Abstract
Clathrin-coated vesicles (CCVs) facilitate the transport of cargo between the trans-Golgi network, endosomes, and the plasma membrane. This study presents the first comparative proteomics investigation of CCVs. A CCV-enriched fraction was isolated from HeLa cells and a “mock CCV” fraction from clathrin-depleted cells. We used a combination of 2D difference gel electrophoresis and isobaric tags for relative and absolute quantification (iTRAQ) in conjunction with mass spectrometry to analyze and compare the two fractions. In total, 63 bona fide CCV proteins were identified, including 28 proteins whose association with CCVs had not previously been established. These include numerous post-Golgi SNAREs; subunits of the AP-3, retromer, and BLOC-1 complexes; lysosomal enzymes; CHC22; and five novel proteins of unknown function. The strategy outlined in this paper should be widely applicable as a means of distinguishing genuine organelle components from contaminants.
Introduction
Clathrin-coated vesicles (CCVs) are important transport intermediates in all eukaryotic cells. Their coats consist of two major components: clathrin, which provides a stabilizing scaffold, and heterotetrameric adaptor protein (AP) complexes, which attach the clathrin to the membrane and select the vesicle cargo (Robinson, 2004). There are at least two AP complexes associated with CCVs: AP-1, which functions in transport between the TGN and endosomes (although there is some question about directionality), and AP-2, which functions in clathrin-mediated endocytosis (Traub, 2005; Bonifacino and Rojas, 2006; Foote and Nothwehr, 2006).
Although it was originally assumed that clathrin and AP complexes were all that was necessary to make a CCV, it is now apparent that CCV formation is much more complex. New components of the machinery are continually being discovered, including various “alternative adaptors” and proteins that contribute to other stages of the CCV cycle (Traub, 2005). Thus, although the most abundant components of CCVs are known, many questions remain regarding the initiation of vesicle formation, cargo selection, budding, scission, uncoating, and transport. Clearly, a complete knowledge of the protein composition of CCVs would greatly advance our understanding of clathrin-mediated trafficking.
In recent years, organelle proteomics has emerged as a powerful tool to guide cell biological research (McDonald and Yates, 2000; Andersen and Mann, 2006; Dunkley et al., 2006), and two studies have so far been published on the CCV proteome. Blondeau et al. (2004) and Girard et al. (2005) prepared CCV-enriched fractions from rat brain and liver, respectively, and identified proteins by tandem mass spectrometry (MS/MS). In both studies, an impressive degree of CCV enrichment was achieved (73–89% vesicle homogeneity, as judged by electron microscopy), and numerous proteins were identified. However, neither study could distinguish which of the identified proteins were true constituents of CCVs and which were copurifying contaminants.
Because it is impossible to prepare completely pure CCVs, the challenge becomes finding unbiased criteria that allow one to identify genuine CCV components. With such criteria at hand, the purity of the preparation is no longer critical. Here, we introduce a novel criterion: the dependence of a protein on clathrin to be present in a CCV fraction. By pairing cell biological tools with state-of-the-art quantitative proteomics techniques, we develop a strategy for identifying bona fide CCV proteins from human tissue culture cells.
Results and discussion
Unbiased proteomic analysis of CCVs
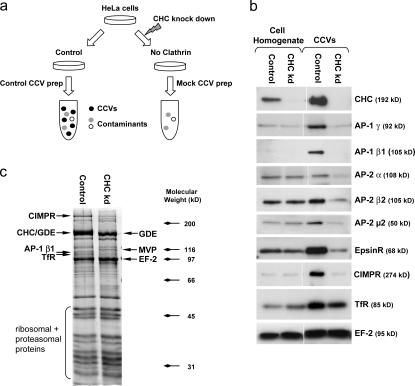
A CCV-enriched fraction was prepared from HeLa cells using an established protocol (Hirst et al., 2004), and the same procedure was performed on cells that had been depleted of clathrin heavy chain (CHC) by siRNA knockdown. Such cells have been shown to contain no detectable clathrin-coated budding profiles or vesicles (Motley et al., 2003), so the prediction is that the “mock CCV” fraction from these cells should be devoid of CCVs but should still contain proteins that contaminate CCV preparations from control cells. Thus, a comparison between the two fractions should reveal genuine CCV proteins as those present only (or mainly) in the control CCV fraction and contaminants as those equally present in both fractions (Fig. 1 a).
Figure 1.
Unbiased comparative proteomics of CCVs. (a) Schematic representation of the experimental approach. (b) Control and mock CCV fractions were prepared and analyzed by Western blotting (∼7.5 μg protein/lane for homogenates and ∼2 μg for CCV fractions). (c) Comparison of control and mock CCV fractions by 1D SDS-PAGE. Proteins were identified by mass spectrometry. EF, elongation factor; GDE, glycogen debranching enzyme; MVP, major vault protein; TfR, transferrin receptor.
To validate the approach, control and mock CCV fractions were analyzed by Western blotting (Fig. 1 b). All of the known CCV components that we tested, including coat proteins (e.g., CHC, AP-1 and AP-2 subunits, and epsinR) and cargo proteins (e.g., cation-independent mannose 6-phosphate receptor [CIMPR] and transferrin receptor), were enriched in our CCV preparations compared with whole cell homogenate (the homogenate lanes contain approximately four times as much protein as the CCV lanes) and were absent or depleted from mock CCVs. In contrast, known contaminants (e.g., elongation factor 2) were equally present in control and mock CCVs. Thus, our comparative proteomics approach should allow us to distinguish bona fide CCV constituents from irrelevant proteins that copurify. Interestingly, we consistently observed differences in the degree of enrichment and depletion of AP-1 and AP-2 subunits. AP-1 subunits were more highly enriched in CCVs relative to cell homogenate, and they were also more strongly depleted from the mock CCVs, suggesting that our preparation favors intracellular, nonendocytic CCVs.
As a first step toward analyzing the CCV proteome, control and mock CCV fractions were separated by 1D SDS-PAGE and stained with Coomassie blue (Fig. 1 c). The two fractions have similar protein patterns, indicating that CCV-enriched preparations from control cells are heavily contaminated with non-CCV material. However, four high molecular weight proteins appear to be depleted from mock CCVs (Fig. 1 c, arrows). These proteins were analyzed by MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight) and identified as CHC (which comigrates with glycogen debranching enzyme), CIMPR, the β1 subunit of AP-1, and transferrin receptor. Thus, the 1D SDS-PAGE data confirm the results of the Western blot, but they offer insufficient resolution to analyze more minor constituents.
Analysis of CCVs by 2D difference gel electrophoresis (DIGE)
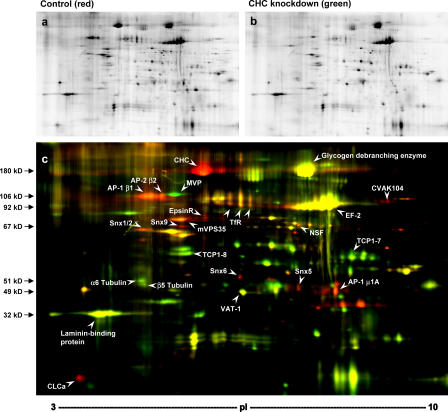
To identify additional CCV components, control and mock CCV fractions were compared by 2D DIGE (Borner et al., 2003). The two fractions were labeled with different fluorescent dyes, pooled, and analyzed in single 2D gels. A representative gel is shown in Fig. 2, with the control CCV fraction in red and the mock CCV fraction in green. The spot pattern was highly reproducible and allowed a clear distinction between proteins present in both fractions (yellow spots) and proteins depleted from the mock CCVs (red spots), which are likely to be genuine CCV components. Proteins were excised from the gels and identified by liquid chromatography (LC)–MS/MS (Fig. 2 c). Several prominent red spots were found to correspond to known clathrin coat components (clathrin heavy and light chains, β1, μ1A, β2, epsinR, and CVAK104) and fusion machinery (N-ethylmaleimide–sensitive fusion protein [NSF]), lending further support to the validity of the approach. Again, we consistently observed more AP-1 than AP-2, as indicated by the relative intensities of the β1 and β2 spots.
Figure 2.
2D DIGE analysis of CCVs. A CCV-enriched fraction (control) and a mock CCV fraction (CHC knockdown) were labeled with Cy3 and Cy5, respectively, pooled, and analyzed in a single 2D gel. (a) Control CCV fraction. (b) CHC-knockdown mock CCV fraction. (c) Overlay of a and b with control CCVs in red and mock CCVs in green. Yellow spots correspond to proteins with similar abundance in both fractions. White arrowheads indicate proteins that were identified by mass spectrometry. EF, elongation factor; MVP, major vault protein; TfR, transferrin receptor.
Several other proteins implicated in membrane traffic were also identified as red spots. These include five sorting nexins, Snx1, Snx2, Snx5, Snx6, and Snx9, none of which had been identified in either of the two previous proteomic analyses of CCVs. Snx1 and Snx2 are putative components of the mammalian retromer complex, which functions in retrograde traffic from endosomes to the TGN (Seaman, 2004). Another retromer component, mVPS35, also appears as a red spot. mVPS35 was also found in the proteomic analyses of brain and liver CCVs (Blondeau et al., 2004; Girard et al., 2005), but at the time, the physiological relevance of this observation was unclear.
The majority of the spots on the 2D gel are yellow, indicating that they are equally abundant in control and mock CCVs. Several of the yellow spots were identified by LC-MS/MS and found to correspond to irrelevant proteins. Thus, the 2D DIGE method enabled us to distinguish between genuine CCV components and contaminants and to identify 15 proteins that are largely depleted from the mock CCVs, including several novel CCV proteins.
Analysis of CCVs by quantitative mass spectrometry
An inherent disadvantage of 2D gels is the poor resolution of hydrophobic proteins. Hence, it is likely that integral membrane proteins, including CCV cargo, are strongly underrepresented in our DIGE analysis. To overcome this limitation, as well as to increase the sensitivity of our screen, we used iTRAQ (isobaric tags for relative and absolute quantification), a gel-free quantitative proteomics method (Ross et al., 2004). Control and mock CCVs were digested with trypsin and labeled with iTRAQ tags, which bind to free amines. The labeled peptides were then pooled into a single sample, and the peptide mixture was fractionated by cation exchange chromatography and analyzed by LC-MS/MS. The tags are chemically similar and isobaric (i.e., have the same mass), so identical peptides that originally came from different samples and therefore have different tags coelute from the LC and are simultaneously analyzed by precursor ion scanning. However, fragmentation of the iTRAQ tags during MS/MS results in different signature peaks. Integration of these peaks allows the determination of the relative abundance of a given peptide in the original samples. In turn, this allows the quantification of the parent protein from which the peptide was derived.
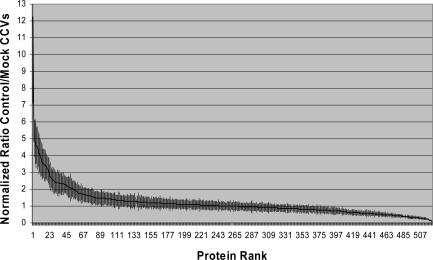
Using this approach, 522 proteins were identified and quantified from control and mock CCV fractions (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200607164/DC1). To determine which proteins were depleted from mock CCVs, the mean relative abundance of proteins in control and mock CCVs was calculated and expressed as a ratio (control/mock CCVs). High ratios correspond to proteins depleted from mock CCVs and thus to candidate CCV proteins. Proteins were ranked according to this ratio in descending order.
A plot of normalized ratio over rank (Fig. 3) reveals that the majority of proteins did not change substantially between mock and control CCVs; in fact, 378 proteins have ratios between 0.5 and 1.5 (i.e., changes in relative abundance of 50% or less). However, ∼10% of the proteins were significantly depleted from the mock CCVs (ratios between ∼2.0 and 12.2; Fig. 3, left). Although the differences between control and mock CCVs are often not as great as predicted by our other data, the curve has the expected biphasic shape, and the figure of 10% depleted proteins agrees well with our 1D and 2D gels.
Figure 3.
iTRAQ quantification of control and mock CCV fractions. Fractions were tagged with iTRAQ reagents, and the relative abundance of identified proteins in control and mock CCVs was ascertained by quantifying the iTRAQ tags. The 522 proteins were sorted by decreasing ratios of control/mock CCVs. The figure shows a normalized plot of ratio over rank. Error bars indicate standard deviations from means of two technical replicates.
To determine a useful (albeit arbitrary) cutoff, we chose the lowest ranking (i.e., least depleted) AP-1 or AP-2 subunit, as APs are known CCV proteins that show clear depletion from mock CCVs. This was the α subunit of AP-2 (rank 53; ratio 2.03). By prediction, all proteins with higher ranks (i.e., ranks 1–52) should correspond to bona fide CCV proteins (Table I). Indeed, out of the top 53 proteins, about half are established CCV components, including clathrin heavy and light chains, subunits of the AP-1 and AP-2 complexes, cargo molecules, alternative adaptors, and other machinery. Of the remaining proteins, four could be classified as likely false positives, based on their known functions in RNA binding or metabolism. The others are novel candidate CCV components.
Table I.
Results of iTRAQ analysis
| Rank | Protein ID | Protein name | Previously identified? |
Established CCV protein? |
Mascot score | Peptides quantified |
Control/mock ratio | SD | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Top 53 proteins | ||||||||||||||||
| 1 | gi|30582397 | AP-1 σ1A | ✓ | ✓ | 96 | 1 | 12.24 | 5.13 | ||||||||
| 2 | gi|32451593 | CHC17 | ✓ | ✓ | 1826 | 30 | 6.27 | 1.46 | ||||||||
| 3 | gi|30582933 | DNase II (lysosomal) | new | new | 63 | 1 | 5.01 | 1.09 | ||||||||
| 4 | gi|1363934 | Dynamin-2 | new | ✓ | 569 | 11 | 4.83 | 1.31 | ||||||||
| 5 | gi|41016993 | EpsinR/Enthoprotin | ✓ | ✓ | 478 | 10 | 4.59 | 1.31 | ||||||||
| 6 | gi|17028334 | AP-1 μ1A | ✓ | ✓ | 379 | 8 | 4.59 | 1.08 | ||||||||
| 7 | gi|56203491 | TPD52L1 | new | new | 104 | 1 | 4.45 | 1.24 | ||||||||
| 8 | gi|12643391 | AP-1 γ | ✓ | ✓ | 540 | 18 | 4.37 | 1.05 | ||||||||
| 9 | gi|3294548 | Cathepsin Z (lysosomal) |
new | new | 102 | 2 | 4.10 | 1.19 | ||||||||
| 10 | gi|38570101 | Unknown protein (RAB-GAP domain) |
new | new | 45 | 3 | 4.05 | 0.98 | ||||||||
| 11 | gi|1335854 | Clathrin heavy chain homologue(CHC22) |
✓ | new | 558 | 1 | 3.84 | 1.09 | ||||||||
| 12 | gi|17402231 | Clathrin light chain a | ✓ | ✓ | 231 | 9 | 3.81 | 1.02 | ||||||||
| 13 | gi|2143260 | PI 3-kinase C2α | ✓ | ✓ | 822 | 11 | 3.62 | 1.03 | ||||||||
| 14 | gi|55662275 | CI-Mannose 6- phosphate receptor |
✓ | ✓ | 668 | 14 | 3.53 | 0.85 | ||||||||
| 15 | gi|70608172 | TPD52 | new | new | 160 | 1 | 3.52 | 0.85 | ||||||||
| 16 | gi|25090897 | CALM | ✓ | ✓ | 258 | 5 | 3.43 | 0.94 | ||||||||
| 17 | gi|21903712 | Carboxypeptidase D | ✓ | ✓ | 406 | 7 | 3.41 | 0.93 | ||||||||
| 18 | gi|116505 | Clathrin light chain b | ✓ | ✓ | 171 | 9 | 3.39 | 0.77 | ||||||||
| 19 | gi|2226273 | TGN46 | new | ✓ | 163 | 1 | 3.30 | 0.75 | ||||||||
| 20 | gi|33150596 | AP-1 σ1B | new | ✓ | 49 | 1 | 3.20 | 0.74 | ||||||||
| 21 | gi|55663531 | Syntaxin 6 | ✓ | new | 121 | 1 | 3.15 | 0.78 | ||||||||
| 22 | gi|13477131 | Sorting nexin 9 | new | ✓ | 260 | 6 | 3.07 | 0.79 | ||||||||
| 23 | gi|56205909 | Rab4A | ✓ | ✓ | 319 | 4 | 2.78 | 0.71 | ||||||||
| 24 | gi|62287155 | NECAP-1 | ✓ | ✓ | 66 | 1 | 2.75 | 0.86 | ||||||||
| 25 | gi|182516 | Ferritin light subunit | ✓ | ✓ | 131 | 3 | 2.67 | 0.57 | ||||||||
| 26 | gi|14043007 | AP-1 β1 | ✓ | ✓ | 1020 | 8 | 2.60 | 0.73 | ||||||||
| 27 | gi|2827434 | Sorting nexin 2 | new | new | 403 | 4 | 2.58 | 0.54 | ||||||||
| 28 | gi|9716092 | Sortilin | new | ✓ | 125 | 2 | 2.53 | 0.77 | ||||||||
| 29 | gi|66932909 | Pumilio 1 | RNA binding | — | 105 | 1 | 2.46 | 0.74 | ||||||||
| 30 | gi|8546849 | CD-Mannose 6-phosphate receptor |
✓ | ✓ | 152 | 5 | 2.44 | 0.63 | ||||||||
| 31 | gi|67477390 | Inositolpolyphosphate 5-phosphatase OCRL-1 |
new | ✓ | 581 | 8 | 2.40 | 0.61 | ||||||||
| 32 | gi|2337920 | Syntaxin 7 | ✓ | new | 220 | 3 | 2.38 | 0.61 | ||||||||
| 33 | gi|47125326 | Ferritin heavy chain | ✓ | ✓ | 84 | 5 | 2.37 | 0.77 | ||||||||
| 34 | gi|55958410 | Argininosuccinate synthetase | metabolism | — | 176 | 5 | 2.37 | 0.62 | ||||||||
| 35 | gi|8922952 | Cappuccino | new | new | 73 | 1 | 2.36 | 0.79 | ||||||||
| 36 | gi|47086495 | BLOC-1, subunit 3 | new | new | 96 | 3 | 2.35 | 0.58 | ||||||||
| 37 | gi|56205243 | Auxilin | ✓ | ✓ | 150 | 3 | 2.33 | 0.63 | ||||||||
| 38 | gi|7920147 | NSF | ✓ | ✓ | 63 | 1 | 2.33 | 0.64 | ||||||||
| 39 | gi|4557469 | AP-2 β2 | ✓ | ✓ | 831 | 7 | 2.32 | 0.66 | ||||||||
| 40 | gi|57162630 | AP-3 μ3A | new | new | 90 | 1 | 2.31 | 0.46 | ||||||||
| 41 | gi|15489411 | AP-2 μ2 | ✓ | ✓ | 121 | 4 | 2.31 | 0.67 | ||||||||
| 42 | gi|54695838 | Rab5C | ✓ | ✓ | 311 | 5 | 2.29 | 0.55 | ||||||||
| 43 | gi|62751805 | D19 | new | new | 88 | 2 | 2.29 | 0.47 | ||||||||
| 44 | gi|15214676 | Unknown protein (putative Rab interactor) |
new | new | 62 | 1 | 2.28 | 0.50 | ||||||||
| 45 | gi|4433649 | Syntaxin 8 | ✓ | new | 122 | 2 | 2.21 | 0.52 | ||||||||
| 46 | gi|13543973 | IMP (inosine monophosphate dehydrogenase) 2 | metabolism | — | 510 | 15 | 2.17 | 0.55 | ||||||||
| 47 | gi|9557955 | Sorting nexin 5 | new | new | 206 | 3 | 2.15 | 0.61 | ||||||||
| 48 | gi|17375734 | cyclin G–associated kinase/auxilin2 |
✓ | ✓ | 288 | 3 | 2.12 | 0.64 | ||||||||
| 49 | gi|15214696 | Glucosamine (N-acetyl)- 6-sulfatase (lysosomal) |
new | new | 162 | 4 | 2.11 | 0.58 | ||||||||
| 50 | gi|13431563 | Huntingtin-interacting protein 1 related (Hip1R) |
✓ | ✓ | 210 | 1 | 2.09 | 0.81 | ||||||||
| 51 | gi|1184699 | tyrosyl-tRNA synthetase | RNA-binding | — | 47 | 1 | 2.06 | 0.61 | ||||||||
| 52 | gi|30582345 | Snapin | new | new | 48 | 1 | 2.05 | 0.53 | ||||||||
| 53 | gi|19913414 | AP-2 α | ✓ | ✓ | 388 | 7 | 2.03 | 0.46 | ||||||||
| Selected additional proteins | ||||||||||||||||
| 88 | gi|23512245 | AP-3 β3A | new | new | 438 | 9 | 1.47 | 0.34 | ||||||||
| 94 | gi|12654697 | Transferrin receptor | ✓ | ✓ | 953 | 20 | 1.46 | 0.38 | ||||||||
| 114 | gi|32880009 | AP-3 σ3A | new | new | 57 | 1 | 1.33 | 0.46 | ||||||||
| 155 | gi|5442368 | AP-4 σ4 | — | — | 76 | 1 | 1.20 | 0.29 | ||||||||
| 192 | gi|37675283 | AP-4 ɛ | — | — | 90 | 3 | 1.11 | 0.22 | ||||||||
| 196 | gi|6580116 | Glycogen-debranching enzyme |
metabolism | — | 1898 | 98 | 1.10 | 0.26 | ||||||||
| 197 | gi|56204938 | AP-4 β4 | — | — | 69 | 2 | 1.10 | 0.26 | ||||||||
| 252 | gi|976227 | 26S proteasome subunit p45 |
proteasome | — | 905 | 28 | 1.01 | 0.24 | ||||||||
| 270 | gi|31108 | EF-2 | ribosome | — | 544 | 13 | 0.98 | 0.22 | ||||||||
| 279 | gi|16306837 | TCP1, subunit 5 (ɛ) | TCP complex | — | 689 | 26 | 0.95 | 0.21 | ||||||||
| 520 | gi|15990478 | Major vault protein (MVP) |
Vault complex | — | 838 | 20 | 0.15 | 0.03 | ||||||||
Control and mock CCV fractions were prepared as in Fig. 1 and analyzed by iTRAQ (Fig. 3). Identified proteins were ranked based on their relative abundance in control and mock CCVs; a high rank corresponds to a high ratio of control/mock CCVs and, thus, to candidate CCV proteins. Shown are the top-ranking 53 proteins (ratio >2.0), some representative contaminants (ratios near 1), and some other proteins of interest. The fourth column indicates whether a protein had been previously identified by CCV proteomics in Blondeau et al. (2004) or Girard et al. (2005). The Mascot score reflects the confidence with which a protein was identified; a score >35 indicates >95% confidence of identification. “Peptides quantified” indicates how many iTRAQ-labelled peptides were used for the quantification. “Control/mock ratio” corresponds to the relative abundance of a protein in control and mock CCVs; the ratio is the mean of two technical replicates. The proteins can be found in the National Center for Biotechnology Information database. Likely false positives among high ranking proteins are in italics. See Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200607164/DC1) for a complete list of the proteins identified.
Evaluation of the comparative proteomics approach
Because all of the known CCV-associated proteins that we identified in our proteomics analyses were significantly depleted in the mock CCV fraction, it is likely that other proteins that behave in a similar manner are also genuinely CCV associated. Nevertheless, there are at least two possible sources of false positives that need to be considered. First, expression levels of other proteins may be decreased by clathrin knockdown, causing them to appear depleted from the mock CCVs. Although none of the proteins that we analyzed by Western blotting were significantly depleted from whole cell homogenates, this may account for the four obvious false positives identified by iTRAQ. Second, we cannot exclude the possibility that blocking CCV-mediated trafficking may interfere with other pathways. For example, if clathrin knockdown prevents the formation of another type of transport intermediate, which is normally also present in the CCV fraction, it will appear depleted from mock CCVs. However, there is currently no evidence to support this possibility.
The iTRAQ analysis may also have produced a few false negatives. The control/mock ratios of known CCV proteins are generally lower than we expected from our other data, suggesting that there may be additional genuine CCV components among proteins with ranks between 54 and ∼100. However, because of overlap with background proteins, such proteins were only included in the list of identified CCV proteins (Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200607164/DC1) when they showed clear depletion from mock CCVs by Western blotting and/or 2D DIGE.
How do our data compare with the two other proteomic analyses of CCVs? The studies on brain and liver both focused on optimizing the CCV preparation method to minimize the number of false positive identifications (Blondeau et al., 2004; Girard et al., 2005). In both cases, the yield and purity of the CCV fractions were higher than we achieved using HeLa cells, but because neither study used unbiased criteria to identify genuine CCV proteins, it is difficult to compare their results quantitatively with ours. Nevertheless, as expected, the CCVs from all three sources show an overlapping composition, including clathrin heavy and light chains, adaptors, and abundant accessory factors such as NSF and cyclin G–associated kinase (Traub, 2005). Some of the promising candidate CCV proteins identified in the two previous studies are not depleted from our mock CCV fraction, including myoferlin, various annexins, and Vac14, suggesting that they may in fact be contaminants (Table S1). There are also several hits from the two previous studies that are clearly physiologically relevant, such as epsin, Eps15, Numb, and the asialoglycoprotein receptor, which we did not find in the present study, either because they are cell type specific or because they are associated with endocytic CCVs, which are underrepresented in the HeLa cell CCV fraction.
AP complexes
One notable feature of the HeLa cell CCV preparation is the high AP-1/AP-2 ratio. AP-1 is more enriched over whole cell homogenate than AP-2, it is more abundant than AP-2, and it is more strongly depleted from the mock CCV fraction than AP-2. There are probably at least two reasons for this phenomenon. First, although HeLa cells appear to contain at least as many plasma membrane as intracellular CCVs (based on immunofluorescence and electron microscopy; Motley et al., 2003), we have found that most of the plasma membrane–associated clathrin remains tenaciously attached to cell remnants upon homogenization (Hirst et al., 2004). Thus, it is likely that the majority of the AP-2–containing CCVs are discarded in our first low-speed centrifugation step. Second, many of the smooth vesicles that contaminate our CCV fraction are derived from the plasma membrane (e.g., the transferrin receptor is a major component of our mock CCVs), and because AP-2 is recruited onto the plasma membrane even in the absence of clathrin (Motley et al., 2003), it is likely to be associated with these contaminating vesicles. Together, these observations probably explain why AP-2 subunits cluster near the lower end of the top-ranking proteins in our iTRAQ analysis (Table I, ranks 39, 41, and 53), whereas subunits of the AP-1 complex are clustered among the highest-ranking proteins (ranks 1, 6, 8, 20, and 26), showing similar ratios to clathrin heavy and light chains (ranks 2, 12, and 18).
This differential depletion of AP-1 and AP-2 can be exploited to interpret the results of the iTRAQ analysis and to assign other proteins in Table I to either the AP-1 or the AP-2 pathway. The highest-ranking component of AP-2 is β2 (rank 39). Thus, as a rough approximation, proteins with ranks higher than 39 should be on the AP-1 pathway, and those that fall betweens ranks 39 and 53 should be on the AP-2 pathway. Supporting this notion, essentially all proteins known to function with AP-1 that were identified here are among the top 35 ranking proteins, including epsinR, PI 3-kinase C2α, and both mannose 6-phosphate receptors. The results are less clear-cut for the AP-2 cluster because it partially overlaps with the AP-3 cluster (see below).
The clustering approach predicts that other high-ranking proteins in Table I, such as syntaxins 6 and 7, are also associated with TGN/endosomal CCVs. It suggests that some proteins with known functions in endocytosis, such as dynamin-2 and Snx9, may also function in intracellular CCV trafficking, a possibility that is supported by immunofluorescence studies (Soulet et al., 2005). The presence of two lysosomal enzymes, DNase II and cathepsin Z, in the AP-1 cluster may provide some insights into the directionality of AP-1–mediated trafficking. Lysosomal enzymes travel from the TGN to lysosomes via endosomal intermediates and are not recycled. Because DNase II and cathepsin Z cocluster with AP-1, our results support a role for AP-1 in forward transport.
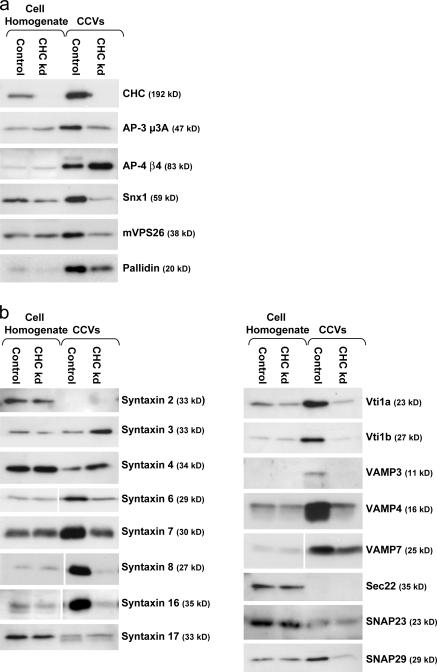
AP-1 and AP-2 belong to a family that includes two other complexes, AP-3 and AP-4, neither of which are enriched in CCVs purified from brain (Simpson et al., 1996; Hirst et al., 1999), although several studies suggest that AP-3 participates in clathrin-mediated trafficking (Dell'Angelica et al., 1998; Peden et al., 2002, 2003). Subunits of both AP-3 and AP-4 complexes were identified in the iTRAQ analysis, with consistently higher ratios for the AP-3 subunits than for the AP-4 subunits (Table I). Western blots of control and mock CCVs show that both complexes are enriched in CCV fractions, but that only AP-3 is depleted from the mock CCVs (Fig. 4 a). Thus, it appears that AP-3 indeed plays a role in CCV trafficking, whereas AP-4 is a contaminant. This example illustrates that neither the presence nor the enrichment of a protein in a CCV preparation is sufficient evidence for its specific association with CCVs and further highlights the discriminating power of our comparative proteomics approach.
Figure 4.
Western blots of selected proteins. Blots of control and mock CCV fractions were probed with antibodies against AP-3, AP-4, retromer, and BLOC-1 subunits (a) or with various SNARE antibodies (b). Loading was as in Fig. 1.
Other CCV components
Subunits of two other protein complexes involved in membrane traffic, retromer and BLOC-1, were also identified as candidate CCV components. Western blots probed with antibodies against the retromer-associated sorting nexin Snx1 and the retromer subunit mVPS26 show similar levels of enrichment in the CCV fraction over the whole cell homogenate and similar levels of depletion from the mock CCV fraction, confirming the iTRAQ data (Fig. 4 a). BLOC-1 consists of at least eight subunits (Dell'Angelica, 2004), seven of which were found in the iTRAQ analysis, with ratios ranging from 1.25 to 2.36. Western blots probed with an antibody against the eighth subunit, pallidin, show that this protein is also moderately depleted from mock CCVs (Fig. 4 a). Thus, all eight of the known BLOC-1 subunits behave like bona fide CCV components. BLOC-1 has recently been shown to interact both physically and genetically with AP-3 (Di Pietro et al., 2006), and the similar ratios of BLOC-1 and AP-3 subunits in the iTRAQ analysis suggest that BLOC-1 may depend on AP-3 for its association with CCVs.
Among the membrane proteins that showed more than twofold depletion in the mock CCV fraction are three post-Golgi SNAREs: syntaxins 6, 7, and 8. SNAREs are essential for the fusion of vesicles with their target organelles, and they are frequently used as markers to define membrane compartments, but little is known about how they traffic through the secretory and endocytic pathways. Peden et al. (2001) showed by Western blotting that several post-Golgi SNAREs are highly enriched in CCVs from rat liver, and the identification of three syntaxins in our iTRAQ analysis suggested that many SNAREs may use clathrin-mediated transport. To test this hypothesis, Western blots of control and mock CCV fractions were screened with a panel of 16 SNARE antibodies (Fig. 4 b). SNAREs involved in traffic between the ER and Golgi (syntaxin 17 and Sec22) or in exocytosis (syntaxins 2, 3, and 4 and SNAP23) showed little or no enrichment in the CCV fraction and were unaffected by CHC knockdown. However, all of the SNARES that have been localized to the TGN, endosomes, and/or lysosomes, including syntaxins 6, 7, 8, and 16; vti1 a and b; VAMPs 3, 4, and 7; and SNAP29, were found to be enriched in control CCVs and depleted from mock CCVs.
One of the most strongly depleted proteins in our iTRAQ analysis of mock CCVs is CHC22, a homologue of conventional CHC that is predominantly expressed in muscle (Liu et al., 2001). The two CHC isoforms are 85% identical, but it is unlikely that CHC22 would be targeted by our 21-base siRNA, because its mRNA contains eight mismatches. Although it has been proposed that CHC22 is not associated with CCVs (Liu et al., 2001), our observations suggest that it is in fact a CCV component.
The proteomic analysis also identified five novel proteins of unknown function in the CCVs. Features of these proteins suggest that they too are bona fide CCV components. Two of the proteins, gi38570101 and gi15214676, have domains predicted to interact with rabs, which are known regulators of membrane traffic. Another protein, D19, is an SH3 domain–containing protein related to intersectin, a protein involved in endocytosis. The other two proteins, TPD52L1 and TPD52, are members of the D52 family of tumor proteins, which has been implicated in the secretory and endocytic pathways (Boutros et al., 2004). Thus, all of these proteins may perform important regulatory roles in clathrin-mediated trafficking, and we are currently investigating their functions.
Outlook: comparative subcellular proteomics
We have introduced an unbiased criterion to identify CCV proteins. This criterion is easily applicable to novel proteins, even outside a proteomic investigation. Simple Western blotting can be used to probe control and mock CCVs for a protein of interest to determine whether it is a bona fide CCV component.
Apart from identifying novel CCV proteins, we have developed a method whose scope can be expanded to investigate the function of individual CCV components. For example, one could knock down specific adaptors to see which cargo proteins are lost from CCVs (Hirst et al., 2004). Similarly, knockdown or overexpression of accessory proteins may affect CCV composition, and this could be monitored by iTRAQ. Such an approach is not limited to studies on CCVs. Other organelles could also be analyzed by 2D DIGE or iTRAQ, using either siRNA knockdowns or HRP-induced compartment ablation to generate mock organelle fractions. Thus, the comparative proteomics approach described in the present study could be used to gain insights not only into CCVs but into other parts of the cell as well.
Materials and methods
RNA interference and CCV preparations
siRNA duplexes against target cDNAs were purchased from Dharmacon. CHC knockdown was performed using a custom-made duplex described by Motley et al. (2003). HeLa cells were transfected using Oligofectamine (Invitrogen) in the presence of 10% fetal calf serum; the final concentration of siRNA was 10 nM. For effective knock down, two sequential transfections were performed on days 1 and 3. Experiments were performed 2 d after the second transfection (day 5). CCV-enriched fractions and mock CCVs were prepared from control and CHC knockdown HeLa cells as described by Hirst et al. (2004), with minor modifications to the protocol.
Gels, mass spectrometry, and iTRAQ
SDS-PAGE and Western blotting were performed according to standard protocols. Detailed descriptions of 2D DIGE analysis, iTRAQ analysis, mass spectrometric identification of proteins, and a list of antibodies used in this study are provided in the supplemental text and Table S3 (available at http://www.jcb.org/cgi/content/full/jcb.200607164/DC1).
Online supplemental material
The supplemental text provides a detailed explanation of iTRAQ and DIGE and a protocol for the preparation of CCVs. Table S1 provides a complete list of proteins identified and quantified by iTRAQ. Table S2 provides a summary of CCV proteins identified in this study by different methods. Table S3 provides a list of antibodies used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200607164/DC1.
Supplementary Material
Acknowledgments
We thank Matthew Seaman, Esteban Dell'Angelica, and Andrew Peden for antibodies, and Paul Luzio, John Kilmartin, and members of the Robinson laboratory for helpful discussions.
This work was supported by grants from the Wellcome Trust and the Medical Research Council.
Abbreviations used in this paper: AP, adaptor protein; CCV, clathrin-coated vesicle; CHC, clathrin heavy chain; DIGE, difference gel electrophoresis; iTRAQ, isobaric tags for relative and absolute quantification; LC, liquid chromatography; MS/MS, tandem mass spectrometry; NSF, N-ethylmaleimide–sensitive fusion protein.
References
- Andersen, J.S., and M. Mann. 2006. Organellar proteomics: turning inventories into insights. EMBO Rep. 7:874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondeau, F., B. Ritter, P.D. Allaire, S. Wasiak, M. Girard, N.K. Hussain, A. Angers, V. Legendre-Guillemin, L. Roy, D. Boismenu, et al. 2004. Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl. Acad. Sci. USA. 101:3833–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J.S., and P. Rojas. 2006. Retrograde transport from endosomes to the trans-Golgi network. Nat. Rev. Mol. Cell Biol. 7:568–579. [DOI] [PubMed] [Google Scholar]
- Borner, G.H.H., K.S. Lilley, T.J. Stevens, and P. Dupree. 2003. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132:568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros, R., S. Fanayan, M. Shehata, and J.A. Byrne. 2004. The tumor protein D52 family: many pieces, many puzzles. Biochem. Biophys. Res. Commun. 325:1115–1121. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C. 2004. The building BLOC(k)s of lysosomes and related organelles. Curr. Opin. Cell Biol. 16:458–464. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., J. Klumperman, W. Stoorvogel, and J.S. Bonifacino. 1998. Association of the AP-3 adaptor complex with clathrin. Science. 280:431–434. [DOI] [PubMed] [Google Scholar]
- Di Pietro, S.M., J.M. Falcon-Perez, D. Tenza, S.R. Setty, M.S. Marks, G. Raposo, and E.C. Dell'angelica. 2006. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol. Biol. Cell. 17:4027–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley, T.P., S. Hester, I.P. Shadforth, J. Runions, T. Weimar, S.L. Hanton, J.L. Griffin, C. Bessant, F. Brandizzi, C. Hawes, et al. 2006. Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA. 103:6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote, C., and S.F. Nothwehr. 2006. The clathrin adaptor complex 1 directly binds to a sorting signal in Ste13p to reduce the rate of its trafficking to the late endosome of yeast. J. Cell Biol. 173:615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard, M., P.D. Allaire, P.S. McPherson, and F. Blondeau. 2005. Non-stoichiometric relationship between clathrin heavy and light chains revealed by quantitative comparative proteomics of clathrin-coated vesicles from brain and liver. Mol. Cell. Proteomics. 4:1145–1154. [DOI] [PubMed] [Google Scholar]
- Hirst, J., N.A. Bright, B. Rous, and M.S. Robinson. 1999. Characterization of a fourth adaptor-related protein complex. Mol. Biol. Cell. 10:2787–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst, J., S.E. Miller, M.J. Taylor, G.F. von Mollard, and M.S. Robinson. 2004. EpsinR is an adaptor for the SNARE protein Vti1b. Mol. Biol. Cell. 15:5593–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.H., M.C. Towler, E. Chen, C.Y. Chen, W. Song, G. Apodaca, and F.M. Brodsky. 2001. A novel clathrin homolog that co-distributes with cytoskeletal components functions in the trans-Golgi network. EMBO J. 20:272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, W.H., and J.R. Yates III. 2000. Proteomic tools for cell biology. Traffic. 1:747–754. [DOI] [PubMed] [Google Scholar]
- Motley, A., N.A. Bright, M.N.J. Seaman, and M.S. Robinson. 2003. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162:909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden, A.A., G.Y. Park, and R.H. Scheller. 2001. The di-leucine motif of vesicle-associated membrane protein 4 is required for its localization and AP-1 binding. J. Biol. Chem. 276:49183–49187. [DOI] [PubMed] [Google Scholar]
- Peden, A.A., R.E. Rudge, W.W.Y. Lui, and M.S. Robinson. 2002. Assembly and function of AP-3 complexes in cells expressing mutant subunits. J. Cell Biol. 156:327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden, A.A., V. Oorschot, B.A. Hesser, C.D. Austin, R.H. Scheller, and J. Klumperman. 2004. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 164:1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.S. 2004. Adaptable adaptors for coated vesicles. Trends Cell Biol. 14:167–174. [DOI] [PubMed] [Google Scholar]
- Ross, P.L., Y.L.N. Huang, J.N. Marchese, B. Williamson, K. Parker, S. Hattan, N. Khainovski, S. Pillai, S. Dey, S. Daniels, et al. 2004. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 3:1154–1169. [DOI] [PubMed] [Google Scholar]
- Seaman, M.N. 2004. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J. Cell Biol. 165:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, F., M.A. West, N.A. Bright, L.S. Newman, R. Darnell, and M.S. Robinson. 1996. A novel adaptor-related protein complex. J. Cell Biol. 133:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulet, F., D. Yarar, M. Leonard, and S.L. Schmid. 2005. SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol. Biol. Cell. 16:2058–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub, L.M. 2005. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim. Biophys. Acta. 1744:415–437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.