Abstract
Many cargoes destined for nuclear import carry nuclear localization signals that are recognized by karyopherins (Kaps). We present methods to quantitate import rates and measure Kap and cargo concentrations in single yeast cells in vivo, providing new insights into import kinetics. By systematically manipulating the amounts, types, and affinities of Kaps and cargos, we show that import rates in vivo are simply governed by the concentrations of Kaps and their cargo and the affinity between them. These rates fit to a straightforward pump–leak model for the import process. Unexpectedly, we deduced that the main limiting factor for import is the poor ability of Kaps and cargos to find each other in the cytoplasm in a background of overwhelming nonspecific competition, rather than other more obvious candidates such as the nuclear pore complex and Ran. It is likely that most of every import round is taken up by Kaps and nuclear localization signals sampling other cytoplasmic proteins as they locate each other in the cytoplasm.
Introduction
The presence of a nucleus requires that a diverse set of macromolecules must be efficiently transported across the nuclear envelope (NE). The sole mediators of this exchange are nuclear pore complexes (NPCs), which span pores in the NE to connect the nuclear and cytoplasmic compartments. Transport of macromolecules across the NPC depends on dynamic interactions between transport cargoes, their cognate-soluble transport factors, and NPCs (Macara, 2001). Many transport factors belong to a related family collectively termed karyopherins (Kaps; also called importins, exportins, and transportins). Kaps bind to specific import (NLS) or export (NES) signals in their cargoes (Mosammaparast and Pemberton, 2004). Unexpectedly, few Kaps are essential, as there appears to be a significant degree of functional redundancy amongst family members (Wozniak et al., 1998). On import, a Kap–NLS cargo complex diffuses from the cytoplasm to the NPC; transient binding and unbinding with a particular set of NPC proteins (FG-Nups) is central to all proposed models for how transport complexes traverse the NPC (Rout et al., 2003; Suntharalingam and Wente, 2003).
Once in the nucleus, import Kap–cargo complexes are dissociated by RanGTP. Ran is maintained in its GTP-bound form in the nucleus by a nuclear GDP/GTP exchange factor, RanGEF. Conversely, in the cytoplasm, RanGTP is hydrolyzed to RanGDP by a cytoplasmically restricted GTPase-activating protein, RanGAP. In this way, cells maintain Ran in its GTP-bound form in the nucleus and limit its GDP-bound form to the cytoplasm. This RanGTP/RanGDP gradient is an essential indicator for the directionality of nucleocytoplasmic transport, and possibly the only directional cue for many Kap-mediated transport pathways. With the cargo delivered, Kaps and Ran are then recycled via a nested series of reactions and translocations (for review see Macara, 2001).
Most kinetic studies of nuclear transport have been performed in vitro using permeabilized cell systems. More recently, the interplay between Kaps, NLS-bearing cargoes, and Ran has been modeled in silico from data collected in vivo from mammalian cells. Expanding on an earlier study (Smith et al., 2002), Riddick and Macara (2005) fitted import rate data using a systems analysis including >60 separate parameters. Their findings suggest that the maximum flux of the NPCs in the cell was ∼500 molecules/NPC/s (at least for the flux of Ran across the NE), and that the NPC is not the rate-limiting factor for nuclear transport.
The yeast Saccharomyces cerevisiae represents an excellent organism with which to examine the mechanism of nuclear translocation, as it is possible to make systematic alterations in components of its nucleocytoplasmic transport machinery in vivo. However, we have been limited in our ability to study nucleocytoplasmic transport quantitatively in yeast by two factors: the lack of a method to accurately quantitate import rates in single living yeast cells, and the inability to accurately quantitate the concentrations of key players in the import reaction in those individual cells. Therefore, we defined a model import pathway (Kap123p-mediated import of ribosomal proteins), and devised high-resolution quantitative single-cell assays to measure the effectiveness of that import pathway. We determined the import rate of Kap123p (as well as other Kaps), as a function of intracellular concentrations of Kap123p and its cargo. Our results indicate that simple concentration and binding-constant relationships between Kap123p, its cargo, and NPCs determine the rate of import; surprisingly, it is the inefficient formation of the Kap–cargo complex in the cytoplasm, rather than limitations in the NPC or the Ran gradient, that restricts import rates in vivo.
Results
A quantitative assay of nuclear import in yeast
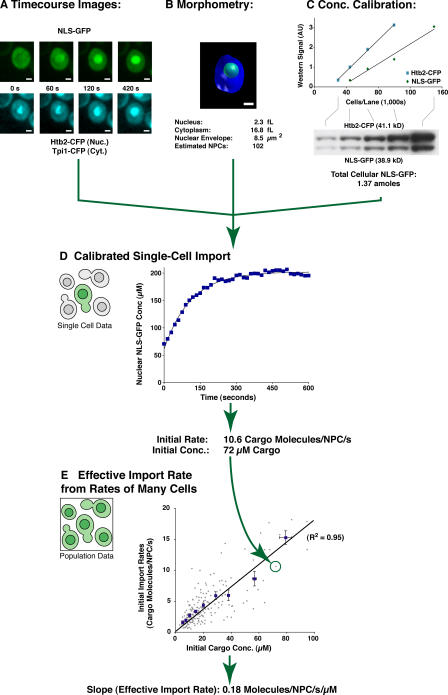
To quantitate Kap-mediated import, we developed a nuclear import assay (based, in part, on a previous method; Shulga et al., 1996) that facilitates rapid, semiautomated cell-by-cell quantitation of import with high spatial and temporal resolution (Fig. 1; for a detailed description of these methods see Leslie et al., 2006). Our model cargoes were NLSs fused to either GFP or a GFP carrying a C-terminal copy of a single PrA repeat. These fusion proteins were small enough to diffuse rapidly across the NPC; hence, in the absence of active import, they equilibrate between the nucleus and cytoplasm within minutes (Shulga et al., 1996). Transport was stopped by the addition of metabolic energy poisons, which destroy the RanGTP/GDP gradient (Schwoebel et al., 2002). Re-import of NLS-GFP was observed seconds after a sample of cells had been washed free of poison and resuspended in glucose-containing media on a microscope slide, allowing the Ran gradient to reform. Import assays were performed in a strain background that contained CFP-tagged Htb2p (a histone) and CFP-tagged Tpi1p (a glycolysis enzyme), which demarked the nucleoplasm and cytoplasm, respectively, whereas Htb2-CFPp also served as an internal calibrant for NLS-GFP concentration. An automated spinning-disk microscopy system was used to acquire confocal images at 15-s intervals over a 10-min time course, by which time the NLS-GFP had fully returned to its steady-state distribution. At each time point, images were taken at three focal points throughout the cells of both the GFP and CFP signals (Fig. 1 A).
Figure 1.
A quantitative measurement of nuclear import rates in vivo. (A) Fluorescence images of a single cell showing NLS-GFP being re-imported over time and of the CFP-tagged marker proteins. (B) Subcellular morphometry obtained from 3D confocal images of the cell in A. (C) Quantitative Western blots of the importing cell population from which the cell in A was taken, which allowed us to calculate the amount of fluorescent protein in that cell. (D) These data were combined to calculate the import curve of nuclear NLS-GFP concentration over time for the cell in A. Import curves for many such importing cells in the population were calculated. (E) Each cell's import rate was plotted against its initial NLS-GFP concentration (gray dots). This population data was split into several statistical bins of increasing cargo concentration, and the mean import rate and concentration in each bin was calculated (blue squares with error bars of the SD of the mean for each mean). A linear regression analysis of this data revealed a strong linear relationship between import rate and cargo concentration, the slope of which we term effective import rate (±95% confidence limits of the slope-fit coefficient). Bars, 1 μm.
The NLS-GFP concentration in a population of importing cells was calibrated using quantitative Western blotting on a sample from this population, which compared the abundance of NLS-GFP to that of the internal standard, Htb2-CFPp (Fig. 1 C). Once the quantitative blotting had determined the mean NLS-GFP molar amount for cells in an import assay and correlated this to the mean cellular NLS-GFP fluorescence in that same assay, we could then use a given cell's actual fluorescence to calculate the abundance of NLS-GFP cargo in that cell (Leslie et al., 2006). We estimated the probable error of these single-cell cargo abundance values to be ±30%.
We needed to measure the size and shape of the assayed cells because a cell with a smaller nucleus would appear to import faster (simply because it has less volume to fill), just as a cell with a greater number of NPCs could potentially import cargo faster. To measure each cell's volumetric statistics, a confocal image series was acquired through the cells, immediately after the time course. From these data, nuclear and cytoplasmic volumes could be directly measured, whereas the area of the nuclear envelope was interpolated from the 3D data using standard isosurface location algorithms (Fig. 1 B). From similar volumetric measurements of commercially prepared spherical beads (2.5 μm diam), we estimated the uncertainty in our volume measurements to be ±10% and in our surface area measurements to be ±6%. Because it has been shown in yeast that the density of NPCs is a relatively constant 12 NPCs/μm2 throughout the cell cycle (Winey et al., 1997), we could convert the surface area of the nuclear envelope to an estimate of the number of NPCs in each cell.
These fluorescence microscopy, concentration calibration, and cellular morphometric measurements were combined to give plots of cargo import over time in single cells, which fit single exponential relationships (Fig. 1 D; average R2 for all single-cell fits, 0.95). The initial rate of cargo import (i.e., import rate at t = 0, where net passive diffusion of naked NLS-GFP across the NPC is negligible) could be estimated in units of cargo molecules/NPC/s for each and every cell assayed. Each time course assayed ∼30 cells within a single microscope field, and multiple assays were repeated until the data from ∼100 cells had been collected.
Import rates were spread over wide ranges because the NLS-GFP expression constructs were cloned in multiple copy vectors, conferring a random number of copies of the gene to each cell with a commensurate random expression level of the fusion protein. We used this variability to examine the relationship between cargo concentration and import rate (Fig. 1 E). Each cell's initial import rate measurement and initial cytoplasmic NLS-GFP concentration is displayed as a single point in a scatter plot (gray dots). To better visualize trends in this population dataset, the cargo concentration range was split into several statistical bins, and the mean import rate and cargo concentration within each bin was calculated (blue squares; error bars are the SD of the mean). This moving-average analysis determined that initial import rates of NLS-GFP cargoes followed a simple linear relationship in respect to the available cytoplasmic concentrations of the fluorescent cargo (Fig. 1 E). We term the slope of this line an effective import rate, being a measure of how quickly a given cargo is imported by its available transport pathways, in units of cargo molecules per NPC per second per micromolar concentration of cargo.
Import of the model ribosomal import cargo Rpl25NLS-GFP-PrAp is largely dependent on Kap123p
We began our investigations with one particular model transport pathway, that of Kap123p, chosen for several reasons. First, it is the most abundant yeast Kap (Rout et al., 1997; Ghaemmaghami et al., 2003; this study), mediating the import of ribosomal proteins (Rout et al., 1997; Sydorskyy et al., 2003), histones (Mosammaparast et al., 2002; Glowczewski et al., 2004), and the mRNA export factor Yra1p (Zenklusen et al., 2001). Second, deletion of Kap123p leads only to a mild growth defect, so we can work with Δkap123 yeast without deleterious effects on cell growth and nuclear transport as a whole. Third, it is partially redundant with other Kaps (Rout et al., 1997; Zenklusen et al., 2001; Sydorskyy et al., 2003), which accounts for its nonessential nature, and is a matter of considerable interest, being typical of the partial redundancy often observed between import pathways.
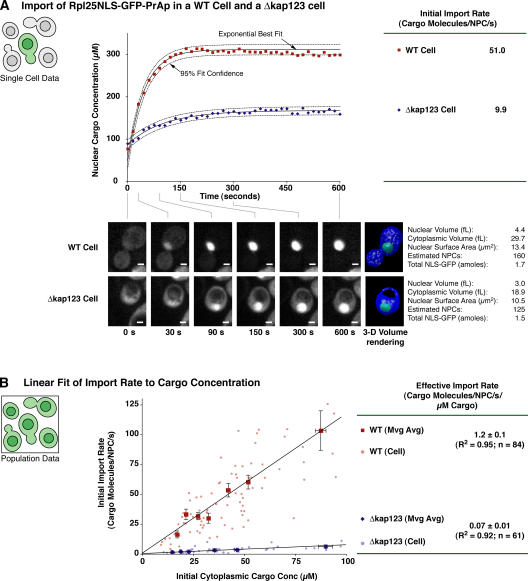
The NLS of the ribosomal protein Rpl25p (Schaap et al., 1991) has been used previously as the model cargo for Kap123p (Rout et al., 1997). To test whether this NLS is largely dependent on Kap123p, we used our import assay to quantitatively compare import of the Rpl25NLS between wild-type and Δkap123 cells (Fig. 2). In wild-type cells, these initial import rates ranged from 5 to 200 cargo molecules/NPC/s/cell, with a mean of 62, whereas these rates varied from 1 to 11 cargo molecules/NPC/s/cell with a significantly lower mean of 3 in Δkap123 cells. By normalizing for the different concentrations of NLS-GFP cargo in each cell (as previously described; Fig. 2 B), we determined that Rpl25NLS cargo was imported at a rate of 1.2 ± 0.1 cargo molecules/NPC/s/μM cargo in wild-type cells; a rate that was ∼17 times more rapid than in the absence of Kap123p. Thus, in wild-type cells, Rpl25NLS-GFP-PrAp is primarily imported by Kap123p, with only a residual 6% of its import proceeding through alternative pathways. Hence, in cells with Kap123p present, we could reasonably ignore this low residual transport component.
Figure 2.
Almost all the import of Rpl25NLS-GFP-PrAp is through Kap123p. (A) Import rate of Rpl25NLS-GFP-PrAp in Δkap123 and wild-type cells was quantitated using the import assay (Fig. 1; see Materials and methods). Import in one cell from each population, each containing similar quantities of Rpl25NLS-GFP-PrAp, are compared. The images, 3D renderings, and subsequent morphometric measurements of each single cell are shown below their individual calibrated import curves, each of which fit well to a single exponential relationship. The rates measured from the initial time points of import are shown to the right of the graph. (B) Many such import rate measurements were taken from single-cells in each population (dots). The population relationships between initial import rate and the initial concentration of Rpl25NLS-GFP-PrAp were analyzed by taking a moving average of the relational data over a wide range of initial concentrations (as in Fig. 1). The SEM for each moving average is expressed as error bars. The slope of a linear fit to these averaged data for each population provided effective import rate measurements (±95% confidence limits for the fit coefficients).
To test whether other NLSs that are recognized by Kap123p behave the same as Rpl25p, we compared the import of Rpl25NLS-GFPp with GFP carrying the NLSs from other Kap123p import cargoes. First, we identified numerous Kap123p-binding proteins, all of which were known to be targeted to the nucleus, and thus likely to contain NLSs (Supplemental materials and methods and Figs. S1 and S2, available at http://www.jcb.org/cgi/content/full/jcb.200608141/DC1); as expected, the majority of these were either ribosomal proteins or proteins involved in the ribosomal assembly process (Table S3; Rout et al., 1997; Sydorskyy et al., 2003). We constructed NLS-GFP fusion proteins from five proteins of varied function chosen amongst this Kap123p-binding group (Table I). The distributions of all but one NLS-GFP fusion protein was significantly affected by Kap123p deletion, but most were still strongly localized to the nucleus, indicating that a significant proportion of their import likely goes through pathways mediated by Kaps other than Kap123p (Fig. S3). One of the NLSs identified was that of another ribosomal protein (Rps1b), which was as profoundly mislocalized in the absence of Kap123p as Rpl25NLS-GFPp. Throughout this study, Rps1bNLS-GFPp transport was quantitated in addition to Rpl25NLS-GFPp and demonstrated essentially identical behavior in both wild-type and Δkap123 yeast (Fig. 3). Thus, Rpl25p (and likely other ribosomal proteins) is an appropriate model cargo for our studies.
Table I.
NLSs cloned as GFP fusion proteins for this study
| Protein | Function | NLS Sequence | Import Kapa | Reference |
|---|---|---|---|---|
| Rpl25p | Ribosome | MAPSAKATAAKKAVVKGTNGKKALKVRTSATFRLPKTLKLARAPK (aa 1–45) | Kap123p | Schaap et al., 1991 |
| Rps1bp | Ribosome | MAVGKNKRLSRGKKGLKKKVVDPFTRKEWFDIKAPSTFENRNVGKTLVNK(aa 1–50) | Kap123p | This study |
| Mak16p | Ribosome assembly | NKNSAKRRKKGTSAKTKRPKV(aa 267–287) | Unknown | This study |
| Nug1p | Ribosome assembly | MRVRKRQSRRTSTKLKEGIKKKASAHRKKEKKMAKKDVTWRSRSKKDPGIPSNFPYKAKIL (aa 1–61) | Kap123p/ Unknown | This study |
| Yil096cp | Ribosome assembly | MARKLKGKIGSKGLKGALLRHKAKVKLVRNIESKQKHELRKKNSSANNKTVKRNQEFQK- LNQGKVMP (aa 1–67) | Kap123p/ Unknown | This study |
| Yra1p | mRNA export | GSNKAGSNRARVGGTRGNGPRRVGKQVGSQRRSLPNRRGPIRKNTRAPPNAVARVAK (aa 14–70) | Kap121p/ Kap123p | Zenklusen et al., 2001; this study |
| Nab2p | mRNA binding | DNSQRFTQRGGGAVGKNRRGGRGGNRGGRNNNSTRFNPLAKALGMAGESN (aa 201–250) | Kap104p | Lee and Aitchison, 1999 |
| Pho4p | Phosphate regulation | ANKVTKNKSNSSPYLNKRRGKPGPDSATSLFELPDSVIPTPKPKPKPKQYPKVILP (aa 141–196) | Kap121p | Kaffman et al., 1998; this study |
According to references; also see Fig. 2, Fig. 3, and Fig. S3 (available at http://www.jcb.org/cgi/content/full/jcb.200608141/DC1).
Figure 3.
Rps1bNLS-GFPp is also a cargo of Kap123p and imported similarly to Rpl25NLS-GFPp. (A) Import of the Rps1bNLS-GFPp cargo was compared with that of Rpl25NLS-GFPp at steady-state in wild-type and Δkap123 cells. The average cellular N/C fluorescence ratio for each strain was calculated from images of several cells, with uncertainty values of the SD of the mean. Bars, 3 μm. (B) Import assays for Rps1bNLS-GFPp and Rpl25NLS-GFPp in wild-type cells were performed and analyzed as previously described (Fig. 1). Shown are the moving average plots for each importing population of initial rate against initial cargo concentration. Linear regression of this data produced trend lines with nearly identical slopes, from which the effective import rates of each cargo are stated to the right of the graph (±95% confidence limits for the fit coefficients).
Kap123p imports cargoes more rapidly than either Kap121p or Kap104p at their wild-type abundances
We compared the import of ribosomal NLS-GFP cargoes with that of similarly sized GFP cargoes carrying other published NLSs (Table I); the NLSs of Nab2p, which is carried predominantly by Kap104p (Lee and Aitchison, 1999), and of Pho4p, which is a cargo of Kap121p (Kaffman et al., 1998). Individual cell data from these two other cargoes also produced smooth import curves (Fig. 4 A), which again gave rise to simple linear relationships between import rate and cargo concentration (Fig. 4 B). These linear relationships suggested that the import system was never saturated with NLS-GFP cargo, even at cargo concentrations exceeding 100 μM, which is some 20-fold higher than the estimated natural cargo concentrations (Riddick and Macara, 2005). Import of Rpl25NLS by Kap123p was found to be significantly faster than that of the two other pathways, being ∼5-fold faster than Nab2NLS/Kap104p and ∼10-fold faster than Pho4NLS/Kap121p. Kap123p, thus, appears to be a significantly more effective importer than the other Kaps tested here.
Figure 4.
Quantitative comparison of the import rates of model cargoes for various karyopherins. (A) Import in three representative wild-type cells, each containing similar amounts of three different import cargoes: Rpl25NLS, Nab2NLS, or Pho4NLS. The Kap mainly responsible for the transport of each cargo is indicated. Import assays were analyzed as previously described, and the single-cell data is displayed as in Fig. 2. Bars, 1 μm. (B) As before, initial rate and concentration measurements were collected from many cells importing each cargo. Rate–concentration pairs for each cell in a population are indicated with gray dots. Moving averages were calculated along the complete concentration range of this population dataset (colored markers), and a best fit line was fitted to these averages with the indicated R 2 values. As discussed, the slope of this line provides effective import rate measurements (±95% confidence limits for the fit coefficients).
Kap123p and Kap121p share identical import kinetics and saturation points in respect to Rpl25NLS-YFP
Based on in vitro data, it has been suggested that a large variety of Kaps can replace Kap123p for import of ribosomal proteins (Jakel and Gorlich, 1998). Indeed, the results in Fig. 2 indicate that Kaps other than Kap123p are able to import Rpl25NLS-bearing cargoes, although not nearly as rapidly. Therefore, we tested whether other Kaps could substitute for Kap123p's import of Rpl25NLS cargo in vivo if expressed at comparable levels (Fig. 5). Rpl25NLS-GFPp–expressing Δkap123 cells were transformed with vectors overexpressing HA tagged versions of the four Kaps examined in this study. From quantitative Western blots (Rout et al., 2000; Cross et al., 2002) performed on strains containing genomically tagged versions of these Kaps, we measured the natural abundances of Kap95p, Kap104p, Kap121p, and Kap123p to be 60,000, 12,000, 18,000 and 100,000 copies/cell, respectively. These data are consistent with codon bias data indicating that Kap123p is the most highly expressed (i.e., abundant) of all Kaps. Although Western blot analysis demonstrated that all four HA-tagged Kaps were expressed at significantly higher levels than endogenous Kap123p, only overexpression of Kap121-HAp (1.8-fold above the natural Kap123p level) was able to compensate for loss of Kap123p in the import of Rpl25NLS-GFPp (Fig. 5), in agreement with previous studies showing that Kap123p was partially redundant with Kap121p (Rout et al., 1997; Zenklusen et al., 2001; Sydorskyy et al., 2003). Thus, our results confirm that, in vivo, ribosomal proteins are not cargos general to most Kaps, but, instead, are only imported by particular Kaps. This contrasts with results obtained for homologous mammalian Kaps in vitro (Jakel and Gorlich, 1998).
Figure 5.
Kap121p overexpression specifically compensated for import of Rpl25NLS-GFPp in the absence of Kap123p. Rpl25NLS-GFPp and HA-tagged versions of Kap95p, Kap104p, Kap121p, or Kap123p were expressed from separate overexpression plasmids in Δkap123 cells. Distributions of the Rpl25NLS-GFPp in response to overexpression of these Kaps are shown in the four rightmost images. Control images are of Rpl25NLS-GFPp in either wild-type or Δkap123 cells with an empty Kap expression vector. Bars, 3 μm. Quantitation of the expression levels of the HA-tagged Kaps from Western blots is stated below each image, relative to genomically expressed Kap123p levels.
As increasing amounts of Kap121p augmented the import rate of the model ribosomal cargo, we tested the hypothesis that Kap123p's effective import rate may be caused by its high cellular abundance. To do this, we manipulated the concentration of either Kap123p or Kap121p and measured how such concentration changes affected Rpl25NLS import. Either Kap123p or Kap121p was expressed as a CFP-tagged fusion protein in Δkap123 cells coexpressing Rpl25NLS-YFPp to allow simultaneous quantitation of both Kap and cargo in individual cells (Fig. 6 A). Because KAP-CFP and RPL25NLS-YFP expression plasmids were separate 2μ multiple-copy vectors, each cotransformed cell produced random amounts of the fluorescently labeled Kap and cargo, presumably proportionate to the amount of each plasmid they received. The Kap-CFPp and Rpl25NLS-YFPp quantities were found to vary independently of each other, resulting in cells with a wide range of either protein (Fig. 6 A). With two independent concentration variables, each potentially affecting import-rate, we needed to analyze larger numbers of cells than was practical with the quantitative import assay. Hence, we chose to measure import rate using the nuclear-to-cytoplasmic ratio (N/C) of cargo, as this value could be obtained relatively rapidly. We analyzed dependence of the steady-state N/C ratio of Rpl25NLS-YFPp on the cytoplasmic concentrations of either Kap121-CFPp or Kap123-CFPp.
Figure 6.
Measurement of karyopherin-dependent saturation kinetics in vivo. (A) Rpl25NLS-YFPp and either Kap121- or Kap123-CFPp were expressed from separate random-copy plasmids in Δkap123 yeast. Images of cells simultaneously expressing different quantities of each fluorescently tagged protein were acquired. Bars, 3 μm. Fluorescence was calibrated to concentration units as previously described for import assays (see Materials and methods). Thus, for several hundred individual cells, Kap-CFPp concentrations and Rpl25NLS-YFPp N/C ratios were measured. (B) The population concentration ranges of Kap123- and Kap121-CFPp were divided into small bins that each contained the data of many cells. The mean Rpl25NLS-YFPp N/C ratio and Kap-CFPp concentration of the data in each bin was calculated and plotted with error bars of the corresponding SDs of the mean for each average. The dashed lines illustrate the normal expression levels of each Kap in wild-type cells, calculated from genomically tagged Kap123-GFPp–expressing yeast using the same concentration calibration method previously mentioned.
Around the physiological concentration of Kap123p (∼5 μM), we observed an approximately linear relationship between its cytoplasmic concentration and the import of Rpl25NLS-YFPp (Fig. 6 B). At concentrations of Kap123-CFPp in excess of 15 μM, the N/C ratio of Rpl25NLS-YFPp plateaued with a half-maximal Kap123p concentration of ∼7 μM.
Interestingly, Kap121-CFPp displayed a relationship in respect to Rpl25NLS import that was practically indistinguishable from Kap123p-CFPp (Fig. 6 B). These highly similar import curves were observed despite findings that Kap123p and Kap121p prefer a significantly different subset of Nups to mediate their exchange across the NPC (Rout et al., 1997; Marelli et al., 1998; Seedorf et al., 1999; Denning et al., 2001).
A reduction in NLS-Kap affinity proportionally reduces import rate
To further examine the apparent equivalence of Kap121p or Kap123p's import kinetics when normalized for concentration, we examined the strength of binding to their common cargo, Rpl25NLS. If these Kaps recognize their common cargo with similar affinities, this would result in equivalent cytoplasmic concentrations of Kap–cargo import complex, which would be indicative of comparable import rates across the NPC.
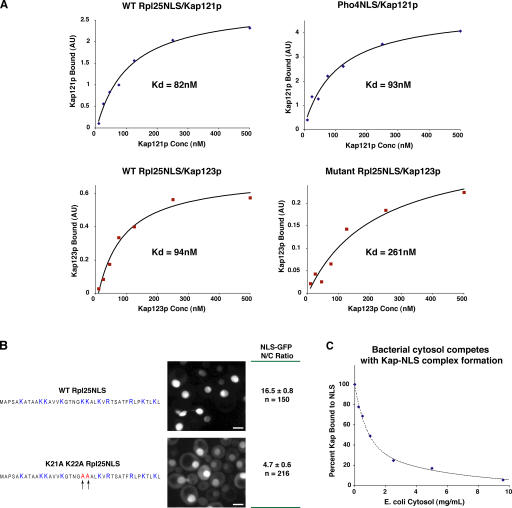
Hence, we measured the dissociation binding constants (K d) between Kaps and NLSs, using recombinant purified proteins and an in vitro binding assay (Fig. 7 and Table II; see Materials and methods). Binding of Kap121p and Kap123p to Rpl25NLS was found to be essentially indistinguishable, with K d values of 82 and 94 nM, respectively; this is consistent with both Kaps having similar import kinetics. We also measured binding between Pho4NLS and Kap121p to be similar to Rpl25NLS binding, at 93 nM, whereas Nab2NLS binding to Kap104p was significantly tighter at 17 nM. Cleavage of the GST from the Kaps had no significant effect on the measured K d values (unpublished data). Binding controls found no quantifiable binding between nonspecific Kap–cargo pairs, such as Pho4NLS-Kap123p (unpublished data).
Figure 7.
Measurement and manipulation of NLS-Kap dissociation constants. (A) K ds between NLSs and their Kap partners were determined by binding assays using purified, recombinantly expressed proteins (see Materials and methods). Results of four typical experiments are shown, between Kap121p and either Rpl25NLS or Pho4NLS, and between Kap123p and either wild-type or mutagenized Rpl25NLS. These binding curves quantitate the amount of Kap bound to the immobilized NLS-GFP-HIS, at the indicated concentrations of Kap, and were used to calculate the K d values of each interaction; stated K ds are the averages of at least three measurements. (B) The Rpl25NLS was mutated to alanines at lysines 21 and 22 using site directed mutagenesis. The effect that these mutations had on import was quantitated by comparing the steady-state N/C distributions of wild-type or mutant NLS-GFP fusion proteins (± the SD of the mean). Bars, 3 μm. The difference between these N/C ratios was statistically significant to a P value of <0.001 by the t test. (C) The competition of nonspecific cytosolic proteins with the formation of Kap–NLS complexes was measured by incubating 100 nM Kap121p and immobilized Pho4NLS with increasing concentrations of E. coli cytosol, using a method otherwise identical that used to measure K d. The amount of Kap bound to NLS was measured from gels and is plotted as a percentage of that bound with no cytosol present.
Table II.
Summary of measured import statistics
| Cargo | Rpl25NLS-GFP-PrA | Rpl25NLS-GFP-PrA | Pho4NLS-GFP-PrA | Nab2NLS-GFP-PrA |
|---|---|---|---|---|
| Cell type | Wild type | Δkap123 | Wild type | Wild type |
| Primary Kap | Kap123 | Kap121 | Kap121 | Kap104 |
| Mean nuclear volume (fL)a | 3.8 ± 1.4 | 3.2 ± 1.5 | 2.6 ± 1 | 2.9 ± 1.2 |
| Mean cytoplasmic volume (fL)a | 36 ± 17 | 35 ± 19 | 27 ± 12 | 30 ± 14 |
| Mean number of NPCsa | 142 ± 37 | 127 ± 42 | 110 ± 28 | 121 ± 33 |
| Initial NLS-GFP concentration range (μM)b | 7.2–109 | 6–104 | 3.6–68 | 4–58 |
| Mean Kap concentration (μM) | 5.3 | 1.0 | 1.0 | 0.6 |
| Effective import rate (cargo molecules NPC−1 s−1 μM−1 cargo)c | 1.2 ± 0.1 | 0.07 ± 0.01 | 0.10 ± 0.01 | 0.18 ± 0.02 |
| K d Kap–NLS (nM) | 94 | 82 | 93 | 17 |
| Kap half saturation (μM) | 7 | 7 | n.d. | n.d. |
| Estimated bulk cargo concentration (μM; 20 nM K d) | 5 | 5 | 5 | 0 |
| Estimated nonspecific competitors concentration (μM; 500 nM K d ) e | 1,000 | 1,000 | 1,000 | 1,000 |
| k' (cargo molecules μM−1 NPC−1 s−1)c | 44 ± 8 | 24 ± 8d | 39 ± 5 | 22 ± 4d |
| p (cargo molecules μM−1 NPC−1 s−1)c | 0.05 ± 0.01 | 0.06 ± 0.01 | 0.06 ± 0.01 | 0.064 ± 0.007 |
| k (cargo molecules NPC−1 s−1) | 300 | 300 | n.d. | n.d. |
| KS (μM) | 1 | 1 | n.d. | n.d. |
± SD.
5th to 95th interquartile range.
± 95% confidence interval for mean.
Highly significant difference from Rpl25NLS-GFP-PrA in wild-type cells.
See discussion.
To test whether these binding affinities were important in determining import rates, we manipulated the K d of the Rpl25NLS interaction with Kap123p and Kap121p. We mutagenized lysine residues 21 and 22 in the Rpl25NLS sequence to alanines, which reduced the binding strength of the NLS–Kap interaction with both Kap121p and Kap123 ∼2.8-fold, and consequently reduced the steady-state NLS-GFP N/C distributions by ∼3.5-fold (Fig. 7, A and B). This indicates that an increase in K d between Kap and cargo gives a proportionate decrease in cargo import.
Nonspecific binding effectively competes with NLSs for Kap binding
Certain observations from our studies led us to propose that binding between NLS-GFP cargo and Kaps is highly competitive with its cytoplasmic environment (see Discussion). To test this hypothesis, we examined the effect of adding bacterial cytosol, absent of any natural Kap cargoes, to the NLS–Kap interactions. Increasing concentrations of bacterial cytosol were found to effectively compete with the binding of Pho4NLS to Kap121p (Fig. 7 C). 1 mg/ml cytosol disrupted ∼50% of the specific Kap–NLS binding, and 10 mg/ml cytosol was sufficient to disrupt >90% of the import complexes. This result is unlikely to be a consequence of specific competition, because there are no NLSs in Escherichia coli, which is a prokaryote.
Import is fitted well by a pump–leak model
We showed that import fits well to single exponential relationships, the initial rates of which are linearly related to cytoplasmic cargo concentration (Fig. 4). Around physiological levels, the Kap concentration is linearly related to the import rates. However, at still higher concentrations of Kap the import rates ultimately saturate, following Michaelis–Menten–like curves (Fig. 6). In addition, we find that reducing the affinity of a cargo for its Kap proportionally reduced its import (Fig. 7 B). Considering these observations, nuclear import of our small NLS-GFP cargoes appears to be well represented by a pump–leak model, such as those used to describe a variety of biological pumps (e.g., ion channels). These pumps actively transport their cargo across a membrane, against a tendency for that cargo to leak back across the membrane through channels (Hodgkin and Keynes, 1955). We therefore fitted our import data to a simple pump–leak model (Fig. 8). From this model, we obtained quantitative estimates of the kinetic parameters of nuclear import in living yeast cells.
Figure 8.
Import through the NPC was modeled as a saturable first-order process balanced against a passive leak. (A) Diagram of the seven basic processes considered in our pump–leak model of import: (i) the passive leak of naked NLS-GFP cargo across the NPC; (ii) binding of NLS-GFP cargo to its cognate Kap; (iii) binding with unlabeled cognate NLS-cargoes of that same Kap; (iv and v) nonspecific binding of cytoplasmic proteins with Kap and NLS-GFP cargo; (vi) active import of Kap–cargo complexes by the NPC. Shown below the diagram are the mathematical relationships that were used to represent the component reactions in our pump–leak model. See text for explanation. (B) The kinetic, morphological and concentration parameters for a particular representative cell were substituted into the model's differential equation set and solved over time using standard ordinary differential equation solution algorithms. Thus, the model predicts cargo import to follow a single exponential curve (black line) that closely fits the measured single-cell import data (red diamonds). (C) The relationship between NLS-GFP cargo concentration and initial import rates was simulated at several concentrations of nonspecific competitors. At low concentrations of these competitors (light-colored curves), import is predicted to be more rapid, but saturate with increasing cargo concentrations. However, at a 1,000-μM concentration of nonspecific proteins, this simulated import relationship (black curve) is close to linear, which is similar to what we observed in vivo (Fig. 4).
In building our model, we assumed a constant Kap concentration in the cytoplasm, consistent with our observations that Kap distributions were not altered by even the highest obtainable Kap or cargo concentrations, and that Kap distributions did not change during our import assays (unpublished data). This assumption was further supported by the reported rapid translocation of naked Kaps (Ribbeck and Gorlich, 2001). Thus, the recycling system for Ran and Kaps, as previously modeled (Smith et al., 2002; Riddick and Macara, 2005), was regarded as an essential but imperturbable and very rapid background process. We also assumed instantaneous mixing in the nucleus and cytoplasm, justified by the small size of yeast cells and the estimated 100-ms intracellular diffusion rates (Wachsmuth et al., 2000; Smith et al., 2002), as compared with the several minutes each cargo required to return to steady-state (Fig. 4).
Fig. 8 A illustrates the components of our pump–leak model. In this model, NLS-GFP cargo binds its cognate Kap (Fig. 8 A, ii), competing with other unlabeled or “bulk” cargoes (iii) and nonspecific cytosolic proteins (iv and v). Because it was not possible to quantitate the degree of nonspecific competition (see Discussion), we assumed a value for this nonspecific competition that was common to all Kap–cargo pairs; namely, an interaction of 500 nM K d with 1-mM competitors. The magnitude of this competition is the minimum capable of simulating the linearity of the observed relationships (Fig. 8 C), but is physiologically reasonable, given the high concentration of total protein in the cytoplasm ( Zimmerman and Minton, 1993). Also, we cannot state whether this competition is indicative of binding to the NLS-GFP, to the Kap, or to both, but the resulting import complex concentrations will be the same in either case (unpublished data); hence, we ignore nonspecific protein binding to NLS-GFP (Fig. 8 A, v). We adopted an estimate for unlabeled NLS-bearing cargo (B) of 5 μM for each Kap (Riddick and Macara, 2005) and assumed a mean K d of 20 nM for these cargoes (Catimel et al., 2001), except for Kap104p, for which we assumed a competitor concentration of 0 μM, as it was observed to bind very few cargos (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200608141/DC1). All three binding processes (Fig. 8 A, ii–iv) were assumed to be at equilibrium. The three equilibrium binding expressions, of the general form K d = [A][B]/[AB], were solved for [AB] in terms of total Kap and binding protein concentrations (denoted by subscript “T” in their variable names), resulting in the quadratic equations shown in Fig. 8 A (bottom). Thus, the concentration of NLS-GFP–Kap import complexes (KC) was calculated by simultaneous solution of these equations, using the measured and estimated values for the dissociation constants and the total cytoplasmic abundances of each reactant, as listed in Table II.
Next, we considered the import processes. Import through NPCs and subsequent nuclear dissociation of import complexes was modeled as a saturable first-order process (Fig. 8 A, vi) because we had observed saturation of import by Kap that followed a Michaelis–Menten–like relationship (Fig. 6). Here, the kinetic constant k is analogous to the Michaelis–Menten's VMAX, and the saturation constant KS is analogous to KM. Because nonspecific proteins likely interact only transiently with Kaps, we assumed such complexes (KU) were not involved in import. Saturation of import was assumed to be caused by the total concentration of import-competent complexes (i.e., KB + KC), although at nonsaturating conditions (i.e., low Kap concentrations) we modeled import of complexes as a pseudolinear system. Pseudolinear rate constants (k') were calculated for each cell importing each model import cargo in Figs. 2 and 4 from their initial import rates and their expected intracellular NLS-GFP–Kap concentration at that time point (see values in Table II).
Lastly, we considered the relevant passive diffusion processes. Naked cargo is free to leak through NPCs via diffusion, where the rate of passive flux is proportional to the difference between the nuclear and cytoplasmic concentrations of cargo, with an NPC permeability constant of p (Fig. 8 A, i). At steady-state, cargo maintains a constant N/C ratio, which can be shown to be proportional to the ratio k'/p + 1 (Shulga et al., 2000). Having estimated k' as described in the previous paragraph, we calculated p, simply using the final N/C ratio for each cell in these import assays. Estimated k' and p values were combined with the measurements and estimates of the other cellular and biochemical parameters listed in Table II, creating a parameter set that was used to simulate import of NLS-GFP cargoes in single cells over time. These simulated curves closely fit the measured import data (Fig. 8 B).
Discussion
We have used our single-cell assay to answer several key questions concerning the nature of import, namely, whether different import pathways have inherently different rates; whether import by the NPC is saturable in vivo; why so many parallel partially redundant import pathways exist; and why high cargo concentrations are unable to saturate their import systems in the cytoplasm. We fitted the results of our studies to a simple pump–leak model of the import system to examine how these data combine to provide the overall measured import rates.
Import rate measurements in vivo in single yeast cells
Note that in contrast to methods where cells are permeabilized or microinjected, our approach can only measure import of model cargoes whose passive NPC permeability allows them to equilibrate in a few minutes after active import is inhibited. Moreover, it is not possible to add exogenous transport factors to these cells or to conduct these experiments using a minimal contingent of defined transport factors. Nevertheless, import rates in yeast determined with our poison-based assay (0.07–1.2 cargo molecules/s/NPC/μM; Table II) are in the same range as in vivo measurements for nuclear import in metazoan cells and rate measurements for single Xenopus laevis NPCs in vitro (∼0.5 cargo molecules/s/NPC/μM; Goldfarb et al., 1986; Keminer et al., 1999; Riddick and Macara, 2005). Most researchers also found no indication of saturation of import by cargo, although none of these measurements were made with NLS-GFP concentrations as high as in this study. Prior measurements in permeabilized cells yielded import rates that were significantly greater than those measured in vivo (28–50 cargo molecules/s/NPC/μM; Ribbeck and Gorlich, 2001; Yang et al., 2004), likely because many competitive factors discussed here had been washed away after disruption of the plasma membrane.
Saturability of the NPC/Ran system in vivo
Kap123p is the most abundant of all 15 yeast Kaps, and is approximately fivefold more abundant than Kap121p (at ∼1 μM). Kap121p is responsible for most residual import of Rpl25NLS-GFPp in Δkap123 cells. Thus, we tested the hypothesis that Kap123p's greater abundance than Kap121p is the cause of its greater import rate of ribosomal cargo. For these experiments, we followed import of cargo quantitatively by the steady-state cargo N/C ratio, which is approximately proportional to the import rate (Shulga et al., 2000). Our results demonstrated that cells expressing similar amounts of either Kap121-CFPp or Kap123-CFPp yielded equivalent N/C ratios of Rpl25NLS-YFPp cargo (Fig. 6). Notably, at their physiological concentrations, both Kap121p and Kap123p are well below their saturating concentrations, where changes in the amount of either Kap would be expected to proportionately affect the import rate of their cargo. Thus, we deduce that the high concentration of Kap123p is the key cause of its rapid import properties.
We also found that the import rates saturated to yield a maximal cargo N/C ratio at concentrations of Kaps higher than physiological (∼15 μM; Fig. 6). This Michaelis–Menten–like relationship is likely caused by saturation of either the NPC, or the RanGTP transport complex dissociation machinery, or both. This relationship is different from that observed by Riddick and Macara (2005), who found that elevated concentrations of importin-β retarded import rates, but is consistent with the recent single-molecule observations of Yang and Musser (2006).
Further investigation of Rpl25NLS-cargo recognition by Kap121p or Kap123p revealed that each Kap bound their common cargo with equivalent ∼90 nM K d (Fig. 7). Thus, similar concentrations of either Kap would produce similar quantities of Kap–cargo import complex (which we observed to be imported at approximately equal rates), saturating the import system in an indistinguishable manner. Therefore, Kap121p and Kap123p carry their cargoes across the NPC at similar rates, despite apparently recognizing different subsets of FG-Nups within the NPC (Rout et al., 1997; Marelli et al., 1998; Seedorf et al., 1999; Allen et al., 2000). It will be interesting to determine whether other Kaps are “tuned” to transport at similar rates, regardless of the FG-Nups that they recognize.
Meeting the import requirements of specific cargoes: Kap123p is a rapid importer for a set of abundant cargoes
Because Kap concentration appears to be a crucial determinant of import-rates, it is possible that Kap123p is the most rapid importer of all Kaps simply because it is the most abundant. Indeed, we found Kap123p import of Rpl25NLS-cargo to be 7- and 12-fold more rapid than the import of the cognate cargoes of Kap104p or Kap121p, respectively, at their natural concentrations (Fig. 4).
Why is the most rapidly importing Kap, whose substrates are a set of proteins required for an essential process (ribosomal biogenesis), not itself essential? To understand this, we compared the published rates of ribosomal biogenesis with our measured rates of import. A wild-type yeast must make ∼4,000 ribosomal subunits/min (Warner, 1999), requiring the import of at least ∼0.4 copies of each ribosomal (r)-protein/NPC/s. The import rate data in Table II suggest that Kap123p could import at this rate with a driving cytoplasmic concentration of only ∼0.3 μM for each r-protein. However, less abundant Kaps, such as Kap121p, would need unincorporated r-proteins to build up to concentrations >6 μM to achieve the import rates required for wild-type growth; with the known short half-life of r-proteins, this could expose them to proteolysis. This may explain the slow growth of Δkap123 yeast (Rout et al., 1997), which have only less effective import pathways available to them.
Our results suggest that Kap123p is a transport factor that has specialized in the import of small abundant cargoes, particularly ribosomal proteins. It has done so by being able to be expressed to relatively high concentrations without detriment to cell. Meanwhile, Kap121p has specialized in the import of cargo whose entry needs to be regulated more than it needs to be particularly rapid (Kaffman et al., 1998; Leslie et al., 2002; Makhnevych et al., 2003). The continuing recognition of ribosomal cargo by Kap121p may represent a vestigial remnant of this evolutionary diversification, while perhaps also providing a small additional boost to ribosomal import rates. Similar specializations probably drove the diversification of the 15 different yeast Kaps, several of which may be partially redundant in respect to some cargoes, such as that described between Kap121p and Kap104p (Leslie et al., 2004).
Cargo saturability: Kap–NLS interactions are subject to nonspecific competition in vivo
Certain observations from these studies of import led us to propose that the binding between NLS-GFP cargo and Kaps occurs in a highly competitive cytoplasmic environment.
First, Kaps were found to be carrying NLS-GFP molecules into the nucleus at quite low frequencies, compared with the measured rapidity of binding between Kap and NLS (Catimel et al., 2001), the speed of intracellular diffusion (Riddick and Macara, 2005), and the relatively short residence time of import complexes at the NPC (Yang et al., 2004; Kubitscheck et al., 2005). For example, ∼5 μM Kap123p required ∼200 s to import most of the >100 μM Rpl25NLS-GFP-PrAp into the nucleus (Fig. 2). This implies that each Kap123p molecule had carried a net of ∼20 Rpl25NLS-GFP-PrAp molecules into the nucleus, with ∼10 s between each cycle. However, accounting for the known rates of transport reactions, we might expect each cycle to take <1 s.
Second, we found no indication of saturation of import rates by the NLS-GFP cargo of any Kap (Fig. 4 B), despite NLS-GFP cargoes (cytoplasmic concentrations from 1 to >100 μM) being over 20-fold more abundant than their Kaps (0.5–5.3 μM) in many cells; we might have expected saturation of import-rates, given the 20–100 nM K d values measured between cognate Kap-NLS pairs in vitro (Fig. 7).
Third, a reduction in the binding affinity of the mutant Rpl25NLS for Kap123p resulted in a proportionate reduction in the import rate of this cargo. In the absence of competition, this small reduction in binding affinity would not be expected to significantly affect the quantity of import complexes formed nor their subsequent import rates, as both Kap and cargo concentrations in vivo are well above the Kd value for their interaction (Fig. 7 B).
The simplest possibility that could account for all of these observations is in vivo competition for the NLS–Kap interaction. In living cells, the NLS-GFP cargoes are competing with the natural unlabeled cargoes, thus reducing the ability of NLS-GFP to saturate the Kap. A Kap can be expected to have hundreds of different cargoes. However, the total amount of competing natural cargoes in the cytoplasm needed to reproduce the linear import relationships we observed would be ∼6 mg/ml for each Kap, which is physiologically unreasonable.
These considerations led us to postulate that binding between Kaps and NLSs is indeed competitive, but with nonspecific cytosolic proteins. We envisage two nonexclusive possibilities for how such competition could occur. First, Kaps could be sampling NLS-like sequences in the cytosol, which contains many sequences partially homologous to those of their actual cargoes. Second, the highly charged NLSs might be adhering to cytosolic proteins nonspecifically. In these paradigms, Kaps and NLSs sample a large number of potential partners in the cytoplasmic milieu, in an effort to form true long-lived Kap–cargo complexes.
Severe nonspecific competition may be the penalty paid for Kaps and NLSs using large binding sites with loose consensus sequences (Conti et al., 1998; Lee et al., 2006). It might seem that the import system could overcome such competition by selecting for Kaps and NLSs binding to each other more tightly. However, studies of export found that stronger Kap–cargo associations (<100 nM) retarded export rates (Engelsma et al., 2004), presumably because efficient transport requires rapid cargo release. Finally, it is difficult to conceive how evolution could have selected amongst ∼2,000 nuclear proteins for anything other than the loose consensus NLSs that we observe, many of which perform dual service as functional domains.
Our finding that E. coli whole-cell lysate competes effectively with the Kap–NLS interaction (Fig. 7 C) supports our hypothesis that nonspecific competition strongly inhibits import rates. It is noteworthy that the total cytoplasmic protein concentration in yeast is estimated to be on the order of 200–300 mg/ml (Zimmerman and Minton, 1993), whereas we saw significant effects at an order of magnitude of lower concentrations; it seems probable that nonspecific competition of cytosolic proteins for Kaps and cargo, rather that the NPC/Ran system itself, is the key step that limits the efficiency of nuclear import.
A simple pump–leak model of the import system
On the basis of our results and our modeling, we can describe the kinetic behavior of the NPC/Ran system. At the start of an import assay, when nuclear and cytoplasmic cargo concentrations are equal, the increase in nuclear cargo concentration is solely caused by active import of Kap–cargo complexes; the net passive flux of molecules across the NPC is zero when there is no significant concentration differential across the NE. Therefore, from initial import rates we calculate the pump characteristics of the NPC/Ran system. In many of our assays, neither Kap nor cargo were saturating (Fig. 4 and Fig. 6), allowing us to calculate a pseudolinear import rate constant, k', from the initial import rates. These rate constants (Table II) reinforce the observed similarity of the rates of the Kap121p and Kap123p pathways, which gave statistically indistinguishable values for import of Pho4NLS and Rpl25NLS, respectively. The fitted k' value for Kap104p import was significantly less (i.e., slower) than those for the other Kaps, as a result of its significantly tighter cargo-binding. This relatively strong binding may be part of a postulated system for Kap104p to take Nab2p directly to its target site within the nucleus (Lee and Aitchison, 1999).
Over the course of an import assay, cargo builds up in the nucleus, resulting in an ever-increasing rate of leak outwards, while diminishing cytoplasmic cargo concentrations simultaneously reduce the rate of its import. Therefore, simulated import follows a single exponential function. These simulated curves fit well to our measured import data (Fig. 8 B), proceeding until a steady-state is reached when the import-rate equals the rate of leak. As would be expected for cargoes of similar sizes, their passive permeability constants (p) were found to be similar to each other (Table II). Because the permeabilities of our reporter cargoes are the same, their steady-state N/C ratios can be used as a measure of the import rate itself (Fig. 6). Our model accurately reproduces our observed population relationships between Kap and cargo concentrations and import rates.
Lastly, we fitted the import saturation data of Kap123p and Kap121p for Rpl25NLS cargo (Fig. 6) to the model, calculating best-fit values for the kinetic parameters of the NPC/Ran system for each pathway. We calculated values of k, the maximum import rate of the NPC/Ran system for each pathway, and analogous to VMAX in Michaelis–Menten kinetics. Both Kap121p and Kap123p gave a k of ∼300 cargo molecules/NPC/s (Table II), which is consistent with in vitro measurements (Ribbeck and Gorlich, 2001), and reflecting the high potential transport flux capacity of each NPC in vivo. We also calculated the values for KS, the K d between import complexes and the NPC/Ran system (and analogous to KM in Michaelis–Menten kinetics), to be ∼1 μM for each of the two pathways (Table II). These similar k and KS values for Kap123p and Kap121p underscore the similar kinetic behavior of these two different pathways. Moreover, these KS values calculated from in vivo experiments are, again, consistent with in vitro measurements (Ribbeck and Gorlich, 2001), and are also in the same range as certain estimates of Kap to FG-Nup binding (Bayliss et al., 1999, 2002; Ben-Efraim and Gerace, 2001; Pyhtila and Rexach, 2003). Our in vivo KS values (∼1 μM) support the idea that nucleocytoplasmic transport is dependent on relatively weak interactions between the NPC and Kaps, as suggested in various NPC mechanistic models (Macara, 2001; Rout et al., 2003; Suntharalingam and Wente, 2003).
Concluding remarks
An advantage of our current methodology is that these quantitative measurements can be made in single cells of a genetically tractable organism in vivo. Future use of this assay will, thus, examine the quantitative effect of key alterations in the transport machinery, such as deletions in particular subsets of NPC Kap docking sites (Strawn et al., 2004). The challenge now is to use these tools to look into the “black box” of the NPC/Ran machinery, manipulating those components expected to be important and measuring their differential effect on import rates and NPC permeability.
Our conclusion that nonspecific binding limits nuclear transport rates has significant ramifications. First, FG-Nups also interact with Kaps via weakly interacting large binding sites, and so may also experience the effects of nonspecific competition. Second, it is conceivable that many other cellular processes may be limited by nonspecific competition in the crowded environment of the cell. Thus, such macromolecular crowding effects may need to be considered in any analysis of cellular processes.
Materials and methods
Plasmids and strains
All plasmids used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200608141/DC1) and were constructed by standard recombinant DNA methods (Sambrook and Russell, 2001) using the restriction sites described in the Supplemental materials and methods. All strains listed in Table S2 were made in DF5 S. cerevisiae (Finley et al., 1987), with the exception of Kap-GFP strains (Ghaemmaghami et al., 2003).
Imaging yeast at steady-state
Cells transformed with NLS-GFP plasmids were, for some experiments, simultaneously transformed with KAP-HA or -CFP expression plasmids. The trace amounts of copper in synthetic selective minimal media (US Biological) facilitated the CUP1 promoter of the Kap expression plasmids to express their fusion proteins several fold above their wild-type levels (as measured from Western blots that compared the signals from the plasmid-borne Kaps with those of genomically tagged versions). Yeast were grown to mid-log phase in selective minimal media and harvested by centrifugation. Cell pellets were reserved for concentration calibration (see import assay). Confocal image sections of several fields of cells in growth media were acquired at 0.4-μm increments, using the microscope setup described in Microscopy. Cell-by-cell image analysis measured subcellular volumes, nuclear envelope area, and fluorescent fusion protein concentrations for a statistically significant number of cells, essentially as described for import assays.
Quantitative import assay
Cells were grown as described for steady-state analysis and then treated essentially as described by Shulga et al. (1996), using metabolic energy poisons to stop import, before measuring the rate of re-import of fluorescent cargoes. Modifications were made to data collection and analysis of this yeast import assay, such that import-rates in single cells could be quantitated in units of cargo molecules/NPC/s. First, advanced automated microscopy was used to observe import in single cells over time, while 3D reconstruction of confocal images was used to measure subcellular volumes. Second, a quantitative Western blotting technique was used to calibrate fluorescence measurements to actual subcellular concentrations of NLS-GFP cargo. For a detailed description of these methods, see our recent survey of nuclear import methods in yeast (Leslie et al., 2006).
Microscopy
Images were collected at room temperature (air conditioned to ∼23°C) with a cooled charge-coupled device camera (Orca ER; Hamamatsu) attached to a microscope (Axiovert 200; Carl Zeiss MicroImaging, Inc.) fitted with a spinning disk (UltraView; Perkin-Elmer) confocal imaging head and using a 100× objective lens (NA 1.45). CFP and YFP were excited with HeCad 442-nm or Argon 514-nm lasers, respectively, and separate images of each were obtained using a standard CFP/YFP dichroic with separate excitation/emission filter sets (Chroma Corp.). These same CFP/YFP optics were used for import assays where NLS-GFP needed to be imaged along with Htb2-CFPp and Tpi1-CFPp; no bleed-through of fluorescence was seen between GFP and CFP image channels. For other experiments, GFP was excited with the 488-nm line of a Krypton–Argon laser, using a dedicated 488-nm dichroic and standard GFP excitation/emission filters (Chroma Corp.). The system was controlled with MetaMorph imaging software (Universal Imaging Corporation). All images were background subtracted and controlled for uniform field illumination before analysis. For presentation, a maximum intensity projection of all focused image planes is shown, and noise in the CFP images was reduced with the adaptive Weiner filter (MatLab).
Recombinant expression and purification of proteins
Nab2NLS-GFPp, Pho4NLS-YFPp, and Rpl25NLS-GFPp were expressed as 6xHis, C-terminal fusion proteins in BL21DE3 Gold E. coli (Novagen). Protein production was induced at OD600 of 0.6–0.8 with 1 mM IPTG (Roche) for 4–5 h at 30°C. Cells were harvested at 4°C in PBS (50 mM NaH2PO4, pH 7.0, and 300 mM NaCl), containing the protease inhibitors 0.1 mM EDTA, 1/100 Solution P (0.5 mg/ml Pepstatin A and 20 mg/ml PMSF in ethanol), 1/500 Protease Inhibitor Cocktail (Sigma-Aldrich), and 1/100 Protease Arrest (G-Biosciences). Cells were lysed by four passages through a microfluidizer (Microfluidics). Cell debris was removed by centrifugation, and fusion proteins were bound, washed, and eluted from TALON resin (BD Biosciences) via their 6xHIS tags at 4°C, as per the manufacturer's recommendations (buffers used after binding did not contain either EDTA or the Protease Arrest cocktail). Purified fusion proteins were concentrated with 10,000 MWCO centrifugal concentrators (Millipore). Protein concentrations were determined by the Bradford method (Bradford, 1976).
GST-Kap104p, GST-Kap121p, and GST-Kap123-HAp were expressed in BL21DE3 pLYS E. coli. Proteins were expressed and purified exactly as previously described (Leslie et al., 2004). When removal of the GST was required, the Thrombin Cleavage Capture kit (Novagen) was used according to the manufacturer's instructions.
Measurement of affinities by resin-binding assays
Binding assays were performed in TB-T (20 mM Hepes-KOH, pH 7.5, 110 mM KOAc, 2 mM MgCl2, 0.1% Tween-20, 1 mM DTT, and 1/100 Solution P). A custom high-titer anti-GFP rabbit polyclonal antibody (Cristea et al., 2005) was affinity purified and conjugated to Sepharose by standard methods (Harlow and Lane, 1988). For each experiment, 0.5 μg of purified NLS-GFP-HIS was incubated per microliter (bed volume) of anti-GFP–conjugated Sepharose, generating immobilized NLS-GFP-HIS resin. 10-μl aliquots of this resin were dispensed into 10–25 ml of TB-T plus 2.5% milk, containing a particular concentration of purified Kap; volumes were adjusted so that the Kap was always in molar excess to the NLS. NLSs were allowed to reach binding equilibrium with the Kap overnight at 4°C, with mixing. The resin, with bound Kap, was harvested and transferred to minicentrifugation columns. Binding buffer was removed by centrifugation, and the resin was washed with 500 μl of ammonium acetate buffer (0.1 M NH4OAc, 0.1 mM MgCl2, and 0.02% Tween-20). All proteins were eluted from the resin by incubation with ammonium hydroxide solution (0.5 M NH4OH and 0.5 mM EDTA). Evaporated protein samples were prepared for SDS-PAGE, and proteins were observed via Coomassie blue staining. The total background-subtracted pixel intensities of bands from scanned gels were measured using OpenLab (Improvision) image analysis software. Dissociation constants were calculated from the resulting binding curves by fitting the data to the predicted simple bimolecular equilibrium relationship.
Online supplemental material
Provided as online supplemental material are descriptions of the plasmids and strains used throughout this work. Also provided, are the raw data of overlay assays, performed to identify Kap123p-binding proteins, and the identification and distribution of NLS-GFP fusion proteins created from these Kap123p binding proteins. Fig. S1 shows Kap123p interacts with a distinct subset of small nuclear proteins. Fig. S2 shows that the identities of Kap123p-interacting proteins were determined from overlay assays. Fig. S3 shows that steady-state distributions of ribosomal NLS-GFP cargoes are significantly altered in Δkap123 yeast. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200608141/DC1.
Supplementary Material
Acknowledgments
We would like to thank Alison North (Director) and Dan Elreda of the Bio-Imaging Resource Center of The Rockefeller University for their continual help and assistance with the collection and analysis of all microscopy images. Protein sequence and amino acid analyses were provided by The Rockefeller University Proteomics Resource Center, which is supported in part by National Institutes of Health shared instrumentation grants and by funds provided by the U.S. Army and Navy for purchase of equipment. We also thank members of the Rout and Chait laboratories for much help and advice, and for sharing their protocols and reagents. We would also like to thank Susan Wente, Marcelo Magnasco, and David Gadsby for their insight and advice.
This work was supported by an Irma T. Hirschl Career Scientist Award, a Sinsheimer Scholar Award, and a grant from the Rita Allen Foundation to M.P. Rout, and grants from the National Institutes of Health to M.P. Rout (GM062427, GM071329) and B.T. Chait (RR00862). J. Novatt is a Howard Hughes Medical Institute Predoctoral Fellow.
Note added in proof. Hodel et al. (2006) recently found a monotonic relationship between Kap–cargo affinity and nuclear accumulation of cargo at steady-state. These data support and expand upon our findings reported in this study, that import rates in vivo are largely determined by the amount of Kap–cargo complexes that can be formed in the cytoplasm.
W. Zhang's present address is Columbia University, Institute for Cancer Genetics, New York, NY 10032.
Abbreviations used in this paper: N/C, nuclear-to-cytoplasmic; NE, nuclear envelope; NPC, nuclear pore complex.
References
- Allen, T.D., J.M. Cronshaw, S. Bagley, E. Kiseleva, and M.W. Goldberg. 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113:1651–1659. [DOI] [PubMed] [Google Scholar]
- Bayliss, R., K. Ribbeck, D. Akin, H.M. Kent, C.M. Feldherr, D. Gorlich, and M. Stewart. 1999. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 293:579–593. [DOI] [PubMed] [Google Scholar]
- Bayliss, R., S.W. Leung, R.P. Baker, B.B. Quimby, A.H. Corbett, and M. Stewart. 2002. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 21:2843–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin β for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152:411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. [DOI] [PubMed] [Google Scholar]
- Catimel, B., T. Teh, M.R. Fontes, I.G. Jennings, D.A. Jans, G.J. Howlett, E.C. Nice, and B. Kobe. 2001. Biophysical characterization of interactions involving importin-alpha during nuclear import. J. Biol. Chem. 276:34189–34198. [DOI] [PubMed] [Google Scholar]
- Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 94:193–204. [DOI] [PubMed] [Google Scholar]
- Cristea, I.M., R. Williams, B.T. Chait, and M.P. Rout. 2005. Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics. 4:1933–1941. [DOI] [PubMed] [Google Scholar]
- Cross, F.R., V. Archambault, M. Miller, and M. Klovstad. 2002. Testing a mathematical model of the yeast cell cycle. Mol. Biol. Cell. 13:52–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, D., B. Mykytka, N.P. Allen, L. Huang, B. Al, and M. Rexach. 2001. The nucleoporin Nup60p functions as a Gsp1p–GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154:937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsma, D., R. Bernad, J. Calafat, and M. Fornerod. 2004. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 23:3643–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, D., E. Ozkaynak, and A. Varshavsky. 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 48:1035–1046. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami, S., W.K. Huh, K. Bower, R.W. Howson, A. Belle, N. Dephoure, E.K. O'Shea, and J.S. Weissman. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741. [DOI] [PubMed] [Google Scholar]
- Glowczewski, L., J.H. Waterborg, and J.G. Berman. 2004. Yeast chromatin assembly complex 1 protein excludes nonacetylatable forms of histone H4 from chromatin and the nucleus. Mol. Cell. Biol. 24:10180–10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb, D.S., J. Gariepy, G. Schoolnik, and R.D. Kornberg. 1986. Synthetic peptides as nuclear localization signals. Nature. 322:641–644. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 726 pp.
- Hodel, A.E., M.T. Harreman, K.F. Pulliam, M.E. Harben, J.S. Holmes, M.R. Hodel, K.M. Berland, and A.H. Corbett. 2006. Nuclear localization signal receptor affinity correlates with in vivo localization in Saccharomyces cerevisiae. J. Biol. Chem. 281:23545–23556. [DOI] [PubMed] [Google Scholar]
- Hodgkin, A.L., and R.D. Keynes. 1955. Active transport of cations in giant axons from Sepia and Loligo. J. Physiol. 128:28–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel, S., and D. Gorlich. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17:4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman, A., N.M. Rank, and E.K. O'Shea. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12:2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keminer, O., J.P. Siebrasse, K. Zerf, and R. Peters. 1999. Optical recording of signal-mediated protein transport through single nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 96:11842–11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitscheck, U., D. Grunwald, A. Hoekstra, D. Rohleder, T. Kues, J.P. Siebrasse, and R. Peters. 2005. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J. Cell Biol. 168:233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.J., A.E. Cansizoglu, K.E. Suel, T.H. Louis, Z. Zhang, and Y.M. Chook. 2006. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 126:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.C., and J.D. Aitchison. 1999. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J. Biol. Chem. 274:29031–29037. [DOI] [PubMed] [Google Scholar]
- Leslie, D.M., B. Grill, M.P. Rout, R.W. Wozniak, and J.D. Aitchison. 2002. Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol. Cell. Biol. 22:2544–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, D.M., W. Zhang, B.L. Timney, B.T. Chait, M.P. Rout, R.W. Wozniak, and J.D. Aitchison. 2004. Characterization of karyopherin cargoes reveals unique mechanisms of Kap121p-mediated nuclear import. Mol. Cell. Biol. 24:8487–8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, D.M., B. Timney, M.P. Rout, and J.D. Aitchison. 2006. Studying nuclear protein import in yeast. Methods. 39:291–308. [DOI] [PubMed] [Google Scholar]
- Macara, I.G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych, T., C.P. Lusk, A.M. Anderson, J.D. Aitchison, and R.W. Wozniak. 2003. Cell cycle regulated transport controlled by alterations in the nuclear pore complex. Cell. 115:813–823. [DOI] [PubMed] [Google Scholar]
- Marelli, M., J.D. Aitchison, and R.W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast, N., Y. Guo, J. Shabanowitz, D.F. Hunt, and L.F. Pemberton. 2002. Pathways mediating the nuclear import of histones H3 and H4 in yeast. J. Biol. Chem. 277:862–868. [DOI] [PubMed] [Google Scholar]
- Mosammaparast, N., and L.F. Pemberton. 2004. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14:547–556. [DOI] [PubMed] [Google Scholar]
- Pyhtila, B., and M. Rexach. 2003. A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J. Biol. Chem. 278:42699–42709. [DOI] [PubMed] [Google Scholar]
- Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick, G., and I.G. Macara. 2005. A systems analysis of importin-α–β mediated nuclear protein import. J. Cell Biol. 168:1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., G. Blobel, and J.D. Aitchison. 1997. A distinct nuclear import pathway used by ribosomal proteins. Cell. 89:715–725. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, M.O. Magnasco, and B.T. Chait. 2003. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 13:622–628. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D.W. Russell. 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schaap, P.J., J. van't Riet, C.L. Woldringh, and H.A. Raue. 1991. Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. J. Mol. Biol. 221:225–237. [DOI] [PubMed] [Google Scholar]
- Schwoebel, E.D., T.H. Ho, and M.S. Moore. 2002. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J. Cell Biol. 157:963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf, M., M. Damelin, J. Kahana, T. Taura, and P.A. Silver. 1999. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 19:1547–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga, N., P. Roberts, Z. Gu, L. Spitz, M.M. Tabb, M. Nomura, and D.S. Goldfarb. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga, N., N. Mosammaparast, R. Wozniak, and D.S. Goldfarb. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149:1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.E., B.M. Slepchenko, J.C. Schaff, L.M. Loew, and I.G. Macara. 2002. Systems analysis of Ran transport. Science. 295:488–491. [DOI] [PubMed] [Google Scholar]
- Strawn, L.A., T. Shen, N. Shulga, D.S. Goldfarb, and S.R. Wente. 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6:197–206. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., and S.R. Wente. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 4:775–789. [DOI] [PubMed] [Google Scholar]
- Sydorskyy, Y., D.J. Dilworth, E.C. Yi, D.R. Goodlett, R.W. Wozniak, and J.D. Aitchison. 2003. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell. Biol. 23:2042–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth, M., W. Waldeck, and J. Langowski. 2000. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298:677–689. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437–440. [DOI] [PubMed] [Google Scholar]
- Winey, M., D. Yarar, T.H. Giddings Jr., and D.N. Mastronarde. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 8:2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, R.W., M.P. Rout, and J.D. Aitchison. 1998. Karyopherins and kissing cousins. Trends Cell Biol. 8:184–188. [DOI] [PubMed] [Google Scholar]
- Yang, W., J. Gelles, and S.M. Musser. 2004. Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. USA. 101:12887–12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W., and S.M. Musser. 2006. Nuclear import time and transport efficiency depend on importin β concentration. J. Cell Biol. 174:951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen, D., P. Vinciguerra, Y. Strahm, and F. Stutz. 2001. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol. 21:4219–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, S.B., and A.P. Minton. 1993. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 22:27–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.