 µM Δ-[Ru(bpy)2eilatin]2+ (2), 8 µM ethidium bromide (4) and 0.1 µM eilatin (3).
| Nucleic acid | Λ-[Ru(bpy)2eilatin]2+ (1) | Δ-[Ru(bpy)2eilatin]2+(2) | Eilatin (3)b | Ethidium bromide (4)c |
|---|---|---|---|---|
| CT DNA | 18 | 27 | 3 | 20 |
| Poly d(A)–Poly d(T) | 13 | 23 | 1 | 105 |
| Poly d(AT)–Poly d(AT) | 9 | 8 | 5 | 13 |
| Poly d(G)–Poly d(C) | 15 | 13 | 5 | 22 |
| Poly d(GC)–Poly d(GC) | 18 | 55 | 8 | 16 |
| Poly r(A)–Poly r(U) | 13 | 22 | 12 | 18 |
| Poly r(G)–Poly r(C) | 125 | 85 | 20 | 400 |
| Poly r(I)–Poly r(C) | 200 | 180 | 150 | 170 |
| Poly r(U) | 8 | 8 | >2000 | >6000 |
| Poly r(C) | 52 | 75 | >1000 | >4000 |
| Poly r(A) | 14 | 13 | 70 | 2500 |
| Poly r(G) | 18 | 20 | 4 | 240 |
| Poly r(I) | 13 | 10 | 0.2 | 950 |
| Oligo r(I)15 | 68 | 53 | n.d. | n.d. |
| Poly d(A) | 20 | 200 | n.d. | n.d. |
| Oligo d(A)15 | 200 | 70 | n.d. | n.d. |
| Oligo d(T)15 | 21 | 21 | n.d | n.d. |
aC50 values for duplexes are reported per base pair. The values for single-stranded polymers are reported per base. The approximate experimental deviation for all values is less than or equal to ±35% of the reported C50 value.
bCompared to 1, 2 and 4, a much lower concentration of 3 was used for spectroscopic measurements (8.0 versus 0.1 µM, respectively); the C50 values for 3 cannot, therefore, be directly compared to those of 1, 2 and 4.
cChanges in UV-vis absorption monitored for duplex nucleic acids, and changes in emission monitored for single-stranded nucleic acids.
Table 2. Approximate Kd values (µM) for Λ-[Ru(bpy)2eilatin]2+ (1), Δ-[Ru(bpy)2eilatin]2+ (2), eilatin (3) and ethidium bromide (4) as calculated from C50 values (Table 1)a.
| Nucleic acid | Λ-[Ru(bpy)2eilatin]2+(1) | Δ-[Ru(bpy)2eilatin]2+(2) | Eilatin (3) | Ethidium bromide (4) |
|---|---|---|---|---|
| CT DNA | 5 | 9.5 | 1.5 | 2.7 |
| Poly d(A)–Poly d(T) | 2.5 | 7.5 | 0.5 | 31 |
| Poly d(AT)–Poly d(AT) | 0.5 | <0.5 | 2.5 | 0.33 |
| Poly d(G)–Poly d(C) | 3.5 | 2.5 | 2.5 | 3.3 |
| Poly d(GC)–Poly d(GC) | 5 | 24 | 4 | 1.3 |
| Poly r(A)–Poly r(U) | 2.5 | 7 | 6 | 2 |
| Poly r(G)–Poly r(C) | 60 | 40 | 10 | 130 |
| Poly r(I)–Poly r(C) | 100 | 86 | 75 | 53 |
| Poly r(U) | <0.5 | <0.5 | >1000 | >2000 |
| Poly r(C) | 22 | 34 | >500 | >1300 |
| Poly r(A) | 3 | 2.5 | 35 | 830 |
| Poly r(G) | 5 | 6 | 2 | 76 |
| Poly r(I) | 2.5 | 1 | 0.05 | 310 |
| Oligo r(I)15 | 30 | 23 | n.d. | n.d. |
| Poly d(A) | 6 | 100 | n.d. | n.d. |
| Oligo d(A)15 | 96 | 31 | n.d. | n.d. |
| Oligo d(T)15 | 6.5 | 6.5 | n.d | n.d. |
aThe Kd values for 1 and 2 are estimated as Kd = [(C50/2) – 4]. The Kd values for 3 are estimated as Kd = [(C50/2) – 0.05]. The Kd values for 4 are estimated as Kd = [(C50/3) – 4]. These values serve as estimates only, as differences in binding stoichiometries and cooperativity will affect the calculated values.
The binding isotherms of 1 and 2 for both single-stranded and duplex polymeric nucleic acids suggest positive cooperativity. Titrations using poly r(U) conducted at both 4 and 8 µM of either 1 and 2 fit well to association isotherms with Hill coefficients of n = 2 (Fig. 6). Duplex nucleic acids also show positive cooperativity in their binding of 1 and 2. For example, poly r(A)–r(U) also has a Hill coefficient of n = 2 for binding to both 1 and 2 (Fig. 6). All of the ‘higher affinity’ interactions with polymeric nucleic acids (C50 < 30 µM, Table 1) show some degree of positive cooperativity in their binding of 1 and 2 (Hill coefficients ranging from ∼1.5 to 2.2). These cooperative-binding interactions may explain why, for the most part, the short single-stranded oligos exhibit a lower apparent binding affinity as compared to the corresponding polymers. Poly r(I), for example, has a 20-fold higher affinity to 1 and 2 as compared to oligo r(I)15 (Tables 1 and 2). 1 and 2 bind to poly r(I) cooperatively (Hill coefficient = 2), but show much less cooperativity when binding oligo (I)15 (Hill coefficient = 1.3). Large differences between the two enantiomers (1 versus 2) are observed for poly d(A) versus oligo r(A)15 (Tables 1 and 2). This may reflect secondary structure formation in the single-stranded nucleic acids and/or higher order aggregates for some of these interactions.
Binding studies: 1 and 2 to non-polymeric nucleic acids
None of the non-polymeric nucleic acids tested (RREJW, TAR31 and tRNAPhe) shows significant cooperativity in their binding of 1 and 2 (Hill coefficients range from 1.1 to 1.4, Fig. 6). Scatchard plots can, therefore, be used to examine the average affinity and stoichiometry of 1 and 2 for binding to these three RNAs (Fig. 7 and Table 3). Non-whole numbers for the binding stoichiometry are obtained for some of these interactions (Table 3). This suggests that these RNAs have multiple non-equivalent binding sites that contribute to the binding isotherms. The reported affinities (Kd), therefore, represent the average affinity of all the contributing binding sites. The apparent binding stoichiometry of these titrations can change depending upon the absolute concentration of the ‘fixed’ species (in this case the small molecule) relative to the affinities of the interactions being studied (34,35). Despite these ambiguities in data analysis, similar trends in binding affinities have been identified using indirect methods (4,17). Consistent with Rev peptide and ethidium displacement experiments (4), 1 and 2 have a higher affinity to the RRE as compared to the TAR (Table 3). Consistent with results from a novel solid-phase assay (17) 1 and 2 have a higher affinity to the RRE as compared to tRNA (Table 3). Unfortunately, due to the limited sensitivity of monitoring changes in UV-vis absorption, it is difficult to probe the highest affinity binding site(s) of 1 and 2 on the RRE. Since these compounds are not luminescent, low µM concentrations are needed to observe their UV-vis absorbance changes upon binding nucleic acids. For example, even if the RRE possesses a 10 nM binding site for 1, it cannot be characterized under these conditions, since the metal complex is at a much higher concentration relative to this theoretical affinity. This limitation is also reflected in some of the C50 values reported in Table 1. Since the concentration of 1 and 2 used for these experiments is 8 µM, and the binding stoichiometry is approximately one ligand per every 2 bases (or base pairs), the theoretical minimum C50 value for all polymeric nucleic acids is ∼8 µM (Table 1). We can only report, therefore, that the interaction between poly r(U) and both 1 and 2 exhibit affinities of Kd < 0.5 µM (Table 2). This is, however, an unusually high affinity for an intercalating agent binding to single-stranded polymeric nucleic acid, and is in stark contrast to ethidium bromide.
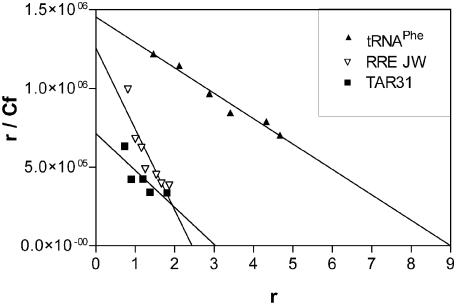
Figure 7.
Scatchard analysis of the interaction between 1 and the RRE JW, tRNAPhe and TAR31. The slope of the graph = 1/Kd(ave), and the r value at r/Cf = 0 is the number of apparent binding sites.
Table 3. Summary of the binding stoichiometries and average affinity of 1 and 2 for RREJW, tRNAPhe and TAR31.
| Nucleic acid | Binding sites for 1 | Average Kd (µM) for 1 | Binding sites for 2 | Average Kd (µM) for 2 |
|---|---|---|---|---|
| RREJW | 2.4 | 1.9 | 3.8 | 1 |
| tRNAPhe | 9 | 5.6 | 10 | 5.6 |
| TAR31 | 3 | 4.6 | 4 | 9 |
Binding studies: ethidium
The classic intercalating agent ethidium bromide (4) has been evaluated for nucleic acid specificity by monitoring the changes in its UV-vis absorption or emission spectrum upon titration of polymeric nucleic acids (36,37). The C50 values for ethidium are summarized in Table 1 and have been used to estimate Kd values (summarized in Table 2). These Kd estimates assume a binding stoichiometry of 3 molecules of ethidium per 10 bases (or base pairs) of polymeric nucleic acid. This assumption is supported by previous studies that have shown that ethidium binds to many different duplex nucleic acids with a stoichiometry of 3 ± 1 molecules ethidium per 10 base pairs (31). Tables 1 and 2 represent, to the best of our knowledge, the most comprehensive study of ethidium bromide and its affinity to different nucleic acids. Bresloff and Crothers (31) used equilibrium dialysis to study the binding of ethidium to 12 different nucleic acids, five of which appear in Table 2. Their studies were conducted using equilibrium dialysis in 1 M NaNO3, while our studies are conducted under more physiological conditions. Despite these differences, we report a very similar trend in ethidium affinity for all five nucleic acids (31). At much lower ionic strengths (0.010 M), Baguley and Falkenhaug (38) have also reported a very similar trend for all comparable nucleic acids. Interestingly, at lower ionic strengths, somewhat smaller differences in sequence selectivity are observed (38).
Ethidium is sometimes regarded as a non-specific intercalating agent (39,40). Our studies, however, confirm that at physiological ionic strength, ethidium is very sensitive to both the sequence and the composition of polymeric nucleic acids. For example, ethidium has a 100-fold higher affinity to poly d(AT)–poly d(AT) as compared to poly d(A)–poly d(T) (Tables 1 and 2). Ethidium exhibits a preference for the alternating purine–pyrimidine tract of poly d(GC)–poly d(CG) as compared to poly d(G)–poly d(C) (Tables 1 and 2) (38). Ethidium is sometimes regarded as being selective for G/C base pairs (41). Ethidium does, in fact, exhibit a 10-fold higher affinity to poly d(G)–d(C) over poly d(A)–d(T), but the opposite trend is revealed for duplex RNA, where it has over a 50-fold higher affinity to poly r(A)–r(U) as compared to poly r(G)–r(C) (Table 2).
Binding studies: eilatin (3)
To examine how the nucleic acid specificity of an intercalating agent changes upon its incorporation into a ruthenium bypyridyl complex, the nucleic acid specificity of ‘free’ eilatin (3) was also investigated. Unlike the eilatin-containing metal complexes (1 and 2), eilatin itself (3) is highly fluorescent and shows dramatic changes in its emission spectrum upon addition of nucleic acids (Fig. 8). For all nucleic acids tested, a decrease in its emission intensity is observed (similar to Fig. 8), allowing C50 values to be measured (summarized in Table 1). Unlike the metal complexes 1 and 2, no evidence for the cooperative binding of eilatin (3) to polymeric nucleic acids has been found. The Hill coefficients for 3 (as for ethidium bromide) are equal to 1.0 for all titrations conducted. The C50 values measured for eilatin (3) are not comparable with those determined for 1, 2 and 4 (Table 1). Due to the limited water solubility of 3, the concentration of 3 used for the fluorescence experiments is 80-fold lower than the concentrations used for the UV-vis titrations of 1, 2 and 4 (Table 1). To allow for a more direct comparison of these values, the binding constants of eilatin (3) have been estimated from C50 values by assuming its binding stoichiometry to the polymeric nucleic acids is similar to those measured for 1 and 2 (at one molecule per every 2 bases or base pairs). The limited solubility of eilatin (3) in water (<3 µM at pH 7.5) makes the independent validation of binding stoichiometry difficult to attain. Nonetheless, the estimated Kd values presented in Table 2 allow for a much more direct comparison of eilatin (3) to compounds 1, 2 and 4. Any deviation in binding stoichiometry from the estimated values will have a roughly linear effect upon the calculated affinities. This, combined with the cooperative binding of 1 and 2 to polymeric nucleic acids, makes these Kd values approximations only (Table 2). Despite this, many interesting trends in nucleic acid specificity emerge.
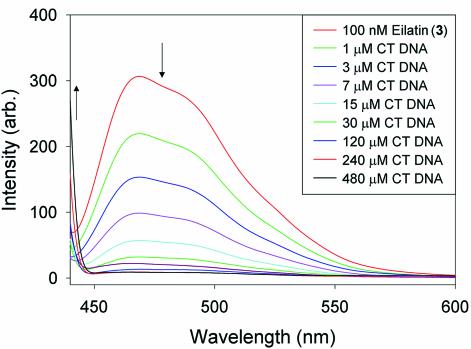
Figure 8.
Changes in the fluorescence emission spectrum of a 0.1 µM solution of eilatin (3) upon addition of CT DNA. Excitation is at 417 nm. Nearly identical changes are seen for all other nucleic acids tested. Eilatin’s limited water solubility (<3 µM at pH 7.5) prevents titrations that would be directly comparable to those conducted for 1 and 2 (Figs 4–6).
Compounds 1–4 compared
The eilatin-containing metal complexes (1 and 2) bind to duplex RNA and DNA with an affinity range similar to ethidium bromide, but exhibit some differences in sequence selectivity (Tables 1 and 2). For example, 2 has an ∼10-fold higher affinity for poly d(G)–d(C) as compared to poly d(GC)–d(GC). This is the opposite selectivity as exhibited by ethidium (Table 2). Similar to ethidium, 1 and 2 have a higher affinity to poly d(AT)–d(AT) as compared to poly d(A)–d(T) (Table 1). Interestingly, all four compounds (1–4) have a much higher affinity to poly r(A)–r(U) as compared to both poly r(I)–r(C) and r(G)–r(C). This may indicate that some duplex nucleic acids have, in general, a low affinity for any intercalating agent. Some nucleic acids, on the other hand, are highly sensitive to the identity of the intercalating agent. For example, poly d(A)–d(T), has a low affinity for ethidium (4), a modest affinity for the metal complexes (1 and 2) and a very high affinity for the free eilatin ligand (3) (Tables 1 and 2). All four compounds exhibit a moderate selectivity for duplex DNA over duplex RNA. Each compound, for example, has an ∼10-fold higher affinity for poly d(G)–d(C) versus poly r(G)–r(C) (Table 2). We conclude that the trends in the nucleic acid selectivity of 1–4 are, for the most part, similar for all the duplex nucleic acids tested. We interpret this as additional evidence that 1–3 bind to duplex nucleic acids via intercalation.
The trends observed for single-stranded versus double-stranded nucleic acids are strikingly different for each compound (Tables 1 and 2). In many cases, the eilatin-containing metal complexes exhibit a higher affinity to single-stranded versus duplex polymers (Tables 1 and 2). Very few examples of intercalating agents that exhibit such a high affinity to single-stranded nucleic acids can be found in the literature (42,43). Classic intercalating agents, like ethidium bromide, typically exhibit a 1000-fold lower affinity for most single-stranded nucleic acids as compared to duplex nucleic acids (Table 2). Both eilatin and the eilatin-containing metal complexes, on the other hand, bind to single-stranded nucleic acids with moderate to high affinity. In addition, these compounds exhibit very different trends in specificity. Eilatin (3) exhibits a high degree of differentiation among the three homopurine single-stranded nucleic acids [poly r(I) > poly r(G) > poly r(A)]. This suggests a preference for electron-poor purines. Interestingly, both ‘free’ eilatin (3) and ethidium (4) show similar preferences for single-stranded purines, while the eilatin-containing metal complexes 1 and 2 exhibit the highest affinity to the single-stranded pyrimidine poly r[U]. Indeed, the incorporation of eilatin (3) into the complexes 1 and 2 increases its affinity to poly r[U] by >2000-fold (Table 2). The ability of the metal complexes to bind to poly r[U] cooperatively may explain their high apparent affinity (Kd < 0.5 µM). On the other hand, no evidence for the cooperative binding of 3 or 4 by any of the nucleic acids tested is apparent by Hill analysis.
Tables 1 and 2 contain other examples of how the nucleic acid specificity of an intercalating ligand can change upon its incorporation into an octahedral metal complex. For the most part, eilatin (3) has a higher affinity to both DNA and RNA duplexes as compared to 1 and 2 (Table 2). A notable exception is poly d(AT)–poly d(AT), to which 1 and 2 have an ∼5-fold higher affinity as compared to 3 (Table 2). Interestingly, the eilatin-containing metal complexes (1 and 2) have a higher affinity for poly d(AT)–poly d(AT) as compared to poly d(A)–poly d(T), while eilatin (3) has the opposite selectivity (Table 2). It appears, therefore, that the sequence specificity of an intercalating agent is fairly plastic and can be changed by its incorporation into an octahedral metal complex.
To the best of our knowledge, only one other group has examined the nucleic acid affinity of eilatin (3), and they reported very different results compared to ours (14). Ethidium bromide displacement experiments from CT DNA were conducted, but only a low affinity to 3 was suggested (IC50 > 100 µM) (14). The limited solubility of eilatin (<3 µM in water) may explain this result. The same report also used fluorescence-based experiments to monitor the changes of a solution of 3 as a function of CT DNA (14). They showed an apparent increase in the fluorescence intensity of eilatin upon addition of CT DNA (14). We found, however, a dramatic decrease for all nucleic acids tested (Fig. 8). In the previous study, the sample was both excited and monitored at 520 nm (14). Eilatin (3) shows no absorption at this wavelength (11). In addition, since the sample was excited and monitored at the same wavelength, the ‘increase in emission’ from the eilatin solution was most likely due to increased light scattering upon addition of CT DNA (14). This is a well-documented effect (44), and appears in our data as a shoulder below 450 nm (Fig. 8), which exhibits a non-saturatable increase with each addition of CT DNA.
Interestingly, we have found that eilatin (3) exhibits a high apparent affinity to the RRE (Kd = 130 nM), but it does not displace the Rev peptide from the RRE (through its solubility limit in water) (35). 1 and 2, on the other hand, are freely soluble in water, bind to the RRE and displace Rev with inhibitory activities that are similar to their RRE binding constants (Table 3) (35). It is possible that the steric bulk provided by the Ru(bpy)2 moiety is needed to displace Rev. Alternatively, 1 and 2 may bind to the RRE at different locations than 3. There is some evidence for the latter possibility. In terms of RNA selectivity, 1 and 2 have a higher apparent affinity for single-stranded RNAs as compared to duplex polymeric RNAs, while 3 has the opposite selectivity. Since most natural RNAs, including the RRE, have bulged regions and other structural features with single-stranded characteristics, it is possible that 1 and 2 preferentially bind to these regions. 1 and 2 may, therefore, bind to the internal bulge of the RRE, which serves as the Rev binding site (Fig. 3). This G-rich bulge is known to possess single-stranded characteristics in the absence of Rev (45). Indeed, preliminary experiments indicate that the mutation of the bases in this bulge affects the affinity of 1 and 2 to the RRE (46). Taken together, this may indicate the internal bulge of the RRE is the preferred binding site of 1 and 2, while 3 prefers duplex regions of the RRE.
CONCLUSION
Eilatin-containing metal complexes represent a new family of compounds with unusual nucleic acid binding specificity. The eilatin ligand is essential for nucleic acid binding and the anti-HIV activity of these compounds, but the metal-free ligand exhibits dramatically different trends in nucleic acid specificity as compared to the eilatin-containing coordination complexes. Due to their unusual trends in enantiomeric selectivity, high single-stranded nucleic acid binding affinity and diverse sequence specificity, the eilatin-containing metal complexes are two of the most unusual metallo intercalators characterized to date.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
Financial support was provided, in part, by the National Institutes of Health (AI 47673 and GM 58447 to Y.T.). D.G. and M.K. are grateful to the Israel Science Foundation and the Israel Academy of Sciences and Humanities for support. J.S.H. thanks the Beckman Scholars Program for an undergraduate research fellowship.
REFERENCES
- 1.Erkkila K.E., Odom,D.T. and Barton,J.K. (1999) Recognition and reaction of metallointercalators with DNA. Chem. Rev., 99, 2777–2796. [DOI] [PubMed] [Google Scholar]
- 2.Cheng C.C., Huang-Fu,W.C., Hung,K.C., Chen,P.J., Wang,W.C. and Chen,Y.I. (2003) Mechanistic aspects of CoII(HAPP)(TFA)2 in DNA bulge-specific recognition. Nucleic Acids Res., 31, 2227–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Gijte O. and Mesmaeker,A.K.-D. (1999) The dinuclear ruthenium(II) complex [{Ru(Phen)2}2(HAT)]4+ (HAT = 1,4,5,8,9,12-hexaazatriphenylene), a new photoreagent for nucleobases and photoprobe for denatured DNA. J. Chem. Soc., Dalton Trans., 951–956. [Google Scholar]
- 4.Luedtke N.W., Hwang,J.S., Glazer,E.C., Gut,D., Kol,M. and Tor,Y. (2002) Eilatin Ru(II) complexes display anti-HIV activity and enantiomeric diversity in the binding of RNA. ChemBioChem., 3, 766–771. [DOI] [PubMed] [Google Scholar]
- 5.Novakova O., Kasparkova,J., Vrana,O., van Vliet,P.M., Reedijk,J. and Brabec,V. (1995) Correlation between cytotoxicity and DNA binding of polypyridyl ruthenium complexes. Biochemistry, 34, 12369–12378. [DOI] [PubMed] [Google Scholar]
- 6.Mishra L., Sinha,R., Itokawa,H., Bastow,K.F., Tachibana,Y., Nakanishi,Y., Kilgore,N. and Lee,K.H. (2001) Anti-HIV and cytotoxic activities of Ru(II)/Ru(III) polypyridyl complexes containing 2,6-(2′-benzimidazolyl)-pyridine/chalcone as co-ligand. Bioorg. Med. Chem., 9, 1667–1671. [DOI] [PubMed] [Google Scholar]
- 7.Kumar C.V., Barton,J.K. and Turro,N.J. (1985) Photophysical properties of ruthenium complexes bound to double helical DNA. J. Am. Chem. Soc., 107, 5518–5523. [Google Scholar]
- 8.Dandliker P.J., Nunez,M.E. and Barton,J.K. (1998) Oxidative charge transfer to repair thymidine dimers and damage guanine bases in DNA assemblies containing tethered metallointercalators. Biochemistry, 37, 6491–6502. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya P.K. and Barton,J.K. (2001) Influence of intervening mismatches on long-range guanine oxidation in DNA duplexes. J. Am. Chem. Soc., 123, 8649–8656. [DOI] [PubMed] [Google Scholar]
- 10.Odom D.T., Dill,E.A. and Barton,J.K. (2000) Robust charge transport in DNA double crossover assemblies. Chem. Biol., 7, 475–481. [DOI] [PubMed] [Google Scholar]
- 11.Rudi A., Benayahu,Y., Goldberg,I. and Kashman,Y. (1988) Eilatin, a novel alkaloid from the marine tunicate Eudistoma sp. Tetrahedron Lett., 29, 6655–6656. [Google Scholar]
- 12.Shochet N.R., Rudi,A., Kashman,Y., Yaacov,H., El-Maghrabi,M.R. and Spector,I. (1993) Novel marine alkaloids from the tunicate Eudistoma sp. are potent regulators of cellular growth and differentiation and affect cAMP-mediated processes. J. Cell. Physiol., 157, 481–492. [DOI] [PubMed] [Google Scholar]
- 13.Einat M., Lishner,M., Amiel,A., Nagler,A., Yarkorli,S., Rudi,A., Kashman,Y., Markel,D. and Fabian,I. (1995) Eilatin: a novel marine alkaloid inhibits in vitro proliferation of progenitor cells in chronic myeloid leukemia patients. Exp. Hematol., 23, 1439–1444. [PubMed] [Google Scholar]
- 14.McDonald L.A., Eldredge,G.S., Barrows,L.R. and Ireland,C.M. (1994) Inhibition of topoisomerase II catalytic activity by pyridoacridine alkaloids from a Cystodytes sp. ascidian: a mechanism for the apparent intercalator-induced inhibition of topoisomerase II. J. Med. Chem., 37, 3819–3827. [DOI] [PubMed] [Google Scholar]
- 15.Rudi A., Kashman,Y., Gut,D., Lellouche,F. and Kol,M. (1997) Ruthenium complexes of eilatin: Face selectivity in octahedral geometry; Synthesis of [Ru(bpy)2(eilatin)]2+ and [Ru(phen)2(eilatin)]2+. Chem. Commun., 1, 17–18. [Google Scholar]
- 16.Gut D., Rudi,A., Kopilov,J., Goldberg,I. and Kol,M. (2002) Pairing of propellers: dimerization of octahedral ruthenium(II) and osmium(II) complexes of eilatin via pi-pi stacking featuring heterochiral recognition. J. Am. Chem. Soc., 124, 5449–5456. [DOI] [PubMed] [Google Scholar]
- 17.Luedtke N.W. and Tor,Y. (2000) A novel solid-phase assembly for identifying potent and selective RNA ligands. Angew. Chem. Int. Ed. Engl., 39, 1788–1790. [DOI] [PubMed] [Google Scholar]
- 18.Proudfoot E.M., Mackay,J.P. and Karuso,P. (2001) Probing site specificity of DNA binding metallointercalators by NMR spectroscopy and molecular modeling. Biochemistry, 40, 4867–4878. [DOI] [PubMed] [Google Scholar]
- 19.Barton J.K., Danishefsky,A.T. and Goldberg,J.M. (1984) Tris(phenanthroline) Ru(II): stereoselectivity in binding to DNA. J. Am. Chem. Soc., 106, 2172–2176. [Google Scholar]
- 20.Haq I., Lincoln,P., Suh,D., Norden,B. Chowdhry,B.Z. and Chaires,J.B. (1995) Interaction of delta- and lambda-[Ru(phen)2 DPPZ] with DNA—A calorimetric and equilibrium binding study. J. Am. Chem. Soc., 117, 4788–4796. [Google Scholar]
- 21.Collins J.G., Aldrich-Wright,J.R., Greguric,I.D. and Pellegrini,P.A. (1999) Binding of the delta- and lambda-enantiomers of [Ru(dmphen)2+ to the hexanucleotide d(GTCGAC)2. Inorg. Chem., 38, 5502–5509. [DOI] [PubMed] [Google Scholar]
- 22.Ji L.-N., Zou,X.-H. and Liu,J.-G. (2001) Shape- and enantioselective interaction of Ru(II)/Co(III) polypyridyl complexes with DNA. Coord. Chem. Rev., 216, 513–536. [Google Scholar]
- 23.Satyanarayana S., Dabrowiak,J.C. and Chaires,J.B. (1992) Neither delta- nor lambda tris(phenanthroline) ruthenium(II) binds to DNA by classical intercalation. Biochemistry, 31, 9319–9324. [DOI] [PubMed] [Google Scholar]
- 24.Coggan D.Z., Haworth,I.S., Bates,P.J., Robinson,A. and Rodger,A. (1999) DNA binding of ruthenium Tris(1,10-phenanthroline): evidence for the dependence of binding mode on metal complex concentration. Inorg. Chem., 38, 4486–4497. [DOI] [PubMed] [Google Scholar]
- 25.Liu J.G., Zhang,Q.L., Shi,X.F. and Ji,L.N. (2001) Interaction of [Ru(dmp)2(dppz)]2+ and [Ru(dmb)2(dppz)]2+ with DNA: effects of the ancillary ligands on the DNA-binding behaviors. Inorg. Chem., 40, 5045–5050. [DOI] [PubMed] [Google Scholar]
- 26.Lerman L.S. (1961) Structural considerations in the interaction of DNA and acridines. J. Mol. Biol., 3, 18–30. [DOI] [PubMed] [Google Scholar]
- 27.Cohen G. and Eisenberg,H. (1969) Viscosity and sedimentation study of sonicated DNA-proflavin complexes. Biopolymers, 8, 45–55. [Google Scholar]
- 28.LePecq J.-B. and Paoletti,C.J. (1967) A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J. Mol. Biol., 27, 87–106. [DOI] [PubMed] [Google Scholar]
- 29.Jin E., Katritch,V., Olson,W.K., Kharatisvili,M., Abagyan,R. and Pilch,D.S. (2000) Aminoglycoside binding in the major groove of duplex RNA: the thermodynamic and electrostatic forces that govern recognition. J. Mol. Biol., 298, 95–110. [DOI] [PubMed] [Google Scholar]
- 30.Yielding L.W., Yielding,K.L. and Donoghue,J.E. (1994) Ethidium binding to deoxyribonucleic acid: spectrophotometric analysis of analogs with amino, azido, and hydrogen substituents. Biopolymers, 23, 83–110. [DOI] [PubMed] [Google Scholar]
- 31.Bresloff J.L. and Crothers,D.M. (1981) Equilibrium studies of ethidium-polynucleotide interactions. Biochemistry, 20, 3547–3553. [DOI] [PubMed] [Google Scholar]
- 32.Barton J.K. and Lippard,S.J. (1979) Cooperative binding of a platinum metallointercalation reagent to poly(A)-poly(U). Biochemistry, 18, 2661–2668. [DOI] [PubMed] [Google Scholar]
- 33.Kubota Y. and Motoda,Y. (1980) Nanosecond fluorescence decay studies of the deoxyribonucleic acid-9-aminoacridine and deoxyribonucleic acid-9-amino-10-methylacridinium complexes. Biochemistry, 19, 4189–4197. [DOI] [PubMed] [Google Scholar]
- 34.Klotz I.R. (1997) Ligand-Receptor Energetics. John Wiley & Sons, Inc., New York, NY. [Google Scholar]
- 35.Luedtke N.W. and Tor,Y. (2003) Fluorescence-based methods for evaluating the RNA affinity and specificity of HIV-1 Rev-RRE inhibitors. Biopolymers, 70, 103–119. [DOI] [PubMed] [Google Scholar]
- 36.Warring M.J. (1965) Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol., 13, 269–282. [DOI] [PubMed] [Google Scholar]
- 37.LePecq J.B. and Paoletti,C. (1967) A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J. Mol. Biol., 27, 87–106. [DOI] [PubMed] [Google Scholar]
- 38.Baguley B.C. and Falkenhaug,E.M. (1978) The interaction of ethidium with synthetic double-stranded polynucleotides at low ionic strength. Nucleic Acids Res., 5, 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boger D.L., Fink,B.E., Brunette,S.R., Tse,W.C. and Hedrick,M.P. (2001) A simple, high-resolution method for establishing DNA binding affinity and sequence selectivity. J. Am. Chem. Soc., 123, 5878–5891. [DOI] [PubMed] [Google Scholar]
- 40.Gabelica V., De Pauw,E. and Rosu,F. (1999) Interaction between antitumor drugs and a double-stranded oligonucleotide studied by electrospray ionization mass spectrometry. J. Mass Spectr., 34, 1328–1337. [DOI] [PubMed] [Google Scholar]
- 41.Montalbano A., Diana,P., Barraja,P., Lauria,A., Cirrincione,E., Dattolo,G. and Almerico,A.M. (2002) Pyrimido [5,4-c] pyrrolo [2,1-a] isoquinoline: a new potential DNA-interactive ring system. Arkivoc. (pt.II), 264–273. [Google Scholar]
- 42.Piantanida I., Palm,B.S., Zinic,M. and Schneider,H-J. (2001) A new 4,9-diazapyrenium intercalator for single- and double-stranded nucleic acids; distinct differences from related diazapyrenium compounds and ethidium bromide. J. Chem. Soc., Perkin Trans., 2, 1808–1816. [Google Scholar]
- 43.Juranovic I., Meic,Z., Piantanida,I., Tumir,L.M. and Zinic,M. (2002) Interactions of phenanthridium-nucleobase conjugates with polynucleotides in aqueous media. Recognition of poly U. Chem. Commun., 13, 1432–1433. [Google Scholar]
- 44.Fishman D.M. and Patterson,G.D. (1996) Light scattering studies of supercoiled and nicked DNA. Biopolymers, 38, 535–552. [DOI] [PubMed] [Google Scholar]
- 45.Battiste J.L., Tan,R., Frankel,A.D. and Williamson,J.R. (1994) Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry, 33, 2741–2747. [DOI] [PubMed] [Google Scholar]
- 46.Luedtke N.W. (2003) RNA Affinity and Specificity of Modified Aminoglycosides, Metal Complexes, and Intercalating Agents That Target the HIV-1 Rev Response Element. PhD Thesis, University of California, San Diego, CA.
- 47.Battiste J.L., Mao,H., Rao,N.S., Tan,R., Muhandiram,D.R., Kay,L.E., Frankel,A.D. and Williamson,J.R. (1996) Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science, 273, 1547–1551. [DOI] [PubMed] [Google Scholar]
- 48.Weeks K.M., Ampe,C., Schultz,S.C., Steitz,T.A. and Crothers,D.M. (1990) Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science, 249, 1281–1285. [DOI] [PubMed] [Google Scholar]
- 49.Teeter M.M. Quigley,G.J. and Rich,A. (1980) Metal ions and transfer RNA. In Spiro,T.G. (ed.) Nucleic Acid—Metal Ion Interactions. Wiley-Interscience: New York, NY, pp. 145–177. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.