Abstract
The functional specificity conferred by glycophosphatidylinositol (GPI) anchors on certain membrane proteins may arise from their occupancy of specific membrane microdomains. We show that membrane proteins with noninteractive external domains attached to the same carcinoembryonic antigen (CEA) GPI anchor, but not to unrelated neural cell adhesion molecule GPI anchors, colocalize on the cell surface, confirming that the GPI anchor mediates association with specific membrane domains and providing a mechanism for specific signaling. This directed targeting was exploited by coexpressing an external domain-defective protein with a functional protein, both with the CEA GPI anchor. The result was a complete loss of signaling capabilities (through integrin–ECM interaction) and cellular effect (differentiation blockage) of the active protein, which involved an alteration of the size of the microdomains occupied by the active protein. This work clarifies how the GPI anchor can determine protein function, while offering a novel method for its modulation.
Introduction
Many cell surface proteins are attached to the membrane by a glycophosphatidylinositol (GPI) anchor, which consists of a conserved central structure (Low, 1989) with variable carbohydrate and lipid peripheral components (Homans et al., 1988). GPI anchors can determine protein functional specificity, just as switching a transmembrane (TM) domain for a GPI anchor can result in novel function caused by association with new signaling elements located in a shared membrane microdomain (Shenoy-Scaria et al., 1992, 1993).
Membrane rafts, originally defined by their insolubility in cold, nonionic detergents such as Triton X-100 (Simons and Ikonen, 1997), are small, heterogeneous aggregations of cholesterol and sphingolipids on the cell surface (Pralle et al., 2000; Pike, 2004) that concentrate GPI-anchored proteins, but also contain other proteins. Although the existence of membrane rafts in vivo has been questioned (Munro, 2003), recent studies using a variety of methods have provided evidence for raftlike membrane microdomains (Friedrichson and Kurzchalia, 1998; Varma and Mayor, 1998; Pralle et al., 2000; Dietrich et al., 2002; Gaus et al., 2003; Sharma et al., 2004). Such microdomains may act as signaling scaffolds, determining the identity of a subset of signaling elements, as proteomic analyses have found a high concentration of such proteins in purified rafts (von Haller et al., 2001; Foster et al., 2003), with GPI-anchored proteins involved in activating this signaling (Robinson, 1997; Solomon et al., 1998). The existence of heterogeneous raft populations has been inferred from studies showing that different GPI-anchored proteins exist in separate rafts (Madore et al., 1999; Wang et al., 2002; Li et al., 2003). External rafts with different proteins may each have a defined set of associated cytoplasmic proteins, whereby aggregation of GPI-anchored proteins by external domain self-binding or by multivalent ligand binding could cluster specific rafts, resulting in downstream signaling (Harris and Siu, 2002).
Carcinoembryonic antigen (CEA), and the closely related CEACAM6, are GPI-anchored, cell surface glycoproteins that block cellular differentiation (Eidelman et al., 1993) and inhibit the apoptotic process of anoikis (Ordonez et al., 2000; Duxbury et al., 2004b), effects that appear to be caused by the activation of specific integrins (Duxbury et al., 2004a; Ordonez et al., 2006). CEA is up-regulated in many human malignancies (Hinoda et al., 1991; Ilantzis et al., 1997), implying a similar role in human cancer, whereas the TM-anchored CEACAM1 (CC1) may act as a tumor suppressor (Kunath et al., 1995; Luo et al., 1997).
Most CEA family members mediate intercellular adhesion by antiparallel self-binding (Zhou et al., 1993), which, together with parallel binding on the same cell surface (Taheri et al., 2003), may result in clustering of rafts containing CEA (Benchimol et al., 1989). Deletion of the last two thirds of the CEA N-terminal domain (ΔNCEA) abrogates its adhesive ability, which leads to a loss of differentiation-blocking activity (Eidelman et al., 1993). The method of membrane anchorage determines CEA family member activity, as genetically fusing the GPI anchor of CEA to CC1's external domain creates a differentiation-blocking molecule, whereas a chimera consisting of the external domain of CEA attached to the TM domain of CC1 does not block differentiation (Screaton et al., 2000). The fact that GPI-anchored neural cell adhesion molecule (NCAM) does not block differentiation, but can be converted to a differentiation-blocking molecule, denoted NCB (previously “NC blunt”), by swapping its GPI anchor for that of CEA, suggests that the CEA GPI anchor harbors the specificity for the differentiation-blocking function and that the external domains merely function to cluster the molecules, and thus, the associated rafts (Screaton et al., 2000).
Based on the aforementioned model, it should be possible to inhibit the biological functions of CEA (and, by implication, that of any GPI-anchored molecule whose function is regulated by a similar mechanism) by interfering with clustering. This has been achieved for CEA by mutating regions in its N-terminal external domain responsible for self-binding or by the addition of peptides or monovalent mAbs that target these regions (Taheri et al., 2003). We test a second strategy which exploits the specificity of the CEA GPI anchor; if “shank-defective” or “shankless” CEA GPI anchors that were incapable of self-association and clustering were introduced, they could occupy the same rafts as CEA, and thus, possibly interfere with its clustering. We show that nonfunctional ΔNCEA inhabits the same membrane microdomains as NCB, as both have the same GPI anchor, but not those of NCAM, and is capable of completely inhibiting NCB's CEA-like differentiation-blocking activity.
Results
Design and construction of experiments
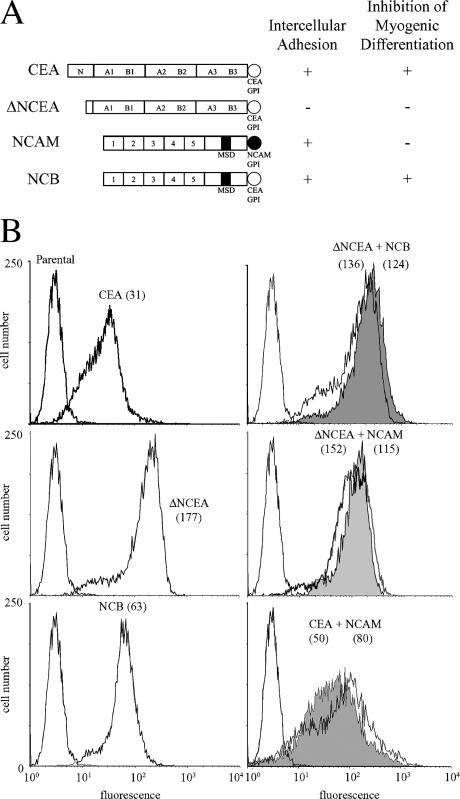
To test the hypothesis that the functional specificity of GPI anchors could be exploited to specifically inhibit the activity of GPI-anchored proteins, cells expressing a functional GPI- anchored protein were supertransfected with a shank-defective molecule with the same GPI anchor, and assessed for effects on function. The former functioning molecule was NCB, which has NCAM self-binding external domains linked to the CEA GPI anchor (Screaton et al., 2000); the defective molecule was ΔNCEA, which has the same GPI anchor, but external domains that are defective in self-binding (Fig. 1 A; Eidelman et al., 1993). Because ΔNCEA cannot bind to the external NCAM domain of NCB (Zhou et al., 1990), this combination allowed a study focused on the potential interaction between their GPI anchors.
Figure 1.
Surface expression of CEA and NCAM proteins on L6 myoblasts. (A) Schematic representation of proteins used in this study, with their ability to mediate intercellular adhesion and to inhibit myogenic differentiation shown (as + or −). (B) FACS profiles, after staining with mAbs D14 (anti-CEA) or 123C3 (anti-NCAM) and FITC-conjugated secondary antibody, demonstrate cell surface levels of indicated proteins with fluorescence means in parentheses. In coexpressing cells, D14 staining is shown by shaded profiles.
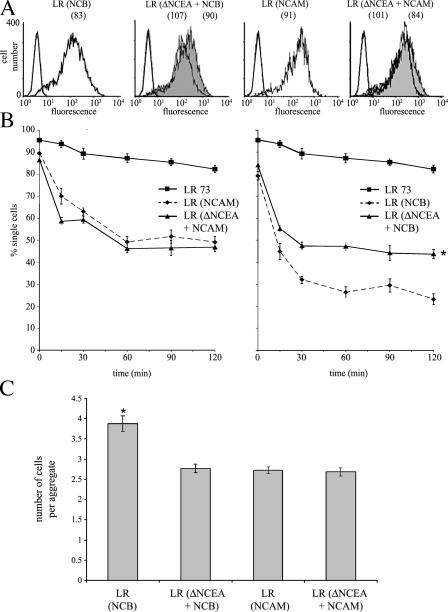
ΔNCEA was stably cotransfected into NCB transfectants of rat L6 myoblasts, which are blocked for myogenic differentiation because of the expression of NCB. ΔNCEA was present on the cell surface of the double transfectants at slightly higher levels than NCB, as seen by FACS (Fig. 1 B) and Western blot (unpublished data). As a control for specificity of effects, double transfectants stably expressing molecules with different GPI anchors were used, i.e., ΔNCEA or CEA with CEA GPI anchors, and NCAM with the NCAM GPI anchor. Similar expression levels were also obtained for these transfectants (Fig. 1 B).
ΔNCEA and NCB exist in close proximity
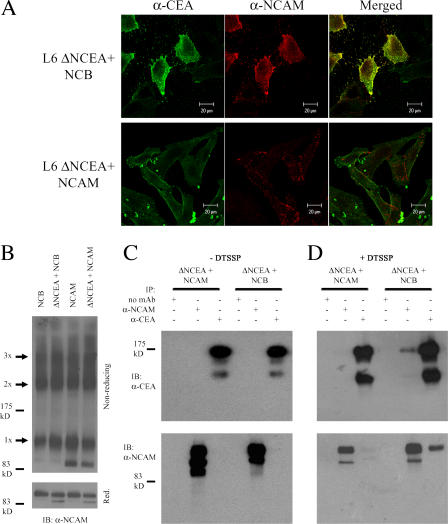
CEA and NCAM appear to exist in separate membrane regions, potentially explaining their opposite biological effects (Screaton et al., 2000). If the GPI anchor alone determines cell surface localization, then molecules with the same GPI anchor should exist in close proximity, whereas those with different anchors should not. Thus, NCAM and NCB would be expected to have different cell surface distributions, with NCB showing a distribution similar to that of CEA. To test this hypothesis, we examined whether ΔNCEA existed in close proximity on the cell surface to NCB, but not to NCAM, using confocal microscopy to examine the cell surface localization of these proteins. The relative surface distribution of NCAM and NCB compared with ΔNCEA was determined after indirect immunofluorescent staining. ΔNCEA showed substantial, although incomplete, colocalization with NCB, whereas ΔNCEA and NCAM showed essentially no colocalization (Fig. 2 A). Because the incubations were performed at room temperature, the antibodies used for detection may have caused partial clustering of the proteins. This, however, should not affect the heterophilic association in question, as clustering of rafts containing both proteins should not change the final amount of colocalization seen. Indeed, fixing the cells before antibody incubation to avoid clustering resulted in very similar patterns of colocalization to what is shown in Fig. 2 A (not depicted). This therefore suggests that the GPI anchor of CEA is sufficient to determine cell surface localization of a protein.
Figure 2.
ΔNCEA exists in close proximity to NCB, but not to NCAM. (A) ΔNCEA colocalizes with NCB, but not with NCAM. Indirect immunofluorescence was visualized by confocal microscopy, using mouse anti-NCAM and rabbit anti-CEA primary mAbs, followed by Cy2-conjugated goat anti–rabbit (left) and rhodamine-conjugated goat anti–mouse secondary antibodies (center). Merged figures (right) demonstrate considerable ΔNCEA colocalization with NCB, but not NCAM, as shown by the yellow regions. (B) Cross-linking with DTSSP resulted in a similar pattern of NCB or NCAM high molecular weight bands (with monomer [1x], dimer [2x], and trimer [3x] being the predominant bands) on a nonreducing acrylamide gel (top), with similar protein levels seen on a reducing (Red.) gel (bottom). (C) IP of these samples was then undertaken. IP in the absence of DTSSP cross-linking showed specific antibody binding, as no coIP occurred. (D) IP of DTSSP cross-linked samples demonstrated coIP of ΔNCEA with NCB and vice-versa, but not with NCAM, demonstrating a specific, close association of ΔNCEA and NCB. IB, immunoblot.
To verify these results, L6 cotransfectants were treated at 4°C (to limit protein diffusion) with the chemical cross-linker DTSSP. Nonreducing Western blots demonstrated similar cross-linking patterns for NCB and NCAM, consisting of dimers, trimers, and higher molecular weight complexes, both alone and in the presence of ΔNCEA (Fig. 2 B). To determine the cellular distribution of ΔNCEA relative to NCAM and NCB, immunoprecipitation (IP) studies of extracts from cross-linked cells expressing similar amounts of these proteins were performed. The cross-linking approach was undertaken, rather than using detergent lysis because of the potential effects of detergents on membrane raft structure. IP of extracts from untreated cells did not result in any coIP (Fig. 2 C), confirming the expected antibody specificity. However, IP with an anti-CEA mAb of extracts of DTSSP-treated cotransfectants resulted in the coIP of a considerable amount of NCB, but, importantly, not of NCAM (Fig. 2 D). Similarly, IP with an anti-NCAM mAb of extracts of cross-linked cotransfectants showed coIP of ΔNCEA only in the case of NCB, but not of NCAM (Fig. 2 D). The low proportion of coimmunoprecipitated protein can likely be explained by the lack of interaction between the external NCAM and CEA protein domains, the requirement for close (<12 Å) apposition to be cross-linked, the presence of large levels of monomeric proteins even after cross-linking (Fig. 2 B), and the incomplete colocalization seen by confocal microscopy (Fig. 2 A). These results demonstrate that proteins with GPI anchors of the same type can exist in close proximity, providing a rationalization for specific interference with protein function.
ΔNCEA restores differentiation to NCB-expressing cells
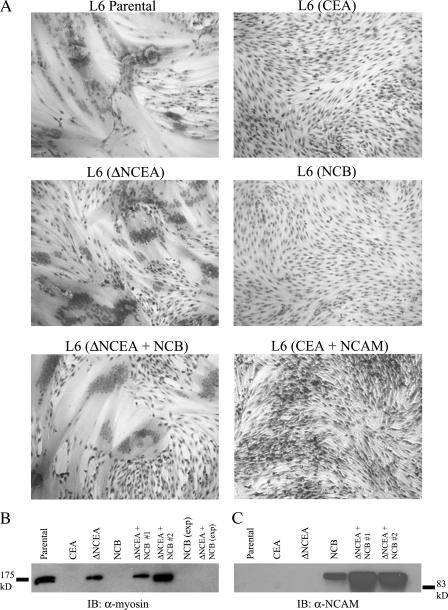
Having demonstrated specific colocalization of ΔNCEA and NCB on the cell surface, the effect of this defective protein on NCB's ability to block differentiation was examined. NCB levels in the ΔNCEA coexpressing L6 transfectants were actually higher than those in NCB-only transfectants, thus, validating comparisons between NCB alone and in the presence of ΔNCEA (Fig. 1 B; FACS means of 124 vs. 63, respectively). NCB completely blocked differentiation, whereas coexpression of ΔNCEA with NCB resulted in an almost complete restoration of differentiation, with a fusion index of 78% of that seen for ΔNCEA alone (Fig. 3 A and Table I; P < 0.0001). As a control, coexpressing NCAM had no effect on the differentiation block imposed by CEA (Fig. 3 A), despite the differentiation-enhancing effects of NCAM (Dickson et al., 1990). To confirm this result, up-regulation of myosin, a biochemical differentiation marker, was examined. ΔNCEA induced myosin production in two independent populations of NCB-expressing cells, as shown by Western blot, whereas NCB alone showed no myosin expression (Fig. 3 B), confirming the previous results. Because of the length of the differentiation assay (10 d total), it was possible that a loss of NCB expression caused the differentiation restoration in these cotransfectants. However, no decrease in NCB levels was seen in differentiated ΔNCEA + NCB cultures, as Western blots showed higher expression levels in both cell populations than the NCB-alone transfectants for which differentiation was blocked (Fig. 3 C). Thus, ΔNCEA expression interfered markedly with the differentiation-blocking function of NCB, presumably via their common feature, the GPI anchor.
Figure 3.
ΔNCEA restores differentiation to NCB-expressing cells. (A) Photomicrographs of L6 transfectants tested for myogenic differentiation after a 7-d incubation in DM. Parental and ΔNCEA cells fused readily, whereas NCB and CEA completely blocked differentiation; coexpression of ΔNCEA with NCB restored differentiation. (B) Up-regulation of the differentiation marker myosin, as determined by Western blot with mAb 47A on 10 μg of total lysate, was seen only in parental, ΔNCEA, and two separate populations of ΔNCEA + NCB. No myosin expression was seen in exponentially growing (exp) cultures of NCB or ΔNCEA + NCB transfectants. (C) NCB expression in differentiated cultures. Western blots of 10 μg of total cellular lysate demonstrated that NCB expression remained higher in coexpressing cells than in NCB-alone transfectant cells after 7 d in DM.
Table I.
Differentiation of L6 transfectants
| Cell line | Fusiona |
|---|---|
| % | |
| L6 parental | 71 |
| L6 (CEA) | 0 |
| L6 (ΔNCEA) | 64 |
| L6 (NCB) | 0 |
| L6 (ΔNCEA + NCB) | 50 |
| L6 (CEA + NCAM) | 0 |
Fusion index was measured as the number of cells containing three or more nuclei divided by total nuclei in the field and expressed as a percentage. Values are the mean of three different experiments, with three fields scored from each experiment.
Effects of CEA-like proteins on binding to ECM
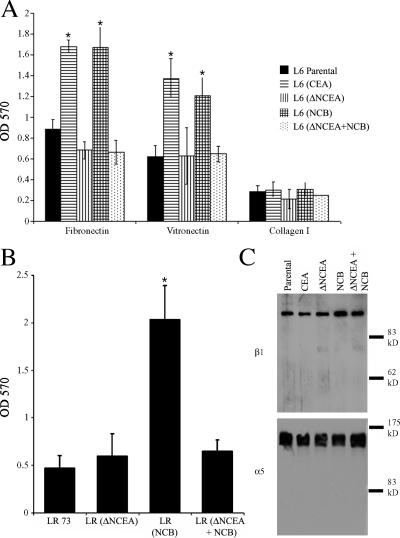
ΔNCEA releases NCB's block of differentiation, suggesting that it is interfering with downstream signaling by NCB. CEA signaling has been found to involve activation of the integrin α5β1 in rat myoblast and human colonic cell lines (Ordonez et al., 2006) and the integrin αvβ3 in neuronal cells (unpublished data). We assessed NCB signaling by incubating single-cell suspensions, prepared from exponential cultures, with plates coated with the ECM components fibronectin (Fn), vitronectin (Vn), and collagen I. Either CEA or NCB expression increased binding to both Fn and Vn, relative to L6 parental cells (Fig. 4 A; P < 0.004). Cells expressing ΔNCEA + NCB (and ΔNCEA alone) showed no such increase, demonstrating a complete loss, in the presence of ΔNCEA, of the NCB-mediated increase in ECM binding (P < 0.0001). As a control, no difference in binding to collagen I was seen between any of these cell lines (Fig. 4 A). In addition, LR-73 (LR) transfectants were tested for binding to Fn, and these cells showed a similar loss of NCB-mediated effects upon coexpression of ΔNCEA (Fig. 4 B; P < 0.01). The total cell levels of α5 and β1 integrins in the L6 transfectants were assessed by Western blot (Fig. 4 C), and cell surface levels were assessed by FACS (α5 only; not depicted) and showed only minor differences between transfectants, confirming that the ability of cells to adhere to Fn, rather than changes in integrin surface expression level, was the source of the observed difference (Ordonez et al., 2006; unpublished data). Thus, the ability of ΔNCEA to interfere with the NCB-mediated differentiation blockage is correlated with interference of enhanced integrin–ECM interaction promoted by NCB.
Figure 4.
Increased ECM binding by NCB transfectants is lost in the presence of ΔNCEA. (A) L6 (CEA) and L6 (NCB) transfectants showed increased binding, compared with parental L6 cells, to immobilized Fn and Vn, whereas the presence of ΔNCEA in NCB transfectants abrogated this increase. No difference in collagen I binding was seen between parental and transfectant lines. Mean values are ± the SD from three independent experiments are shown (*, P < 0.004). (B) LR transfectants showed a similar pattern of binding to Fn, with an increase seen for NCB, but not for ΔNCEA + NCB transfectants (*, P < 0.003). (C) No significant alteration in integrin β1 (top) or α5 (bottom) expression was seen by Western blots on 5 μg of lysate from each L6 cell line.
NCB raft association is unaltered by the presence of ΔNCEA
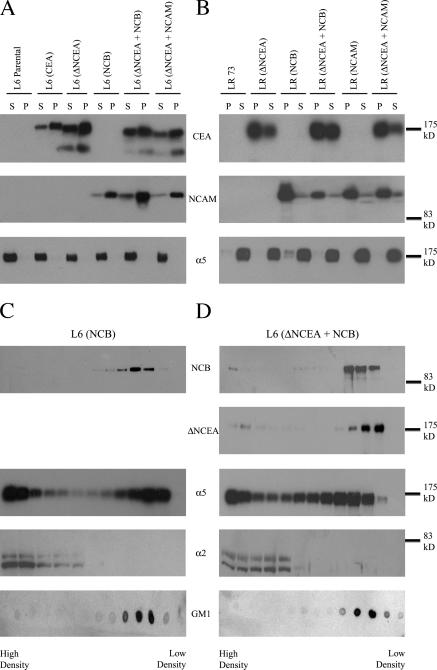
Signaling by GPI-anchored proteins requires intact membrane rafts (Stulnig et al., 1997). One possible mechanism for the effects of ΔNCEA on NCB functional properties could be by expulsion of NCB from rafts. When this possibility was examined, however, NCB remained primarily insoluble in cold Triton X-100 after coexpression of ΔNCEA in either L6 or LR cells (Fig. 5, A and B, respectively). Complete cellular lysis was demonstrated by the fact that the integrin α5 chain, an integral membrane protein, was localized in the soluble fractions. As confirmation, isopycnic sucrose gradient ultracentrifugation, where raft-associated proteins migrate to the lower density regions of the gradient, was performed on cold Brj-98 lysates of L6 transfectants. Again, no obvious difference was noted between NCB alone and NCB coexpressed with ΔNCEA (Fig. 5, C and D), as almost all of the NCB was present in the low-density fractions in both cases. Under these conditions, the α5 integrin chain showed partial raft association for both transfectants, demonstrating that an alteration of α5 localization was not responsible for the lack of NCB function. The distribution of ΔNCEA was also found to be essentially the same as that of NCB, as expected for two GPI-anchored proteins. As controls, the α2 integrin chain was found solely in higher density fractions, whereas the raft lipid GM1 was entirely in the low- density fractions. Thus, NCB retained membrane raft association in the presence of ΔNCEA, so that this could not explain the loss of NCB function.
Figure 5.
NCB membrane raft association is unaltered in the presence of ΔNCEA. Triton X-100 solubility assays of L6 (A) and LR (B) cells at 4°C showed no alteration in membrane raft association of NCB ± ΔNCEA transfectants with the majority of the protein in the insoluble (P), i.e., raft-associated, fraction. As a control for efficiency of lysis, the integrin α5 chain showed essentially no insolubility, with all protein found in the soluble (S) fraction for both L6 and LR parental and transfectant cell lines. Isopycnic sucrose density gradient ultracentrifugation showed similar membrane raft association, as seen by flotation at lower density fractions of NCB in both L6 (NCB; C) and L6 (ΔNCEA + NCB; D) transfectants. Integrin α5 had partial raft association, whereas integrin α2 was not found in lower density fractions. The raft marker GM1 was found exclusively in lower density fractions, whereas ΔNCEA showed similar distribution to NCB.
Effect of expression of ΔNCEA on NCB-mediated intercellular adhesion
The proteins of the CEA family mediate intercellular adhesion, as does NCAM (Soroka et al., 2003), by external domain self-binding. Such self-binding is required for the differentiation-inhibitory activity of CEA, presumably to affect raft clustering (Eidelman et al., 1993; Taheri et al., 2003). If ΔNCEA interfered with NCB clustering, one might predict a reduction in the ability of NCB to mediate intercellular adhesion. NCAM was used as a control, as it inhabits different rafts from ΔNCEA (Fig. 2 C). NCAM and NCB were expressed at very similar levels, with and without ΔNCEA, on the surface of LR cells, thus allowing for quantitative comparisons in adhesion between populations (Fig. 6 A). A significant reduction in the strength of NCB-mediated adhesion occurred in the presence of ΔNCEA, as shown by a reproducible decrease of ∼20% in the number of aggregated cells in suspension after 2 h (P < 0.001), a difference that was not seen for NCAM-mediated adhesion (Fig. 6 B). This was accompanied by a decrease in the size of aggregates in NCB-expressing cells as a result of ΔNCEA coexpression (P < 0.0001), which, again, was not seen for NCAM (Fig. 6 C). Thus, introducing the same functional GPI anchor with a defective shank led to a specific reduction in the strength of intercellular adhesion by NCB. Effective intercellular adhesion by GPI-anchored proteins is believed to involve the formation of large, zipperlike structures through the aggregation of multiple proteins and rafts, creating stabilized platforms (Harris and Siu, 2002). The ability of ΔNCEA to interfere with NCB-mediated adhesion is thus consistent with models invoking interference with NCB clustering.
Figure 6.
ΔNCEA interferes with NCB-mediated intercellular adhesion. (A) LR surface expression (fluorescence means are shown in parenthesis) of NCAM-like proteins (unshaded profiles) and ΔNCEA (shaded profiles) after staining with mAbs 123C3 and D14, respectively. (B) Adhesion assays of LR (NCB) and LR (ΔNCEA + NCB; right) and LR (NCAM) and LR (ΔNCEA + NCAM; left). Note the 20% decrease (*, P < 0.001) in NCB-mediated adhesion in the presence of ΔNCEA, which is not seen in NCAM versus NCAM + ΔNCEA transfectants. Values represent the mean ± the SEM for four independent experiments. (C) Aggregate assays of cells from B, showing that the size of multicellular aggregates created by NCB was significantly reduced in the presence of ΔNCEA. The number of cells per multicellular aggregate was determined, after 1 h of incubation, in four separate fields per experiment. Data represent the mean ± SEM of three independent experiments (*, P < 0.0001).
ΔNCEA alters the size of NCB-containing rafts
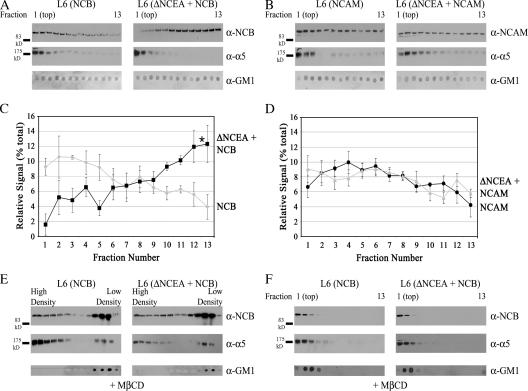
One mechanism whereby ΔNCEA could interfere with NCB clustering is by altering the structure of the rafts it is associated with. Therefore, the size of the rafts that NCB occupied was approximated by lysing the cells under conditions identical to those used for isopycnic separation on sucrose density gradients and separating the lysate by velocity sedimentation through a uniform 12.5% sucrose solution. Under these conditions, NCB was almost entirely raft associated (Fig. 5); therefore, this technique should provide a measure of the size of the rafts inhabited by these proteins. The fractions, which were collected from the top (fraction #1), were assessed by Western blot for protein localization, using equal volumes of each fraction. NCB was found to be shifted to fractions farther from the top when ΔNCEA was coexpressed, indicating that it was present in larger complexes under these conditions (Fig. 7 A). The distribution of NCAM, on the other hand, was found to be similar whether ΔNCEA was present or not (Fig. 7 B), demonstrating that the size of the NCAM complexes was not altered in the presence of ΔNCEA. The distribution of ΔNCEA was very similar to that of NCB in shifting to larger complexes when coexpressed with NCB, while remaining in smaller complexes when coexpressed with NCAM (unpublished data). A significant (P < 0.05) difference in NCB distribution (Fig. 7 C) relative to NCAM distribution (Fig. 7 D) upon coexpression with ΔNCEA was demonstrated by densitometric analysis of three independent experiments. This suggests that the presence of ΔNCEA specifically alters the rafts containing NCB. To confirm that this was a raft-specific effect, cells were pretreated with methyl-β-cyclodextrin (MβCD) to sequester cholesterol and disrupt raft structure. Initially, sucrose gradient ultracentrifugation was performed on lysates of these treated cells, to confirm the disruption of the rafts. The distribution of NCB demonstrated that this treatment partially disrupted the rafts, as a portion of the NCB was now present in higher density fractions (Fig. 7 E; compare to Fig. 5). When these samples were tested for the size of the complexes that NCB was localized to, it was found that NCB, both alone and coexpressed with ΔNCEA, remained in the first few fractions after velocity sedimentation (Fig. 7 F). Thus, treatment with MβCD abrogated the difference seen for NCB complex size after ΔNCEA coexpression, confirming that the difference seen was a raft-mediated effect. Although cellular lysis with detergents at low temperatures can affect raft structure (Shogomori and Brown, 2003), the fact that the ΔNCEA-dependent sedimentation difference is seen for NCB, but not for NCAM transfectants, suggests that it represents a valid increase in raft size. This would indicate a dilution of the NCB concentration in membrane rafts, as an increase in the size of a raft containing the same number of NCB molecules would cause a relative concentration decrease. This would thus reduce the incidence of cis-interactions between the proteins, which are necessary for clustering, explaining the decrease in intercellular adhesion (Fig. 6 B), and the loss of biological function (Figs. 3 A and 4 A).
Figure 7.
ΔNCEA increases the size of NCB-containing rafts. Cellular lysates were assessed for membrane raft size by velocity centrifugation through a 12.5% sucrose column. Fractions collected from the top were probed for protein localization; note that only the first 13 of 25 fractions are shown, as no NCB/NCAM protein was located in lower fractions. (A) Fractions of L6 (NCB) and L6 (ΔNCEA + NCB) were analyzed for NCB, integrin α5, and GM1 distribution by Western blot. (B) Similar analysis of L6 (NCAM) and L6 (ΔNCEA + NCAM) lysates was performed. Densitometric quantitation of NCB (C) and NCAM (D) localization was then performed for these fractions. Values represent the mean ± the SEM of 3 independent experiments (*, P < 0.05). (E) Effect of raft disruption with MβCD on sucrose gradient flotation of NCB. Note the shift of NCB distribution into higher density fractions, when compared with Fig. 5. (F) Treatment with MβCD results in a complete loss of the effect of ΔNCEA on NCB complex size. Fractions of cells pretreated with MβCD were collected as in A, and probed for protein localization. Raft disruption caused NCB to be found solely in the highest fractions, whether ΔNCEA was present or not.
Antibody cross-linking restores function of NCB inhibited by ΔNCEA
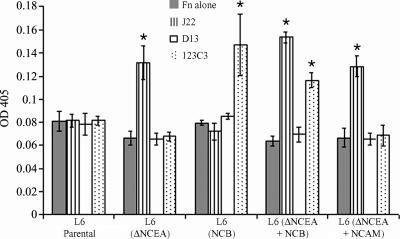
As the mechanism of inhibition of NCB function by ΔNCEA appears to involve interference with clustering, NCB function should be restored by artificial clustering with antibodies. Antibody cross-linking of cell surface proteins induces signaling events, including restoring the defective differentiation-blocking function of ΔNCEA (Taheri et al., 2003) through integrin activation manifested by increased cellular binding of Fn (unpublished data). ΔNCEA and NCB coexpressed with ΔNCEA both appear to be nonfunctional because of defects in protein clustering, so clustering of NCB with antibodies should have a similar effect to what has previously been seen for ΔNCEA. To test if NCB retained the potential to modulate ECM binding, in spite of the deactivating effects of coexpressed ΔNCEA, cells in monolayer culture were treated with mAbs directed against the NCAM external domains of NCB, along with secondary antibodies to enhance clustering, and binding of soluble Fn was measured. Several mAbs were used, including J22, which binds to internal CEA domains and, as such, remains capable of clustering ΔNCEA; D13, which is a control mAb that has an epitope in the region deleted from ΔNCEA; and 123C3, which binds to the NCAM external domains of both NCAM 125 and NCB. As expected, cross-linking ΔNCEA with J22, but not with D13, resulted in a significant increase in bound Fn (Fig. 8). Similarly, cross-linking of NCB, alone and in the presence of ΔNCEA, increased bound Fn (Fig. 8). Cross-linking NCAM, which does not normally modulate integrins, with the NCAM-specific antibody did not lead to an increase in bound Fn levels, demonstrating the specificity of this effect. The lack of difference in Fn binding between NCB and parental cells, unlike that seen in Fig. 4, in monolayer culture is likely caused by the intact ECM surrounding the cells in this assay. This would provide the ligands for integrins that have previously been activated, so that these integrins would not bind to the Fn added to the culture medium. Thus, NCB, in the presence of ΔNCEA, remained capable of altering Fn interaction after antibody cross-linking, which is consistent with the hypothesis that a defect in NCB clustering is created upon introduction of ΔNCEA.
Figure 8.
Antibody cross-linking restores integrin activation. Clustering induced by anti-CEA mAb J22 and anti-NCAM mAb 123C3, plus secondary anti–mouse antibodies, of ΔNCEA and NCB, respectively, leads to an increase in binding of Fn from the culture media by ΔNCEA- and NCB-expressing cells. A nonbinding antibody, D13, as well as cross-linking of NCAM, did not lead to such an increase. Values shown are the means ± the SD for a representative experiment (*, P < 0.005).
Discussion
The GPI anchor of CEA contains specific information determining protein function (Screaton et al., 2000) when attached to an adhesive extracellular protein domain. We investigated the hypothesis that interference with the GPI anchor could cause specific inhibition of function using a defective CEA GPI anchor–bearing molecule, ΔNCEA, coexpressed with NCB, a chimera of the NCAM external domain attached to the CEA GPI anchor. Coexpression of ΔNCEA with NCB resulted in a complete loss of the latter's biological function.
Determination of specific protein function by GPI anchors
TM- and GPI-anchored NCAM isoforms exhibit different effects on myoblast fusion (Peck and Walsh, 1993). Genetically attaching the CEA GPI anchor to the external domains of CC1 and NCAM, two adhesive cell surface molecules that do not inhibit cellular differentiation, results in chimeras that block differentiation, demonstrating functional anchor specificity (Screaton et al., 2000). Anchors may determine associated signaling proteins, as GPI-anchored CD59 can signal through kinases after having been exogenously administered to cells (van den Berg et al., 1995). Attaching this anchor to GFP also recreates the same signaling events upon antibody cross-linking (Hiscox et al., 2002). Membrane rafts concentrate a wide variety of different proteins on both sides of the plasma membrane; thus, GPI anchor-mediated targeting of a protein to rafts containing specific signaling molecules, in conjunction with clustering through external domain interactions, would explain how the CEA anchor can determine specific protein function.
Existence of distinct membrane raft domains
To have functionally distinct membrane rafts, these domains should contain different molecules; indeed, the segregation of gangliosides GM1 and GD3 into different domains has been observed (Gomez-Mouton et al., 2001, 2004; Vyas et al., 2001). This may help determine signaling specificity, as a close and specific association occurs between some gangliosides and certain signaling molecules (Kasahara et al., 1997; Iwabuchi et al., 1998). Similarly, GPI anchors are heterogeneous, as differences in both hydrophobic and hydrophilic GPI anchor regions have been documented (Ferguson et al., 1988; Armesto et al., 1996). These raft and anchor variations may be sufficient to create GPI anchor–specific membrane domains with different protein repertoires. In support of this idea, human folate receptor and placental AP do not exist in close proximity (Wang et al., 2002), whereas Thy-1 and prion protein associate with different domains (Madore et al., 1999) that show major differences in lipid composition (Brugger et al., 2004). CEA and NCAM do not copatch on the cell surface (Screaton et al., 2000), which was confirmed by IP and colocalization results presented in this study (Fig. 2). Replacing the GPI anchor signal sequence of NCAM for that of CEA, however, produces a chimera (NCB) partially located in close proximity to ΔNCEA (Fig. 2). This incomplete colocalization is likely a result of the small size of membrane rafts before activation, as it has been suggested that under resting conditions GPI-anchored proteins are primarily found as monomers, with a proportion found in small clusters (Sharma et al., 2004). NCB affects cell–Fn interaction similarly to CEA (Fig. 4 A), but unlike NCAM, suggesting that this altered localization affects signaling. Different Ras isoforms inhabit separate cytoplasmic leaflet compartments (Prior et al., 2003), showing that differences in protein distribution on the cell surface may be mirrored on the cell interior. We suggest that the C-terminal primary amino acid sequence can direct the addition of a specific type of GPI anchor, which determines both membrane localization and function through associated signaling elements.
Role of membrane raft clustering in signaling
Intense study of membrane raft dynamics has been undertaken in T cells, where these domains have a key role in signaling upon activation of the T cell receptor. Costimulation of T cells by CD28 is caused by raft redistribution, which alters internal tyrosine phosphorylation patterns (Viola et al., 1999), whereas antibody cross-linking of raft lipids replicates the membrane rearrangements that occur upon receptor activation (Janes et al., 1999). CEA mediates intercellular adhesion by antiparallel binding (Zhou et al., 1993) and differentiation inhibition by both antiparallel and parallel binding (Taheri et al., 2003), both of which may cause clustering of the rafts inhabited by CEA. The discovery of CEA mutants that retain intercellular adhesive ability but do not block differentiation indicates the key role, in conjunction with antiparallel adhesion, of parallel interactions in CEA's biological function (Taheri et al., 2003). NCAM-mediated adhesion involves intermolecular cis- and trans-interactions (Rao et al., 1992; Soroka et al., 2003), so at a threshold cell surface density, CEA and NCAM could create large cell surface raft aggregates leading to signaling (Harris and Siu, 2002). CEACAM7, a weakly adhesive member of the CEA family, poorly activates integrin α5β1; however, attaching its anchor to the stronger adhesive external domain of CC1 results in increased integrin activation (unpublished data). Previous work has demonstrated that treatment of CEA-expressing cells with fragment antigen-binding antibody fragments (Fabs) to reduce protein clustering is sufficient to release the CEA block of differentiation (Taheri et al., 2003). Similarly, reducing initial cis-clustering, through the addition of molecules with defective nonadhesive extracellular domains attached to the same GPI anchor, could also interfere with effective protein and raft clustering, causing substantial effects on downstream signaling. ΔNCEA coexpression resulted in a reproducible decrease in intercellular adhesive strength of NCB-expressing cells (Fig. 6 B) and an observed loss of cell–Fn interaction (Fig. 4 A). ΔNCEA expression increased the size of the membrane rafts containing NCB (Fig. 7 A), which would dilute the NCB molecules within the raft compartment. Thus, a larger raft with the same number of NCB molecules would lead to less efficient cis-clustering, decreasing the resulting adhesive strength. Consistent with this model, mimicking the clustering seen upon adhesion by antibody-mediated cross-linking of the NCB molecules restored its effects on integrin–Fn interaction (Fig. 8). Therefore, it would appear that treatment with Fabs or nonfunctional GPI anchors leads to similar effects on clustering and protein activity.
Modulation of raft lipid content
The function of membrane proteins can be altered by modulating membrane lipid content. Depletion of cellular cholesterol inhibits signaling from membrane raft domains (Incardona and Eaton, 2000), whereas administration of exogenous gangliosides displaces GPI-anchored proteins from membrane rafts (Simons et al., 1999). Treatment of T cells with polyunsaturated fatty acids interferes with tyrosine kinase activation and calcium release upon stimulation (Stulnig et al., 1998), which appears similar to this study, where signaling by NCB is inhibited via its GPI anchor. The key difference lies in the GPI-anchor specificity because exogenous administration of lipids likely causes a global alteration in raft structure, whereas GPI anchors contain information that targets specific subdomains. Thus, ΔNCEA is not found in close proximity to NCAM, but changing the anchor of NCAM for that of CEA alters the localization of this protein, such that it now colocalizes with ΔNCEA (Fig. 2). This colocalization explains how ΔNCEA, which shows no effect on differentiation, can restore differentiation to NCB-expressing cells, whereas NCAM, which accelerates differentiation, does not restore fusion to CEA-expressing cells (Fig. 3 A). It is intriguing to note that ΔNCEA expression has such a significant effect on NCB function despite the fact that the proteins are found on the cell surface at similar levels. This suggests that once a threshold number of proteins have been inhibited, the biological function is completely lost, as any remaining functional proteins may be incapable of clustering sufficiently to lead to effective signaling activation.
We therefore suggest that the GPI anchor plays a key role in protein function by directing localization to a specific subset of membrane rafts, which determines the associated signaling molecules. Exploiting this biological specificity by competition with functionally specific GPI anchors attached to nonfunctional external protein domains results in a complete loss of biological activity. This therefore confirms the key biological role of the GPI anchor, and suggests a novel method for the manipulation GPI-anchored proteins.
Materials and methods
Constructs and antibodies
ΔNCEA is a CEA deletion mutant that has the last 75 amino acids of the N domain deleted, such that it is no longer biologically active (Zhou et al., 1993). The NCAM splice variant used in this study, p125, is a human GPI-anchored NCAM isoform containing the muscle-specific domain (Barton et al., 1988). NCB is a chimera of the NCAM p125 external domain genetically fused to the CEA GPI anchor signal sequence (Screaton et al., 2000). The mAbs J22 and D14 bind to internal CEA domains (Zhou et al., 1993), whereas the epitope of D13 is in the portion of the CEA N domain that is deleted in ΔNCEA, and rabbit polyclonal anti-CEA binds to all CEA external protein domains. The mAb 123C3 (Santa Cruz Biotechnology, Inc.) recognizes human NCAM, whereas antibodies H-293, H-104, and M-106 (Santa Cruz Biotechnology, Inc.) recognize the α2, α5, and β1 integrins, respectively. The mAb 47A (De Giovanni et al., 1993) binds to myosin. C20 is a goat polyclonal anti-Fn antibody (Santa Cruz Biotechnology, Inc.).
Cell culture and differentiation assay
Cells were grown attached to tissue culture plastic surfaces (Nunc), as previously described (Screaton et al., 2000). In brief, CHO-derived LR-73 fibroblasts were grown in α-MEM with 10% FBS. Rat L6 myoblasts were grown in DME containing 10% FBS (GM), and were subcultured before reaching confluency to avoid selecting for nonfusing variants. Cell concentrations were determined using a particle counter (Beckman Coulter). For myoblast differentiation, 104 L6 cells/cm2 were seeded in 60-mm dishes. After 3 d, the media was switched to DME with 2% horse serum (DM). 4–7 d later, cultures were assessed for differentiation by hematoxylin (Sigma-Aldrich) staining and microscopic examination (Screaton et al., 1997), or by lysing and assessing myosin levels by Western blotting.
Transfections
100-mm dishes were seeded with 2 or 4 × 105 cells/plate for LR or L6, respectively. 24 h later, cells were cotransfected by calcium phosphate coprecipitation with 5 μg of cDNA, 0.5 μg of pSV2(neo), and 10 μg of carrier DNA isolated from LR-73 cells. Double transfections were performed in the same manner, either by cotransfecting both cDNAs at once (for LR cells) with pSV2(neo) or with 0.5 μg of pBabe(puro) to supertransfect L6 transfectants; transfectants were isolated by selection with 400–600 μg/ml neomycin (G418; Invitrogen) or 1 μg/ml puromycin (Sigma-Aldrich). After 10–14 d, resistant clones were pooled and sorted for high expression by FACS using mAbs J22 or 123C3. Although pooled populations of many clones were used, two independent transfections of L6 cells were performed to ensure no clonal variation occurred, with identical assay results. Note that the L6 (NCB) cells were pooled colonies resistant to both G418 and puromycin, and that although data from L6 and LR-73 parental cells is shown, no difference between these cells and pooled G418-resistant clones transfected with the pSV2(neo) alone has been noted (Ordonez et al., 2006).
FACS analysis
Cells were collected with PBS-citrate containing 4 mM EDTA (PBSCE) for NCAM-expressing transfectants (because of the sensitivity of the NCAM external domain to trypsinization; Screaton et al., 2000) or 0.063% trypsin in PBS-citrate for CEA transfectants. 2.5 × 105 cells were resuspended in ice-cold PBS with 2% FBS (PBSF). Cells were incubated for 30 min with mAb at a dilution of 1:50–100, washed with PBSF, and incubated with FITC-conjugated goat anti–mouse antibody (Jackson ImmunoResearch Laboratories) diluted 1:100. After an additional 30 min, cells were pelleted, resuspended in PBSF, and analyzed using a FACScan instrument (Becton Dickinson).
Binding to purified ECM components
Cells were seeded at 104 cells/cm2 on day 0. On day 2, subconfluent cultures were collected with PBSCE, resuspended in GM, and incubated at 37°C for 30 min. Cells were washed with serum-free media and resuspended in serum-free media at 4 × 105 cells/ml. 100 μl/well of this suspension was added to wells from a 96-well plate coated with Fn, Vn, or collagen I (CHEMICON International, Inc.) and incubated for 1 h at 37°C. Wells were washed with PBS containing Mg2+, and adherent cells were stained with crystal violet. Wells were washed again with PBS, and the bound stain was solubilized with 0.05 M NaH2PO4, pH 4.5, plus 25% ethanol. Staining was quantified with a microplate reader (Bio-Tek Instruments) at 570 nm. Statistical significance was determined using a t test (http://www.physics.csbsju.edu/stats/t-test_bulk_form.html).
Immunoblotting
Proteins in cellular lysates were resolved by SDS-PAGE and transferred electrophoretically to a 0.45-μm PVDF membrane (Millipore). Immunoblotting was performed as previously described (Screaton et al., 2000), with antibody binding detected using ECL Plus reagent (GE Healthcare).
Triton X-100 solubility and isopycnic sucrose gradient ultracentrifugation
Triton X-100 solubility was determined as previously described (Screaton et al., 2000). In brief, subconfluent cell cultures were rinsed with PBS, collected with PBSCE, and rendered single-cell suspensions by passing through a 27-gauge needle. 107 cells/ml were resuspended in ice-cold lysis buffer containing 1% Triton X-100 and the protease inhibitors aprotonin (Roche), leupeptin (Roche), and PMSF (Sigma-Aldrich). Lysates were syringed with a 27-gauge needle, incubated on ice for 15 min, and centrifuged at 13,500 g for 20 min at 4°C. Soluble fractions were removed, and the pellets were resuspended in 0.9 vol of lysis buffer and 0.1 vol 10% SDS. The relative amounts of soluble versus pellet protein were determined by immunoblotting. For sucrose gradient ultracentrifugation, two T175 flasks were seeded with 104 cells/cm2. 2 d later, cells were collected with PBSCE, and lysed with 1 ml of 1% Brij-98 (Sigma-Aldrich) in sucrose gradient buffer (10 mM Tris, pH 8.0, and 140 mM NaCl) containing aprotonin, leupeptin, and PMSF for 30 min at 4°C. 1 ml of ice-cold 80% sucrose was added to this lysate and overlayed successively with 2 ml of 35% sucrose and 1 ml of 5% sucrose. Lysates were centrifuged with a rotor (SW55; Beckman Coulter) for 19 h at 45,000 RPM at 4°C. 400-μl fractions were collected from the top of the gradient, and equal volumes of each fraction were assessed by immunoblotting.
Velocity sedimentation
L6 cells from 2–4 T175 flasks were collected with PBSCE, pooled, and lysed with 500 μl of 1% Brj-98 in sucrose gradient buffer for 30 min on ice. The lysate was then added on top of 11 ml of 12.5% sucrose, and centrifuged for 1 h at 12,300 RPM (∼18,700 g) in an SW41 rotor (Beckman Coulter). 25 460-μl fractions were collected from the top, and assayed by immunoblotting. In certain cases, cells were pretreated with mβCD (Sigma-Aldrich) for 15 min at 37°C before Brj-98 lysis, to disrupt membrane rafts.
Chemical cross-linking and IP
4 × 105 L6 cells were seeded in three 100-mm dishes for each transfectant. 2 d later, cells were washed with PBS and incubated, with gentle rocking at 4°C, with either 1 ml of 1 mM DTSSP (Pierce Chemical Co.) in PBS or with PBS alone. After 1 h, unconjugated DTSSP was neutralized with 100 mM Tris, pH 7.4. Cells were lysed with 400 μl/plate of 60 mM n-Octyl β-D glucopyranoside (Sigma-Aldrich) in lysis buffer containing protease inhibitors. Lysates were pooled and syringed to reduce viscosity. 1.2 ml of each lysate was precleared by rotation with 75 μl of Protein A/G Plus–Agarose beads (Santa Cruz Biotechnology, Inc.) for 3 h at 4°C. Precleared lysates were then diluted with an equal amount of lysis buffer, and divided into three aliquots, receiving no antibody, 5 μg 123C3, or 5 μg J22. Samples were rotated overnight at 4°C, and then 75 μl of Protein A/G Plus–Agarose beads were added. 3 h later, the beads were washed five times with lysis buffer and resuspended in 75 μl 1× Laemmli sample buffer for analysis by Western blotting
Immunofluorescence and confocal microscopy
L6 transfectants were seeded in 8-well Lab-Tek Permanox chamber slides (Nunc) at a density of 104 cells/well. 2 d later, cells were washed with PBSF and incubated with primary antibodies 123C3 (at a dilution of 1:100 in PBSF) and rabbit polyclonal anti-CEA (1:2,000 dilution) for 30 min at RT. Cells were washed with PBSF, and then incubated at RT for 30 min, in the dark, with a 1:250 dilution of both Cy2-conjugated goat anti–rabbit and rhodamine-conjugated goat anti–mouse secondary antibodies. Cells were then washed twice with PBSF, and fixed by incubation with 4% formaldehyde for 10 min at 4°C, followed by 100% methanol for 20 min at 4°C. Samples were then mounted using fluorescent mounting medium (DakoCytomation). Localization of stained proteins was observed using a LSM 510 Axiovert 100M confocal microscope with a Plan-Achromat 63×/1.4 NA oil differential interference contrast objective (both Carl Zeiss MicroImaging, Inc.).
Adhesion and aggregate size assays
Adhesion assays were performed as previously described (Zhou et al., 1993). In brief, 106 LR cells were seeded in 80-cm2 tissue culture flasks (Nunc), and collected 2 d later by incubation with PBSCE. 3 × 106 cells were resuspended in 3 ml α-MEM containing 0.8% FBS and 10 μg/ml DNase I (Roche), syringed with a 27-gauge needle to obtain single-cell suspensions, and allowed to aggregate at 37°C with stirring at 100 rpm using a small magnetic stirring bar. Aliquots were removed at the indicated times, and the cells were counted with a hemocytometer to determine the percentage of single cells. For the aggregate assay, cells were prepared as for adhesion assays, but the number of cells present in each of ∼50 multicellular aggregates was scored after 1 h in suspension.
Soluble Fn-binding assay
On day 0, 104 cells/well were seeded in a 96-well plate. 2 d later, cells were washed with PBSF and incubated for 30 min at 37°C in DME with 50 μl of 10 μg/ml human Fn (BD Biosciences), along with, where indicated, 5 μg/ml primary mAb and 30 μg/ml donkey anti–mouse secondary antibody (Jackson ImmunoResearch Laboratories) to further cross-link CEA or NCAM constructs. Cells were then washed three times with PBSF, and fixed with 4% formaldehyde. Bound Fn was determined by incubation with anti-Fn antibody, C20, at a dilution of 1:100 in 3% BSA (Sigma-Aldrich) in PBS (PBSB) for 90 min at RT, having blocked nonspecific binding by incubation for 1 h at RT with PBSB. Cells were washed with PBSB, and incubated with HRP-conjugated rabbit anti–goat secondary antibody (Jackson) at a dilution of 1:2,500 in PBSB for 1 h. After incubation with a hydrogen peroxide solution containing ABTS (Sigma-Aldrich), bound Fn was determined with a microplate reader at 405 nm, with a reference wavelength of 490 nm.
Acknowledgments
We thank Jacynthe Laliberté for technical assistance with confocal microscopy.
This work was supported by the Cancer Research Society and by a research contract to C.P. Stanners from Novothera Biotechnologies, Inc. T.B. Nicholson was the recipient of a Natural Sciences and Engineering Research Council of Canada scholarship.
Abbreviations used in this paper: CEA, carcinoembryonic antigen; CC1, CEA-related cell adhesion molecule CEACAM1; Fn, fibronectin; GPI, glycophosphatidylinositol; IP, immunoprecipitation; MβCD, methyl-β-cyclodextrin; NCAM, neural cell adhesion molecule; TM, transmembrane; Vn, vitronectin.
References
- Armesto, J., E. Hannappel, K. Leopold, W. Fischer, R. Bublitz, L. Langer, G.A. Cumme, and A. Horn. 1996. Microheterogeneity of the hydrophobic and hydrophilic part of the glycosylphosphatidylinositol anchor of alkaline phosphatase from calf intestine. Eur. J. Biochem. 238:259–269. [DOI] [PubMed] [Google Scholar]
- Barton, C.H., G. Dickson, H.J. Gower, L.H. Rowett, W. Putt, V. Elsom, S.E. Moore, C. Goridis, and F.S. Walsh. 1988. Complete sequence and in vitro expression of a tissue-specific phosphatidylinositol-linked N-CAM isoform from skeletal muscle. Development. 104:165–173. [DOI] [PubMed] [Google Scholar]
- Benchimol, S., A. Fuks, S. Jothy, N. Beauchemin, K. Shirota, and C.P. Stanners. 1989. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 57:327–334. [DOI] [PubMed] [Google Scholar]
- Brugger, B., C. Graham, I. Leibrecht, E. Mombelli, A. Jen, F. Wieland, and R. Morris. 2004. The membrane domains occupied by glycosylphosphatidylinositol-anchored prion protein and Thy-1 differ in lipid composition. J. Biol. Chem. 279:7530–7536. [DOI] [PubMed] [Google Scholar]
- De Giovanni, C., P.L. Lollini, R. Dolcetti, L. Landuzzi, G. Nicoletti, E. D'Andrea, K. Scotland, and P. Nanni. 1993. Uncoupling of growth inhibition and differentiation in dexamethasone-treated human rhabdomyosarcoma cells. Br. J. Cancer. 67:674–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, G., D. Peck, S.E. Moore, C.H. Barton, and F.S. Walsh. 1990. Enhanced myogenesis in NCAM-transfected mouse myoblasts. Nature. 344:348–351. [DOI] [PubMed] [Google Scholar]
- Dietrich, C., B. Yang, T. Fujiwara, A. Kusumi, and K. Jacobson. 2002. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 82:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury, M.S., H. Ito, S.W. Ashley, and E.E. Whang. 2004. a. c-Src-dependent cross-talk between CEACAM6 and alpha(v)beta(3) integrin enhances pancreatic adenocarcinoma cell adhesion to extracellular matrix components. Biochem. Biophys. Res. Commun. 317:133–141. [DOI] [PubMed] [Google Scholar]
- Duxbury, M.S., H. Ito, M.J. Zinner, S.W. Ashley, and E.E. Whang. 2004. b. CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene. 23:465–473. [DOI] [PubMed] [Google Scholar]
- Eidelman, F.J., A. Fuks, L. DeMarte, M. Taheri, and C.P. Stanners. 1993. Human carcinoembryonic antigen, an intercellular adhesion molecule, blocks fusion and differentiation of rat myoblasts. J. Cell Biol. 123:467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, M.A., S.W. Homans, R.A. Dwek, and T.W. Rademacher. 1988. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science. 239:753–759. [DOI] [PubMed] [Google Scholar]
- Foster, L.J., C.L. De Hoog, and M. Mann. 2003. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. USA. 100:5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichson, T., and T.V. Kurzchalia. 1998. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 394:802–805. [DOI] [PubMed] [Google Scholar]
- Gaus, K., E. Gratton, E.P. Kable, A.S. Jones, I. Gelissen, L. Kritharides, and W. Jessup. 2003. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. USA. 100:15554–15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mouton, C., J.L. Abad, E. Mira, R.A. Lacalle, E. Gallardo, S. Jimenez-Baranda, I. Illa, A. Bernad, S. Manes, and A.C. Martinez. 2001. Segregation of leading-edge and uropod components into specific lipid rafts during T cell polarization. Proc. Natl. Acad. Sci. USA. 98:9642–9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mouton, C., R.A. Lacalle, E. Mira, S. Jimenez-Baranda, D.F. Barber, A.C. Carrera, A.C. Martinez, and S. Manes. 2004. Dynamic redistribution of raft domains as an organizing platform for signaling during cell chemotaxis. J. Cell Biol. 164:759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, T.J., and C.H. Siu. 2002. Reciprocal raft-receptor interactions and the assembly of adhesion complexes. Bioessays. 24:996–1003. [DOI] [PubMed] [Google Scholar]
- Hinoda, Y., H. Takahashi, T. Higashide, T. Nakano, Y. Arimura, M. Yoshimoto, M. Tsujisaki, K. Imai, and A. Yachi. 1991. Correlated expression of mRNAs of carcinoembryonic antigen and nonspecific cross-reacting antigen genes in malignant and nonmalignant tissues of the colon. Jpn. J. Clin. Oncol. 21:75–81. [DOI] [PubMed] [Google Scholar]
- Hiscox, S., M.B. Hallett, B.P. Morgan, and C.W. van den Berg. 2002. GPI-anchored GFP signals Ca2+ but is homogeneously distributed on the cell surface. Biochem. Biophys. Res. Commun. 293:714–721. [DOI] [PubMed] [Google Scholar]
- Homans, S.W., M.A. Ferguson, R.A. Dwek, T.W. Rademacher, R. Anand, and A.F. Williams. 1988. Complete structure of the glycosyl phosphatidylinositol membrane anchor of rat brain Thy-1 glycoprotein. Nature. 333:269–272. [DOI] [PubMed] [Google Scholar]
- Ilantzis, C., S. Jothy, L.C. Alpert, P. Draber, and C.P. Stanners. 1997. Cell-surface levels of human carcinoembryonic antigen are inversely correlated with colonocyte differentiation in colon carcinogenesis. Lab. Invest. 76:703–716. [PubMed] [Google Scholar]
- Incardona, J.P., and S. Eaton. 2000. Cholesterol in signal transduction. Curr. Opin. Cell Biol. 12:193–203. [DOI] [PubMed] [Google Scholar]
- Iwabuchi, K., S. Yamamura, A. Prinetti, K. Handa, and S. Hakomori. 1998. GM3-enriched microdomain involved in cell adhesion and signal transduction through carbohydrate-carbohydrate interaction in mouse melanoma B16 cells. J. Biol. Chem. 273:9130–9138. [DOI] [PubMed] [Google Scholar]
- Janes, P.W., S.C. Ley, and A.I. Magee. 1999. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. J. Cell Biol. 147:447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, K., Y. Watanabe, T. Yamamoto, and Y. Sanai. 1997. Association of Src family tyrosine kinase Lyn with ganglioside GD3 in rat brain. Possible regulation of Lyn by glycosphingolipid in caveolae-like domains. J. Biol. Chem. 272:29947–29953. [DOI] [PubMed] [Google Scholar]
- Kunath, T., C. Ordonez-Garcia, C. Turbide, and N. Beauchemin. 1995. Inhibition of colonic tumor cell growth by biliary glycoprotein. Oncogene. 11:2375–2382. [PubMed] [Google Scholar]
- Li, R., T. Liu, F. Yoshihiro, M. Tary-Lehmann, M. Obrenovich, H. Kuekrek, S.C. Kang, T. Pan, B.S. Wong, M.E. Medof, and M.S. Sy. 2003. On the same cell type GPI-anchored normal cellular prion and DAF protein exhibit different biological properties. Biochem. Biophys. Res. Commun. 303:446–451. [DOI] [PubMed] [Google Scholar]
- Low, M.G. 1989. Glycosyl-phosphatidylinositol: a versatile anchor for cell surface proteins. FASEB J. 3:1600–1608. [DOI] [PubMed] [Google Scholar]
- Luo, W., C.G. Wood, K. Earley, M.C. Hung, and S.H. Lin. 1997. Suppression of tumorigenicity of breast cancer cells by an epithelial cell adhesion molecule (C-CAM1): the adhesion and growth suppression are mediated by different domains. Oncogene. 14:1697–1704. [DOI] [PubMed] [Google Scholar]
- Madore, N., K.L. Smith, C.H. Graham, A. Jen, K. Brady, S. Hall, and R. Morris. 1999. Functionally different GPI proteins are organized in different domains on the neuronal surface. EMBO J. 18:6917–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S. 2003. Lipid rafts: elusive or illusive? Cell. 115:377–388. [DOI] [PubMed] [Google Scholar]
- Ordonez, C., R.A. Screaton, C. Ilantzis, and C.P. Stanners. 2000. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res. 60:3419–3424. [PubMed] [Google Scholar]
- Ordonez, C., A.B. Zhai, P. Camacho-Leal, L. DeMarte, M.Y. Fan, and C.P. Stanners. 2006. GPI-anchored CEA family glycoproteins CEA and CEACAM6 mediate their biological effects through enhanced integrin α5β1-fibronectin interaction. J. Cell. Physiol. In press. [DOI] [PubMed]
- Peck, D., and F.S. Walsh. 1993. Differential effects of over-expressed neural cell adhesion molecule isoforms on myoblast fusion. J. Cell Biol. 123:1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike, L.J. 2004. Lipid rafts: heterogeneity on the high seas. Biochem. J. 378:281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralle, A., P. Keller, E.L. Florin, K. Simons, and J.K. Horber. 2000. Sphingolipid–cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148:997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior, I.A., C. Muncke, R.G. Parton, and J.F. Hancock. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160:165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, Y., X.F. Wu, J. Gariepy, U. Rutishauser, and C.H. Siu. 1992. Identification of a peptide sequence involved in homophilic binding in the neural cell adhesion molecule NCAM. J. Cell Biol. 118:937–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, P.J. 1997. Signal transduction via GPI-anchored membrane proteins. Adv. Exp. Med. Biol. 419:365–370. [DOI] [PubMed] [Google Scholar]
- Screaton, R.A., L.Z. Penn, and C.P. Stanners. 1997. Carcinoembryonic antigen, a human tumor marker, cooperates with Myc and Bcl-2 in cellular transformation. J. Cell Biol. 137:939–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton, R.A., L. DeMarte, P. Draber, and C.P. Stanners. 2000. The specificity for the differentiation blocking activity of carcinoembryonic antigen resides in its glycophosphatidyl-inositol anchor. J. Cell Biol. 150:613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, P., R. Varma, R.C. Sarasij, Ira, K. Gousset, G. Krishnamoorthy, M. Rao, and S. Mayor. 2004. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 116:577–589. [DOI] [PubMed] [Google Scholar]
- Shenoy-Scaria, A.M., J. Kwong, T. Fujita, M.W. Olszowy, A.S. Shaw, and D.M. Lublin. 1992. Signal transduction through decay-accelerating factor. Interaction of glycosyl-phosphatidylinositol anchor and protein tyrosine kinases p56lck and p59fyn 1. J. Immunol. 149:3535–3541. [PubMed] [Google Scholar]
- Shenoy-Scaria, A.M., L.K. Gauen, J. Kwong, A.S. Shaw, and D.M. Lublin. 1993. Palmitylation of an amino-terminal cysteine motif of protein tyrosine kinases p56lck and p59fyn mediates interaction with glycosyl-phosphatidylinositol-anchored proteins. Mol. Cell. Biol. 13:6385–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogomori, H., and D.A. Brown. 2003. Use of detergents to study membrane rafts: the good, the bad, and the ugly. Biol. Chem. 384:1259–1263. [DOI] [PubMed] [Google Scholar]
- Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- Simons, M., T. Friedrichson, J.B. Schulz, M. Pitto, M. Masserini, and T.V. Kurzchalia. 1999. Exogenous administration of gangliosides displaces GPI-anchored proteins from lipid microdomains in living cells. Mol. Biol. Cell. 10:3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, K.R., M.A. Mallory, K.A. Hanify, and R.W. Finberg. 1998. The nature of membrane anchorage determines kinase association and detergent solubility of CD4. Biochem. Biophys. Res. Commun. 242:423–428. [DOI] [PubMed] [Google Scholar]
- Soroka, V., K. Kolkova, J.S. Kastrup, K. Diederichs, J. Breed, V.V. Kiselyov, F.M. Poulsen, I.K. Larsen, W. Welte, V. Berezin, et al. 2003. Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion. Structure (Camb). 11:1291–1301. [DOI] [PubMed] [Google Scholar]
- Stulnig, T.M., M. Berger, T. Sigmund, H. Stockinger, V. Horejsi, and W. Waldhausl. 1997. Signal transduction via glycosyl phosphatidylinositol-anchored proteins in T cells is inhibited by lowering cellular cholesterol. J. Biol. Chem. 272:19242–19247. [DOI] [PubMed] [Google Scholar]
- Stulnig, T.M., M. Berger, T. Sigmund, D. Raederstorff, H. Stockinger, and W. Waldhausl. 1998. Polyunsaturated fatty acids inhibit T cell signal transduction by modification of detergent-insoluble membrane domains. J. Cell Biol. 143:637–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri, M., H.U. Saragovi, and C.P. Stanners. 2003. The adhesion and differentiation-inhibitory activities of the immunoglobulin superfamily member, carcinoembryonic antigen, can be independently blocked. J. Biol. Chem. 278:14632–14639. [DOI] [PubMed] [Google Scholar]
- van den Berg, C.W., T. Cinek, M.B. Hallett, V. Horejsi, and B.P. Morgan. 1995. Exogenous glycosyl phosphatidylinositol-anchored CD59 associates with kinases in membrane clusters on U937 cells and becomes Ca2+-signaling competent. J. Cell Biol. 131:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, R., and S. Mayor. 1998. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 394:798–801. [DOI] [PubMed] [Google Scholar]
- Viola, A., S. Schroeder, Y. Sakakibara, and A. Lanzavecchia. 1999. T lymphocyte costimulation mediated by reorganization of membrane microdomains. Science. 283:680–682. [DOI] [PubMed] [Google Scholar]
- von Haller, P.D., S. Donohoe, D.R. Goodlett, R. Aebersold, and J.D. Watts. 2001. Mass spectrometric characterization of proteins extracted from Jurkat T cell detergent-resistant membrane domains. Proteomics. 1:1010–1021. [DOI] [PubMed] [Google Scholar]
- Vyas, K.A., H.V. Patel, A.A. Vyas, and R.L. Schnaar. 2001. Segregation of gangliosides GM1 and GD3 on cell membranes, isolated membrane rafts, and defined supported lipid monolayers. Biol. Chem. 382:241–250. [DOI] [PubMed] [Google Scholar]
- Wang, J., W. Gunning, K.M. Kelley, and M. Ratnam. 2002. Evidence for segregation of heterologous GPI-anchored proteins into separate lipid rafts within the plasma membrane. J. Membr. Biol. 189:35–43. [DOI] [PubMed] [Google Scholar]
- Zhou, H., A. Fuks, and C.P. Stanners. 1990. Specificity of intercellular adhesion mediated by various members of the immunoglobulin supergene family. Cell Growth Differ. 1:209–215. [PubMed] [Google Scholar]
- Zhou, H., A. Fuks, G. Alcaraz, T.J. Bolling, and C.P. Stanners. 1993. Homophilic adhesion between Ig superfamily carcinoembryonic antigen molecules involves double reciprocal bonds. J. Cell Biol. 122:951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]