Abstract
LAPTM5 is a lysosomal transmembrane protein expressed in immune cells. We show that LAPTM5 binds the ubiquitin-ligase Nedd4 and GGA3 to promote LAPTM5 sorting from the Golgi to the lysosome, an event that is independent of LAPTM5 ubiquitination. LAPTM5 contains three PY motifs (L/PPxY), which bind Nedd4-WW domains, and a ubiquitin-interacting motif (UIM) motif. The Nedd4–LAPTM5 complex recruits ubiquitinated GGA3, which binds the LAPTM5-UIM; this interaction does not require the GGA3-GAT domain. LAPTM5 mutated in its Nedd4-binding sites (PY motifs) or its UIM is retained in the Golgi, as is LAPTM5 expressed in cells in which Nedd4 or GGA3 is knocked-down with RNAi. However, ubiquitination-impaired LAPTM5 can still traffic to the lysosome, suggesting that Nedd4 binding to LAPTM5, not LAPTM5 ubiquitination, is required for targeting. Interestingly, Nedd4 is also able to ubiquitinate GGA3. These results demonstrate a novel mechanism by which the ubiquitin-ligase Nedd4, via interactions with GGA3 and cargo (LAPTM5), regulates cargo trafficking to the lysosome without requiring cargo ubiquitination.
Introduction
Ubiquitin, especially monoubiquitin, has been a well-documented sorting signal for both endocytosed plasma membrane (PM) proteins and intracellular resident proteins trafficking from the TGN to endosomes or lysosomes (Hicke, 2001). This has been extensively studied in yeast, where ubiquitination serves an important sorting signal to determine protein targeting to either the lumen or the limiting membrane of the vacuole (lysosome; Helliwell et al., 2001; Katzmann et al., 2001; Soetens et al., 2001; Blondel et al., 2004). The role of ubiquitin in sorting proteins from the Golgi to endosomes/lysosomes in mammalian cells is not as clear.
Either blocking ubiquitin modification or adding a single ubiquitin to cargo proteins diverts them away from their normal cellular destination. For example, blocking ubiquitination of the vacuolar carboxypeptidase S or adding a ubiquitin to the hydrolase dipeptidylaminopeptidase B in yeast diverts these proteins to the limiting membrane or lumen of the vacuole, respectively (Katzmann et al., 2001, 2002). Likewise, the ubiquitination status of the yeast GAP1 and Fur4 permeases determines their fate during sorting at the Golgi (Blondel et al., 2004; Helliwell et al., 2001; Soetens et al., 2001). In both yeast and mammalian cells, most of the ubiquitinated proteins that reach the vacuole/lysosome interior are either PM proteins, which then undergo degradation (Levkowitz et al., 1998; Hicke, 2001; Rocca et al., 2001), or hydrolytic enzymes activated inside the vacuolar lumen (Odorizzi et al., 1998). If proteins are not ubiquitinated, they are either recycled back to the PM (Levkowitz et al., 1998) or delivered to the limiting membrane of multivesicular bodies (MVBs) or the vacuole/lysosome (Katzmann et al., 2001). Therefore, mono- or multimonoubiquitination plays a critical role in targeting cargo proteins to their proper cellular destination (Terrell et al., 1998; Shih et al., 2000; Mosesson et al., 2003), hence deciding their fate (degradation vs. recycling).
The monoubiquitin sorting signal can be recognized and transmitted by several proteins involved in both the endocytic and biosynthetic pathways through direct physical interaction. These proteins often possess ubiquitin-binding motifs/domains (“ubiquitin receptors”), such as ubiquitin-interacting motif (UIM; Hofmann and Falquet, 2001), GAT (GGA and Tom homologue), UBA, UEV, VHS, and CUE (Hicke et al., 2005). The UIMs in epsin and eps15/eps15R play an important role in internalization and the early stages of endocytosis of PM proteins, such as the EGFR (Oldham et al., 2002; Polo et al., 2002), whereas the UIMs of Vps27/Hrs and Hse1/STAM are associated with sorting function at the early and late endosome (Bilodeau et al., 2002; Raiborg et al., 2002; Shih et al., 2002). Interestingly, several UIM-containing endocytic proteins (e.g., epsin, eps15, and Hrs) are themselves ubiquitinated outside of the UIM sequence itself (Katz et al., 2002; Polo et al., 2002; Miller et al., 2004). At least in some cases, this ubiquitination appears to involve Nedd4 family members. Nedd4 is a ubiquitin ligase comprised of a C2 domain, 3–4 WW domains that bind PY motifs (L/PPxY) on target proteins, and a ubiquitin ligase Hect domain (Staub and Rotin, 2006). However, it is not yet clear how Nedd4 proteins are recruited to these endocytic proteins, which do not possess PY motifs. Moreover, the precise role of ubiquitination of these UIM-containing endocytic proteins in cargo sorting and transport is not known.
The Golgi-localized, γ-ear–containing, ADP ribosylation factor–binding proteins (GGAs) are primarily associated with the TGN, and play a role in lysosomal and endosomal sorting (Dell'Angelica et al., 2000; Bonifacino, 2004; Pelham, 2004). For example, they sort mannose 6-phosphate receptors from the Golgi to the lysosome by binding to the cytoplasmic dileucine motifs of the receptor (Puertollano et al., 2001; Zhu et al., 2001; Misra et al., 2002). GGA3 was reported to also function as an adaptor to sort the internalized EGFR to endosomes (Puertollano and Bonifacino, 2004; Kametaka et al., 2005). In yeast, it was shown that GGA binds to the ubiquitinated Gap1 permease, diverting it from the TGN to the vacuole/lysosome (Soetens et al., 2001; Scott et al., 2004). In both mammalian and yeast cells, ubiquitin was shown to bind to the GAT domain of the GGAs (Puertollano and Bonifacino, 2004; Scott et al., 2004; Shiba et al., 2004). Thus, monoubiquitin is a universal sorting signal not only at the cell surface or in endosomes but also at the Golgi.
Lysosomal-associated protein transmembrane 5 (LAPTM5) is a multispanning transmembrane protein that resides in the late endosome/lysosome and is expressed primarily in hematopoietic cells (Adra et al., 1996; Scott et al., 1996). We isolated LAPTM5 in a screen for Nedd4-WW domain–interacting proteins, and detected three PY motifs and a UIM in that protein (unpublished data). The function of this lysosomal protein is so far unknown. LAPTM5 is related to LAPTM4 (also known as MTP), a lysosomal protein that is widely expressed in many cell types (Hogue et al., 1996, 1999; Cabrita et al., 1999). LAPTM4 was shown to confer multidrug resistance by transporting a range of small molecules, including nucleosides, nucleobase analogues, antibiotics, anthracyclines, ionophores, and steroid hormones (Cabrita et al., 1999; Hogue et al., 1999). It is believed to function in sequestration of these small molecules into the lysosome, thus, protecting the cell from their harmful effects. LAPTM4 has two splice isoforms, LAPTM4α and -β (Hogue et al., 2002). LAPTM4α was shown to possess two Tyr-based motifs (YxxΠ) that are responsible for its localization to the lysosome (Hogue et al., 2002). How LAPTM5 is sorted to the lysosome has not been elucidated and was the focus of our studies.
We show that LAPTM5 is sorted from the Golgi to the lysosome by association via its PY motifs with Nedd4. The Nedd4–LAPTM5 complex recruits GGA3, and ubiquitinated GGA3 binds the UIM of LAPTM5. The Nedd4-triggered LAPTM5–GGA3 association then promotes translocation of LAPTM5 from the Golgi to the lysosome. Interestingly, this translocation does not require LAPTM5 ubiquitination.
Results
Interaction of LAPTM5 with Nedd4 is necessary for its sorting to the lysosomes
Our earlier work identified LAPTM5 in a screen for proteins that interact with the second WW domain of Nedd4-1 (Nedd4; unpublished data). The intracellular domains of LAPTM5 contain three PY (L/PPxY) motifs; an LPAY motif located between transmembrane (TM) domains 3 and 4, and LPSY and PPPY motifs in the cytoplasmic C terminus (Fig. 1). We also identified a UIM in this protein (Fig. 1). The presence of the three PY motifs in LAPTM5, our identification of Nedd4 as a binding partner for LAPTM5, and the observation that LAPTM5 is localized to late endosomes (LEs) or lysosome (Adra et al., 1996), prompted us to investigate whether Nedd4 may be involved in sorting of LAPTM5 to the LE/lysosomes.
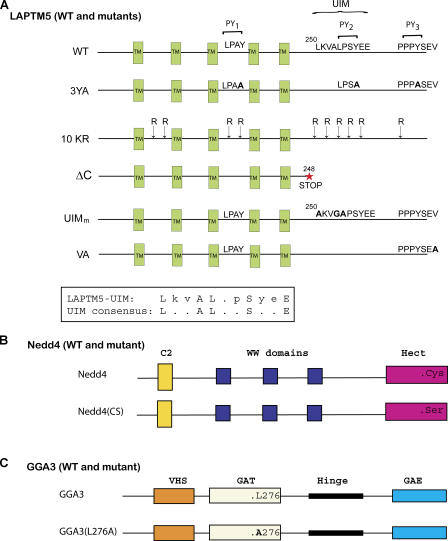
Figure 1.
Schematic representation of LAPTM5, Nedd4, and GGA3, and their mutants used in the study. (A) WT LAPTM5 showing its predicted TM domains (TM), the three PY motifs (PY1, PY2, PY3), the UIM (also depicted in the boxed sequence at the bottom), and the C-terminal SxV (putative PDZ-binding) motif. The various mutants of LAPTM5 used in the study are shown as well. (B) Nedd4 (Nedd4-1) and its catalytically inactive Nedd4(CS) bearing a Cys®Ser mutation in the Hect domain. (C) GGA3 and its L276A mutant in the GAT domain that cannot bind ubiquitin.
To test whether these three PY motifs are responsible for Nedd4 binding, all three Tyr residues in the PY motifs were mutated to alanines (3YA; Fig. 1), and wild-type (WT) LAPTM5 or its 3YA mutant were tested for binding to a catalytically inactive Nedd4(CS) in a coimmunoprecipitation (coIP) assay. A catalytically inactive Nedd4(CS) was used to prevent a possible degradation of LAPTM5 in the event that it is a substrate for Nedd4. As shown in Fig. 2 A (top), after ectopic expression in human embryonic kidney 293T (HEK293T) cells, LAPTM5(WT) was able to coIP with Nedd4(CS), and this binding was lost in the LAPTM5-3YA mutant. Of the three PY motifs, the third one (PY3) appears most important for binding to Nedd4, with the first PY motif (PY1), but not PY2, contributing to the interaction as well (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200603001/DC1). These results demonstrate that Nedd4 binds to the PY motifs of LAPTM5.
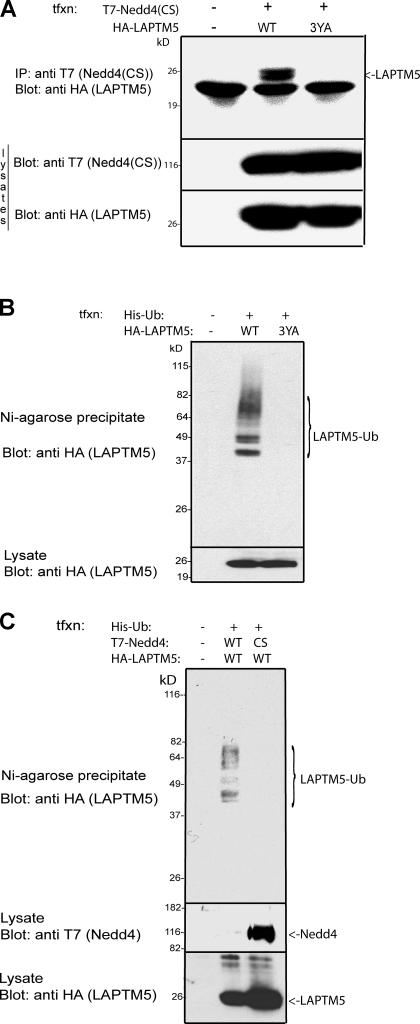
Figure 2.
Interactions between Nedd4 and LAPTM5 and Nedd4-mediated ubiquitination of LAPTM5. (A) coIP of WT, or the 3YA mutant of LAPTM5 (3YA), with Nedd4. T7-tagged Nedd4 was immunoprecipitated from HEK293T cells coexpressing HA-tagged LAPTM5 and the precipitate immunoblotted with anti-HA antibodies to detect coimmunoprecipitated LAPTM5. (B) Ubiquitination of LAPTM5 is dependent on the presence of its PY motifs (Nedd4 binding sites). HEK293T cells were cotransfected with HA-LAPTM5 (WT or the 3YA mutant) and His-tagged ubiquitin (His-Ub), and then the cells were lysed and the lysate was boiled in SDS. Ubiquitinated (histidinated) proteins were then precipitated with Ni–Agarose2+ (Ni–Agarose) beads and blotted with anti-HA antibodies to detect ubiquitinated LAPTM5. (C) LAPTM5 ubiquitination is mediated by Nedd4. The experiment was performed as in B, only His-Ub and HA-LAPTM5 were cotransfected with either WT or a catalytically inactive Nedd4(CS). Bottom sections of all blots represent lysate controls for protein expression.
To examine whether Nedd4 binding leads to ubiquitination of LAPTM5, lysates from HEK293T cells expressing HA-tagged LAPTM5 together with His(x8)-tagged ubiquitin were boiled in SDS (to dissociate other putative interacting proteins), precipitated with Ni2+–Agarose beads (which bind His), and subjected to immunoblotting with anti-HA antibodies to detect ubiquitinated LAPTM5. As seen in Fig. 2 B, LAPTM5(WT) was ubiquitinated in cells (that express endogenous Nedd4), and this ubiquitination was dependent on the presence of its PY motifs, as it was eliminated in the LAPTM5-3YA mutant. In agreement with the binding data (Fig. S1 A), PY3 appears most important for LAPTM5 ubiquitination (Fig. S1 B). These results suggest that LAPTM5 ubiquitination is mediated by Nedd4. Indeed, overexpression of a catalytically inactive Nedd4(CS), together with LAPTM5, inhibited this ubiquitination (Fig. 2 C), likely in a dominant-negative fashion. Most of the LAPTM5 in these assays appeared to be either mono- or diubiquitinated. Collectively, these results indicate that Nedd4 is likely the ubiquitin ligase that ubiquitinates LAPTM5.
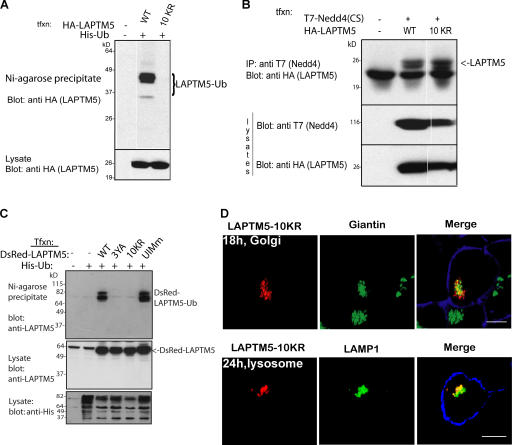
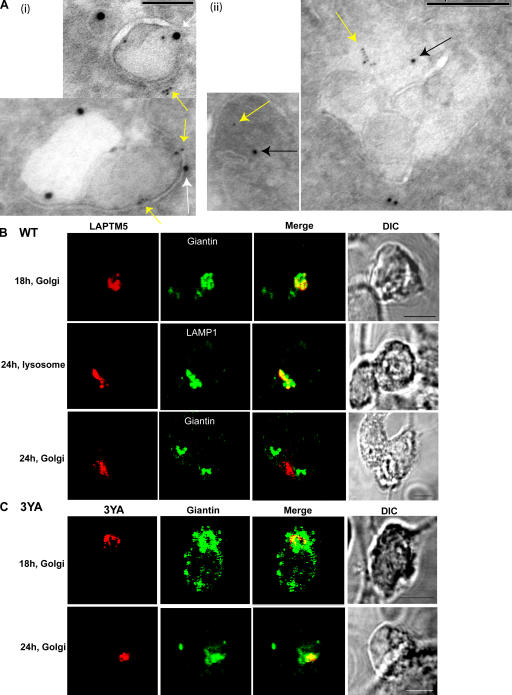
Because LAPTM5 was previously shown to localize to lysosomes (Adra et al., 1996), we next tested whether this localization is dependent on Nedd4-mediated interaction with LAPTM5. Thus, DsRed-tagged LAPTM5(WT) was transfected into HEK293T cells, and its localization was assessed by confocal microscopy. As seen in Fig. 3 A, at 14 h after transfection, LAPTM5(WT) was localized to the Golgi, as determined by colocalization with giantin. By 18 h, some of it has translocated to vesicles that costain with the lysosomal marker LAMP1 (Fig. 3 B), and by 24 h, most of it was localized to the lysosome (Fig. 3 C, Table I, and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200603001/DC1). This was further validated by immunoEM analysis (Fig. 3 D), which demonstrates localization of LAPTM5 in LAMP2-containing vesicles (lysosomes). In contrast, the mutant LAPTM5-3YA, which cannot bind Nedd4, was retained in the Golgi and appeared unable to translocate to the lysosome even after 24 (Fig. 3, A–C and E, Table I, and Table S1) or 48 h (not depicted) after transfection. It was also not localized to EEA1- (early endosomes), Hrs- (sorting endosomes), or Rab11 (recycling endosome)-containing vesicles (Table I and Fig. S2). The ability of the LAPTM5-3YA mutant to localize early after transfection to the Golgi suggests that this mutant is properly folded and able to enter the Golgi from the ER (Fig. 3 E, immunoEM), much like the WT LAPTM5. Rather, it is the sorting step from the Golgi to the lysosome that is defective in the 3YA mutant, most likely caused by its inability to bind Nedd4.
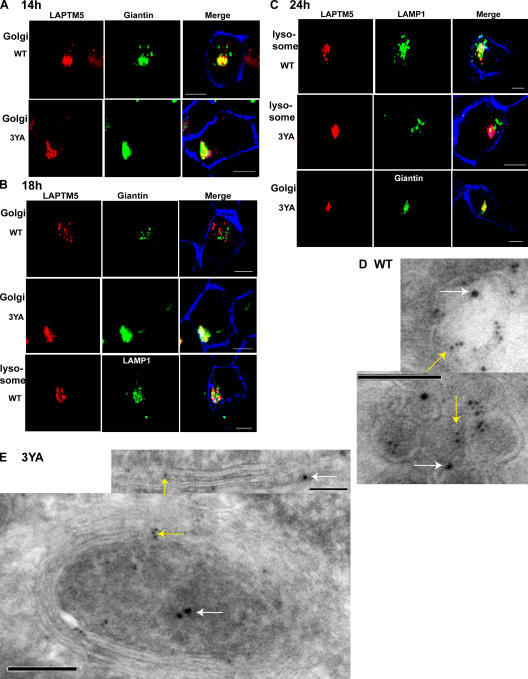
Figure 3.
Golgi-to-lysosome sorting of LAPTM5 is dependent on its PY motifs. (A–C) Immunofluorescent (confocal) analysis of Golgi versus lysosomal localization of LAPTM5 and its 3YA mutant; HEK293T cells were transfected with DsRed-LAPTM5(WT) or the DsRed-LAPTM5(3YA) mutant and costained with either the Golgi marker giantin (green) or the lysosomal marker LAMP1 (green). 14 h after transfection (A), 18 h after transfection (B), and 24 h after transfection (C). Concanavalin A (ConA) was used as a marker for the plasma membrane (blue). Images represent confocal analysis. (D and E) ImmunoEM analysis of lysosomal or Golgi localization of WT-LAPTM5 and its 3YA mutant expressed in HEK293T cells. (D) Colocalization of HA-tagged WT LAPTM5 (6-nm gold particles, yellow arrow) with the LE/lysosomal marker LAMP2 (10-nm gold particles, white arrow). (E) Colocalization of HA-tagged LAPTM5-3YA mutant (6-nm gold particles, yellow arrow) with the Golgi markers giantin or furin (10-nm gold particles, white arrow). Bars: (A–C) 5 μm; (D and E) 100 nm.
Table I.
Quantitative analysis of colocalization of LAPTM5 with organelle markers
| Cell type | Organelle | Pearson's coefficient |
|---|---|---|
| HEK293T | ||
| WT | Lysosomes | 0.819, r(20), P < 0.01 |
| Golgi | 0.296, r(20), P > 0.10 | |
| 3YA | Lysosomes | 0.241, r(20), P > 0.10 |
| Golgi | 0.809, r(20), P < 0.01 | |
| Recycling e | 0.118, r(15), P > 0.10 | |
| Sorting e | 0.103, r(15), P > 0.10 | |
| Early e | 0.104, r(15), P > 0.10 | |
| 10KR | Lysosomes | 0.733, r(20), P < 0.01 |
| Golgi | 0.419, r(20), P > 0.05 | |
| Recycling e | 0.151, r(15), P > 0.10 | |
| Sorting e | 0.165, r(15), P > 0.10 | |
| Early e | 0.157, r(15), P > 0.10 | |
| UIMm | Lysosomes | 0.410, r(20), P > 0.05 |
| Golgi | 0.680, r(20), P < 0.01 | |
| Recycling e | 0.165, r(15), P > 0.10 | |
| Sorting e | 0.125, r(15), P > 0.10 | |
| Early e | 0.133, r(15), P > 0.10 | |
| WT + Nedd4 RNAi | Lysosomes | 0.359, r(20), P > 0.10 |
| Golgi | 0.783, r(20), P < 0.01 | |
| WT + GGA3 RNAi | Lysosomes | 0.318, r(20), P > 0.10 |
| Golgi | 0.748, r(20), P < 0.01 | |
| DC2.4 | ||
| WT | Lysosomes | 0.712, r(20), P < 0.01 |
| Golgi | 0.218, r(20), P > 0.10 | |
| 3YA | Lysosomes | 0.398, r(25), P < 0.05 |
| Golgi | 0.453, r(25), P < 0.05 | |
| Recycling E | 0.165, r(15), P > 0.10 | |
| Sorting E | 0.167, r(15), P > 0.10 | |
| Early E | 0.162, r(15), P > 0.10 |
Quantification on individual randomly selected dsRed-LAPTM5–transfected cells was performed using LSM510 and Volocity 3.7.0. The extent of colocalization is indicated as Pearson's coefficient, corresponding to the degrees of freedom (r) and level of significance (P). The following markers of organelles were used in the analysis: LAMP1 (lysosomes), giantin (Golgi), Rab11 (recycling endosomes), Hrs (sorting endosomes), and EEA1 (early endosomes).
To further test for the role of Nedd4 in the Golgi to lysosome sorting of LAPTM5, we knocked down endogenous Nedd4 in HEK293T cells with RNAi (Fig. 4 A). As seen in Fig. 4 B, Table I, and Table S1, knockdown of endogenous Nedd4 resulted in retention of LAPTM5 in the Golgi. Interestingly, this impaired Golgi to lysosomal sorting was observed despite normal expression of endogenous Nedd4-2, which was unaffected by our knockdown of endogenous Nedd4 (Nedd4-1; Fig. 4 A). This suggest that Nedd4, rather than Nedd4-2, is involved in lysosomal targeting of LAPTM5.
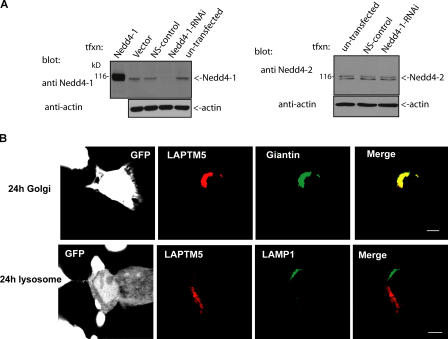
Figure 4.
Knockdown of Nedd4 inhibits Golgi-to-lysosome sorting of LAPTM5. (A) Expression of GFP-Nedd4-1-RNAi (Nedd4-RNAi) in HEK293T cells leads to knockdown of endogenous Nedd4-1 (left), but not Nedd4-2 (right). Affinity-pure antibodies that preferentially recognize Nedd4-1 (anti-Hect antibodies, left) or Nedd4-2 (right) were used to detect the respective Nedd4 proteins after transfection with pSUPER-GFP-Nedd4-1-RNAi (Nedd4-1-RNAi), pSUPER-GFP (vector), pSUPER-GFP–nonspecific control (NS-control), or Nedd4-1. Actin controls are shown as well. (B) Cells coexpressing GFP-Nedd4-RNAi (white) and DsRed-LAPTM5 (red) were costained with anti-giantin (green) or -LAMP1 (green) antibodies 24 h after transfection of LAPTM5. GFP-Nedd4-RNAi (in pSUPER vector) was transfected 48 h before the transfection of LAPTM5, to ensure sufficient time for knockdown of endogenous Nedd4 (i.e., Nedd4-1). Bar, 5 μm.
Collectively, these results suggest that Nedd4 binding to LAPTM5 is necessary for sorting of LAPTM5 from the Golgi to the lysosome.
Ubiquitination of LAPTM5 is not necessary for translocation of LAPTM5 to the lysosomes
Ubiquitination has been well documented as a sorting signal for delivery of biosynthetic proteins to the lumen of the yeast vacuole (Katzmann et al., 2002). Its role in sorting mammalian transmembrane proteins is less clear. Because we found that Nedd4 binding to LAPTM5 is required for its sorting to the lysosome (Fig. 3), and LAPTM5 is ubiquitinated by Nedd4 (Fig. 2), we next tested if LAPTM5 ubiquitination is involved in its sorting to the lysosome. We, thus, generated a mutant LAPTM5 with all of its 10 cytoplasmic Lys residues mutated to Arg (10KR; Fig. 1) to render it ubiquitination-impaired. The ubiquitination status of this mutant was then analyzed after its cotransfection with His-tagged ubiquitin, as described in Fig. 2. As expected, the LAPTM5-10KR mutant failed to become ubiquitinated (Fig. 5 A), although it was still able to bind Nedd4 (Fig. 5 B) because its PY motifs are intact.
Figure 5.
Loss of LAPTM5 ubiquitination does not block its lysosomal sorting. (A) HA-tagged LAPTM5 or LAPTM5(10KR) were coexpressed with His-Ub in HEK293T cells. Cells were lysed, and then boiled lysates were incubated with Ni2+–Agarose beads to precipitate ubiquitinated (Histidinated) proteins, and immunoblotted with HA antibodies to detect ubiquitinated LAPTM5 or its 10KR mutant, as described in Fig. 2. The majority of WT LAPTM5 appears as a diubiquitinated species. (B) T7-Nedd4 and HA-LAPTM5 (WT or the 10KR mutant) were cotransfected into HEK293T cells and lysates immunoprecipitated with T7 antibodies (Nedd4), and blotted with HA antibodies (LAPTM5) to detect coIP of LAPTM5 with Nedd4. A and B (bottom) represent lysate controls. (C) DsRed-LAPTM5(10KR) is not ubiquitinated; the experiment was performed as in A, except that transfected LAPTM5 or its mutants were tagged with DsRed instead of HA. (D) Cells expressing DsRed-LAPTM5 (WT or the 10KR mutant, red) were stained with anti-giantin (green) or -LAMP1 (green) antibodies, as in Fig. 3, to follow Golgi to lysosomal sorting of the LAPTM5-10KR mutant. Images are confocal pictures. The plasma membrane is stained with ConA (blue). Bar, 5 μm.
To analyze the requirement of LAPTM5 ubiquitination for its sorting from the Golgi to the lysosome, we tested the ability of the ubiquitination-impaired LAPTM5-10KR mutant to translocate to the lysosome from the Golgi. We first verified that LAPTM5-10KR fused to DsRed is not itself ubiquitinated because of the added DsRed moiety (Fig. 5 C), and then we used it for immunolocalization studies. As seen in Fig. 5 D and quantified in Tables I and S1, most of the LAPTM5-10KR mutant was localized to the Golgi at 18 h after transfection and to the lysosomes at 24 h after transfection, quite similar (although not identical) to WT LAPTM5 (Fig. 3 and Table I). Moreover, HA-tagged LAPTM5-10KR was also able to sort to the lysosome (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200603001/DC1). These results suggest that ubiquitination of LAPTM5 is not necessary for its translocation to the lysosome.
Together, these data suggest that binding to Nedd4, rather than ubiquitination, may function as a lysosomal sorting signal for LAPTM5.
GGA3 interacts with LAPTM5 in the presence of Nedd4
We next tested the possibility that binding of Nedd4 to LAPTM5 may unravel an as yet unidentified sorting signal within LAPTM5, permitting lysosomal targeting. Because the 3YA mutant of LAPTM5, which failed to bind to Nedd4, was retained in the Golgi, we focused our studies on Golgi sorting.
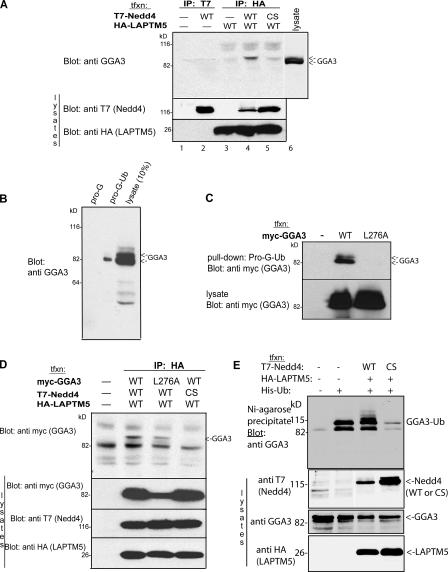
The GGA proteins bind mannose 6-phosphate receptors at the Golgi and play an essential role in lysosomal sorting (Bonifacino, 2004; Pelham, 2004). Therefore, we first tested whether GGA3 could bind to LAPTM5. Cell lysates expressing either T7-tagged Nedd4 alone or HA-tagged LAPTM5 were immunoprecipitated and subjected to immunoblotting with anti- GGA3 antibody to detect if endogenous GGA3 could coimmunoprecipitate in the complex. As shown in Fig. 6 A, neither protein alone could bind GGA3 (lanes 2 and 3). However, GGA3 was immunoprecipitated in a complex with LAPTM5(WT) and Nedd4(WT), but only poorly with the catalytically inactive Nedd4(CS; Fig. 6 A, lane 4 and 5). Furthermore, only a higher form of endogenous GGA3 was found in the complex, although a doublet is seen in the lysate (Fig. 6 A, lane 6). Immunoprecipitating endogenous GGA3 followed by immunoblotting with anti-ubiquitin antibody demonstrated that this upper band was ubiquitinated GGA3 (not depicted), in accord with earlier work, which reported that GGA3 undergoes monoubiquitination (Shiba et al., 2004). Collectively, these data suggest that GGA3 is found in a complex with LAPTM5 and Nedd4 (but not Nedd4[CS]), and that GGA3 in this complex is most likely the ubiquitinated form of the protein.
Figure 6.
GGA3 associates with the Nedd4–LAPTM5 complex and can be ubiquitinated by Nedd4. (A) Lysates of HEK293T cells expressing HA-tagged LAPTM5 and T7-tagged Nedd4 (WT or CS) were immunoprecipitated with HA (LAPTM5) and blotted with GGA3 antibodies. Note the CoIP of GGA3 with LAPTM5 only when active Nedd4 is present. (B) HEK293T cell lysate (which expresses endogenous GGA3) was incubated with ubiquitin immobilized on protein G beads (pro-G-Ub), the beads were washed, and GGA3 was bound to the immobilized ubiquitin detected with anti-GGA3 antibodies. (C) As in B, only cells were transfected with Myc-tagged WT or the L276A mutant of GGA3 (which cannot bind ubiquitin), and binding to pro-G-Ub was subsequently tested. Note in B and C that the lower band of GGA3 (likely nonubiquitinated) is the one that more strongly binds ubiquitin. (D) Cells were transfected with T7-Nedd4 (WT or CS mutant), Myc-GGA3 (WT or the L276A mutant that cannot bind ubiquitin), and HA-LAPTM5, and their lysate was subjected to IP with HA (LAPTM5) antibodies and immunoblotting for Myc (GGA3). (E) Nedd4 is able to ubiquitinate GGA3; HEK293T cells were cotransfected (or not) with LAPTM5, His-Ub, T7-Nedd4(WT), or T7-Nedd4(CS). Lysates of these cells were incubated with Ni2+–Agarose beads to precipitate histidinated (ubiquitinated) proteins, and with proteins immunoblotted with anti-GGA3 antibodies, to detect ubiquitination of endogenous GGA3. Bottom portions of all blots represent lysate controls for protein expression.
The GGA proteins contain various domains that are involved in protein–protein interactions (Fig. 1; Bonifacino, 2004; Pelham, 2004). Recently, the GAT domain of GGA3 was reported to bind ubiquitin (Puertollano and Bonifacino, 2004; Shiba et al., 2004). Thus, we tested whether ubiquitinated LAPTM5 can bind to the GGA3-GAT domain. We first confirmed that endogenous GGA3 binds ubiquitin by demonstrating that G protein–conjugated ubiquitin (prot-G-Ub) is able to precipitate GGA3 (Fig. 6 B). To confirm that the GGA3-GAT domain is responsible for ubiquitin binding, we expressed wt and GAT domain mutant GGA3 (GGA3-L276A) in HEK293T cells and repeated the precipitation assay with prot-G-Ub. As seen in Fig. 6 C, GGA3-L276A was no longer able to bind ubiquitin, as previously reported (Puertollano and Bonifacino, 2004; Shiba et al., 2004). However, it is interesting that more of the lower band of GGA3 binds to ubiquitin (Fig. 6 C), which is different from the form that is in a complex with LAPTM5 and Nedd4 (Fig. 6 A).
To understand how GGA3 forms a complex with LAPTM5 and Nedd4, we expressed WT and the GGA3-L276A mutant, together with a combination of WT and mutant LAPTM5 and Nedd4, and subjected the lysate to coIP with LAPTM5. If ubiquitinated LAPTM5 binds to the GAT domain of GGA3, only WT GGA3 should complex with LAPTM5 and Nedd4. However, both WT GGA3 and the GGA3-L276A mutant were able to bind to the Nedd4–LAPTM5 complex (Fig. 6 D), suggesting that GAT function as a ubiquitin-binding motif is not necessary for the association with this complex. Furthermore, it suggests that ubiquitination of LAPTM5 may not contribute to the formation of the complex. This is consistent with our data (Fig. 5 and Table I) that show efficient sorting of LAPTM5-10KR to the lysosome.
Because GGA3 can coprecipitate with the Nedd4–LAPTM5 complex, we tested the possibility that GGA3 itself may become ubiquitinated by Nedd4. Fig. 6 E demonstrates that, indeed, endogenous GGA3 can become ubiquitinated by coexpressed Nedd4, but not by the catalytically inactive Nedd4(CS).
LAPTM5 has a UIM that is responsible for GGA3 binding and lysosomal targeting
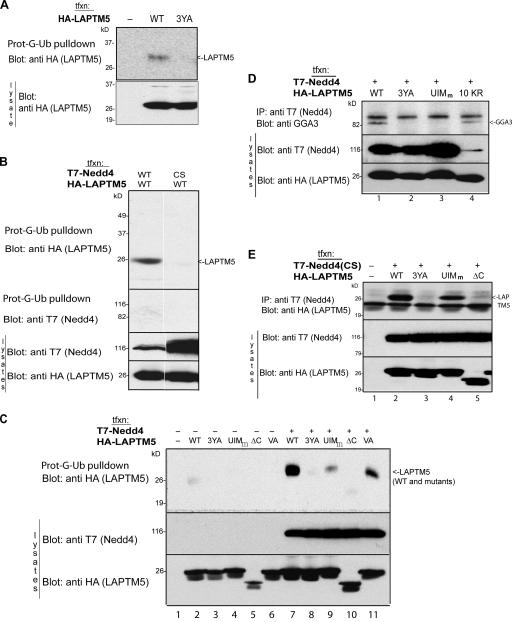
As mentioned above, only the higher form of GGA3 was immunoprecipitated in a complex with LAPTM5 and Nedd4 (Fig. 6 A), suggesting that GGA3 in that complex is most likely the ubiquitinated form. LAPTM5 contains a putative UIM at its C terminus (Fig. 1). To test if this putative UIM binds ubiquitin and ubiquitinated GGA3, cell lysates expressing either HA-tagged LAPTM5 or T7-tagged Nedd4 were precipitated with prot-G-Ub. Nedd4 did not bind to this immobilized ubiquitin (Fig. 7 B). However, LAPTM5 was able to bind ubiquitin (Fig. 7, A–C), binding that was not seen in its 3YA mutant (Fig. 7, A and C). These results suggest that binding of Nedd4 to LAPTM5 affects the ability of LAPTM5 to bind ubiquitin. To further test this, we incubated cell lysates expressing LAPTM5 together with either WT or catalytically inactive Nedd4(CS) with prot-G-Ub, and immunoblotted the precipitate with antibodies to LAPTM5 or Nedd4 (i.e., to their tags). Fig. 7 B (top) shows that ubiquitin was able to bind to LAPTM5 only in the presence of WT Nedd4, but not Nedd4(CS). This suggests that active Nedd4 is needed to allow binding of LAPTM5 to ubiquitin.
Figure 7.
LAPTM5 binds ubiquitin and GGA3 via its UIM, and active Nedd4 is required for this binding. (A) LAPTM5 (WT or the 3YA mutant) was transfected into HEK293T cells and lysates incubated with pro-G-Ub to test for its ability to bind ubiquitin, as in Fig 6. (B) WT-LAPTM5 was transfected with WT or catalytically inactive Nedd4(CS) and lysate incubated with pro-G-Ub, as in A. (C) WT or the indicated mutants of LAPTM5 were transfected in the presence or absence of cotransfected WT Nedd4 and cell lysate incubated with pro-G-Ub to detect binding of the mutants to ubiquitin. (D) WT-Nedd4 and the indicated mutants of LAPTM5 were transfected into cells and the coprecipitation of endogenous GGA3 with the complex analyzed by immunoblotting with anti-GGA3 antibodies. (E) WT or the indicated mutants of LAPTM5 were cotransfected with catalytically inactive Nedd4(CS), Nedd4(CS) immunoprecipitated from cell lysates and coimmunoprecipitated LAPTM5, or its mutants analyzed by immunoblotting with anti-HA antibodies. Bottom portions of all blots represent lysate controls for protein expression.
We next tested the role of the LAPTM5-UIM in ubiquitin binding, as well as the dependency of binding on the levels of Nedd4 expressed in cells. As shown in Fig. 7 C, LAPTM5 was able to bind ubiquitin substantially better when Nedd4 was overexpressed (compare lane 2 to 7). Mutant LAPTM5 truncated at amino acid 248 at the C-tail (ΔC, Fig. 1) almost completely lost its ubiquitin-binding capacity (Fig. 7 C, lanes 5 and 10), suggesting that the UIM, which resides in this region, may be important for binding to ubiquitin. Because this C-terminal region also contains a potential PDZ-binding sequence (SxV), we made two independent mutants of LAPTM5; a UIM mutant (UIMm; Fig. 1) known to be defective in ubiquitin binding (Shekhtman and Cowburn, 2002), and a Val to Ala substitution of the last residue of LAPTM5 (LAPTM5-VA; Fig. 1), which should preclude binding to PDZ domains. We then tested the ability of these mutants to bind ubiquitin. Our results show that the VA mutant had a slightly reduced ubiquitin-binding ability (Fig. 7 C, lane 6 and 11). The UIMm mutant, however, revealed a dramatic decrease in ubiquitin binding (Fig. 7 C), suggesting that the UIM of LAPTM5 plays a key role in binding ubiquitin.
As detailed above, we observed that the LAPTM5-10KR and -3YA mutants, both unable to become ubiquitinated, showed a different pattern of subcellular distribution 24 h after transfection; the 10KR mutant was primarily translocated to the lysosome (similar to WT LAPTM5), whereas the PY motif mutant was retained in the Golgi (Figs. 3 and 5). We thus tested whether these two mutants showed a difference in GGA3 binding. Cell lysates expressing Nedd4 together with a combination of either WT or mutant LAPTM5 were immunoprecipitated with Nedd4 antibodies and subjected to immunoblotting with anti-GGA3 antibody to detect if endogenous GGA3 coIPs with the complex. As seen in Fig. 7 D, the LAPTM5-10KR mutant was able to complex with GGA3 in the presence of Nedd4, much like WT LAPTM5 (Fig. 7 D, lane 1 and 4). However, the LAPTM5-3YA mutant was unable to bind GGA3 (Fig. 7 D, lane 2), suggesting that interactions with Nedd4, rather than the ubiquitination status of LAPTM5, is important for GGA3 binding. Furthermore, the UIM of LAPTM5 was responsible for GGA3 interaction with the complex because GGA3 binding was completely abolished in the UIM mutant, LAPTM5-UIMm (Fig. 7 D, lane 3). Importantly, this same UIM mutant that cannot bind ubiquitin or GGA3 was also unable to sort LAPTM5 from the Golgi to the lysosome (Fig. 8 A, Table I, and Table S1). Because the mutations in the UIM were close to the second PY motif (LPsY) of LAPTM5, we tested the ability of this UIMm mutant to bind Nedd4. As seen in Fig. 7 E, the UIMm mutant was still able to bind Nedd4, similar to WT LAPTM5 (lane 2 and 4), suggesting that Nedd4 binding to LAPTM5 does not require the UIM. Collectively, these data suggest that binding of Nedd4 to LAPTM5 promotes the UIM of LAPTM5 to bind ubiquitinated GGA3. This then allows LAPTM5 to translocate from the Golgi to the lysosome.
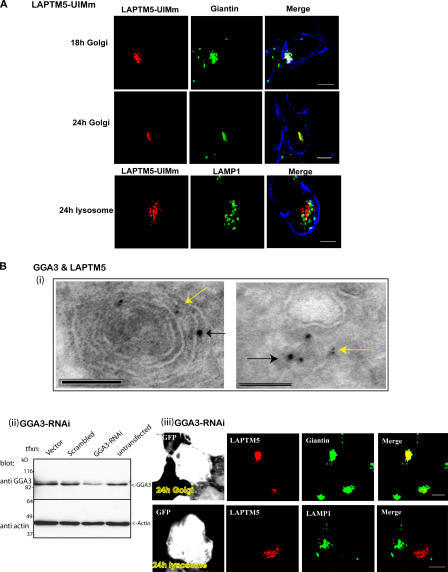
Figure 8.
The LAPTM5-UIM and GGA3 are required for Golgi-to-lysosome sorting of LAPTM5. (A) HEK293T cells were transfected with the UIM mutant DsRed-LAPTM5-UIMm (red), and its localization to the Golgi and lysosome 18 and 24 h after transfection was analyzed by staining with giantin or LAMP1, respectively (green). The PM is marked with ConA (blue) staining. (B; i) ImmunoEM analysis reveals colocalization of LAPTM5 (6-nm gold particles, yellow arrow) and endogenous GGA3 (10-nm gold particles, black arrow) in what appears to be a lysosome (left) and vesicles (right). (ii) Expression of GFP-tagged GGA3-RNAi in HEK293T cells leads to knockdown of endogenous GGA3. GGA3 antibodies were used to detect knockdown of GGA3 after (or not) transfection of pSUPER-GFP (vector), pSUPER-GFP–scrambled sequence (scrambled), and pSUPER-GFP-GGA3-RNAi (GGA3-RNAi). Actin controls are shown as well. (iii) Cells coexpressing GFP-GGA3-RNAi (white) and DsRed–LAPTM5 (red) were stained with anti-giantin (green) or anti-LAMP1 (green) antibodies 24 h after transfection of LAPTM5. GFP-GGA3-RNAi was transfected twice (at 0 and 48 h), followed by transfection with DsRed–LAPTM5 24 h later. Bars: (A) 5 μm; (B, i) 100 nm; (B, iii) 5 μm.
GGA3 is required for LAPTM5 translocation from the Golgi to lysosome
The ability of the LAPTM5-UIM to bind ubiquitinated GGA3 (Figs. 6 and 7), and the inability of the LAPTM5-UIMm mutant (which cannot bind ubiquitinated GGA3) to translocate to the lysosome (Fig. 8 A), suggest that GGA3 may be responsible for LAPTM5 sorting and translocation. To test this, RNAi for GGA3 was used to knockdown endogenous GGA3 in HEK293T cells cotransfected with LAPTM5 (and expressing endogenous Nedd4). As seen in Fig. 8 B (ii and iii) and Tables I, and S1, knockdown of GGA3 led to a substantial retention of LAPTM5 in the Golgi and impairment of lysosomal sorting.
To further substantiate these results, we performed immunoEM analysis to test for colocalization of LAPTM5 with GGA3. As seen in Fig. 8 B (i), LAPTM5 and GGA3 are detected together in vesicles that emanate from the TGN and lysosomes.
Golgi to lysosome translocation of LAPTM5 in dendritic DC2.4 cells
The aforementioned studies were performed in HEK293T cells. Although these cells express endogenous Nedd4 and GGA3, they do not express endogenous LAPTM5, which is primarily a hematopoietic protein. To ensure that the Nedd4/GGA3-dependent sorting of LAPTM5 from the Golgi to the lysosome was not an artifact of ectopic expression of LAPTM5 in HEK293T cells, we tested this sorting in DC2.4 cells, a cell line derived from dendritic cells of the immune system (Shen et al., 1997). As seen in Fig. S4 (available at http://www.jcb.org/cgi/content/full/jcb.200603001/DC1), DC2.4 cells express small amounts of endogenous LAPTM5. They also express endogenous Nedd4 (unpublished data). We next tested Golgi to lysosomal transport in these cells after transfection of DsRed-tagged WT or the 3YA mutant of LAPTM5. Fig. 9 B demonstrates that WT LAPTM5 expressed in DC2.4 cells is sorted from the Golgi to the lysosome, as seen in HEK293T cells. This is also validated by demonstrating colocalization of LAPTM5 with LAMP1 at the membrane of lysosomes using immunoEM (Fig. 9 A, i). In contrast, this sorting is attenuated in mutant LAPTM5 that cannot bind Nedd4 (the 3YA mutant); by 24 h after transfection, the proportion of cells translocated from the Golgi to the lysosome was much greater in the WT relative to the 3YA mutant ( Tables I and S1). These results suggest that, much like our observation with HEK293T cells, lysosomal sorting of LAPTM5 is also seen in dendritic cells that normally express endogenous LAPTM5, and this sorting likely involves Nedd4 binding as well. Moreover, the segregation of LAPTM5 to GGA3-containing TGN/vesicles and MVBs is also seen in the DC2.4 cells (Fig. 9 A, ii).
Figure 9.
LAPTM5 expressed in dendritic DC2.4 cells is also sorted from the Golgi to the lysosome in a PY motif–dependent manner. (A; i) ImmunoEM analysis reveals colocalization of HA-tagged LAPTM5 (6-nm gold particles, yellow arrow) with LAMP1 (15-nm gold particles, white arrow)-containing vesicles (LE/lysosomes) in DC2.4 cells. (ii) ImmunoEM analysis in DC2.4 cells demonstrates colocalization of HA-tagged LAPTM5 (6-nm gold particles, yellow arrow) with endogenous GGA3 (10-nm gold particles, black arrow) in a vesicle and in MVBs. (B and C) DC2.4 cells were transfected with DsRed-WT-LAPTM5 (B, red) or DsRed-LAPTM5-3YA mutant (C, red), and cells were immunostained with anti-giantin or LAMP1 antibodies (green) to detect Golgi to lysosomal sorting at the indicated times after transfection. Bars: (A, i) 100 nm; (A, ii) 200 nm; (B and C) 5 μm.
Discussion
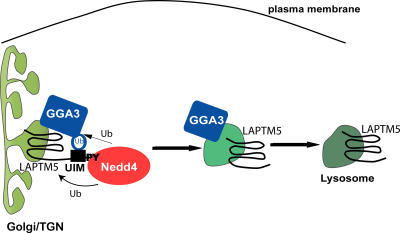
In this paper, we demonstrate that the transmembrane lysosomal protein LAPTM5 is sorted from the Golgi to the lysosome with the aid of Nedd4 and GGA3, a process not requiring LAPTM5 ubiquitination. We propose that LAPTM5, Nedd4, and GGA3 form a complex, whereby ubiquitinated GGA3 binds the UIM of LAPTM5, facilitating transport of LAPTM5 from the Golgi to lysosomes (Fig. 10). It is possible that Nedd4 dissociates from this complex once GGA3 is ubiquitinated (possibly by Nedd4) and able to bind LAPTM5, allowing the GGA3–LAPTM5 complex to be sorted into GGA3-containing vesicles that ultimately arrive at the MVBs/lysosomes (Fig. 10).
Figure 10.
A model depicting the role of Nedd4 and GGA3 in sorting of LAPTM5 to the lysosome from the Golgi. LAPTM5, via its PY motifs, binds (to the WW domains of) Nedd4, permitting the UIM of LAPTM5 to bind ubiquitinated GGA3. Nedd4 may contribute to the ubiquitination of GGA3. Thus, Nedd4 promotes the recruitment of GGA3 to LAPTM5, allowing LAPTM5 translocation from the Golgi to the lysosome with the aid of GGA3. After Nedd4-mediated ubiquitination, Nedd4 may dissociate, leaving the GGA3–LAPTM5 complex to sort into GGA3-containing vesicles traveling to the lysosome.
LAPTM5 and LAPTM4α and -β belong to a family of lysosomal membrane proteins (Adra et al., 1996; Hogue et al., 2002). Although the ubiquitously expressed LAPTM4 proteins are able to transport small molecules, such as nucleosides, into the lysosome (Hogue et al., 1996, 1999), the function of the hematopoietic-expressed LAPTM5 is not yet known. Moreover, the two family members (LAPTM4 and LAPTM5) may use different sorting signals for lysosomal targeting; LAPTM4α and -β have two Tyr-based sorting signals (YxxΦ) at their C termini, which, together with a di-Leu motif, are responsible for lysosomal targeting (Hogue et al., 2002). This is similar to other lysosome-associated membrane proteins, in which Tyr-based or di-Leu sorting signals were shown to interact with adaptor protein complexes (AP-1, -3, and -4; Huizing et al., 2001; Janvier et al., 2003; Janvier and Bonifacino, 2005; Theos et al., 2005). As we show, LAPTM5 has adapted a different sorting strategy; it utilizes its PY motifs (which bind Nedd4-WW domains) and UIM region (which binds ubiquitinated GGA3) to facilitate its sorting to lysosomes. It is interesting that two of the three PY motifs in LAPTM5 are also conserved in LAPTM4α and -β, but it is not yet known whether they also contribute to lysosomal targeting in these proteins.
Based on our work here, we propose that Nedd4 plays a crucial role in targeting LAPTM5 to lysosomes because a mutant that cannot bind Nedd4 (LAPTM5-3YA) is retained in the Golgi, and knockdown of Nedd4 leads to the same Golgi-retention defect. Interestingly, although Nedd4 can ubiquitinate LAPTM5, LAPTM5 ubiquitination is not essential for its sorting because the ubiquitination-impaired LAPTM5-10KR mutant can still bind GGA3 and be sorted to lysosomes. This suggests that LAPTM5 ubiquitination may have other, yet unknown, function, perhaps recruiting other proteins that contain ubiquitin-binding domains, enhancing function of the UIM, or regulating LAPTM5 stability. In contrast, GGA3 cannot bind the LAPTM5-3YA mutant, and this mutant is retained in the Golgi, suggesting that GGA3 can only interact with LAPTM5 in the presence of Nedd4. Moreover, this interaction requires the presence of active Nedd4, because overexpression of a catalytically inactive Nedd4(CS) precludes the association of GGA3 with the Nedd4–LAPTM5 complex. Exactly how Nedd4 regulates the interaction between LAPTM5 and GGA3 is currently unknown, but it is intriguing that the presence of active Nedd4 is required for the LAPTM5-UIM to more strongly bind ubiquitin. Thus, it is possible that Nedd4 binding to LAPTM5 ensures that the UIM of LAPTM5 can properly bind ubiquitinated GGA3 (Fig. 10). Moreover, we show that Nedd4 is also able to ubiquitinate GGA3, thereby further promoting the association of GGA3 with the LAPTM5-UIM.
Based on our observation that GGA3 bearing a mutation in its GAT domain, GGA3-L276A (which we show cannot bind ubiquitin), is still able to interact with LAPTM5, we believe that ubiquitination of LAPTM5 is not required for GGA3 binding, an observation that is in accord with our findings of proper lysosomal sorting of LAPTM5 lacking its ubiquitination sites. Thus, our work describes a novel mode of recruiting GGA3 to a protein complex in the Golgi; an interaction of ubiquitinated GGA3 with the UIM of LAPTM5 (Fig. 10). Moreover, this recruitment involves the ubiquitin ligase Nedd4, placing this E3 ligase in a complex with GGA3. Although we do not know which physiological E3 ligases are responsible for GGA3 ubiquitination, we show here that Nedd4 can ubiquitinate GGA3 in cells, suggesting that Nedd4 may be one such E3 ligase.
Our observation of the abolishment of binding of the GGA3-GAT domain mutant L276A to ubiquitin, although in agreement with previous work (Puertollano and Bonifacino, 2004; Shiba et al., 2004), is at odds with recent reports (Kawasaki et al., 2005; Prag et al., 2005) that demonstrate, using structural determinations, that a second site (site 1) in the GAT domain of GGA3 provides stronger affinity of interactions with ubiquitin than site 2, to which L276 belongs. As discussed elsewhere (Prag et al., 2005), ubiquitin binding is strongly affected by the mode of presentation and by the oligomerization state of its interacting partners, which could provide a possible explanation for the discrepant results.
It had been previously documented that the presence of the UIM in several endocytic proteins (e.g., epsin, eps15, Hrs) not only allows binding of ubiquitin to the UIM itself, but also promotes ubiquitination of these proteins on regions outside the UIM (Katz et al., 2002; Polo et al., 2002). Moreover, this ubiquitination often involves Nedd4 family members. However, it is not known how Nedd4 proteins are recruited to proteins such as epsin, eps15, or Hrs, which do not possess PY motifs. In the case of LAPTM5, we show here that it contains both UIM and PY motifs, allowing it to directly bind Nedd4, to become ubiquitinated by Nedd4, and to bind ubiquitin (attached to GGA3) via its UIM. GGA3 itself was recently shown to become monoubiquitinated (Shiba et al., 2004).
As already stated, the function of LAPTM5 is, thus far, unknown. Given its selective expression in immune cells and its lysosomal localization and similarities to LAPTM4 proteins, it is tempting to speculate that it may be involved in lysosomal movement in immune cells, and possibly in transport of molecules destined to be released from the lysosome once at the plasma membrane.
Materials and methods
Constructs and antibodies
HA-LAPTM5 was inserted into pCDNA3. To generate DsRed-LAPTM5, LAPTM5 cDNA was cloned into the pDsRed C1 vector (Invitrogen) downstream of and in frame with the DsRed fluorescent protein coding sequence. Mutations in LAPTM5, depicted in Fig. 1, were generated by site-directed mutagenesis using PCR, verified by sequencing and subcloned into pCDNA3. GST-WW2 constructs were previously described (Staub et al., 1996). His-tagged ubiquitin construct was obtained from D. Bohmann (University of Rochester Medical Center, Rochester, NY; Treier et al., 1994) and Myc-tagged WT and mutant GGA3 (L276A) were provided by J. Bonifacino (National Institutes of Health, Bethesda, MD). To knock down Nedd4-1, the oligo GATGAAGCCACCATGTATA was synthesized and inserted into the pSUPER-EGFP vector, as previously described (Amsen et al., 2006). For GGA3 knockdown, the oligo AAACGGCTTCCGCATCCTC was used as reported earlier (Puertollano and Bonifacino, 2004) and inserted into the pSUPER-EGFP as previously described (Amsen et al., 2006). Polyclonal antibody against LAPTM5 was generated using GST fusion proteins containing the C terminus of LAPTM5 (amino acid sequence between 225 and 260). Polyclonal anti–Nedd4-Hect domain antibodies (specific for Nedd4-1) were described earlier (Staub et al., 1997). Anti–Nedd4-2 antibodies were generated against a GST fusion protein encompassing a regions that is unique to Nedd4-2. Both anti–Nedd4-1 and -2 antibodies were affinity purified before use. The anti-GGA3 antibody was obtained from Santa Cruz Biotechnology, Inc. Rabbit anti-HA antibody for immunofluorescence was purchased from Sigma-Aldrich.
Cell culture and transfections
HEK293T cells were maintained in DME supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were transfected using the calcium phosphate precipitation method.
The immature murine DC2.4 cells (Shen et al., 1997) provided by K. Rock (University of Massachusetts Medical Center, Worcester, MA) were maintained in complete RPMI 1640 medium containing 10% fetal bovine serum, 2 mM l-glutamine, 100 μM nonessential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM β-mercaptoethanol. Transfections were performed using ESCORT V transfection reagent (Sigma-Aldrich) according to the manufacturer's protocol.
Immunofluorescent confocal microscopy
Autofluorescent DsRed-LAPTM5 was visualized using fluorescent confocal microscopy (excitation maximum, 558 nm; emission maximum, 583 nm). To visualize the plasma membrane, the HEK293T cells were stained with Alexa Fluor 488–conjugated concanavalin A (Invitrogen). The cells were fixed at varying times after transfection using partial Shaudinn's fixative (one part 95% ethanol/two parts saturated mercuric chloride) followed by wash in 70% methanol or 4% paraformaldehyde in PBS, followed by 0.1% Triton X-100. Fixed cells were incubated with the appropriate serum to block nonspecific antibody binding. To visualize Golgi and lysosomes, the cells were stained with polyclonal antibody against giantin (1:750; Covance) and H4A3 monoclonal antibody against LAMP1 (1:1,000; Developmental Studies Hybridoma Bank, under NICHD auspices; University of Iowa), followed by Cy5-conjgugated (Jackson ImmunoResearch Laboratories) or Alexa Fluor 647–conjugated (Invitrogen) anti–rabbit or anti–mouse secondary antibodies, respectively. To visualize early, sorting, and recycling endosomes, we used anti-EEA1 (Abcam), Hrs (H. Stenmark, The Norwegian Radium Hospital, Oslo, Norway), and Rab11 (Zymed) antibodies, respectively (each at 1:250 dilution). Single-plane images were acquired with a LSM 510 confocal microscope under 63×, 1.4 NA, oil-immersion objective (Carl Zeiss MicroImaging, Inc.). Fluorescent intensity was quantified with the LSM510 v3.2 ImagePC software (Carl Zeiss MicroImaging, Inc.). To better visualize colocalization of proteins, the Cy5/Alexa Fluor 647 and Alexa Fluor 488 were reassigned into green and blue colors, respectively.
DC2.4 cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.1% Triton X-100. Fixed cells were incubated with appropriate sera to block nonspecific antibody binding. To visualize Golgi and lysosomes, the cells were stained with polyclonal antibody against giantin (1:250; Covance) and monoclonal rat CD107a antibody against LAMP1 (1:750; BD Biosciences), followed by Cy5-conjugated anti–rabbit or anti–rat secondary antibodies (Jackson ImmunoResearch Laboratories), respectively.
Immunogold labeling and EM
Cryosectioning and immunolabeling were performed as previously described (Folsch et al., 2001), with the following changes. 24 h after transfection, cells were fixed for 1 h in 4% paraformaldehyde, followed by overnight fixation in 8% paraformaldehyde. 75-nm-thick sections were collected on formvar-coated nickel grids, at −120°C. The grids were blocked in a solution of 5% fish skin gelatin for 30 min and incubated with mouse anti-HA antibodies (1:25) and either rat CD107a (1:20) or rabbit LAMP2 (1:20; Sigma-Aldrich), giantin/furin (1:20; Abcam), or GGA3 (1:40; Abcam), followed by five washes in PBS and appropriate combination of 6 nm goat anti–mouse IgG/IgM and 10 nm goat anti–rabbit IgG, or with 15 nm goat anti–rat IgG (1:20; Electron Microscopy Sciences). The images were taken using a transmission EM (Tecnai 20; FEI) at 80 kV equipped with a camera (Dualview; Gatan), and processed using acquisition software (DigitalMicrograph; Gatan).
Pull-down and coIP assays
HEK293T (or DC2.4 cells) cells expressing either WT or mutant LAPTM5 cells were lysed with 1 ml of lysis buffer (50 mM Hepes, pH 7.5, 150 mN NaCl, 1% Triton X-100, 10% glycerol, 1.5 mM MgCl2, 1.0 mM EGTA, 10 mg/ml leupeptin, 10 mg/ml aprotinin, and 1 mM PMSF) and cleared by centrifugation at 14,000 rpm for 10–20 min. The cleared supernatants were used for pull-down and coIP experiments. For pull-down experiments, 500 μg of cell lysates were incubated with 50 μg GST or GST-Nedd4-WW2 (GST-WW2) protein on glutathione–Sepharose beads for 2 h at 4°C. Beads were washed twice with 1 ml HNTG (20 mM Hepes, pH 7.5, 150 mM NaCl, 10% glycerol, and 0.1% Triton X-100) and twice with lysis buffer. Bound proteins were eluted from the bead with 1× SDS-PAGE sample buffer, resolved by SDS-PAGE, and transferred to nitrocellulose membrane. Bound LAPTM5 was identified with anti-HA antibody (1:10,000; Babco), followed by secondary antibodies and ECL detection (GE Healthcare). To analyze the ability of the UIM of LAPTM5 to bind ubiquitin, 500 μg HEK293T cell lysates expressing either WT or mutant LAPTM5 that lacks the UIM (LAPTM5-UIMm) were incubated with ubiquitin immobilized on protein G beads (pro-G-Ub; 20 μg), the beads were washed, and bound LAPTM5 was detected with anti-HA antibody. For coIPs, HEK293T cell lysates expressing transfected WT or mutant HA-LAPTM5 together with T7-Nedd4 (CS; 500 μg each) were incubated with anti-T7 antibody for 1 h at 4°C, followed by the addition of 10 μl of protein G–Sepharose for an additional 1 h. After bound proteins were washed with lysis buffer (two times) and HNTG (three times), the coimmunoprecipitated LAPTM5 proteins were detected with anti-HA antibody (1:10,000). To identify GGA3 in the Nedd4–LAPTM5 complex, HEK293T cells were transfected with T7-Nedd4 (WT or CS mutant), Myc-GGA3 (WT or the L276A mutant that cannot bind ubiquitin) and HA-LAPTM5, and their lysates were subjected to IP with HA (LAPTM5) antibodies and immunoblotting with anti-Myc antibody (1:10,000) to detect bound GGA3.
Ubiquitination assays
HEK293T cells were cotransfected with HA-LAPTM5 or DsRed-LAPTM5 (WT or mutants) and His-tagged ubiquitin (His-Ub), and where indicated, cells were also cotransfected with T7-tagged Nedd4 (WT or catalytically inactive CS mutant). Cells were lysed in lysis buffer supplemented with 50 μM LLnL (N-acetyl-Leu-Leu-norleucinal; Sigma-Aldrich). Ubiquitinated (histidinated) proteins were then precipitated with Ni–Agarose beads for 4 h at 4°C and blotted with anti-HA antibodies to detect ubiquitinated LAPTM5, or where applicable, with anti-GGA3 antibodies to detect ubiquitinated GGA3. To ensure ubiquitination of LAPTM5, and not of associated proteins, the cell lysates were treated with 1% SDS and boiled for 5 min. These boiled lysates were then diluted 11 times with lysis buffer to dilute the SDS before their use in the Ni-bead precipitation step. Complexes with Ni–Agarose bead were washed with lysis buffer (two times) and HNTG (three times) and immunoblotted with anti-HA antibodies to detect LAPTM5, followed by anti–mouse horseradish peroxidase secondary antibodies and ECL detection. Equal amounts of different lysates (∼2 mg) were used for the precipitations.
Online supplemental material
Fig. S1 describes the contribution of the individual PY motifs of LAPTM5 to binding to Nedd4 and to LAPTM5 ubiquitination. Fig. S2 shows lack of colocalization of LATPM5(3YA) with EEA1, Rab11, or Hrs. Fig. S3 shows lysosomal localization of HA-tagged LAPTM5(10KR). Fig. S4 shows endogenous expression of LAPTM5 in DC2.4 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200603001/DC1.
Supplementary Material
Acknowledgments
We thank Dr. K. Rock for DC2.4 cells, Dr. J. Bonifacino for GGA3 cDNA, Dr. H. Stenmark for anti-Hrs antibodies, Dr. D. Bohmann for His-Ub cDNA, and A.Gaulis, A. S-Kotler, R. Bagshaw, M. Tropak, and B. Temkin for technical help.
This work was supported by the National Cancer Institute of Canada with funds from the Canadian Cancer Society, and the Canadian Institutes of Health Research (CIHR; to D. Rotin). Y. Pak was supported by a CIHR Fellowship, and N. Pham was supported by a CIHR Studentship. D. Rotin is supported by a Canada Research Chair (Tier II) award.
Y. Pak and W.K. Glowacka contributed equally to this paper.
Abbreviations used in this paper: HEK, human embryonic kidney; IP, immunoprecipitation; LAPTM, lysosomal-associated protein transmembrane; LE, late endosome; MVB, multivesicular body; Pm, plasma membrane; TM, transmembrane; UIM, ubiquitin-interacting motif; WT, wild type.
References
- Adra, C.N., S. Zhu, J.L. Ko, J.C. Guillemot, A.M. Cuervo, H. Kobayashi, T. Horiuchi, J.M. Lelias, J.D. Rowley, and B. Lim. 1996. LAPTM5: a novel lysosomal-associated multispanning membrane protein preferentially expressed in hematopoietic cells. Genomics. 35:328–337. [DOI] [PubMed] [Google Scholar]
- Amsen, E.M., N. Pham, Y. Pak, and D. Rotin. 2006. The guanine nucleotide exchange factor CNrasGEF regulates melanogenesis and cell survival in melanoma cells. J. Biol. Chem. 281:121–128. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P.S., J.L. Urbanowski, S.C. Winistorfer, and R.C. Piper. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534–539. [DOI] [PubMed] [Google Scholar]
- Blondel, M.O., J. Morvan, S. Dupre, D. Urban-Grimal, R. Haguenauer-Tsapis, and C. Volland. 2004. Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell. 15:883–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J.S. 2004. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 5:23–32. [DOI] [PubMed] [Google Scholar]
- Cabrita, M.A., T.C. Hobman, D.L. Hogue, K.M. King, and C.E. Cass. 1999. Mouse transporter protein, a membrane protein that regulates cellular multidrug resistance, is localized to lysosomes. Cancer Res. 59:4890–4897. [PubMed] [Google Scholar]
- Dell'Angelica, E.C., R. Puertollano, C. Mullins, R.C. Aguilar, J.D. Vargas, L.M. Hartnell, and J.S. Bonifacino. 2000. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsch, H., M. Pypaert, P. Schu, and I. Mellman. 2001. Distribution and function of AP-1 clathrin adaptor complexes in polarized epithelial cells. J. Cell Biol. 152:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, S.B., S. Losko, and C.A. Kaiser. 2001. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153:649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195–201. [DOI] [PubMed] [Google Scholar]
- Hicke, L., H.L. Schubert, and C.P. Hill. 2005. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6:610–621. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and L. Falquet. 2001. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 26:347–350. [DOI] [PubMed] [Google Scholar]
- Hogue, D.L., M.J. Ellison, J.D. Young, and C.E. Cass. 1996. Identification of a novel membrane transporter associated with intracellular membranes by phenotypic complementation in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 271:9801–9808. [DOI] [PubMed] [Google Scholar]
- Hogue, D.L., L. Kerby, and V. Ling. 1999. A mammalian lysosomal membrane protein confers multidrug resistance upon expression in Saccharomyces cerevisiae. J. Biol. Chem. 274:12877–12882. [DOI] [PubMed] [Google Scholar]
- Hogue, D.L., C. Nash, V. Ling, and T.C. Hobman. 2002. Lysosome-associated protein transmembrane 4 alpha (LAPTM4 alpha) requires two tandemly arranged tyrosine-based signals for sorting to lysosomes. Biochem. J. 365:721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing, M., R. Sarangarajan, E. Strovel, Y. Zhao, W.A. Gahl, and R.E. Boissy. 2001. AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol. Biol. Cell. 12:2075–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier, K., and J.S. Bonifacino. 2005. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell. 16:4231–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier, K., Y. Kato, M. Boehm, J.R. Rose, J.A. Martina, B.Y. Kim, S. Venkatesan, and J.S. Bonifacino. 2003. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 γ-σ1 and AP-3 δ-σ3 hemicomplexes. J. Cell Biol. 163:1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka, S., R. Mattera, and J.S. Bonifacino. 2005. Epidermal growth factor-dependent phosphorylation of the GGA3 adaptor protein regulates its recruitment to membranes. Mol. Cell. Biol. 25:7988–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz, M., K. Shtiegman, P. Tal-Or, L. Yakir, Y. Mosesson, D. Harari, Y. Machluf, H. Asao, T. Jovin, K. Sugamura, and Y. Yarden. 2002. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic. 3:740–751. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., M. Babst, and S.D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., G. Odorizzi, and S.D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893–905. [DOI] [PubMed] [Google Scholar]
- Kawasaki, M., T. Shiba, Y. Shiba, Y. Yamaguchi, N. Matsugaki, N. Igarashi, M. Suzuki, R. Kato, K. Kato, K. Nakayama, and S. Wakatsuki. 2005. Molecular mechanism of ubiquitin recognition by GGA3 GAT domain. Genes Cells. 10:639–654. [DOI] [PubMed] [Google Scholar]
- Levkowitz, G., H. Waterman, E. Zamir, Z. Kam, S. Oved, W.Y. Langdon, L. Beguinot, B. Geiger, and Y. Yarden. 1998. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12:3663–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.L., E. Malotky, and J.P. O'Bryan. 2004. Analysis of the role of ubiquitin-interacting motifs in ubiquitin binding and ubiquitylation. J. Biol. Chem. 279:33528–33537. [DOI] [PubMed] [Google Scholar]
- Misra, S., R. Puertollano, Y. Kato, J.S. Bonifacino, and J.H. Hurley. 2002. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature. 415:933–937. [DOI] [PubMed] [Google Scholar]
- Mosesson, Y., K. Shtiegman, M. Katz, Y. Zwang, G. Vereb, J. Szollosi, and Y. Yarden. 2003. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 278:21323–21326. [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., M. Babst, and S.D. Emr. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95:847–858. [DOI] [PubMed] [Google Scholar]
- Oldham, C.E., R.P. Mohney, S.L. Miller, R.N. Hanes, and J.P. O'Bryan. 2002. The ubiquitin-interacting motifs target the endocytic adaptor protein epsin for ubiquitination. Curr. Biol. 12:1112–1116. [DOI] [PubMed] [Google Scholar]
- Pelham, H.R. 2004. Membrane traffic: GGAs sort ubiquitin. Curr. Biol. 14:R357–R359. [DOI] [PubMed] [Google Scholar]
- Polo, S., S. Sigismund, M. Faretta, M. Guidi, M.R. Capua, G. Bossi, H. Chen, P. De Camilli, and P.P. Di Fiore. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 416:451–455. [DOI] [PubMed] [Google Scholar]
- Prag, G., S. Lee, R. Mattera, C.N. Arighi, B.M. Beach, J.S. Bonifacino, and J.H. Hurley. 2005. Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, gamma-ear-containing, ADP-ribosylation-factor-binding proteins. Proc. Natl. Acad. Sci. USA. 102:2334–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano, R., and J.S. Bonifacino. 2004. Interactions of GGA3 with the ubiquitin sorting machinery. Nat. Cell Biol. 6:244–251. [DOI] [PubMed] [Google Scholar]
- Puertollano, R., R.C. Aguilar, I. Gorshkova, R.J. Crouch, and J.S. Bonifacino. 2001. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 292:1712–1716. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., K.G. Bache, D.J. Gillooly, I.H. Madshus, E. Stang, and H. Stenmark. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4:394–398. [DOI] [PubMed] [Google Scholar]
- Rocca, A., C. Lamaze, A. Subtil, and A. Dautry-Varsat. 2001. Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor beta chain to late endocytic compartments. Mol. Biol. Cell. 12:1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, L.M., L. Mueller, and S.J. Collins. 1996. E3, a hematopoietic-specific transcript directly regulated by the retinoic acid receptor alpha. Blood. 88:2517–2530. [PubMed] [Google Scholar]
- Scott, P.M., P.S. Bilodeau, O. Zhdankina, S.C. Winistorfer, M.J. Hauglund, M.M. Allaman, W.R. Kearney, A.D. Robertson, A.L. Boman, and R.C. Piper. 2004. GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat. Cell Biol. 6:252–259. [DOI] [PubMed] [Google Scholar]
- Shekhtman, A., and D. Cowburn. 2002. A ubiquitin-interacting motif from Hrs binds to and occludes the ubiquitin surface necessary for polyubiquitination in monoubiquitinated proteins. Biochem. Biophys. Res. Commun. 296:1222–1227. [DOI] [PubMed] [Google Scholar]
- Shen, Z., G. Reznikoff, G. Dranoff, and K.L. Rock. 1997. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J. Immunol. 158:2723–2730. [PubMed] [Google Scholar]
- Shiba, Y., Y. Katoh, T. Shiba, K. Yoshino, H. Takatsu, H. Kobayashi, H.W. Shin, S. Wakatsuki, and K. Nakayama. 2004. GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J. Biol. Chem. 279:7105–7111. [DOI] [PubMed] [Google Scholar]
- Shih, S.C., D.J. Katzmann, J.D. Schnell, M. Sutanto, S.D. Emr, and L. Hicke. 2002. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4:389–393. [DOI] [PubMed] [Google Scholar]
- Shih, S.C., K.E. Sloper-Mould, and L. Hicke. 2000. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 19:187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetens, O., J.O. De Craene, and B. Andre. 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276:43949–43957. [DOI] [PubMed] [Google Scholar]
- Staub, O., S. Dho, P. Henry, J. Correa, T. Ishikawa, J. McGlade, and D. Rotin. 1996. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle's syndrome. EMBO J. 15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- Staub, O., H. Yeger, P.J. Plant, H. Kim, S.A. Ernst, and D. Rotin. 1997. Immunolocalization of the ubiquitin-protein ligase Nedd4 in tissues expressing the epithelial Na+ channel (ENaC). Am. J. Physiol. 272:C1871–C1880. [DOI] [PubMed] [Google Scholar]
- Staub, O., and D. Rotin. 2006. The role of ubiquitylation in cellular membrane transport. Physiol. Rev. 86:669–707. [DOI] [PubMed] [Google Scholar]
- Terrell, J., S. Shih, R. Dunn, and L. Hicke. 1998. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell. 1:193–202. [DOI] [PubMed] [Google Scholar]
- Theos, A.C., D. Tenza, J.A. Martina, I. Hurbain, A.A. Peden, E.V. Sviderskaya, A. Stewart, M.S. Robinson, D.C. Bennett, D.F. Cutler, et al. 2005. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell. 16:5356–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier, M., L.M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 78:787–798. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., B. Doray, A. Poussu, V.P. Lehto, and S. Kornfeld. 2001. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science. 292:1716–1718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.