Abstract
Inhibition of Notch signaling by Numb is critical for many cell fate decisions. In this study, we demonstrate a more complex relationship between Notch and the two vertebrate Numb homologues Numb and Numblike. Although Numb and Numblike at low levels of Notch signaling negatively regulated Notch, high levels of Notch signaling conversely led to a reduction of Numb and Numblike protein levels in cultured cells and in the developing chick central nervous system. The Notch intracellular domain but not the canonical Notch downstream proteins Hes 1 and Hey 1 caused a reduction of Numb and Numblike. The Notch-mediated reduction of Numblike required the PEST domain in the Numblike protein and was blocked by the proteasome inhibitor MG132. Collectively, these observations reveal a reciprocal negative regulation between Notch and Numb/Numblike, which may be of relevance for stabilizing asymmetric cell fate switches and for tumor development.
Introduction
Asymmetric cell division generates distinct progeny from a single cell division, and the two proteins Notch and Numb are critical for this process. In Drosophila melanogaster, Numb functions as a negative regulator of Notch (for review see Cayouette and Raff, 2002). Numb protein is asymmetrically localized to one daughter cell in cell divisions that generate distinct progeny. The cell receiving high levels of Numb suppresses Notch signaling, whereas the cell with low levels of Numb maintains Notch activity (Frise et al., 1996; Guo et al., 1996). Numb and Notch are evolutionally conserved proteins. Two mammalian Numb homologues, Numb and Numblike, have been identified (Zhong et al., 1996, 1997). Gene targeting in mice reveals partially redundant functions for Numb and Numblike; i.e., the compound knockout of Numb and Numblike has a more severe phenotype than knockouts of each gene alone (Petersen et al., 2002; Li et al., 2003). Data from Drosophila (Frise et al., 1996; Guo et al., 1996) and from adult mouse muscle progenitors (i.e., satellite cells; Conboy and Rando, 2002) support a differentiation-promoting role for Numb/Numblike.
The Notch signaling pathway controls numerous cell fate decisions during development, often by maintaining a more undifferentiated fate. The Notch receptor is a single transmembrane protein that undergoes a complex series of proteolytic processing events. This ultimately leads to the release of the intracellular domain (ICD) of the receptor in response to activation from membrane-tethered ligands of the Delta or Serrate type (Artavanis-Tsakonas et al., 1999). The released Notch ICD translocates to the nucleus, where it interacts with the DNA-binding protein CSL (also termed RBP-Jκ [Furukawa et al., 1992] and suppressor of hairless in Drosophila) to control activation of a specific set of downstream genes, most notably the Hes and Hey family basic helix-loop-helix transcription factor genes (Iso et al., 2003).
Although Numb is known to be a negative regulator of Notch, we describe a more complex relationship between Notch and Numb/Numblike. Unexpectedly, high levels of Notch signaling lead to a reduction of Numb/Numblike protein levels, revealing a reciprocal negative regulation between Notch and Numb/Numblike.
Results and discussion
Numb and Numblike negatively regulate Notch signaling and reduce the level of Notch protein
A dose-dependent reduction of the levels of both a truncated membrane-tethered (Notch 1 ΔE) and an intracellular (Notch 1 ICD) form was observed in response to increasing amounts of Numb (Fig. 1, a and b). The Notch 1 ΔE protein was cleaved in the presence of Numb, although cleavage appeared to be somewhat reduced at higher Numb levels (Fig. 1, a and b). Furthermore, Numb and Numblike negatively regulate Notch signaling both from full-length Notch, Notch 1 ΔE, and Notch 1 ICDd (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200602009/DC1). We next studied whether Numb affected Notch intracellular localization. Transfected Numb-HA immunoreactivity was largely confined to intracellular vesicles (Fig. 1 c), which are likely to be endosomes, based on the codistribution of Numb-HA and Eps15 immunoreactivity (not depicted). In cells where the activation and cleavage of full-length Notch 1 was induced by coculture, the resulting Notch 1 ICD was predominantly localized to the nucleus also in the presence of transfected Numb (Fig. 1 d). In summary, these data indicate that Numb and Numblike negatively regulate Notch signaling and that Numb does not sequester Notch 1 ICD in the cytoplasm, arguing against a function for Numb in excluding Notch 1 ICD from the nucleus (Frise et al., 1996; Wakamatsu et al., 1999; Berezovska et al., 2000).
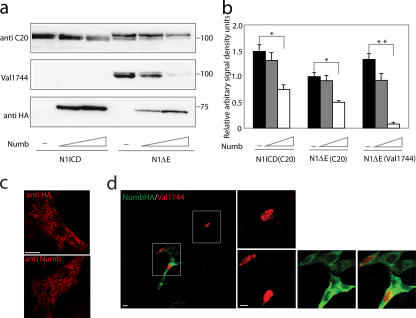
Figure 1.
Numb and Numblike down-regulate Notch protein levels. (a) Western blots of Notch 1 ICD or Notch 1 ΔE protein levels after cotransfection with various amounts of Numb (500 and 1000 ng). The Western blot was stained with antisera recognizing the C terminus of Notch ICD (anti-C20) or the cleaved ICD (Val 1744) and was reprobed with anti-HA antibody to visualize Numb. Note that the Val 1744 antibody does not recognize transfected Notch 1 ICD, which lacks nine amino acids as compared with the cleaved form of Notch 1 ΔE. (b) Densitometric analysis of protein levels in the autoradiogram from panel a and two additional experiments. Error bars represent SD. *, P < 0.05; **, P < 0.01. (c) Intracellular localization of Numb-HA expression visualized by an anti-HA and a new anti-Numb antiserum as indicated. The specificity of the new anti-Numb antiserum is described in Fig. S2 a (available at http://www.jcb.org/cgi/content/full/jcb.200602009/DC1). (d) Visualization of Notch ICD by the Val 1744 antibody after ligand-induced activation of full-length Notch 1 (red) in cells transiently transfected with Numb (green). Cells expressing Notch ICD in the nucleus (boxed areas) are enlarged that are either in the absence (top) or presence (bottom) of transfected Numb. Bars, 10 μm.
Numb promotes differentiation in C2C12 cells at low but not at high levels of Notch signaling
We next analyzed the effects of Numb on cellular differentiation at various levels of Notch signaling in the myogenic cell line C2C12, in which differentiation to myotubes can be blocked by Notch signaling (Dahlqvist et al., 2003; Gustafsson et al., 2005). Different levels of Notch signaling were accomplished by coculturing cells expressing different levels of Notch receptor and ligand. Thus, regular C2C12 cells or C2C12 cells stably expressing Notch 1 (C2C12-N1) cocultured with regular 3T3 cells yielded low levels of Notch signaling as measured by 12XCSL-luc activation, whereas coculture with Jagged-1 (Serrate-1)–expressing 3T3 cells (3T3-J1) yielded high levels of Notch signaling (Fig. 2 a). Transfection of Numb exerted a negative effect on Notch signaling in all combinations, although the remaining level of Notch signaling after Numb transfection was considerably higher after coculture with Jagged-1–expressing cells (Fig. 2 a).
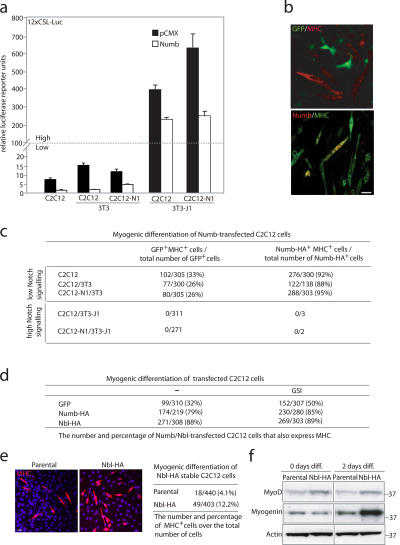
Figure 2.
Numb promotes myogenic differentiation in C2C12 cells at low but not at high levels of Notch signaling. (a) Effects of Numb on 12XCSL-luc activation in various coculture combinations of Notch- and Jagged-expressing cells or in C2C12 cells cultured alone. Error bars represent SD. (b) Visualization of GFP (green) and MHC (red) expression (top) or of Numb-HA (red; anti-HA) and MHC (green) expression (bottom) in differentiated C2C12 cells. (c) A table showing the number and percentage of GFP- or Numb-HA–expressing cells that also express MHC compared with the total number of GFP or Numb-HA–expressing cells after the transfection of GFP or Numb-HA into C2C12 cells under differentiation conditions in various coculture combinations as indicated. (d) A table showing the number and percentage of GFP-, Numb-HA–, or Numblike-HA–expressing cells that also express MHC in the absence or presence (right column) of 400 nM GSI DAPT. (e) MHC-expressing cells (red) in a Numblike stable cell line (Nbl-HA) and the parental cell line 2 d after the induction of differentiation. The percentage of MHC-expressing cells in the Nbl-HA and parental cell lines is shown to the right. (f) Western blot of MyoD and myogenin expression levels in the Nbl-HA and parental cell lines at 0 or 2 d after the induction of differentiation. Bars, 10 μm.
Transfection of Numb into C2C12 cells followed by 3 d under differentiation-promoting conditions resulted in a dramatic increase in the differentiation of Numb-expressing cells at low levels of Notch signaling (Fig. 2 b); 88 and 95% of the Numb-positive cells were also myosin heavy chain (MHC) positive in C2C12/3T3 and C2C12-N1/3T3 cocultures, respectively, and 92% were positive in C2C12 cells cultured alone (Fig. 2 c). Similar values (88%) were obtained for Numblike (Fig. 2 d). The proportion of differentiated cells after Numb and Numblike transfection was considerably higher compared with the blockage of Notch signaling by the γ-secretase inhibitor (GSI) DAPT, which only causes an increase in MHC-positive cells from 32 to 50% (Fig. 2 d). The addition of DAPT did not further enhance the differentiation-promoting effect of Numb and Numblike (Fig. 2 d). This argues for a more instructive role for Numb and Numblike in myogenic differentiation rather than only blocking Notch signaling. In keeping with a differentiation-promoting role, we observed that a C2C12 cell line stably expressing Numblike showed accelerated myogenic differentiation as compared with the parental cell line; 2 d after the switch to prodifferentiation conditions, 12.2% of the cells were MHC positive as compared with only 4.1% in the parental cell line (Fig. 2 e). The stable expression of Numblike was accompanied by somewhat elevated levels of the myogenic differentiation factor MyoD and a robust induction of myogenin during differentiation (Fig. 2 f).
At high levels of Notch signaling (i.e., in C2C12/3T3-J1 and C2C12-N1/3T3-J1 cocultures), we found no MHC-positive cells even after the transfection of Numb (Fig. 2 c). Unexpectedly, Numb-expressing cells were also rare in C2C12/3T3-J1 and C2C12-N1/3T3-J1 cocultures (∼1% as compared with C2C12/3T3 and C2C12-N1/3T3; Fig. 2 c). The combination of Numb expression and elevated Notch signaling did not increase cell death (Fig. S2 b, available at http://www.jcb.org/cgi/content/full/jcb.200602009/DC1). In summary, this indicates that Numb can override the differentiation-inhibiting effects of Notch at lower levels of Notch signaling and promote myogenic differentiation, but, at higher Notch levels, differentiation is blocked, and there is a strong decrease in Numb expression.
High levels of Notch 1 activation reduce Numb protein levels
We examined the apparent loss of Numb expression at high Notch levels in a C2C12 cell line stably expressing HA-tagged Numb (Numb-HA), which was cocultured with 3T3 or 3T3-J1 cells. The Numb protein level was markedly reduced in 3T3-J1 cocultures but not in 3T3 cocultures or in a mixed lysate of Numb-HA and 3T3 or 3T3-J1 cells cultured separately (Fig. 3 a). This effect was dependent on Notch signaling, as addition of the GSI L-685,458 to cocultures of Numb-HA and 3T3-J1 cells blocked Numb down-regulation (Fig. 3 b). In contrast, Numb mRNA levels were not reduced (Fig. S2 c). Transfection of Notch 1 ΔE led to a similar down-regulation of Numb in the cell line stably expressing Numb-HA (Fig. 3 c). To learn whether down-regulation required the activation of Hes 1 and Hey 1, we transiently expressed Notch 1 ICD, Hes 1, or Hey 1 from adenoviral vectors in the stable Numb-HA (Fig. 3 d) and Nbl-HA (Fig. 3 e) cell lines. Robust down-regulation of Numb was observed by Notch 1 ICD but to a much lesser extent by the canonical Notch downstream genes Hes 1 or Hey 1 (Fig. 3, d and e).
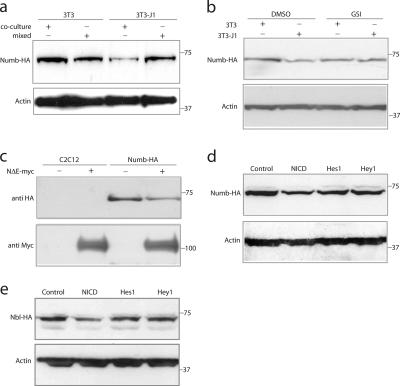
Figure 3.
High levels of Notch activation cause a reduction in Numb protein levels. (a) C2C12 cells stably expressing Numb-HA were cocultured with 3T3 cells, with Jagged-1–expressing 3T3 cells (3T3-J1), or were only mixed with 3T3 or 3T3-J1 cells just before lysis. (b) Cells stably expressing Numb-HA were cocultured with 3T3 or 3T3-J1 cells in the presence or absence of the GSI L-685,458 (1 μM) as indicated. Western blots of cell extracts in panels a and b were stained with an anti-HA antibody and reprobed with an anti–β-actin antibody. (c) Transfection of Notch 1 ΔE-myc (N1ΔE-myc) into C2C12 cells or stable Numb-HA C2C12 cells leads to a reduction of Numb protein levels in the Numb-HA stable cells. The blot was reprobed with anti-myc to visualize Notch expression. (d and e) Western blot of cell extracts from the stable Numb-HA (d) or Numblike-HA (Nbl-HA; e) C2C12 cell lines infected with adenoviruses expressing Notch 1 ICD (NICD), Hes 1, or Hey 1 as indicated.
Importance of the PEST domain for Notch-mediated down-regulation of Numblike
To further investigate the Notch-mediated down-regulation of Numb and Numblike, we speculated that the PEST domain may be of importance, as PEST domains have been shown to be involved in protein turnover and have been implicated in proteasome-mediated degradation (Reverte et al., 2001). The Numb protein contains two PEST domains, and Numblike contains only one, and, as Numblike in all assays behaved similarly to Numb, we generated a Numblike construct lacking the PEST domain (amino acids 260–273; Fig. 4 a) and produced stable cell lines with HA-tagged Numblike (Fig. 3 e) or PEST-deficient Numblike (Nbl-HA and Nbl-HA–ΔPEST, respectively). Like Numb, Nbl-HA protein was reduced by high levels of Notch signaling through coculture with Jagged-1–expressing cells, whereas the mixing of Nbl-HA and Jagged-1 cells immediately before lysis did not reduce Nbl-HA levels (Fig. 4 b). In contrast, Nbl-HA–ΔPEST protein levels did not change after coculture (Fig. 4 b). As for Numb, L-685,458 blocked the 3T3-J1 coculture–induced reduction on Nbl-HA protein levels, whereas Nbl-HA–ΔPEST protein levels were unaffected (Fig. 4 b). No substantial differences in the levels of Nbl-HA or Nbl-HA–ΔPEST mRNA were observed (Fig. S2, d and e), nor was mouse Numb mRNA expression increased by the expression of Notch 1 ICD (Fig. S2 f). Nbl-HA–ΔPEST was equally efficient as Nbl-HA in accelerating C2C12 myogenic differentiation, and 88% of Nbl-HA–ΔPEST-expressing C2C12 cells were MHC positive 3 d after the differentiation switch. Nbl-HA–ΔPEST was more efficient than Nbl-HA in negatively regulating Notch signaling as measured by 12XCSL-luc activation (Fig. S3 a, available at http://www.jcb.org/cgi/content/full/jcb.200602009/DC1). This is in keeping with the increased stability of Nbl-HA–ΔPEST and its resistance to degradation by Notch.
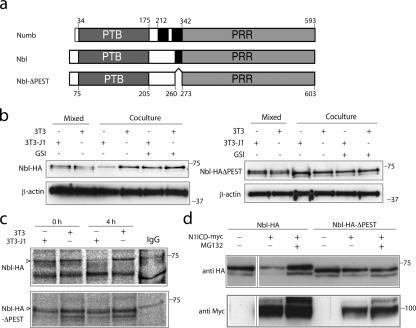
Figure 4.
Notch-induced reduction of Numblike protein levels depends on the PEST domain. (a) A schematic representation of the mouse Numb and Numblike (Nbl) proteins (with the amino acid positions for the phosphotyrosine-binding [PTB] and proline-rich region [PRR] domains [gray] and PEST domains [black] depicted) as well as Numblike-ΔPEST (NblΔPEST), in which the 13–amino acid–long PEST domain has been removed. (b) Cells stably expressing Numblike-HA (Nbl-HA; left) or Nbl-HA–ΔPEST (right) were cocultured with 3T3 cells or with Jagged-1–expressing cells (3T3-J1) as indicated (coculture) in the presence or absence of L-685,458 (GSI) or were only mixed with 3T3 or 3T3-J1 cells just before lysis (mixed). Western blots of cell extracts were stained with an anti-HA antibody. The presence of two bands in the Western blot probably reflects the partial degradation of Numblike, and a similar doublet of bands has previously been observed for Numb (Pece et al., 2004). Loading controls (β-actin) are shown below. (c) The stable Nbl-HA and Nbl-HA–ΔPEST cell lines were pulsed with [35S]methionine for 1 h at 16 h after coculture with Jagged-1–expressing cells and chased for 0 or 4 h as indicated. Note that Nbl-HA but not Nbl-HA–ΔPEST levels are reduced after the 4-h chase. (d) Transfection of myc-tagged Notch 1 ICD (N1ICD) into cell lines stably expressing Nbl-HA or Nbl-HA–ΔPEST in the presence or absence of the proteasome inhibitor MG132. Western blots of cell extracts were incubated with an anti-HA antibody and reprobed with an anti-myc antibody to control for Notch 1 ICD expression.
To address whether the down-regulation of Nbl-HA was a result of increased protein turnover or reduced synthesis, we performed pulse-chase experiments in the stable cell lines. After a 1-h [35S]methionine pulse 16 h after coculture with Jagged-1–expressing cells, there was no difference in the synthesis rate between Nbl-HA and Nbl-HA–ΔPEST (chase = 0 h), but, after 4 h of chase, considerably less Nbl-HA was observed as compared with Nbl-HA–ΔPEST (Fig. 4 c). This suggests that degradation rather than the synthesis rate is affected in the Notch-mediated down-regulation of Nbl-HA. Addition of the proteasome inhibitor MG132 abrogated the Notch 1 ICD–mediated down-regulation of Nbl-HA and, in fact, increased levels to more than what was observed in the absence of Notch signaling (Fig. 4 d, compare the first and third lanes). No effect of MG132 was observed with Nbl-HA–ΔPEST (Fig. 4 d). In conclusion, these experiments argue that the PEST domain is important for the Notch-mediated reduction of Numblike protein levels and that Numblike protein degradation is proteasome dependent.
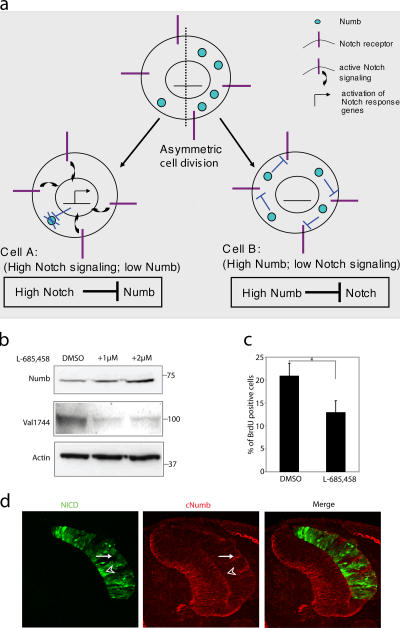
The negative regulation of Numb and Numblike by Notch demonstrated in this study may play a role in stabilizing the cell fate switch by an asymmetric cell division, which generates two distinct cells: one daughter cell that receives high levels of Numb and, therefore, down-regulates Notch signaling and a second daughter cell with no or low Numb and continued Notch signaling (for review see Cayouette and Raff, 2002). In Fig. 5 a, we propose that in the latter cell, down-regulation of Numb/Numblike by Notch may assure that Numb levels are kept low and, thus, safeguard the outcome of the cell fate switch resulting from Numb segregation. Such a mechanism would reduce the requirements on the asymmetric segregation machinery to perfectly distribute Numb to only one daughter cell, as small amounts of Numb segregating to the Notch-signaling cell would be eliminated. It may also be particularly important to reduce Numblike protein levels in this cell, as Numblike appears not to be asymmetrically localized (Zhong et al., 1997).
Figure 5.
Reciprocal negative regulation between Notch and Numb. (a) A model in which cell fate decisions are reinforced and maintained by reciprocal interaction between Notch and Numb. After classical asymmetric cell division, two daughter cells are produced with an asymmetrical inheritance of Numb. In cell A, little Numb protein is present, allowing Notch signaling and consequent cell fate alteration. Degradation of remaining Numb caused by active Notch signaling reinforces this lineage change. Conversely, the inheritance of Numb in cell B acts to inhibit Notch signaling, leading to adoption of the default cell fate. (b) Western blot of protein extracts from the SKOV-3 ovariocarcinoma cell line in the absence or presence (2 d) of L-685,458 (1 and 2 μM). (c) The percentage of BrdU-positive SKOV-3 cells in the absence or presence of L-685,458. Values are significant at *, P < 0.05. Error bars represent SD from three experiments. (d) Expression of Numb after the electroporation of Notch 1 ICD-IRES-GFP in the developing chick spinal cord. Expression of Notch 1 ICD (NICD) and endogenous Numb (cNumb) is visualized by immunohistochemistry on sections. Electroporated side is to the right. The arrow indicates an area with a low level of Notch 1 ICD, which coincides with robust levels of Numb. The arrowhead indicates an area with a high level of Notch 1 ICD, which correlates with reduced Numb levels.
To test the model, we wanted to learn whether the level of Notch signaling was inversely correlated with Numb levels in a tumor cell line because a correlation between reduced Numb protein levels and elevated Notch expression is frequently found in breast tumors (Pece et al., 2004; Stylianou et al., 2006). The human ovarian carcinoma cell line SKOV-3 was selected for analysis because it contains detectable levels of both Notch 1 ICD and Numb (Fig. 5 b). Treatment with L-685,458 resulted in reduced levels of Notch 1 ICD and, importantly, elevated levels of endogenous Numb (Fig. 5 b), suggesting that a reduction of Notch activity leads to enhanced Numb levels. Treatment with L-685,458 also reduced the proliferative rate in the SKOV-3 cells (Fig. 5 c). To test the converse situation (i.e., the experimental elevation of Notch), we introduced Notch 1 ICD by in ovo electroporation into the developing chick central nervous system. Areas of Notch 1 ICD overexpression showed reduced levels of Numb protein (Fig. 5 d, arrowhead), whereas areas of low Notch ICD expression contained higher levels of Numb (Fig. 5 d, arrow). In conclusion, these data support the idea that Notch-mediated down-regulation of Numb can be observed in vivo.
Materials and methods
Cell culture, transfection, and reporter gene analysis
3T3, 293T, and C2C12 cells were grown in DME containing 10% FCS. Transfections were performed using LipofectAMINE Plus reagent (Invitrogen) or FuGene 6 reagent (Roche) according to the manufacturer's instructions. The luciferase assay is described in the supplemental material (available at http://www.jcb.org/cgi/content/full/jcb.200602009/DC1). High-titer stocks of adenoviruses for Notch 1 ICD, Hes 1, and Hey 1 were used for infection of Numb-HA or Nbl-HA stable C2C12 cells, and, in all cases, at least 70% of the cells expressed the protein 24 h after infection with minimal cell toxicity.
Coculture and C2C12 cell differentiation
C2C12 cells grown on 10-cm plates were transfected with 4 μg DNA using LipofectAMINE Plus reagent. For coculture experiments, transfected C2C12 cells were seeded with approximately the same number of 3T3 or 3T3-J1 cells and grown for 24–48 h in DME containing 10% FCS before analysis. Differentiation experiments were performed by plating transfected C2C12 cells at high density onto coverslips precoated with gelatin and laminin in DME containing 10% FCS. Equivalent amounts of 3T3 or 3T3-J1 cells were added several hours later. Culture medium was changed the following day to DME containing 2% horse serum. Cultures were fixed 3 d later in 4% PFA and subjected to immunocytochemistry.
Western blotting and immunoprecipitation
Cultures were washed in PBS, harvested, resuspended in 50–200 μl whole cell extraction buffer (20 mM Hepes, pH 7.8, 420 mM NaCl, 0.5% NP-40, 25% glycerol, 0.2 mM EDTA, 1.5 mM MgCl2, 1 mM DTT, 1 mM PMSF, and complete protease inhibitors [Roche]), and incubated on an end-to-end rotator for 30 min at 4°C. The lysate was centrifuged for 30 min at 12,000 rpm, and protein concentration in the supernatant was determined by Bradford analysis (Bio-Rad Laboratories). Western blotting was performed as described in the supplemental material.
RNA isolation and quantitative RT-PCR
RNA was isolated from cultured cells using the RNeasy Mini Kit (QIAGEN). Reverse transcription was performed on 2.5 μg of total RNA using oligo-dT12–18 and Superscript II reverse transcriptase (Invitrogen). Real-time PCR was performed in accordance with the manufacturer's instructions using a rapid thermal cycler system (LightCycler; Applied Biosystems). A mastermix containing nucleotides, Taq polymerase, SYBR green, and buffer was mixed with primers and cDNA.
Pulse chase
C2C12 cells stably expressing Nbl-HA or Nbl-HA–ΔPEST were cocultured with 3T3-J1 or 3T3 cells. At 16 h of coculture, the growth medium was replaced by serum-free DME without methionine and cysteine containing 30 μCi [35S]methionine. After a 1-h pulse, the cells were either harvested (0 h) or washed, and the medium was replaced with normal growth medium for a 4-h chase. Cells were lysed in radioimmunoprecipitation buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris-HCl, pH 8.0, supplemented with protease inhibitors [Complete; Roche]). Cell lysates were centrifuged 14,000 g for 20 min at 4°C for the removal of insoluble material followed by preclearing with Sepharose G beads. HA-tagged Nbl or Nbl-ΔPEST were immunoprecipitated using a monoclonal HA antibody (HA11; BioSite) and captured by Sepharose G beads. The immunoprecipitated proteins were eluted by Laemmli sample buffer and separated by SDS-polyacrylamide gel electrophoresis.
Online supplemental material
Fig. S1 shows that Numb and Numblike down-regulate Notch signaling. Fig. S2 shows the detection of Numb protein and the role of Notch and Numb for apoptosis. Fig. S3 shows that Numblike-HA–ΔPEST down-regulates Notch-induced reporter gene activation more efficiently than Numblike-HA. Supplemental material provides details about the generation of Numb, Numblike, and Notch DNA constructs as well as generation of the anti-Numb antiserum and the sources of commercial antibodies. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200602009/DC1.
Supplementary Material
Acknowledgments
We thank Drs. J. Muhr and J. Holmberg for chick in ovo electroporation, S. Bergstedt for cell culture work, and B. Joseph, T. Honjo, W. Zhong, D. Ball, P. Dijke, S. Wood, and Y. Wakamatsu for reagents.
This work was supported by grants from the Swedish Foundation for Strategic Research (Center of Excellence in Developmental Biology), the Swedish Cancer Society, the Swedish Research Council, and the European Union project EuroStemCell (to U. Lendahl). G. Chapman was supported by a National Health and Medical Research Council (Australia) CJ Martin Fellowship (No. 158043), and C. Sahlgren was supported by the Academy of Finland and a Sigrid Juselius Foundation Research Fellowship.
G. Chapman and L. Liu contributed equally to this paper.
G. Chapman's present address is Victor Chang Cardiac Research Institute, Darlinghurst NSW 2010, Australia.
Abbreviations used in this paper: GSI, γ-secretase inhibitor; ICD, intracellular domain; MHC, myosin heavy chain.
References
- Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. [DOI] [PubMed] [Google Scholar]
- Berezovska, O., C. Jack, P. McLean, J.C. Aster, C. Hicks, W. Xia, M.S. Wolfe, W.T. Kimberly, G. Weinmaster, D.J. Selkoe, and B.T. Hyman. 2000. Aspartate mutations in presenilin and gamma-secretase inhibitors both impair notch1 proteolysis and nuclear translocation with relative preservation of notch1 signaling. J. Neurochem. 75:583–593. [DOI] [PubMed] [Google Scholar]
- Cayouette, M., and M. Raff. 2002. Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat. Neurosci. 5:1265–1269. [DOI] [PubMed] [Google Scholar]
- Conboy, I.M., and T.A. Rando. 2002. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 3:397–409. [DOI] [PubMed] [Google Scholar]
- Dahlqvist, C., A. Blokzijl, G. Chapman, A. Falk, K. Dannaeus, C.F. Ibanez, and U. Lendahl. 2003. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development. 130:6089–6099. [DOI] [PubMed] [Google Scholar]
- Frise, E., J.A. Knoblich, S. Younger-Shepherd, L.Y. Jan, and Y.N. Jan. 1996. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl. Acad. Sci. USA. 93:11925–11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, T., S. Maruyama, M. Kawaichi, and T. Honjo. 1992. The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell. 69:1191–1197. [DOI] [PubMed] [Google Scholar]
- Guo, M., L.Y. Jan, and Y.N. Jan. 1996. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 17:27–41. [DOI] [PubMed] [Google Scholar]
- Gustafsson, M.V., X. Zheng, T. Pereira, K. Gradin, S. Jin, J. Lundkvist, J.L. Ruas, L. Poellinger, U. Lendahl, and M. Bondesson. 2005. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell. 9:617–628. [DOI] [PubMed] [Google Scholar]
- Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194:237–255. [DOI] [PubMed] [Google Scholar]
- Li, H.S., D. Wang, Q. Shen, M.D. Schonemann, J.A. Gorski, K.R. Jones, S. Temple, L.Y. Jan, and Y.N. Jan. 2003. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 40:1105–1118. [DOI] [PubMed] [Google Scholar]
- Pece, S., M. Serresi, E. Santolini, M. Capra, E. Hulleman, V. Galimberti, S. Zurrida, P. Maisonneuve, G. Viale, and P.P. Di Fiore. 2004. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167:215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, P.H., K. Zou, J.K. Hwang, Y.N. Jan, and W. Zhong. 2002. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 419:929–934. [DOI] [PubMed] [Google Scholar]
- Reverte, C.G., M.D. Ahearn, and L.E. Hake. 2001. CPEB degradation during Xenopus oocyte maturation requires a PEST domain and the 26S proteasome. Dev. Biol. 231:447–458. [DOI] [PubMed] [Google Scholar]
- Stylianou, S., R.B. Clarke, and K. Brennan. 2006. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66:1517–1525. [DOI] [PubMed] [Google Scholar]
- Wakamatsu, Y., T.M. Maynard, S.U. Jones, and J.A. Weston. 1999. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron. 23:71–81. [DOI] [PubMed] [Google Scholar]
- Zhong, W., J.N. Feder, M.M. Jiang, L.Y. Jan, and Y.N. Jan. 1996. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 17:43–53. [DOI] [PubMed] [Google Scholar]
- Zhong, W., M.M. Jiang, G. Weinmaster, L.Y. Jan, and Y.N. Jan. 1997. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development. 124:1887–1897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.