Abstract
Melanocytes reside within the basal layer of the human epidermis, where they attach to the basement membrane and replicate at a rate proportionate to that of keratinocytes, maintaining a lifelong stable ratio. In this study, we report that coculturing melanocytes with keratinocytes up-regulated CCN3, a matricellular protein that we subsequently found to be critical for the spatial localization of melanocytes to the basement membrane. CCN3 knockdown cells were dissociated either upward to the suprabasal layers of the epidermis or downward into the dermis. The overexpression of CCN3 increased adhesion to collagen type IV, the major component of the basement membrane. As the receptor responsible for CCN3-mediated melanocyte localization, we identified discoidin domain receptor 1 (DDR1), a receptor tyrosine kinase that acts as a collagen IV adhesion receptor. DDR1 knockdown decreased melanocyte adhesion to collagen IV and shifted melanocyte localization in a manner similar to CCN3 knockdown. These results demonstrate an intricate and necessary communication between keratinocytes and melanocytes in maintaining normal epidermal homeostasis.
Introduction
During development, melanocyte precursors migrate from the neural crest toward the epidermis, where they are arrested when contacting keratinocytes. Differentiated human melanocytes remain strictly localized at the basement membrane and cannot survive within the upper epidermal layers unless transformed in nevi or melanomas. Keratinocytes control the normal growth of melanocytes to maintain a lifelong stable ratio, and they regulate the expression of cell surface molecules (Valyi-Nagy et al., 1993). They also produce growth factors and cytokines that may act as paracrine factors to regulate the phenotype of melanocytes, including interleukin-1β (IL-1β), TNF-α, stem cell factor (SCF), and EGF (Imokawa, 2004).
CCN3 (nephroblastoma overexpressed) is a matricellular protein that shares, with five other family members, structural modules of insulin-like growth factor–binding domains, von Willebrand factor type C repeats, thrombospondin type 1 repeats, and secreted regulatory factors containing cysteine knot motifs for dimerization (Perbal and Takigawa, 2005). Depending on the cell type and tissue context, matricellular proteins participate in diverse functions, such as cell adhesion, proliferation, differentiation, and survival (Brigstock, 1999).
In a search for the molecular players involved in cross talk between melanocytes and keratinocytes, we compared gene expression profiles of melanocytes grown in monoculture with melanocytes grown under the control of keratinocytes. We found CCN3 protein being up-regulated in melanocytes after coculture with keratinocytes. The CCN3 protein was secreted into the culture medium and affected two fundamental features of melanocytic physiology: it inhibited the proliferation of melanocytes and was required for the 3D organization of the melanocyte network on the basal membrane of human skin equivalent. Furthermore, CCN3 stimulated the adhesion of melanocytes to basal membrane collagen IV but not to dermal collagen I, as confirmed by the siRNA-mediated down-regulation of CCN3 and two-photon (2P) microscopy. We then identified discoidin domain receptor 1 (DDR1), a receptor tyrosine kinase, as being responsible for the adhesive properties of CCN3.
Results and discussion
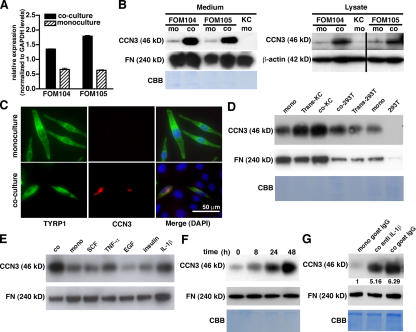
To investigate how keratinocytes change the phenotype of melanocytes, we cocultured pure populations of melanocytes and keratinocytes and conducted global gene expression analyses of monoculture- versus coculture-grown melanocytes. The CCN3 gene was found to be consistently up-regulated in cocultured melanocytes (Fig. 1 A). Keratinocytes in culture or human skin did not express CCN3, whereas melanocytes constitutively expressed it at low levels. After melanocytes were cultured with keratinocytes, CCN3 was strongly expressed in the cytoplasm of melanocytes and was secreted into the culture medium (Fig. 1, B and C). As demonstrated by immunofluorescent staining of normal human skin, CCN3 was expressed at the basal layer of the epidermis, where melanocytes are positioned (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200602132/DC1). Keratinocyte-derived culture supernatants stimulated CCN3 expression, but the up-regulation of CCN3 by melanocytes was strongest when keratinocytes were in direct cell–cell contact (Fig. 1 D, first to third lanes). Coculture with control epithelial cells had no effect on CCN3 secretion by melanocytes (Fig. 1 D, fourth to seventh lanes).
Figure 1.
CCN3 expression in melanocytes. (A) Up-regulated CCN3 mRNA expression in melanocytes in cocultures with keratinocytes as compared with monocultures. Results are from quantitative RT-PCR. Data represent the mean ± SD (error bars). (B) CCN3 protein expression in melanocytes in monoculture (mo) versus coculture (co). Results are from the Western blotting of culture medium (left) with fibronectin (FN) and Coomassie blue staining (CBB) as loading controls and from cell lysates (right) of two melanocyte cell lines (FOM 104 and 105), with β-actin as a loading control. (C) Costaining of melanocytes for TYRP1 (green) and CCN3 (red) shows up-regulation of CCN3 after coculture with keratinocytes. Colocalization is shown in merged photographs of cultures also stained with DAPI for the identification of all nuclei. (D) Immunoblot of conditioned medium from monocultured melanocytes (mono), melanocytes cocultured with keratinocytes while separated by a semiporous Transwell membrane (Trans-KC), cocultured with keratinocytes (coKC), cocultured with HEK 293T cells (co293T), cocultured with HEK 293T cells separated by Transwell membrane (Trans-293T), and monocultured HEK 293T cells (293T). (E) Immunoblot of conditioned medium from melanocytes cocultured with keratinocytes (co), monocultured (mono), or monocultured and stimulated with SCF at 50 ng/ml, TNF-α at 100 ng/ml, EGF at 20 ng/ml, insulin at 10 nM, or IL-1β at 2 ng/ml for 48 h. (F) Immunoblot of conditioned medium from monocultured melanocytes treated with 2 ng/ml IL-1β for the times indicated. (G) Immunoblot of conditioned medium from monocultured melanocytes treated with 2 μg/ml control goat IgG (mono goat IgG), cocultured melanocytes treated with anti–IL-1β antibodies (co anti IL-1β), and cocultured melanocytes treated with 2 μg/ml control goat IgG (co goat IgG). The numbers below the CCN3 blot indicate the relative densities normalized to the fibronectin blot. (D–G) Fibronectin immunoblot and Coomassie blue staining of Bis-Tris gels were performed as loading controls.
Keratinocytes commonly express several growth factors and cytokines that may change the phenotype of melanocytes. TNF-α and IL-1β stimulated the expression of CCN3 (Fig. 1 E). IL-1β was constitutively expressed by keratinocytes (unpublished data), and TNF-α was found to be undetectable in the culture supernatants derived from keratinocytes without stimulation (Lan et al., 2005). Therefore, we chose to further investigate the effect with IL-1β. Melanocytes began to increase CCN3 secretion 8 h after stimulation by IL-1β, and it continued for 48 h (Fig. 1 F). To investigate whether IL-1β produced by keratinocytes contributes to the induction and secretion of CCN3, we performed immunodepletion of IL-1β in coculture medium using neutralizing antibodies (Fig.1 G). The depletion of IL-1β decreased CCN3 in cocultures. However, this inhibiting effect was only partial (20% reduction), suggesting that other keratinocyte-derived factors are involved in the mechanism of CCN3 production by melanocytes.
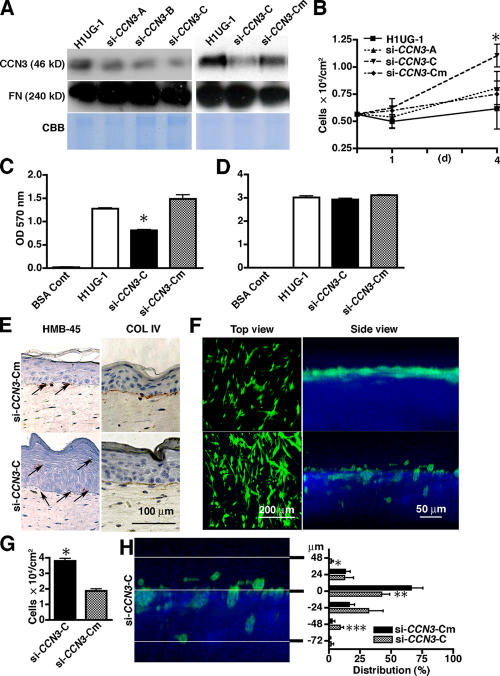
Because CCN3 has antiproliferative activity in fibroblastic, glioma, and Ewing's sarcoma cells (Joliot et al., 1992; Fu et al., 2004; Benini et al., 2005), we sought to determine whether CCN3 inhibits the growth of melanocytes. A lentiviral vector (si-CCN3-C) designed to knockdown CCN3 in melanocytes demonstrated a considerable decrease in protein production compared with an empty vector (H1UG-1), a one-pair mismatch (si-CCN3-Cm), and two related siRNA (si-CCN3-A and -B) vectors in conditioned media (Fig. 2 A) and lysates (not depicted). Melanocytes transduced with si-CCN3-C showed increased growth rates compared with cells transduced with control vectors (Fig. 2 B). The difference in growth rates between CCN3 knockdown (si-CCN3-C) and control cells (si-CCN3-Cm) was significant (P = 0.0095) on day 4 after coculture, when the medium from si-CCN3-Cm contained more CCN3 than si-CCN3-C (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200602132/DC1). They also showed a notable decrease in attachment to collagen type IV, which is present in the basement membrane (Figs. 2 C and S2 B) but not to type I collagen present in the dermis (Fig. 2 D) or laminin, which is another component of the basement membrane (Fig. S2 C, left). This result suggested that CCN3 modulates collagen type IV adhesion of melanocytes.
Figure 2.
CCN3 knockdown increases growth and disturbs the localization of melanocytes on the basement membrane zone in organotypic cultures of human skin. (A) Immunoblot of conditioned medium from CCN3 lentiviral siRNA-transfected melanocytes (si-CCN3-A, -B, and -C). Viral vector alone (H1UG-1) and one base pair mismatch siRNA (si-CCN3-Cm) were used as controls. Fibronectin blot (FN) and Coomassie blue staining (CBB) indicate equal loading. (B) Growth of melanocytes transduced with siRNA for CCN3 in coculture with keratinocytes when seeded at a 1:2 ratio. Cells were counted on days 1 and 4. n = 4. *, P = 0.0095 when compared with si-CCN3-Cm. (C and D) Adhesion on collagen type IV (C) and type I (D) as substrates. Data represent the mean ± SD of triplicates. *, P = 0.00028. (E) Organotypic cultures of human skin. Immunostaining for the melanocyte marker HMB-45 (left; arrows) or the basement membrane protein collagen type IV (COL IV; right). (F) 2P microscopy live images of skin reconstructs at day 14 to visualize melanocytes (green) transduced with control lentiviral vector (si-CCN3-Cm) or siRNA CCN3 (si-CCN3-C). Top view shows x-y view of 3D images, and side view shows x-z views of 3D images. (G) Growth of melanocytes transduced with siRNA for CCN3 in skin reconstructs at day 14. n = 5. *, P = 0.000014. (H) Distribution of melanocytes in skin reconstructs. Level 0 in the y bar indicates the epidermis/dermis border as determined by SHG (blue). Distribution (percentage) = number of melanocytes at each plane/total number of melanocytes × 100. n = 5. *, P = 0.024; **, P = 0.0086; ***, P = 0.014 compared with the control. (B, G, and H) Data represent the mean ± SD (error bars).
The melanocytes in mouse skin are localized in the dermis, suggesting that mouse melanocytes have different regulatory mechanisms from humans. The behavior of melanocytes in the skin reconstructs resembles that in vivo (Haake and Scott, 1991; Meier et al., 2000). We further tested the proliferation and localization of melanocytes within human skin reconstructs (Fig. 2, E–H). To identify melanocytes in sections of skin reconstructs and to determine their localization by immunohistochemistry, we used a melanocyte-specific marker, HMB-45 (Fig. 2 E, left). Staining for collagen IV demonstrated the formation of an intact basement membrane within the 14-d skin formation period (Fig. 2 E, right). Knockdown of CCN3 in melanocytes increased their numbers and changed their localization by migrating either upward to the suprabasal layers of the epidermis or downward into the dermis. To better determine the cell number and localization of melanocytes within the basement membrane zone, we performed 2P microscopy on the skin reconstructs. The lentiviral vectors contained GFP as a marker for melanocytes. Second harmonic generation (SHG) signals served as a sensitive indicator of collagen I to separate the epithelial layer from underlying stroma (Pena et al., 2005). A combination of 2P-excited GFP and SHG imaging of skin reconstructs showed that the knockdown of CCN3 in melanocytes not only increased their numbers within the reconstructs but also shifted their localization (Fig. 2, F–H). Individual melanocytes separated from the basement membrane, migrated into the epidermis, and invaded the dermis (Fig. 2 H). In contrast, melanocytes transduced with control vector remained confined to the basement membrane zone.
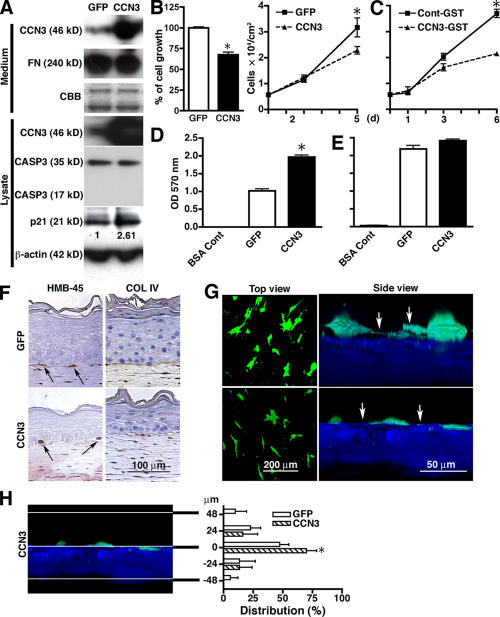
Melanocytes transduced with an adenoviral vector to overexpress the CCN3 protein (Fig. 3 A) were growth inhibited (Fig. 3 B). Although the CCN family members CCN1 and CCN2 have been shown to induce apoptosis (Hishikawa et al., 1999; Todorovicc et al., 2005), apoptosis as measured by caspase 3 levels was not changed in CCN3-overexpressing melanocytes (Fig. 3 A). There was no increase in the sub-G1 population of melanocytes overexpressing CCN3 either as tested by FACS (unpublished data). Moreover, the cyclin kinase inhibitor p21 was up-regulated (Fig. 3 A), suggesting that the decrease in proliferation is caused by growth inhibition and not cell death. Similar inhibition was seen when recombinant CCN3 (Li et al., 2002) protein was added to culture medium (Fig. 3 C). Melanocytes overexpressing CCN3 increased attachment to collagen IV (Fig. 3 D) but not to collagen I (Fig. 3 E) or laminin (Fig. S2 C, right). Melanocytes overexpressing CCN3 were firmly localized to the basement membrane zone of human skin reconstructs. Sections of the reconstructs showed tightly aligned melanocytes at the epidermal/dermal interface as assessed by melanocyte marker HMB-45 and by collagen IV staining (Fig. 3 F). 2P microscopy of skin reconstructs confirmed that the dendrites of control melanocytes remained separate from the basement membrane; however, those overexpressing CCN3 were localized at the border between the epidermis and dermis (Fig. 3, G and H). A limitation of the adenoviral vector system is that it is not integrated into the host genomic DNA, and its gene expression is extinguished through divisions of host cells. Therefore, GFP-positive cells were not observed by 2P imaging as frequently as those using a lentiviral vector system. The number of melanocytes identified by HMB-45 staining decreased when they overexpressed CCN3 (unpublished data). These data demonstrate that melanocyte-derived CCN3 inhibits growth to maintain normal homeostasis and secures the attachment of melanocytes to the basement membrane.
Figure 3.
Overexpression of CCN3 in melanocytes inhibits growth and aligns cells to the basement membrane of skin reconstructs. (A) Immunoblot of conditioned medium and cell lysates from melanocytes transduced with control GFP and CCN3 adenoviral vector. The samples were harvested 72 h after infection. β-actin immunoblot indicates equal loading of lysates. Fibronectin (FN) immunoblot and Coomassie blue staining (CBB) were used as loading controls of conditioned medium. CASP3, caspase 3. The numbers below the p21 blot indicate relative density normalized to the β-actin blot. (B) Growth of melanocytes transduced with either GFP or CCN3 using adenoviral vectors. (left) Cell growth was measured by 3[H]thymidine incorporation assays. n = 4. *, P = 0.00079. (right) Cells were counted on days 2 and 5. n = 4. *, P = 0.012. (C) Growth of melanocytes in the presence of 500 ng/ml CCN3-GST fusion protein or GST control protein. *, P = 0.0001. (D and E) Adhesion on collagen type IV (D) and type I (E) as substrates. n = 3. *, P = 0.015. (F) Immunostaining of human skin reconstructs to identify melanocytes using the HMB-45 marker (left; arrows) and the basement membrane using collagen type IV (COL IV; right). (G) 2P microscopy live images of skin reconstructs to visualize melanocytes (green) transduced with control GFP or CCN3 adenoviral vector. Top view shows x-y view, and side view shows x-z views of 3D images. White arrows indicate dendrites of melanocytes. (H) Distribution of melanocytes in skin reconstructs. Level 0 in the y bar indicates the epidermis/dermis junction as determined by SHG (blue). Distribution (percentage) = number of melanocytes at each level/total number of melanocytes × 100. n = 5. *, P = 0.0027. (B, C, and H) Data represent the mean ± SD (error bars).
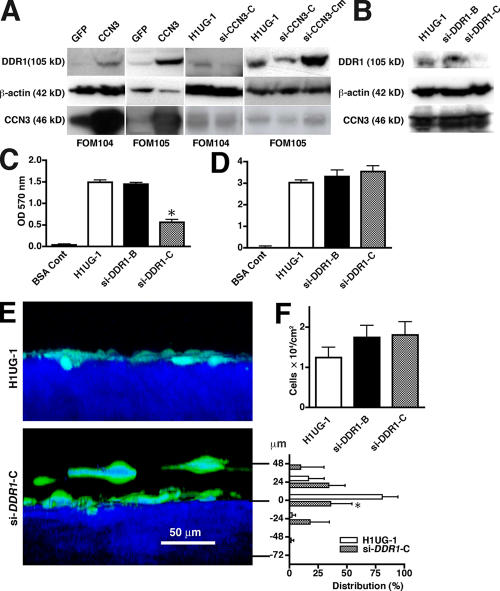
Because matricellular proteins themselves have only weakly adhesive functions (Murphy-Ullrich, 2001), we compared the expression profile of melanocytes overexpressing CCN3 with that of control cells by microarray analysis. DDR1 was up-regulated, as verified by Western blotting in two melanocyte cultures when CCN3 was overexpressed (Fig. 4 A). When melanocytes were transduced with siRNA against CCN3 (si-CCN3-C), DDR1 expression was down-regulated. DDR1 is a tyrosine kinase receptor for several collagens, particularly collagen IV (Vogel, 1999). Down-modulation of DDR1 with an siRNA, as confirmed by Western blotting (Fig. 4 B), showed decreased adhesion to collagen IV (Fig. 4 C) similar to those from siRNA CCN3. Consistently, adhesion to collagen I was not up-regulated (Fig. 4 D). 2P imaging of skin reconstructs showed that the knockdown of DDR1 in melanocytes shifted their localization; the proportion of cells at the basement membrane zone to total cell number was particularly decreased compared with the control (Fig. 4 E). To test whether DDR1 is essential for the regulation of melanocyte adhesion to basement membranes by CCN3, we overexpressed CCN3 in melanocytes transduced with si-DDR1-C. CCN3 overexpression in melanocytes transduced with si-DDR1-C recovered neither adhesion to collagen type IV nor normal localization in skin reconstructs (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200602132/DC1), confirming that up-regulation of the adhesion of melanocytes to the basement membrane by CCN3 is mediated through the collagen IV receptor DDR1. Knockdown of DDR1 in melanocytes did not increase their number in skin reconstructs (Fig. 4 F). Our results suggest that CCN3 regulates melanocyte growth through a mechanism that is distinct from adhesion. It is possible that CCN3 up-regulates DDR1 expression through the activation of p53 because p21 is a downstream target of p53, was up-regulated in CCN3-treated cells, and DDR1 is also one of the transcriptional targets of this tumor suppressor (Ongusaha et al., 2003).
Figure 4.
DDR1 is modulated by CCN3, and its expression determines the adhesion of melanocytes. (A) DDR1 and CCN3 protein expression in melanocytes transduced with GFP or CCN3 for overexpression using an adenoviral vector (left two columns) or siRNA CCN3 for knockdown using a lentiviral vector (right two columns). Results of cell lysates from two cell lines with β-actin as a loading control. (B) DDR1 expression and CCN3 expression in lysates of melanocytes transduced with DDR1 lentiviral siRNA of two different target sequences (si-DDR1-B and -C) and viral vector alone (HIUG-1). β-actin immunoblots indicate equal loading. (C and D) Adhesion of melanocytes transduced with DDR1 siRNA analyzed on collagen type IV (C) and type I (D) as substrates. n = 3. *, P = 0.00075 compared with si-DDR1-B. (E) 2P microscopy x-z views of skin reconstructs at day 14 to visualize the localization of melanocytes (green) transduced with control lentiviral vector (H1UG-1) or siRNA DDR1 (si-DDR1-C). Level 0 in the y bar indicates the epidermis/dermis border as determined by SHG (blue). Distribution (percentage) = number of melanocytes at each plane/total number of melanocytes × 100. n = 5. *, P = 0.0026. (F) Growth of melanocytes transduced with siRNA for DDR1 in skin reconstructs. n = 5. (C–F) Data represent the mean ± SD (error bars).
Melanocytes appear to have a contingency mechanism that is essential for their survival and secures continuous attachment to the basement membrane of the skin. The primary mechanism for attachment was thought to be through integrins (Sonnenberg et al., 1991), of which the laminin-binding integrin α6β1 was the main candidate (Albelda et al., 1990; Hara et al., 1994). Down-modulation of α6 integrin in melanocytes does not alter their localization in skin reconstructs (unpublished data), suggesting that α6 integrin is not essential for anchorage under homeostatic conditions. Because expression of the α6-integrin subunit is down-modulated by ultraviolet irradiation (Krengel et al., 2005), the melanocytes must have developed alternative mechanisms to maintain localization at the basement membrane. Our study indicates that CCN3 production by melanocytes after their contact with keratinocytes up-regulates the DDR1 adhesion receptor for collagen IV and influences melanocyte localization, contributing to the homeostasis in skin. When the proinflammatory cytokine IL-1β produced by keratinocytes up-regulates CCN3 in melanocytes, their normal localization in the skin is secured through DDR1-mediated adhesion to collagen type IV. Knockdown of DDR1 did not affect melanocyte proliferation in skin reconstructs, suggesting that there must be other downstream effectors of CCN3 that are responsible for the growth inhibitory effect of CCN3. Such a mechanism remains to be elucidated. CCN3 can bind to αvβ3 (Lin et al., 2003), a multiligand-binding integrin, but the β3 subunit is not expressed by normal melanocytes (Albelda et al., 1990; Hsu et al., 2000). CCN3 can also bind to Notch (Sakamoto et al., 2002); however, Notch signaling is not activated in melanocytes (unpublished data). In other cell types, CCN3 can be up-regulated by basic FGF (bFGF; Lafont et al., 2002), which stimulates melanocyte growth but does not modulate adhesion. Growth inhibition and basement membrane localization conferred by CCN3 are important, if not essential, functions for maintaining melanocyte homeostasis in the normal skin. Our findings suggest that CCN3 dysregulation may be the first step toward disrupting the normal balance between melanocytes and keratinocytes. Therefore, clarifying CCN3's role in melanocyte pathogenesis and melanoma is an important goal for further work.
Materials and methods
Cell culture
Normal human keratinocytes, melanocytes, and fibroblasts were isolated from neonatal human foreskins. Keratinocytes were cultured in EpiLife medium supplemented with human keratinocyte growth supplement (Cascade Biologics, Inc.). Melanocytes were cultured in MCDB153 (Sigma-Aldrich) supplemented with 2% FBS, 10% chelated FBS, 2 mM glutamine, 20 pM cholera toxin (Sigma-Aldrich), 1.5 nM recombinant human bFGF (Sigma-Aldrich), 100 nM recombinant human endothelin-3 (Peninsula Labs), and 10 ng/ml recombinant human SCF (Sigma-Aldrich). Fibroblasts were cultured in DME with 10% FBS. For cocultures, melanocytes were cultured with keratinocytes at a 1:5 ratio in EpiLife medium for 2 d. As a control, monocultured samples (melanocytes and keratinocytes at a 1:5 ratio) were cultured separately for 2 d. For gene expression comparison of monocultures with cocultures, melanocytes transduced with GFP were cultured with keratinocytes and isolated by FACS. As required, cells were counted or lysed for protein and RNA extraction.
Neutralization of human IL-1β activity
Cocultures were treated with 2 μg/ml human IL-1β–specific goat IgG (R&D Systems) to neutralize IL-1β. Control cultures were treated with 2 μg/ml nonspecific goat IgG.
Viral vectors for overexpression and knockdown
The human CCN3 cDNA was amplified using the primers 5′-GACGGGTACCTGAGCATGCAGAGTGTG-3′ and 5′-CTTGTCTAGAAGGTTACATTTTCCCTCTGG-3′ and were subcloned into pAdTrack-CMV at KpnI–XbaI sites. Recombination between pAdTrack-CMV–CCN3 and pAdEasy was performed in Escherichia coli to generate the CCN3 adenoviral vector (CCN3/Ad5). The mammalian expression vector H1UG-1 derived from the FG12 lentiviral vector (Qin et al., 2003) was used to produce lentiviral RNAi vector. DNA sequences encoding siRNA targeting CCN3 and DDR1 mRNA were cloned into BamH1 and XhoI sites under control of the HuH1 promoter. The original DNA sequences encoding the siRNAs targeting CCN3 mRNA were as follows: si-CCN3-A (5′-GCTGCAAATTCCAGTGCACCT-3′), si-CCN3-B (5′-GTTGAGGTGCCTGGAGAGT-3′), and si-CCN3-C (5′-GTGTCAACTGCATTGAACA-3′). One (si-CCN3-C) out of three siRNA vectors displayed high efficiency CCN3 knockdown in melanocytes and was selected for the creation of a mutated (indicated in bold) control siRNA sequence (si-CCN3-Cm, 5′-GTGTCAACTTCATTGAACA-3′). The DNA sequences encoding the siRNAs targeting DDR1 mRNA were si-DDR1-B (5′-GAGGAGCTGACGGTTCACC-3′) and si-DDR1-C (5′-GATCTGGTTAGTCTTGATT-3′). The lentivirus was produced by cotransfection of human embryonic kidney 293T cells with four plasmids: a packaging defective helper construct, a Rev plasmid, a plasmid coding for a heterologous envelope protein, and the H1UG-1 vector construct harboring the selected siRNA sequence.
RNA extraction and gene chip hybridization
Total RNA was isolated using the total RNeasy kit (QIAGEN). Human Genome U133A arrays were used for mRNA expression profiling according to the manufacturer's instructions (Affymetrix, Inc.). A laser scanner (GeneArray; Hewlett-Packard) was used to analyze gene chips, and the expression levels were calculated using Microarray Suite software (Affymetrix, Inc.). Gene expression values were normalized to the mean value of all genes in each experiment.
Quantitative RT-PCR
Total RNA was reverse transcribed into first-strand cDNA for quantitative RT-PCR. The gene specific primers were designed as CCN3 (5′-GAACCGTCAATGTGAGATGC-3′ and 5′-ACAGAACCTGGGCTTGTAGG-3′) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 5′-ATGGAAATCCCATCACCATCTT-3′ and 5′-CGCCCCACTTGATTTTGG-3′). ABsolute QPCR SYBR Green Mixes (ABgene) were used with 1 ng/ml cDNA and with 70 nM of primers for the evaluation of GAPDH and CCN3 expression. A negative control without the cDNA template was run with each assay. Amplifications were performed in an ABI Prism 7000 Sequence Detection System (Applied Biosystems). Thermal cycler conditions were 95°C for 15 min and 40 cycles of 15 s at 95°C followed by 1 min at 60°C. All experiments were performed in triplicate, and a mean value was used for the determination of mRNA levels. At the end of PCR, baseline and threshold values (C T) for these genes were set using the ABI Prism 7000 software (Applied Biosystems), and the calculated C T values were exported to Excel (Microsoft) for analysis. The relative expression of mRNA was calculated using the comparative C T method according to the manufacturer (Perkin-Elmer). All samples were normalized to the relative levels of GAPDH.
CCN3 protein purification
The CCN3 coding sequence was cloned into the pGEX4T1 vector. Expression of the recombinant GST-CCN3 protein was induced by adding 0.1 mM IPTG to the bacteria cultures when they reached 0.7–0.9 OD at 600 nm. After centrifugation, pellets were resuspended in 50 mM Tris, pH 8.0, 1 mM EDTA, 100 mM NaCl, and proteinase inhibitors (complete cocktail [Roche], 200 mM PMSF, 10 mM TLCK, 200 mM benzamidine, and 10 mM TPCK), and 300 μg/ml lysozyme was added. Lysis was performed for 20 min on ice. Triton X-100 was then added to 1%, and lysates were sonicated on ice. After centrifugation, supernatants were incubated with glutathione–Sepharose beads (GE Healthcare) in PBS for 1 h at 4°C on a rotating wheel. For GST-CCN3, PBS was complemented with 5% fat-free milk and 0.5 mM ATP. Beads were then washed several times with PBS-proteinase inhibitors. Recombinant proteins were recovered by three elutions of 1 h on ice with 20 mM glutathione, 100 mM Tris, pH 8.0, and 120 mM NaCl. Fractions were pooled, dialyzed overnight at 4°C against 10 mM NH4HCO3, and lyophilized. Quantification was performed by SDS-PAGE and Coomassie blue coloration of the gel.
Immunoassays
For Western analyses to detect CCN3 or DDR1 expression, cells were washed with PBS and harvested in radioimmunoprecipitation buffer. To detect secreted CCN3, proteins from conditioned medium corresponding to 2 × 105 melanocytes were collected with heparin–Sepharose beads after overnight incubation at 4°C as described previously (Chevalier et al., 1998). Samples were separated on 4–12% Bis-Tris gels, transferred to polyvinylidene difluoride membranes, and probed with K19M, antifibronectin (Transduction Laboratories), anti–β-actin (Sigma-Aldrich), anti-p21 (BD Biosciences), anticaspase 3 (Cell Signaling Technology), or anti-DDR1 antibodies (C-20; Santa Cruz Biotechnology, Inc.). To detect the signal, HRP-conjugated secondary antibody was added followed by ECL (GE Healthcare). The immunoblot images were acquired with a scanner (Perfection 636U; Epson) and quantitated using Image Beta 4.02 software (Scion Corp.). For immunofluorescence, cells were fixed, permeabilized, and incubated sequentially with mouse anti-TYRP1 (Signet), FITC-conjugated goat anti–mouse IgG, K19M anti-CCN3 rabbit serum, and AlexaFluor594-conjugated goat anti–rabbit IgG. Immunofluorescent staining was also performed on serial sections of human foreskins with K19M and anti-pmel 17/HMB-45 (DakoCytomation) as primary antibodies. For immunohistochemistry, sections were stained with mouse anti–human type IV collagen (Calbiochem) or HMB-45 using standard procedures. Biotin-labeled anti–mouse secondary antibodies were applied, and signal was detected using the ABC kit (Vector Laboratories). After adding 3-amino-9-ethyl carbazole, samples were counterstained with Mayer's hematoxylin (Sigma-Aldrich). The immunostaining images were taken by an upright microscope (E600; Nikon) with 20/40× objectives. A digital camera (SPOT RT Slider; Diagnostic Instruments) was used to acquire the pictures. Photoshop 6.0 software (Adobe) was used for contrast and brightness adjustment.
Cell assays
For cell growth experiments, melanocytes were plated in quadruplicates in 24-well plates at a density of 5.66 × 103 cells/cm2. Cell growth was monitored by counting cells in five random high power fields. For [3H]thymidine uptake assays, recombinant adenovirus-transduced melanocytes were seeded in quadruplicates in 96-well plates at 5,000 cells/well 48 h after transduction and cultured in 200 μl of medium. After 24 h, cells were pulsed with 1 μCi [3H]methyl thymidine and harvested after 12 h for counting.
For adhesion assays, melanocytes were suspended in serum-free MCDB153 (6 × 105 cells/ml) and transferred in triplicate to either CytoMatrix cell adhesion strips coated with human collagen type IV, human laminin (Chemicon), or 96-well plates coated with 10 μg/ml bovine type I collagen (Organogenesis) and incubated for 90 min at 37°C. After washing to remove unattached cells, the attached cells were stained with 0.2% crystal violet. The cell-bound stain was solubilized, and the optical density (570 nm) was determined.
Reconstructions of normal human skin were prepared as previously reported (Berking et al., 2001). 3 ml of fibroblast-containing bovine type I collagen (7.5 × 104 cells/ml) was added to each insert of tissue culture trays (Organogenesis) and allowed to constrict in DME with 10% FBS for 7 d at 37°C. For epidermal reconstruction, keratinocytes were mixed with melanocytes at a ratio of 5:1 in keratinocyte serum-free medium (Invitrogen) containing 2% dialyzed FCS, 60 μg/ml bovine pituitary extract (Invitrogen), 4.5 ng/ml bFGF, 100 nM human endothelin-3, and 10 ng/ml human SCF. A total of 5 × 106 cells were seeded on each contracted collagen gel. Cultures were kept submerged in medium containing 1 ng/ml EGF for 2 d, 0.2 ng/ml EGF for another 2 d, and were raised to the air–liquid interface via feeding from below with high calcium (2.4 mM) medium. After 14 d, skin reconstructs were either directly analyzed using a 2P microscope or fixed with 4% PFA and embedded in paraffin for subsequent sectioning and staining.
2P imaging was performed with an upright multiphoton microscope (Ultima; Prairie Technologies) attached to a microscope (BX-61; Olympus) fitted with 20/40× water immersion objectives (Olympus). This arrangement was combined with a diode pumped wideband mode-locked titanium-sapphire femtosecond laser (Chameleon; Coherent). Components of the extracellular matrix (e.g., collagen) were detected by SHG signals (Pena et al., 2005). In all of the experiments, the samples were exposed to a wavelength of 920 nm. The wavelengths emitted by the GFP (515 nm) and the extracellular matrix (460 nm) were distinguished using a filter cube (Dichroic). Z stacks of a series of x-y planes at a resolution of 2 pixels/μm−1 in step size 2 μm were captured using Photonics photomultiplier tubes (R3896; Hamamatsu) with amplifiers and View acquisition software (Prairie Technologies). Volocity software (Improvision) was used to generate x-z sections and to render 3D reconstructions of the skin. To assess localization of melanocytes in skin reconstructs, five fields (×200) were randomly selected in each reconstruct and scored by counting GFP-positive cells on x-y planes at 24-μm intervals. Distribution (percentage) = number of melanocytes on each plane/total number of melanocytes on all planes × 100. All experiments were performed three times using melanocytes derived from three different donors. The data were analyzed by t test (two-tailed distribution and two-sample unequal variance) and expressed as the mean ± SD. Each figure shows one representative experiment.
Online supplemental material
Fig. S1 shows CCN3 expression in human skin. Fig. S2 shows that adhesion on laminin is not affected by CCN3 modulation. Fig. S3 shows that the overexpression of CCN3 does not restore the localization of melanocytes transduced with si-DDR1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200602132/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Philip, G. Haydon, and Dr. Hajime Takano for helpful discussions and technical assistance.
This work was supported by grants from the National Institutes of Health (CA76674, CA80999, CA47159, CA76674, CA25874, and CA10815) and was partially supported by funds from the Commonwealth Universal Research Enhancement Program (Pennsylvania Department of Health). The work performed in B. Perbal's laboratory was funded by the Ministère de l'Education Nationale de la Recherche et de la Technologie and by a grant from Ligue Nationale Contre le Cancer (Comité du Cher). The work in S.M. Firth's laboratory was supported by the Australian Research Council (Discovery Project DP0345171) and Cancer Institute NSW (Career Development and Support Fellowship). S.M. Firth would like to acknowledge the technical assistance of Xiaolang Yan.
Abbreviations used in this paper: 2P, two photon; bFGF, basic FGF; DDR1, discoidin domain receptor 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IL-1β, interleukin-1β; SCF, stem cell factor; SHG, second harmonic generation.
References
- Albelda, S.M., S.A. Mette, D.E. Elder, R. Stewart, L. Damjanovich, M. Herlyn, and C.A. Buck. 1990. Integrin distribution in malignant melanoma: association of the beta 3 subunit with tumor progression. Cancer Res. 50:6757–6764. [PubMed] [Google Scholar]
- Benini, S., B. Perbal, D. Zambelli, M.P. Colombo, M.C. Manara, M. Serra, M. Parenza, V. Martinez, P. Picci, and K. Scotlandi. 2005. In Ewing's sarcoma CCN3 (NOV) inhibits proliferation while promoting migration and invasion of the same cell type. Oncogene. 24:4349–4361. [DOI] [PubMed] [Google Scholar]
- Berking, C., R. Takemoto, K. Satyamoorthy, R. Elenitsas, and M. Herlyn. 2001. Basic fibroblast growth factor and ultraviolet B transform melanocytes in human skin. Am. J. Pathol. 158:943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigstock, D.R. 1999. The connective tissue growth factor/cysteine-rich 61/nephroblastoma overexpressed (CCN) family. Endocr. Rev. 20:189–206. [DOI] [PubMed] [Google Scholar]
- Chevalier, G., H. Yeger, C. Martinerie, M. Laurent, J. Alami, P.N. Schofield, and B. Perbal. 1998. novH: differential expression in developing kidney and Wilm's tumors. Am. J. Pathol. 152:1563–1575. [PMC free article] [PubMed] [Google Scholar]
- Fu, C.T., J.F. Bechberger, M.A. Ozog, B. Perbal, and C.C. Naus. 2004. CCN3 (NOV) interacts with connexin43 in C6 glioma cells: possible mechanism of connexin-mediated growth suppression. J. Biol. Chem. 279:36943–36950. [DOI] [PubMed] [Google Scholar]
- Haake, A.R., and G.A. Scott. 1991. Physiologic distribution and differentiation of melanocytes in human fetal and neonatal skin equivalents. J. Invest. Dermatol. 96:71–77. [DOI] [PubMed] [Google Scholar]
- Hara, M., M. Yaar, A. Tang, M.S. Eller, W. Reenstra, and B.A. Gilchrest. 1994. Role of integrins in melanocyte attachment and dendricity. J. Cell Sci. 107:2739–2748. [DOI] [PubMed] [Google Scholar]
- Hishikawa, K., B.S. Oemar, F.C. Tanner, T. Nakaki, T.F. Luscher, and T. Fujii. 1999. Connective tissue growth factor induces apoptosis in human breast cancer cell line MCF-7. J. Biol. Chem. 274:37461–37466. [DOI] [PubMed] [Google Scholar]
- Hsu, M.Y., F.E. Meier, M. Nesbit, J.Y. Hsu, P. Van Belle, D.E. Elder, and M. Herlyn. 2000. E-cadherin expression in melanoma cells restores keratinocyte-mediated growth control and down-regulates expression of invasion-related adhesion receptors. Am. J. Pathol. 156:1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imokawa, G. 2004. Autocrine and paracrine regulation of melanocytes in human skin and in pigmentary disorders. Pigment Cell Res. 17:96–110. [DOI] [PubMed] [Google Scholar]
- Joliot, V., C. Martinerie, G. Dambrine, G. Plassiart, M. Brisac, J. Crochet, and B. Perbal. 1992. Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol. Cell. Biol. 12:10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krengel, S., I. Stark, C. Geuchen, B. Knoppe, G. Scheel, P. Schlenke, A. Gebert, L. Wunsch, J. Brinckmann, and M. Tronnier. 2005. Selective down-regulation of the alpha6-integrin subunit in melanocytes by UVB light. Exp. Dermatol. 14:411–419. [DOI] [PubMed] [Google Scholar]
- Lafont, J., M. Laurent, H. Thibout, F. Lallemand, Y. Le Bouc, A. Atfi, and C. Martinerie. 2002. The expression of novH in adrenocortical cells is down-regulated by TGFbeta 1 through c-Jun in a Smad-independent manner. J. Biol. Chem. 277:41220–41229. [DOI] [PubMed] [Google Scholar]
- Lan, C.C., H.S. Yu, C.S. Wu, H.Y. Kuo, C.Y. Chai, and G.S. Chen. 2005. FK506 inhibits tumour necrosis factor-alpha secretion in human keratinocytes via regulation of nuclear factor-kappaB. Br. J. Dermatol. 153:725–732. [DOI] [PubMed] [Google Scholar]
- Li, C.L., V. Martinez, B. He, A. Lombet, and B. Perbal. 2002. A role for CCN3 (NOV) in calcium signalling. Mol. Pathol. 55:250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.G., S.J. Leu, N. Chen, C.M. Tebeau, S.X. Lin, C.Y. Yeung, and L.F. Lau. 2003. CCN3 (NOV) is a novel angiogenic regulator of the CCN protein family. J. Biol. Chem. 278:24200–24208. [DOI] [PubMed] [Google Scholar]
- Meier, F., M. Nesbit, M.Y. Hsu, B. Martin, P. Van Belle, D.E. Elder, G. Schaumburg-Lever, C. Garbe, T.M. Walz, P. Donatien, et al. 2000. Human melanoma progression in skin reconstructs: biological significance of bFGF. Am. J. Pathol. 156:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy-Ullrich, J.E. 2001. The de-adhesive activity of matricellular proteins: is intermediate cell adhesion an adaptive state? J. Clin. Invest. 107:785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongusaha, P.P., J.I. Kim, L. Fang, T.W. Wong, G.D. Yancopoulos, S.A. Aaronson, and S.W. Lee. 2003. p53 induction and activation of DDR1 kinase counteract p53-mediated apoptosis and influence p53 regulation through a positive feedback loop. EMBO J. 22:1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena, A.M., T. Boulesteix, T. Dartigalongue, and M.C. Schanne-Klein. 2005. Chiroptical effects in the second harmonic signal of collagens I and IV. J. Am. Chem. Soc. 127:10314–10322. [DOI] [PubMed] [Google Scholar]
- Perbal, B., and M. Takigawa. 2005. CCN Proteins: a New Family of Cell Growth and Differentiation Regulators. Imperial College Press, London. 324 pp.
- Qin, X.F., D.S. An, I.S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA. 100:183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K., S. Yamaguchi, R. Ando, A. Miyawaki, Y. Kabasawa, M. Takagi, C.L. Li, B. Perbal, and K. Katsube. 2002. The nephroblastoma overexpressed gene (NOV/ccn3) protein associates with Notch1 extracellular domain and inhibits myoblast differentiation via Notch signaling pathway. J. Biol. Chem. 277:29399–29405. [DOI] [PubMed] [Google Scholar]
- Sonnenberg, A., K.R. Gehlsen, M. Aumailley, and R. Timpl. 1991. Isolation of alpha 6 beta 1 integrins from platelets and adherent cells by affinity chromatography on mouse laminin fragment E8 and human laminin pepsin fragment. Exp. Cell Res. 197:234–244. [DOI] [PubMed] [Google Scholar]
- Todorovicc, V., C.C. Chen, N. Hay, and L.F. Lau. 2005. The matrix protein CCN1 (CYR61) induces apoptosis in fibroblasts. J. Cell Biol. 171:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valyi-Nagy, I.T., G. Hirka, P.J. Jensen, I.M. Shih, I. Juhasz, and M. Herlyn. 1993. Undifferentiated keratinocytes control growth, morphology, and antigen expression of normal melanocytes through cell-cell contact. Lab. Invest. 69:152–159. [PubMed] [Google Scholar]
- Vogel, W. 1999. Discoidin domain receptors: structural relations and functional implications. FASEB J. 13:S77–S82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.