Abstract
The purpose of apoptosis in multicellular organisms is obvious: single cells die for the benefit of the whole organism (for example, during tissue development or embryogenesis). Although apoptosis has also been shown in various microorganisms, the reason for this cell death program has remained unexplained. Recently published studies have now described yeast apoptosis during aging, mating, or exposure to killer toxins (Fabrizio, P., L. Battistella, R. Vardavas, C. Gattazzo, L.L. Liou, A. Diaspro, J.W. Dossen, E.B. Gralla, and V.D. Longo. 2004. J. Cell Biol. 166:1055–1067; Herker, E., H. Jungwirth, K.A. Lehmann, C. Maldener, K.U. Frohlich, S. Wissing, S. Buttner, M. Fehr, S. Sigrist, and F. Madeo. 2004. J. Cell Biol. 164:501–507, underscoring the evolutionary benefit of a cell suicide program in yeast and, thus, giving a unicellular organism causes to die for.
Introduction
Apoptosis is an evolutionally conserved cell suicide program used by an organism to selectively eliminate dangerous, superfluous, or damaged cells. The phenomenon of yeast cells undergoing apoptosis has long been controversial, in part because of doubts of whether cell suicide could constitute an evolutionary advantage for unicellular organisms. However, since the first description of apoptosis in a yeast (Saccharomyces cerevisiae) strain carrying a CDC48 mutation (Madeo et al., 1997), several yeast orthologues of crucial mammalian apoptotic proteins have been discovered (Madeo et al., 2002; Fahrenkrog et al., 2004; Wissing et al., 2004; Qiu et al., 2005; Li et al., 2006; Walter et al., 2006), and conserved proteasomal, mitochondrial, and histone-regulated apoptotic pathways have been delineated (Fig. 1; Manon et al., 1997; Ligr et al., 2001; Ludovico et al., 2002; Fannjiang et al., 2004; Ahn et al., 2005a; Gourlay and Ayscough, 2005; Pozniakovsky et al., 2005). By demonstrating that the overexpression of human Bcl-2 in yeast extends chronological life span, Longo et al. (1997) connected yeast aging to conserved mammalian apoptotic pathways. Chronologically aged yeast cells have been used as a valuable model to study oxidative damage and molecularly conserved aging pathways of postmitotic tissues in higher organisms (Bitterman et al., 2003). Recently, both replicative and chronological aging of yeast cells have been demonstrated to culminate in cell death with an apoptotic phenotype (Laun et al., 2001; Fabrizio et al., 2004; Herker et al., 2004). In this paper, we describe physiological scenarios of yeast apoptosis (Fig. 2), suggesting a teleological explanation for this (at first glance useless) behavior of unicellular organisms.
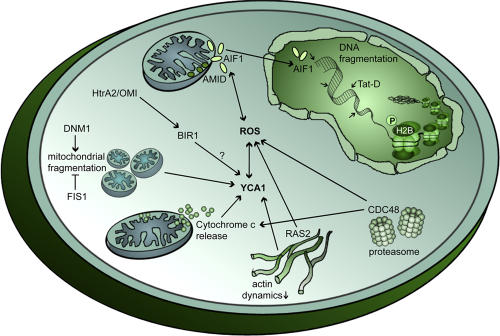
Figure 1.
The basal molecular machinery of yeast apoptosis. Crucial proteins of the basic molecular machinery executing cell death are conserved in yeast, including the yeast caspase YCA1, the mitochondrially located proteins apoptosis-inducing factor 1 (AIF1), HtrA2/Omi (NMA111), and AMID (NDI1), and the antiapoptotic proteins CDC48 and BIR1. In addition, yeast programmed death has been linked to complex apoptotic scenarios such as mitochondrial fragmentation, cytochrome c release, cytoskeletal perturbations, and histone H2B phosphorylation.
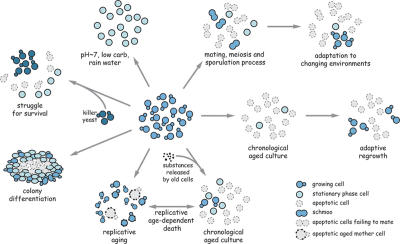
Figure 2.
Physiological scenarios of yeast apoptosis. Wild-type yeast cells die altruistically in times of dwindling resources during chronological aging, after attack by killer toxins from nonclonal enemy strains, and as a result of unsuccessful mating.
Death in times of love: pheromones induce cell death when mating is not successful
Complex social interactions occur both within and between various microbial species and can be either competitive or cooperative. Severin and Hyman (2002) demonstrated that exposure of haploid yeast cells to mating-type pheromones can induce apoptosis in yeast when a suitable mating partner is absent, thus connecting the yeast life cycle to cell death mechanisms. Successful mating prevents apoptosis, suggesting that yeast may use mating factor–induced cell death to eliminate infertile or otherwise damaged cells. Therefore, by accelerating the passage of generations, pheromone-induced apoptosis might ensure evolutionary progress and may favor the diploid state, which is likely to provide an adaptive advantage over the haploid state. This cell death process induced by low doses of pheromone depends on the apoptotic factors cytochrome c and mitochondrial permeabilization (Severin and Hyman, 2002). In contrast, death caused by higher concentrations of pheromone lacked certain hallmarks of apoptosis (Zhang et al., 2006). Zhang et al. (2006) further characterized this death, delineating three genetically and chronologically distinguishable forms of yeast death upon unsuccessful mating attempts, all of which were preceded by an accumulation of reactive oxygen species (ROS). Rapid cell death required cell wall degradation and Fig1p, an integral membrane protein necessary for efficient mating. A second, slower wave of cell death was independent of Fig1p and was dependent on much lower concentrations of pheromones, whereas a third wave of death was shown to be regulated by calcineurin signaling (Zhang et al., 2006). However, death can occur not only before but also after mating; successful mating leads to the generation of diploid cells, which can, as a consequence of scarce nutrition, undergo meiosis and sporulate as a way to stochastically reshuffle and rearrange the genome to increase genetic diversity and, thus, the fitness of the population. This meiosis is coupled to apoptosis, as 20% of cells grown on sporulation media undergo apoptotic cell death, whereas the 80% that survive initiate sporulation (Ahn et al., 2005b; Knorre et al., 2005). This might ensure that only genetic recombinants that are adapted to their surroundings survive.
Death in times of peace: dwindling nutrients trigger the altruistic death of older cells
Various microorganisms tend to cluster together to survive nutrient depletion, forming multicellular communities called biofilms. In such a social community, the benefit of a cellular suicide program seems evident. The self-destruction of virus-infected, damaged, and old cells, which consume dwindling nutrients or spread an infection, contributes to the viability and reproductive success of healthier members of the community harboring similar genomes. S. cerevisiae has been shown to initiate the formation of biofilms in both natural and laboratory environments, particularly when nutrients are depleted (Reynolds and Fink, 2001; Zara et al., 2002).
In the wild, an individual yeast cell landing on a rotting apple will divide and form a colony until all readily utilizable nutrients are exhausted. When the whole fruit is colonized and the next apple is not in sight, cells cease to proliferate and enter a postdiauxic but still metabolic active phase known as chronological aging (Fabrizio and Longo, 2003). To ensure survival of the clone, it seems to make sense for old or damaged cells to undergo cell death instead of consuming vanishing resources in a futile attempt to repair themselves. Chronologically and replicatively aged yeast cells die while exhibiting typical features of apoptosis, accompanied by the accumulation of ROS, which is a crucial factor in aging and apoptosis as well (Laun et al., 2001; Fabrizio et al., 2004; Herker et al., 2004). The question arises of how it is determined exactly which cells are to die in a chronologically aged population. In principle, this could be just a stochastic marker or, alternatively, a selection based on fitness. Notably, however, only replicatively older cells harboring two or more bud scars kill themselves in times of hardship (Allen et al., 2006), demonstrating that during chronological aging, apoptosis selectively removes older individuals from the population. Furthermore, when dying, aged yeast cells actively stimulate the survival of the clone by releasing defined substances into their surrounding (Fabrizio et al., 2004; Herker et al., 2004). Interestingly, Chen and Fink (2006) described specific aromatic alcohols as molecules mediating social interactions between yeast. These alcohols could have putative signaling functions leading to quorum sensing during chronological aging. To summarize, the death of older cells is advantageous for two reasons: first, because it spares nutrients for younger cells; and second, because the older cells release nutrients that can be metabolized by replicatively younger cells.
In addition, apoptosis has been demonstrated to occur during the development of colonies on solid media (Vachova and Palkova, 2005). Aged colonies undergo metabolic changes that induce apoptotic death in a spatially restricted fashion, namely in the center of the colony. These colonies can be considered as multicellular organisms that undergo a sort of differentiation coupled to apoptosis. Interestingly, mechanical deletion of the colony center results in decreased survival of the colony margin. Thus, the outer cells profit from cell death occurring in the colony center, again arguing for an altruistic nature of cell death. This is in accordance with the classical definition of altruism, as this behavior increases the fitness of the group relative to other groups while it decreases the fitness of the altruist compared with others within the group (Wilson and Sober, 1989). Similar to yeast colonies, cells in a metazoan multicellular organism undergo cell death to ensure normal development and survival of the whole organism.
Deletion of the yeast caspase YCA1 gene enhances the resistance against oxidative stress and also delays age-induced cell death (Madeo et al., 2002), although caspase-independent apoptosis occurs in yeast as well (Guscetti et al., 2005; Zhang et al., 2006). The fact that disruption of the apoptotic machinery results in an extended chronological life span is consistent with the idea that apoptosis might have evolved to clean the population of damaged or old cells that would consume resources and, thus, reduce the viability of the clone. However, the existence of a death program may not be advantageous under all conditions. In the wild, a yeast cell washed off a grape by a rain shower gains no advantage by undergoing apoptosis because there is neither food around to economize nor fitter relatives to spare the food for. Consistently, yeast incubated in distilled water have an extended life span as compared with cells cultured in minimal media (Fabrizio et al., 2004). However, it is also possible that nutrient metabolism produces ROS or other molecules that cause apoptosis.
In organisms ranging from yeast to mammals, the link between diet and aging is well established, as calorie restriction extends life span and increases stress resistance (Koubova and Guarente, 2003). Pathways that mediate glucose-dependent signaling have recently been shown to interact with age-induced apoptosis in yeast (Fabrizio et al., 2001). Disruption of SCH9, the yeast orthologue of AKT1 kinase, which is a crucial regulator of glucose-dependent signaling and aging in Caenorhabditis elegans and probably also in humans (Miyauchi et al., 2004), leads to an extended life span and delays apoptosis during chronological aging (Fabrizio et al., 2001, 2004). The decreased activity of Sch9p signals nutrient depletion and might therefore mimic a surrounding of pure water (Fabrizio et al., 2004). In the process of aging, calorie restriction delays apoptotic cell death and prolongs life span.
Death in times of war: competing yeast tribes secrete killer toxins
The production and secretion of virus-encoded killer toxins with antimicrobial activity is a relatively common phenomenon among a great variety of yeast genera. Although these killer yeasts are immune to their own cytotoxic proteins or glycoproteins, they eliminate susceptible microorganisms from the environment (Schmitt and Breinig, 2002). In S. cerevisiae, three different killer toxins (K1, K2, and K28), all encoded by cytosolic double-stranded RNA viruses, have been identified so far. This killer phenomenon might play a major role in competition in fruit communities in which various insects may carry different colonizing yeasts and the fight for nutrients may begin. It is significant that resistant rather than susceptible yeast strains are usually found in fruits and that one quarter of these yeast strains are killers (Starmer et al., 1987). It has recently been shown that the exposure to toxins produced by coexisting killer strains triggers apoptosis in yeast (Ivanovska and Hardwick, 2005; Reiter et al., 2005; Breinig et al., 2006; Schmitt and Breinig, 2006). Reiter et al. (2005) reported that low doses of killer toxins trigger the production of ROS and apoptosis in S. cerevisiae. When exposed to toxin, yeast cells deficient for the antioxidant enzyme GSH1 display an enhanced accumulation of ROS and decreased viability. The increased sensitivity of gsh1 mutants to killer toxins portrays the evident involvement of ROS in toxin-induced cell death. Conversely, a yeast yca1 disruptant exhibits reduced toxin sensitivity, indicating that the presence of yeast caspase is needed for efficient toxin-triggered apoptosis. In contrast, high concentrations of killer toxins induce a necrotic cell death beyond any regulation by Yca1p and ROS. Thus, killer toxins can initiate two modes of cell death: low doses trigger regulated apoptosis, whereas high doses cause unregulated necrosis. In the natural habitat of killer yeasts, where toxin concentration is likely to be low, the induction of apoptosis might play an important role in efficient toxin-mediated cell killing (Reiter et al., 2005).
ROS regulates programmed death and adaptation
Microorganisms have to cope with drastic fluctuations in temperature, nutrient resources, and oxygen tension. Therefore, the ability to adapt to a changing environment ensures optimal fitness. In Escherichia coli, the death of populations maintained in stationary phase is followed by the growth of a better-adapted mutant subpopulation, which rapidly takes over the culture (Zambrano et al., 1993); this phenomenon is known as adaptive regrowth. Similarly, aged yeast populations may favor the growth of a mutant subpopulation (Fabrizio et al., 2004). Monitoring of the age-dependent mutation frequency suggested a role for superoxide and the rate of spontaneous mutations in adaptive regrowth. Higher mutation frequency correlates with shortened life span and improved adaptation to a new environment containing nutrients released by dead cells. Adaptive regrowth hardly occurs in populations of long-lived mutants showing decreased levels of superoxide, whereas almost all cultures of short-lived mutants lacking superoxide dismutase are able to regrow (Fabrizio et al., 2003, 2004). Disruption of the apoptotic machinery via the deletion of YCA1 only initially results in a better survival of aged cultures. An aged yca1-null mutant strain is no longer able to regrow when nutrients become available after a period of starvation, leading to a population with a high percentage of damaged and old cells. In the long run, aging yeast populations benefit from early apoptosis with strains that initiate early death, outlasting long-lived mutants in competition assays (Fabrizio et al., 2004; Herker et al., 2004). In both scenarios, frequent death and growth cycles promote the selection of fitter subpopulations. Under the regime of ROS, programmed aging and the consequent early death together with a relatively high mutation frequency ensure adaptation to changing environments.
Concluding remarks
Accumulating studies indicate that yeast undergo programmed cell death for several good reasons (Fig. 2; Fabrizio et al., 2004; Herker et al., 2004; Knorre et al., 2005; Reiter et al., 2005; Allen et al., 2006). Pheromone signaling leads to the apoptotic death of cells that fail to mate, therefore depleting the population of haploid cells and favoring the survival of diploid cells that increase genetic diversity through meiotic recombination. The early death of old and damaged cells during aging and starvation enhances the chances of the rest of the population to survive and to sporulate, thus increasing the probability that the clone will survive. Moreover, in such conditions, the increased production of ROS enhances the probability of somatic mutations and, thus, the generation of genetic variants that can adapt to continuously changing conditions. Therefore, altruistic cell death via the activation of a highly conserved, self-destructive, enzymatic machinery confers an advantage on the population, enhancing its genetic diversity both via sexual reproduction and somatic mutations. In addition, apoptosis coupled to chronological and replicative aging limits longevity that would maintain ancient genetic variants within the population and, therefore, favor genetic conservatism. As a negative side phenomenon, however, the apoptotic process may be hijacked, at least in laboratory conditions, to facilitate cell killing by competing yeast strains that produce toxins in a tribal war. In summary, the death of old, infertile, or damaged yeast cells may ensure the survival of a colony of yeast cells and introduces the concept of an altruistic aging and death program. Deciphering the components of the upstream signaling and execution pathways of these programs constitutes an ongoing challenge with far-reaching implications for developmental and evolutionary biology.
Acknowledgments
We are grateful to the Austrian Science Fund for grant S-9304-B05 (to F. Madeo, S. Buttner, and D. Carmona-Gutierrez), the Deutsche Forschungsgemeinschaft for grant MA2587 (to F. Madeo), and the European Union for the Trans-Death grant (to G. Kroemer).
Abbreviation used in this paper: ROS, reactive oxygen species.
References
- Ahn, S.H., W.L. Cheung, J.Y. Hsu, R.L. Diaz, M.M. Smith, and C.D. Allis. 2005. a. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 120:25–36. [DOI] [PubMed] [Google Scholar]
- Ahn, S.H., K.A. Henderson, S. Keeney, and C.D. Allis. 2005. b. H2B (Ser10) phosphorylation is induced during apoptosis and meiosis in S. cerevisiae. Cell Cycle. 4:780–783. [DOI] [PubMed] [Google Scholar]
- Allen, C., S. Buttner, A.D. Aragon, J.A. Thomas, O. Meirelles, J.E. Jaetao, D. Benn, S.W. Ruby, M. Veenhuis, F. Madeo, and M. Werner-Washburne. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman, K.J., O. Medvedik, and D.A. Sinclair. 2003. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 67:376–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breinig, F., T. Sendzik, K. Eisfeld, and M.J. Schmitt. 2006. Dissecting toxin immunity in virus-infected killer yeast uncovers an intrinsic strategy of self-protection. Proc. Natl. Acad. Sci. USA. 103:3810–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., and G.R. Fink. 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20:1150–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., and V.D. Longo. 2003. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2:73–81. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., F. Pozza, S.D. Pletcher, C.M. Gendron, and V.D. Longo. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 292:288–290. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., L.L. Liou, V.N. Moy, A. Diaspro, J. Selverstone Valentine, E.B. Gralla, and V.D. Longo. 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 163:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., L. Battistella, R. Vardavas, C. Gattazzo, L.L. Liou, A. Diaspro, J.W. Dossen, E.B. Gralla, and V.D. Longo. 2004. Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae. J. Cell Biol. 166:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog, B., U. Sauder, and U. Aebi. 2004. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell Sci. 117:115–126. [DOI] [PubMed] [Google Scholar]
- Fannjiang, Y., W.C. Cheng, S.J. Lee, B. Qi, J. Pevsner, J.M. McCaffery, R.B. Hill, G. Basanez, and J.M. Hardwick. 2004. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 18:2785–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay, C.W., and K.R. Ayscough. 2005. Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J. Cell Sci. 118:2119–2132. [DOI] [PubMed] [Google Scholar]
- Guscetti, F., N. Nath, and N. Denko. 2005. Functional characterization of human proapoptotic molecules in yeast S. cerevisiae. FASEB J. 19:464–466. [DOI] [PubMed] [Google Scholar]
- Herker, E., H. Jungwirth, K.A. Lehmann, C. Maldener, K.U. Frohlich, S. Wissing, S. Buttner, M. Fehr, S. Sigrist, and F. Madeo. 2004. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska, I., and J.M. Hardwick. 2005. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J. Cell Biol. 170:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorre, D.A., E.A. Smirnova, and F.F. Severin. 2005. Natural conditions inducing programmed cell death in the yeast Saccharomyces cerevisiae. Biochemistry (Mosc.). 70:264–266. [DOI] [PubMed] [Google Scholar]
- Koubova, J., and L. Guarente. 2003. How does calorie restriction work? Genes Dev. 17:313–321. [DOI] [PubMed] [Google Scholar]
- Laun, P., A. Pichova, F. Madeo, J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K.U. Frohlich, and M. Breitenbach. 2001. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39:1166–1173. [PubMed] [Google Scholar]
- Li, W., L. Sun, Q. Liang, J. Wang, W. Mo, and B. Zhou. 2006. Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Mol. Biol. Cell. 17:1802–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligr, M., I. Velten, E. Frohlich, F. Madeo, M. Ledig, K.U. Frohlich, D.H. Wolf, and W. Hilt. 2001. The proteasomal substrate Stm1 participates in apoptosis-like cell death in yeast. Mol. Biol. Cell. 12:2422–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, V.D., L.M. Ellerby, D.E. Bredesen, J.S. Valentine, and E.B. Gralla. 1997. Human Bcl-2 reverses survival defects in yeast lacking superoxide dismutase and delays death of wild-type yeast. J. Cell Biol. 137:1581–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludovico, P., F. Rodrigues, A. Almeida, M.T. Silva, A. Barrientos, and M. Corte-Real. 2002. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, F., E. Frohlich, and K.U. Frohlich. 1997. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139:729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lachelt, M. Herlan, M. Fehr, K. Lauber, S.J. Sigrist, S. Wesselborg, and K.U. Frohlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell. 9:911–917. [DOI] [PubMed] [Google Scholar]
- Manon, S., B. Chaudhuri, and M. Guerin. 1997. Release of cytochrome c and decrease of cytochrome c oxidase in Bax-expressing yeast cells, and prevention of these effects by coexpression of Bcl-xL. FEBS Lett. 415:29–32. [DOI] [PubMed] [Google Scholar]
- Miyauchi, H., T. Minamino, K. Tateno, T. Kunieda, H. Toko, and I. Komuro. 2004. Akt negatively regulates the in vitro lifespan of human endothelial cells via a p53/p21-dependent pathway. EMBO J. 23:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniakovsky, A.I., D.A. Knorre, O.V. Markova, A.A. Hyman, V.P. Skulachev, and F.F. Severin. 2005. Role of mitochondria in the pheromone- and amiodarone-induced programmed death of yeast. J. Cell Biol. 168:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J., J.H. Yoon, and B. Shen. 2005. Search for apoptotic nucleases in yeast: role of Tat-D nucle ase in apoptotic DNA degradation. J. Biol. Chem. 280:15370–15379. [DOI] [PubMed] [Google Scholar]
- Reiter, J., E. Herker, F. Madeo, and M.J. Schmitt. 2005. Viral killer toxins induce caspase-mediated apoptosis in yeast. J. Cell Biol. 168:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, T.B., and G.R. Fink. 2001. Bakers' yeast, a model for fungal biofilm formation. Science. 291:878–881. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.J., and F. Breinig. 2002. The viral killer system in yeast: from molecular biology to application. FEMS Microbiol. Rev. 26:257–276. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.J., and F. Breinig. 2006. Yeast viral killer toxins: lethality and self-protection. Nat. Rev. Microbiol. 4:212–221. [DOI] [PubMed] [Google Scholar]
- Severin, F.F., and A.A. Hyman. 2002. Pheromone induces programmed cell death in S. cerevisiae. Curr. Biol. 12:R233–R235. [DOI] [PubMed] [Google Scholar]
- Starmer, W.T., P.F. Ganter, V. Aberdeen, M.A. Lachance, and H.J. Phaff. 1987. The ecological role of killer yeasts in natural communities of yeasts. Can. J. Microbiol. 33:783–796. [DOI] [PubMed] [Google Scholar]
- Vachova, L., and Z. Palkova. 2005. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 169:711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, D., S. Wissing, F. Madeo, and B. Fahrenkrog. 2006. The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J. Cell Sci. 119:1843–1851. [DOI] [PubMed] [Google Scholar]
- Wilson, D.S., and E. Sober. 1989. Reviving the superorganism. J. Theor. Biol. 136:337–356. [DOI] [PubMed] [Google Scholar]
- Wissing, S., P. Ludovico, E. Herker, S. Buttner, S.M. Engelhardt, T. Decker, A. Link, A. Proksch, F. Rodrigues, M. Corte-Real, et al. 2004. An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 166:969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano, M.M., D.A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 259:1757–1760. [DOI] [PubMed] [Google Scholar]
- Zara, S., F.G. Antonio, M. Budroni, and A.T. Bakalinsky. 2002. HSP12 is essential for biofilm formation by a Sardinian wine strain of S. cerevisiae. Yeast. 19:269–276. [DOI] [PubMed] [Google Scholar]
- Zhang, N.N., D.D. Dudgeon, S. Paliwal, A. Levchenko, E. Grote, and K.W. Cunningham. 2006. Multiple signaling pathways regulate yeast cell death during the response to mating pheromones. Mol. Biol. Cell. 17:3409–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]