Abstract
Skin lies at the interface between the complex physiology of the body and the external environment. This essential epidermal barrier, composed of cornified proteins encased in lipids, prevents both water loss and entry of infectious or toxic substances. We uncover that the transcription factor GATA-3 is required to establish the epidermal barrier and survive in the ex utero environment. Analysis of Gata-3 mutant transcriptional profiles at three critical developmental stages identifies a specific defect in lipid biosynthesis and a delay in differentiation. Genomic analysis identifies highly conserved GATA-3 binding sites bound in vivo by GATA-3 in the first intron of the lipid acyltransferase gene AGPAT5. Skin from both Gata-3−/− and previously characterized barrier-deficient Kruppel-like factor 4−/− newborns up-regulate antimicrobial peptides, effectors of innate immunity. Comparison of these animal models illustrates how impairment of the skin barrier by two genetically distinct mechanisms leads to innate immune responses, as observed in the common human skin disorders psoriasis and atopic dermatitis.
Introduction
Mammalian epidermis functions as a barrier to prevent both water loss to the terrestrial environment and entry of toxic and pathogenic agents into the organism (Elias, 2005; Segre, 2006). Embryonic ectoderm is specified to an epidermal fate at murine embryonic day (E) 8.5, regulated by the p63 transcription factors (Koster and Roop, 2004). At E9.5, this single layer of basal cells express cytokeratin (K) 5 and K14 (Byrne et al., 1994). During midembryogenesis, these basal cells continue to divide as a single-layered epithelium to increase the surface area of the developing embryo. Stratification of the basal epithelium initiates at E12.5 with asymmetric cell divisions perpendicular to the basement membrane (Lechler and Fuchs, 2005). By E15.5, suprabasal cells initiate the differentiation program and express K1/K10 (Byrne et al., 1994). Establishment of the epidermal permeability barrier initiates at E16.5 on the dorsal surface and spreads ventrally to achieve a fully competent barrier by E18 (Hardman et al., 1998). Barrier establishment requires cross-linking of the cells in the upper layer, which then constrains increases to the surface area of the embryo. Although the barrier must be acquired before the typical end of gestation, it is not advantageous to develop a fully competent barrier too early in development because of the need for continued growth.
Interfollicular epidermal cells retain the ability to self-renew under both homeostatic and injured conditions by maintaining mitotically active cells (Blanpain et al., 2004; Morris et al., 2004; Tumbar et al., 2004; Ito et al., 2005; Levy et al., 2005). Terminal differentiation begins when basal cells concomitantly withdraw from the cell cycle and lose adhesion to the basement membrane. In the intermediate spinous layers, the cells assemble a durable cytoskeletal framework that provides mechanical strength to resist physical trauma. In the upper granular layer, a cornified envelope (CE) is assembled directly underneath the plasma membrane by sequential incorporation of precursor proteins. Lipid-containing lamellar bodies fuse with the plasma membrane and attach to the CE scaffold, sealing the now enucleated cells together to create the “bricks and mortar” barrier at the skin surface (Elias, 2005). Recent experimental results have also demonstrated a selective role for tight junctions in establishing the epidermal barrier (Furuse et al., 2002). This process of differentiation from a mitotically active basal cell to a terminally differentiated squame is maintained throughout life as part of epidermal regeneration and maturation.
Mouse models with targeted ablations of genes encoding keratinocyte transcription factors have demonstrated that barrier acquisition is a coordinated and regulated process (Dai and Segre, 2004). Our earlier experiments demonstrated that the transcription factor Kruppel-like factor 4 (Klf4) is necessary to establish the epidermal barrier in utero (Segre et al., 1999). To elucidate further the transcriptional networks regulating this process, we examined the specific role of Gata-3, the most highly expressed member of the GATA family of transcription factors in interfollicular epidermis.
Results
Epidermal-specific deletion of Gata-3 results in perinatal lethality because of selective barrier impairment
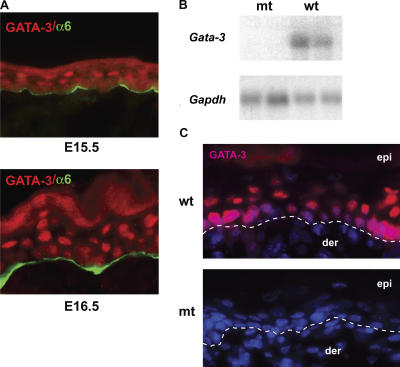
GATA-3 expression in interfollicular epidermis is first detected in the immediate suprabasal layer at E15.5 (Fig. 1 A). At E16.5 and thereafter, GATA-3 is expressed in both basal and immediate suprabasal layers (Fig. 1 A). Gata-3−/− embryos die at E11 (Pandolfi et al., 1995), and pharmacologic rescue of these embryos until E17.5 revealed a role for Gata-3 in development of the inner root sheath of the hair follicle (Kaufman et al., 2003). To elucidate the necessary function of GATA-3 in terminal stages of epidermal differentiation in vivo, we used the cre–loxP system. To generate mice with an epidermal- specific targeted ablation of Gata-3, homozygous floxed Gata-3 (Gata-3 fl/fl) mice were crossed with mice heterozygous for a deletion of Gata-3 (Gata-3 +/−) and expressing the Cre recombinase under the control of the K14 promoter (K14-Cre) to generate mice with a Gata-3 fl/− K14-Cre genotype, hereafter referred to as Gata-3 mutants (Pai et al., 2003; Andl et al., 2004). Quantitative PCR of amplicons both within and outside the Gata-3 locus on mutant and control littermate epidermal genomic DNA demonstrated that >97% of the epidermal cells had deleted the Gata-3 locus (unpublished data). The residual amplification in Gata-3 mutants may be from melanocytes or Langerhans cells, resident in the epidermis. Deletion of GATA-3 mRNA and protein was also demonstrated by Northern and immunohistochemical analysis of Gata-3 mutant newborn skin (Fig. 1, B and C).
Figure 1.
Epidermal-specific deletion of Gata-3. (A) Expression of GATA-3 at E15.5 in immediate suprabasal layer and at E16.5 in both basal and suprabasal layers. α6 marks the basement membrane of the epidermis. (B) Deletion of Gata-3 mRNA, shown by Northern blot of skin mRNA probed with Gata-3 and Gapdh cDNA. wt, wild type; mt, mutant. (C) Immunofluorescence of newborn epidermis, demonstrating deletion of GATA-3 in the epidermis. Nuclei were stained with DAPI. Dotted lines mark the basement membrane. der, dermis; epi, epidermis.
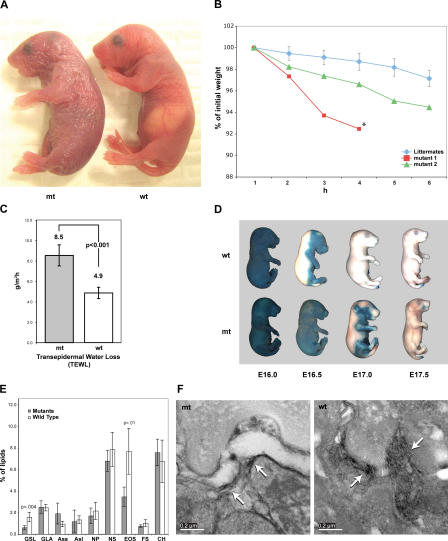
Gata-3 mutants, born at the expected Mendelian ratio, are distinguishable from their littermates at birth by a lack of prominent whiskers. In the perinatal period, Gata-3 mutants do not feed and can be identified by the lack of a typical “milk spot.” Equally striking is the apparent desiccation of the Gata-3 mutant skin during the perinatal period, which takes on a thin, erythemic, wrinkled appearance (Fig. 2 A). During the first 6 h after birth, Gata-3 mutant mice lose an average of 5% of their birth weight. By comparison, littermates who do survive >24 h when unfed do not appear to desiccate and lose significantly less weight during a comparable period (Fig. 2 B). Specifically, individual Gata-3 mutants lose weight at a rate that is >3.5- and 7.8-fold greater than the standard deviation of the control littermates. Because other barrier-deficient animal models display a similar perinatal lethality, we tested this directly (Segre et al., 1999; List et al., 2002).
Figure 2.
Loss of Gata-3 in the epidermis results in perinatal lethality because of a selective barrier impairment. (A) Newborn Gata-3 mutant (mt) mice possess thin, erythemic, wrinkled skin. wt, wild type. (B) Weight loss as a percentage of initial weight over time. The rate of weight loss of mutants 1 and 2 are >3.5 and 7.8 standard deviations from the mean of the control littermates, respectively. The asterisk indicates the point after which mutant 1 expired. (C) Transepidermal water loss assay measured on ventral surface of the newborn. (D) Dye exclusion assay performed on E16.0–E17.5 embryos. (E) Total epidermal lipid component analysis. n = 4 mice per group. GSL, glycosylceramides; GLA, acylglycosylceramides; ASS, ceramide with short α-hydroxyacids (C16) amide linked to sphingosine; ASL, ceramide with long α-hydroxyacids amide linked to sphingosine acylceramides long; NP, ceramide with normal fatty acids amide linked to phytosphingosine; NS, ceramide with normal fatty acids amide linked to sphingosine; EOS, ceramide with long (C30–C34) ω-hydroxyacids amide linked to sphingosine and bearing ester-linked linoleic acid on the ω-hydroxyl group; FS, free sterol; CH, cholesterol. (F) Electron micrographs of a region between a granulocyte and corneocyte displaying distorted lamellar bodies with few disorganized leaflets (arrows) and large, irregular vacuoles in the mutant as compared with densely packed membrane leaflets (arrows) in the wild type. Bars, 0.2 μm.
As compared with control littermates, Gata-3 mutant newborns exhibit a significant increase in the rate of transepidermal water loss across their skin surface (P < 0.001; Fig. 2 C). The increase in the rate of transepidermal water loss and weight loss is of a similar order of magnitude (Fig. 2 B). Gata-3 mutants' impaired skin barrier is unable to retain water in the terrestrial ex utero environment, which results in dehydration and, ultimately, lethality. To complement the studies that measure water loss across the skin surface, we investigated Gata-3 mutants' competence to exclude percutaneous dye penetration. As previously shown, dye exclusion in control littermates (visualized as white areas) initiates on the dorsal surface at approximately E16.5 and spreads ventrally, resulting in complete dye impermeability by E17.5. A transient delay of 0.5 d is observed in both the initiation and completion of the dye exclusion of the Gata-3 mutants (Fig. 2 D). Because the Gata-3 mutants are able to exclude dye penetration before birth, this delay does not explain the increased rate of transepidermal water loss and subsequent lethality. However, the biophysical properties of the skin barrier that regulate the relative permeability of small molecules, infectious agents, water, and gases across this surface are still poorly understood.
To determine which of the three known components of the barrier is disrupted in Gata-3 mutant newborns, we analyzed tight junctions, CEs, and lipid composition. Although total lipid content was similar, Gata-3 mutants exhibit a selective defect in lipid synthesis. Gata-3 mutants have a decreased level of glucosylceramides and its derivative ceramide EOS (Fig. 2 E). Ceramide EOS is one of the precursors of sphingolipids, which interact with free lipids to organize the lipid lamellar structures in the stratum corneum (SC; Wertz and van den Bergh, 1998). Ultrastructural analysis of Gata-3 mutant skin, preserved to maintain lipid structures, revealed a paucity of lamellar bodies in the SC. In addition, these lamellar bodies contain only a few disorganized membrane leaflets and irregular vacuoles (Fig. 2 F). This analysis points to a specific defect in lipid content and organization underlying the selective barrier impairment. In contrast, the other two elements of the barrier appear normal. Specifically, egression of a subcutaneously injected dye halted at occludin-positive structures, indicating that the tight junctions in Gata-3 mutants are fully competent (Furuse et al., 2002; unpublished data). In addition, the CEs of the Gata-3 mutants appear normal: mature, plump, and rigid (unpublished data).
Differentiation defects in GATA-3–deficient skin
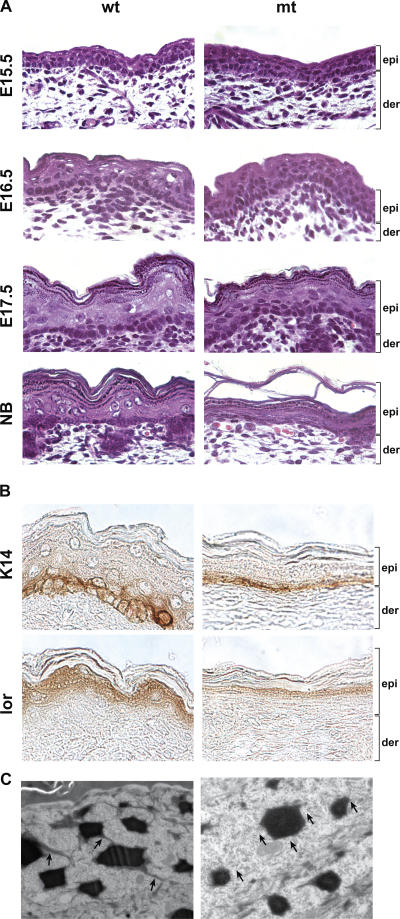
To investigate the etiology of Gata-3 mutants' barrier defect, we examined the histology, differentiation, and proliferation status of embryonic Gata-3 mutant skin. At E1 5.5, when GATA-3 is initially expressed, the histology of Gata-3 mutant skin appears normal (Fig. 3 A). At E16.5, nuclei persisted in the presumptive granular layer of Gata-3–deficient epidermis, consistent with a differentiation defect and delay in barrier acquisition (Fig. 2 D and Fig. 3 A). At E17.5, granular cells of Gata-3–deficient epidermis were properly enucleated and differentiated, again consistent with overcoming the delay in acquiring a selective barrier (Fig. 2 D and Fig. 3 A). Gata-3 mutant newborn epidermis appears thinner with a disorganized basal layer (Fig. 3 A). Immunohistochemical analysis of Gata-3–deficient newborn epidermis demonstrated that the structural proteins K14 (basal) and loricrin (granular) were expressed in the proper cell layer (Fig. 3 B). Ultrastructural analysis of the Gata-3 mutant newborn epidermis revealed the absence of filaments that connect the keratohyalin granules (Fig. 3 C), again suggesting that the terminal differentiation program may be impaired. The rate of proliferation of Gata-3 mutant basal cells is similar to controls, as measured by BrdU immunohistochemistry and cell cycle FACS analysis (unpublished data).
Figure 3.
Differentiation defects in Gata-3–deficient epidermis. (A) Histology of E15.5, E16.5, E17.5, and newborn (NB) dorsal skin. epi, epidermis; der, dermis. (B) Immunohistochemical staining of newborn skin with K14 (basal) and loricrin (granular). (C) EM of the upper granular layer. Arrows highlight the presence/absence of filaments binding and connecting the keratohyalin granules. wt, wild type; mt, mutant.
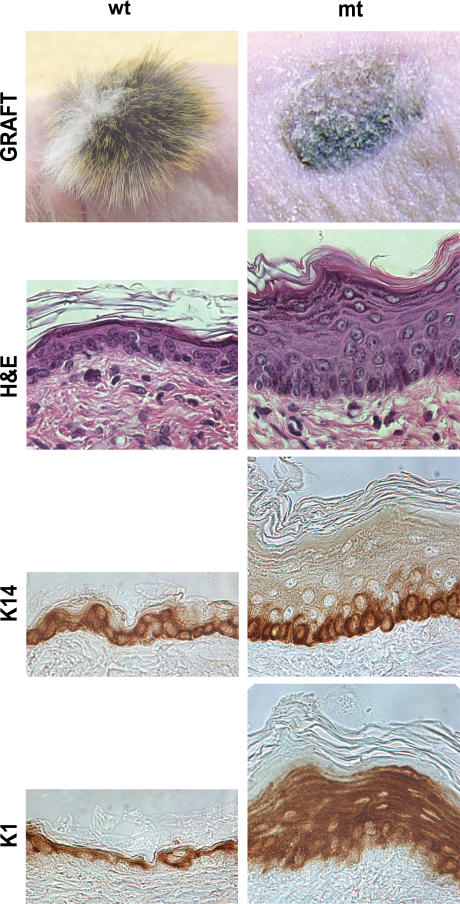
To circumvent the perinatal lethality of Gata-3 mutants and investigate the role GATA-3 plays in epidermal homeostasis, we grafted E18.5 Gata-3 mutant and control littermate skin onto nude mice. The gross morphology of grafted Gata-3 mutant skin confirmed the previously reported role of Gata-3 in hair follicle specification (Fig. 4; Kaufman et al., 2003). Previous studies have established that hyperproliferation and acanthosis (thickened epidermis) are compensatory responses to impaired epidermal barrier (Proksch et al., 1991). Histological analysis of the grafted Gata-3 mutant skin displayed both these hallmark features (Fig. 4). Gata-3 mutant epidermis is ∼10 cell layers thick, with an increase in K1-positive suprabasal cells, whereas both control grafted and hairless nude epidermis are approximately three to four cell layers thick (Fig. 4). Although proliferation was increased in the Gata-3 mutant epidermis, it was restricted to the basal cells (unpublished data). These grafting studies suggest that Gata-3 mutant epidermis retains an inherent barrier defect that extends beyond the perinatal period.
Figure 4.
Differentiation defects and compensatory hyperproliferation in transplanted Gata-3–deficient skin grafts. Histological and immunohistochemical staining with K14 and K1. wt, wild type; mt, mutant; H&E, hematoxylin and eosin.
Epidermal transcriptional pathways regulated by GATA-3 at E15.5, E16.5, and newborn
To identify the pathways of gene expression that are affected by the loss of GATA-3 during development, we analyzed microarray data from Gata-3 mutant and control littermate dorsal skin isolated from three distinct epidermal stages of development: (1) at E15.5, the initial defect in barrier acquisition; (2) at E16.5, the compensatory acquisition of barrier to exclude small molecules; and (3) at newborn, the selective barrier deficiency upon exposure to the terrestrial environment. This tripartite experiment enabled us to query the genes and pathways affected by GATA-3 that led to the delay in epidermal differentiation and the persistent lipid and barrier defect.
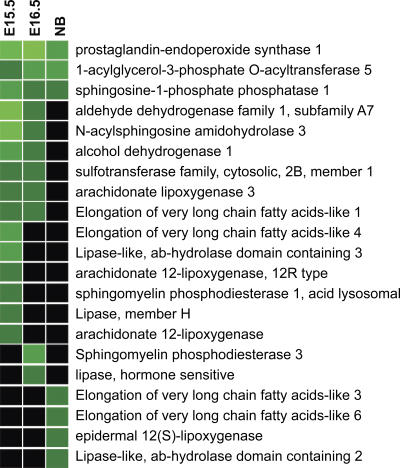
At all developmental stages, lipid synthesis and modification was identified as the most significant and commonly affected pathway in the Gata-3 mutants, consistent with the lipid defect observed in these animals (Fig. 5). Down-regulated at all epidermal developmental stages are prostaglandin-endoperoxide synthase 1 (Ptgs1; greater than threefold), 1-acylglycerol 3 phosphate O-acyltransferase 5 (Agpat5; greater than three- to ninefold), and sphingosine-1-phosphate phosphatase 1 (Sgpp1; greater than two- to fivefold). The family of Elongation of very long fatty acids–like (Elovl) genes, Elovl1, Elovl3, Elovl4, and Elovl6, encoding lipid biosynthetic proteins, is also down-regulated in Gata-3 mutants.
Figure 5.
Lipid synthesis pathway is affected in Gata-3 mutants. Heat map representation of microarray data, which demonstrates down-regulation of genes involved in the lipid synthesis and modification in Gata-3 mutants at E15.5, E16.5, and newborn (NB). Green indicates a more than twofold decrease in mutants, bright green indicates a more than fivefold decrease in mutants, and black indicates no change between mutants and wild type.
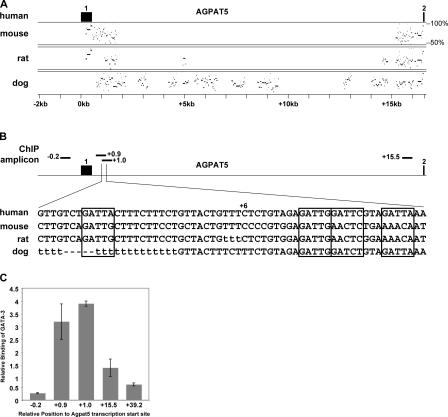
To determine if genes in the lipid biosynthetic pathway are direct targets of GATA-3, we used a genomic approach. First, to identify potential cis-acting regulatory elements in the lipid genes, we performed a multispecies alignment of the human sequences compared with mouse, rat, and dog homologues. Between mouse and human, ∼5% of the genomic sequence is under positive selection; i.e., alignable and conserved (Waterston et al., 2002). Only one third of these regions are predicted to encode an exon of a gene. The other regions of alignment are postulated to encode RNA genes or regulatory elements. An example of the multispecies alignment of the proximal promoter and first intron of AGPAT5 with the program MultiPipMaker is shown in Fig. 6 A (Schwartz et al., 2003). MultiPipMaker identifies two blocks of noncoding sequence conservation: distal to the first exon and proximal to the second exon. To refine this analysis, we used TRANSFAC to query whether GATA-3 binding sites were predicted within these blocks of conserved sequence, with a consensus binding sequence of G A T A/T A/G (Merika and Orkin, 1993; Wingender et al., 2000). Examination of the conservation tracks on the University California Santa Cruz genome web browser enabled us to rapidly determine whether these predicted GATA-3 sites are conserved between species. Examples of two highly conserved GATA-3 sites (GATTA and GATTG) as well as one not conserved (GATTc) and one sequence conserved only with dog (GATTA) are given in Fig. 6 B. Finally, to determine if GATA-3 binds in vivo to these sites, we immunoprecipitated chromatin with a GATA-3–specific antibody. Two overlapping amplicons (+0.9 and +1.0 from AGPAT5 transcription start site), which contain these highly conserved GATA-3 binding sites, were specifically enriched 3.8- and 3.2-fold in the GATA-3 chromatin immunoprecipitated DNA. Sequences in the proximal promoter (−0.2) and more distal in the AGPAT5 gene (+15.5 and +39.2) were not enriched in the GATA-3 chromatin immunoprecipitated DNA (Fig. 6 C). Although a similar genomic analysis of PTGS and SGPP1 were performed, we did not identify multispecies conserved GATA-3 binding sites, which might suggest that the criteria for inclusion were very stringent. In summary, GATA-3 binds in vivo to a region in the first intron of the lipid acyltransferase gene AGPAT5 that contains highly conserved GATA-3 binding sites.
Figure 6.
GATA-3 binds in vivo to sites in the first intron of the lipid acetyltransferase gene AGPAT5. (A) Comparison of human AGPAT5 sequence (−2 kb upstream of transcription start site to second exon) with mouse, rat, and dog Agpat5 sequences. MultiPipMaker calculates the percentage of identity using a Blastz alignment. Percentage of identity (50–100%) is shown on the y axis. Exons 1 and 2 are marked above the sequence identity plot with filled boxes. (B) Location of ChIP amplicons, in proximal promoter (−0.2), in region of sequence conservation distal to exon 1 (+0.9 and +1.0) and in region of sequence conservation proximal to exon 2 (+15.5) with positions relative to AGPAT5 transcription start site. Overlap between +0.9 and +1.0 amplicons (Chr8:6,554,370-6,544,430) contains four GATA-3 binding sites (boxed). (C) C hIP demonstrates specific binding of GATA-3 in vivo to region distal to exon 1 in the first intron of AGPAT5.
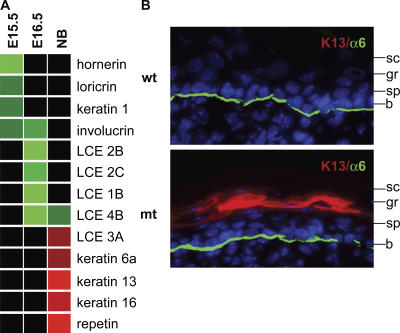
In addition to the defects in lipid synthesis, Gata-3 mutants display a developmental delay in the expression of structural proteins (Fig. 7 A). Specifically at E15.5, genes encoding the cornification proteins hornerin and loricrin, as well as the differentiation proteins K1 and involucrin, are down-regulated in the Gata-3 mutants. At E16.5, expression of late CE genes (LCE 1B, 2B, 2C, 3A, and 4B) are either absent or decreased by more than fivefold in the Gata-3 mutants, demonstrating a continuation of the differentiation delay. Previous work has shown that late CE protein expression immediately precedes in utero dye impermeability in a patterned fashion (Marshall et al., 2001). Therefore, we postulate that this delay in late CE gene expression underlies the delay in barrier acquisition visualized in Fig. 2 D. Because of their temporal expression during development, the genes down-regulated at E15.5 are different than E16.5, but both expression profiles reflect a delay in differentiation. By the newborn stage, Gata-3 mutants express the vast majority of these genes encoding epidermal differentiation and cornification proteins at normal levels. This transcriptional profiling provides the molecular underpinnings to interpret the morphological changes in the Gata-3 mutant skin, observed during development.
Figure 7.
Delayed differentiation during embryonic development and aberrant keratin 13 expression in Gata-3 mutant newborns. (A) Heat map representation of microarray data, demonstrating delayed expression of epidermal differentiation proteins at E15.5 and E16.5. Newborn (NB) Gata-3 mutants display aberrant expression of epidermal structural proteins. Green indicates a more than twofold decrease in mutants, bright green indicates a more than fivefold decrease in mutants, red indicates a more than twofold increase in mutants, bright red indicates a more than fivefold increase in mutants, and black indicates no change between mutants and wild type. LCE, late CE genes. (B) Immunohistochemical staining of K13 in newborn skin. b, basal; sp, spinous; gr, granular.
The transcriptional profile of Gata-3 mutant newborn skin also reflects the pathways invoked to compensate in the ex utero terrestrial environment for an intrinsic barrier defect. Classic studies have shown that barrier deficiency results in increased DNA synthesis and acanthosis (Proksch et al., 1991). Gata-3 mutants express high levels of K6 in the suprabasal layers of the interfollicular epidermis (unpublished data), consistent with many other examples of K6/K16 in hyperproliferative conditions (Wong and Coulombe, 2003). Repetin is a filaggrin-like protein that has been postulated to specifically aggregate K6/K16 filaments, explaining its expression under these conditions (Presland et al., 2006). Unexpectedly, Gata-3 mutants express K13 protein in the spinous layer of the epidermis (Fig. 7 B). K13 is normally expressed only in stratified but not cornified epithelium, such as tongue and esophagus. K13 expression in epidermis has previously only been reported in papillomas at high risk of converting to squamous cell carcinoma (Nischt et al., 1988). The expression of K13 could suggest a role for GATA-3 in squamous cell carcinoma progression. Alternatively, K13 expression could reflect a similar underlying state of the skin that is common to both barrier impairment and tumor progression, such as mounting an inflammatory response.
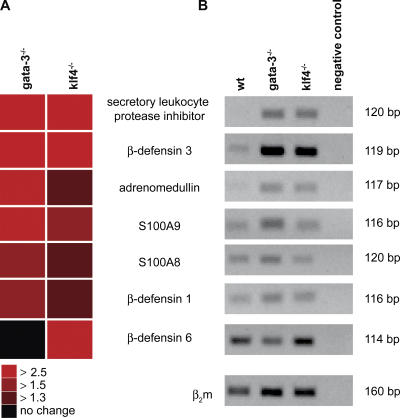
The first line of cutaneous defense against infection by microorganisms is the proteinaceous/lipid skin barrier. Augmenting this physical barrier are both the innate and adaptive immune systems (Zasloff, 2002; Braff et al., 2005; Lehrer, 2005). Antimicrobial peptides, effectors of innate immunity, are expressed by keratinocytes and have distinct but overlapping reactivity against bacteria, fungi, and enveloped viruses (Braff et al., 2005). Antimicrobial peptides are induced to provide a rapid defense, which is particularly important in fetal skin before maturation of immunological memory (Marchini et al., 2002). Epithelial defense is a significantly affected pathway in Gata-3 mutant newborns, including a strong up-regulation of the antimicrobial peptides, secretory leukocyte proteinase inhibitor, adrenomedullin, S100A8, S100A9, and β-defensin 1 and 3 (Fig. 8; Braff et al., 2005; Lehrer, 2005).
Figure 8.
Innate immunity is evoked in both newborn Gata-3 and Klf4−/− mutant skin. (A) Heat map representation of microarray data, demonstrating specific increased expression of genes encoding antimicrobial proteins, effectors of innate immune, in both Gata-3 and Klf4−/− mutant newborn skin. Black indicates no change; dark red indicates >1.3-fold up-regulated; red indicates >1.5-fold up-regulated; and bright red indicates >2.5-fold up-regulated in mutants. (B) Quantitative PCR confirmation of genes encoding antimicrobial peptides up-regulated in Gata-3 and Klf4−/− mutant newborn skin. β2m, β-2-microglobulin for normalization. wt, wild type.
To investigate the specificity of the transcriptional profile of Gata-3 mutant newborn skin, we compared these results with a similar analysis of barrier-impaired Klf4−/− skin (Segre et al., 1999). First, the levels of Klf4 are not altered in Gata-3 mutants and vice versa, suggesting that these transcription factors are not epistatic but distinct in their regulation of epidermal differentiation (unpublished data). Second, the nature of the barrier deficiencies in Klf4−/− and Gata-3 mutants appears completely distinct. Klf4−/− mutants exhibit a specific defect in CE maturation, with normal synthesis but abnormal extrusion of lipids, and a persistent dye penetration even as newborns. Molecularly, Ptgs1 is the only “lipid synthesis pathway” gene with decreased levels in Klf4 mutants. Both mutants do up-regulate genes common to a hyperproliferative state, including K6, K16, and repetin. However, the greatest similarity between the two barrier-deficient mutant mice is an up-regulation of the epithelial defense genes. Klf4−/− and Gata-3 mutants show a similar up-regulation of the innate immune effectors, secretory leukocyte proteinase inhibitor and β-defensin 3 (Fig. 8). However, whereas Klf4−/− mutants show a strong up-regulation of β-defensin 6, Gata-3 mutants more strongly up-regulate adrenomedullin, S100A9, S100A8, and β-defensin 1. Thus, Gata-3 and Klf4 mutants, genetically distinct models of barrier impairment, both activate an innate immune response, but they do so through up-regulation of distinct antimicrobial peptides.
Discussion
These results demonstrate Gata-3's specific role in epidermal barrier acquisition. Gata-3 mutant embryos exhibit a transient delay in differentiation, demonstrated by percutaneous dye penetration. Similar delays in dye exclusion were observed in mice with targeted deletions of genes encoding CE proteins, envoplakin, and loricrin (Koch et al., 2000; Maatta et al., 2001). However, envoplakin- and loricrin-deficient mice survive the perinatal period, perhaps because of the compensatory up-regulation of other structural proteins. In contrast, Gata-3 deficiency in the epidermis results in a perinatal lethality with an inherent barrier defect that extends postnatally, as demonstrated by grafting experiments. Underlying Gata-3 mutant's barrier defect is a severe defect in lipid synthesis, in particular, ceramide EOS and glucosylceramides. The electron micrographs of Gata-3 mutant skin, postfixed to maintain lipid structure, are reminiscent of similar findings in infants with severely affected type 2 Gaucher disease. Mutations in β-glucocerebrosidase, the enzyme that catalyzes the hydrolysis of glucosylceramide to ceramide, underlie Gaucher disease (Sidransky et al., 1992). Type 2 Gaucher disease and a mouse model, with a targeted deletion of β-glucocerebrosidase, manifest at birth with a primary barrier deficiency and display abnormal loosely packed lamellar body–derived sheets in the SC (Holleran et al., 1994). Our findings suggest that GATA-3 may act to regulate this important process of lipid biosynthesis, and genes in this pathway should be tested as a potential modifiers to explain the wide phenotypic variation observed among Gaucher patients (Goker-Alpan et al., 2005). Our studies identified AGPAT5, which catalyzes an essential step in the synthesis of all glycerolipids, as a direct target in vivo for GATA-3 (Lu et al., 2005). Future studies will address the hierarchical transcriptional regulation of lipid synthesis in the skin.
Morphological and transcriptional analyses at distinct developmental stages revealed both GATA-3's regulation of differentiation and lipid synthesis pathways and the compensatory responses to impaired barrier. For example, although the newborn barrier-deficient Gata-3 mutant skin is hypocellular, the grafted Gata-3 mutant skin is acanthotic or hypercellular, as a compensatory response to the impaired barrier in the terrestrial environment (Fig. 3 A and Fig. 4). Analysis of Gata-3 mutants at only one developmental stage would have revealed the specific defect in lipid biosynthesis but would have been refractive to elucidating the transient delay in expression of genes encoding differentiation and cornification proteins. Transcriptional profiling at multiple developmental stages brings clarity to pathways affected by and responding to Gata-3's loss.
This process is remarkably well conserved, as GATA transcription factors are also essential to specify the fate and regulate differentiation of epidermal cells in Caenorhabditis elegans. The cell biology of the C. elegans epidermis closely resembles that of mammals, including intermediate filament networks and cell connections through adherens and tight junctions (Hardin and Lockwood, 2004). GATA transcription factor ELT-1 specifies epidermal cell fate (Page et al., 1997). Subsequently, ELT-5 and -6 (adjacent genes encoding GATA factors) are required throughout development to regulate epidermal cell differentiation (Koh and Rothman, 2001).
Mammalian lung and skin are both epithelia at the interface between the body and the environment that form proteinaceous lipid barriers. Although lung is a branched simple epithelium and the composition of the barriers is distinct, there are remarkable similarities between the systems. At the transcriptional level, corticosteroids and thyroid hormone accelerate barrier maturation in utero of both epidermis and alveoli (Aszterbaum et al., 1993). Just as Klf4 is necessary for the terminal stages of epidermal development, Lklf (Klf2) plays an important role in the terminal stages of lung development (Wani et al., 1999). GATA-6 is the only known GATA factor expressed in the distal epithelium of the developing lung. Expression of a dominant-negative form of GATA-6 in these alveolar cells resulted in a defect in terminal differentiation and proximal airway development. These GATA-6 transgenic mice die perinatally with defects in lipid (surfactant protein) synthesis and decreased expression of Aquaporin 5, a gene encoding a water channel. Similar to GATA-3's role in epidermal barrier, GATA-6 is necessary for maturation of the proteinaceous lipid barrier that regulates alveoli gas exchange (Yang et al., 2002).
Extending the well-established paradigms from hematopoietic cells, it is intriguing to speculate whether GATA-3 will have similar interactions with family members of other transcription factors in the skin. GATA-1 acts upstream of EKLF (KLF1) during erythroid development, and GATA-3 acts upstream of LKLF (KLF2) during lymphocte development (Kuo and Leiden, 1999; Anderson et al., 2000). The expression of GATA-3 and KLF4 in basal and suprabasal cells, respectively, is consistent with GATA-3 acting upstream of KLF4. Klf4 levels are unchanged in Gata-3 mutants, which could reflect compensatory autoregulation or parallel pathways.
Both Klf4 and Gata-3 mutants exhibit an epidermal barrier deficiency, but each activates distinct antimicrobial peptides, effectors of innate immunity. Innate immunity is important before the adaptive immune system mounts a response and particularly during the first year of human life, as the adaptive immune system is maturing. These findings demonstrate that newborn skin can mount a robust activation of an innate immunity. Moreover, an analysis of Gata-3 and Klf4 mutant newborns demonstrates that genetically distinct barrier impairments activate overlapping but distinct innate immune responses. The comparison of Klf4- and Gata-3–deficient newborn epidermis will be very informative to unravel the complex immune response to barrier impairment.
Barrier disruption is a hallmark characteristic of common inflammatory skin disorders, such as atopic dermatitis (more commonly known as eczema) and psoriasis (Segre, 2006). Recent work has examined the distinct innate immune responses of psoriasis and atopic dermatitis (de Jongh et al., 2005). In particular, patients with atopic dermatitis have an increased tendency to develop both disseminated viral skin infection after smallpox vaccine inoculation and recurrent Staphylococcus aureus infections because of inadequate innate immune response (Howell et al., 2006). In contrast, barrier-impaired keratitis-ichthyosis-deafness patients develop recurring Candida albicans yeast infections. Mutations in the epidermal cornification protein filaggrin were recently reported to underlie atopic dermatitis, focusing attention on the role that barrier impairment plays in this disorder (Palmer et al., 2006). Because of the naive state of T and B cells in newborn mice, a full investigation into this complex innate/adaptive immune response requires adult epidermal-specific targeting of Gata-3 and Klf4.
Materials and methods
Generation of Gata-3fl/fl K14-Cre mice and skin grafts
Mice carrying the Gata-3fl allele (Pai et al., 2003) were crossed with mice expressing germline Cre recombinase (Scheel et al., 2003) to generate a Gata-3 − allele. These mice were then crossed onto mice transgenic for human K14-driven Cre recombinase (Andl et al., 2004) to generate Gata-3 +/− K14-Cre. Gata-3 −/fl K14-Cre mice were generated by crossing Gata-3fl/fl mice with Gata-3 +/− K14-Cre mice. Genotyping was done as previously described (Pai et al., 2003; Andl et al., 2004). The morning of the plug was 0.5 d after coitum. E18.5 dorsal skin was grafted onto nude mice in an area that the mice could not scratch, and these mice were individually housed. All animal studies were approved by the National Human Genome Research Institute animal care and use committee, and all mice were housed in our Association for Assessment of Laboratory Animal Care–accredited facility.
Barrier function assays
Dye penetration assays were performed with X-gal at pH 4.5 for 4 h at 37°C as previously described (Hardman et al., 1998). After staining, embryos were photographed under a dissecting scope (MZFLIII; Leica) using a digital camera (AxioCam; Carl Zeiss MicroImaging, Inc.), and images were acquired with OpenLab software (Improvision). Transepiderrmal water loss was measured using a Tewameter (Courage + Khazaka).
Histology and immunohistochemistry
Routine histology and paraffin staining were performed as described previously (Jaubert et al., 2003). For immunofluorescence, frozen sections were fixed in 10% formalin/PBS and stained with primary antibodies: rabbit polyclonal antibodies against GATA-3 (Segre 379-2b; 1:100), K14 (1:1,000; Covance,), K1 (1:1,000; Covance), Loricrin (1:500; Covance), K13 (1:500; a gift from S. Yuspa, National Cancer Institute, Bethesda, MD), and α6 integrin rat polyclonal antibodies (MAB1982; 1:100; Chemicon). Fluorescent secondary antibodies were Alexa 488 goat anti–rabbit (1:400) and Alexa 594 goat anti–rat (1:200). Slides were mounted with DAPI glycerol media, containing SlowFade Gold antifade, to counterstain nuclei (Invitrogen). Fluorescent staining was imaged with a microscope (Axioplot; Carl Zeiss MicroImaging, Inc.) and photographed with a camera (CoolSNAP; Photometrix).
RNA isolation, Northern blot analysis, and microarray
RNA was isolated from the dorsal skin of newborns and embryos, incubated in RNALater (Ambion), snap frozen, homogenized in TRIzol (Invitrogen) using tissue lyser (QIAGEN), and processed according to the manufacturer's instructions. Northern blot was hybridized with probes for Gata-3 and Gapdh. Microarrays were done on independent samples for newborns (n = 4) and E15.5 and E16.5 embryos (n = 3). Control littermates are Gata-3 fl/+ or Gata-3fl/fl, and mutant mice are Gata-3 fl/− K14-Cre. Complimentary RNA was labeled according to the manufacturer's recommendations and hybridized onto Affymetrix 430 2.0 A+B mouse arrays. These arrays contain 45,000 probe sets, representing 34,000 well-substantiated mouse genes. We identified ∼20,000 probes as present in mouse skin during the developmental windows analyzed in these experiments. Microarray results were analyzed by Genesifter using a t test (P < 0.05) and Benjamini and Hochberg correction (VizX Labs). Confirmation of fold changes was made with quantitative PCR on cDNA from Gata-3 and Klf4 mutants on a TaqMan light cycler (Applied Biosystems) with SYBR Green mix (Invitrogen), and primers spanning exon boundaries are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200605057/DC1).
Genomic analysis
Pipmaker and MultiPipmaker were performed with repeat masked sequences (http://pipmaker.bx.psu.edu/pipmaker/) with mouse (Chr8:18,841,481-18,861,523), human (Chr8:6,548,286-6,569,868), rat (Chr16:75,769,607-75,791,434), and dog (Chr16:61,666,596-61,692,310) sequences (Schwartz et al., 2003). Coordinates for blocks 1 and 2 are human (Chr8:6,553,816-6,554,997 and Chr8:6,568,473-6,569,799, respectively). The overlap of amplicon +0.9 and +1.0 in which the GATA-3 conserved sequences are identified is Chr8:6,554,370-6,544,430. Mouse and human sequence coordinates are relative to February 2006 and March 2006 releases, respectively. TRANSFAC was accessed through a National Human Genome Research Institute site license (Wingender et al., 2000).
Chromatin immunoprecipitation (ChIP) studies
ChIP was performed on human MCF-7 cells, an epithelial cell line that expresses GATA-3 and the lipid biosynthetic genes, including AGPAT5. Chromatin was immunoprecipitated with a GATA-3 antibody (SC-9009; Santa Cruz Biotechnology, Inc.) binding to the endogenous protein. Other reagents were provided in the ChIP-IT kit (Active Motif), and we followed the manufacturer's instructions. DNA/GATA-3 antibody complexes were immunoprecipitated with protein G and A beads. DNA was quantified with QuantiTect SYBR Green PCR kit (QIAGEN). Primers are listed in Table S2 (available at http://www.jcb.org/cgi/content/full/jcb.200605057/DC1), and amplification was quantified on a TaqMan light cycler (Applied Biosystems). Binding of GATA-3 to chromatin immunoprecipitated DNA was measured as the change in the number of cycles required to cross a threshold, normalized to sonicated, reverse-cross-linked input DNA.
Ultrastructural and lipid analysis
Whole backskin was removed, placed on a paper towel, and fixed in modified Karnovsky's fixative (2% paraformaldehyde, 2% glutaraldehyde, 0.1 M cacodylate buffer, pH 7.3, and 0.06% CaCl2) overnight at 4°C. Samples were washed twice in 0.1 M cacodylate buffer after fixation before embedding. Lipids were extracted into chloroform: methanol mixtures and analyzed by thin-layer chromatography as previously described (Law et al., 1995). Lipid masses were used to calculate weight percentages. Ruthenium tetroxide transmission EM was performed as previously described (List et al., 2003).
Online supplemental material
Tables S1 and S2 provide the sequences of the primers used for quantitative RT-PCR and ChIP, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200605057/DC1.
Supplementary Material
Acknowledgments
We thank Cherry Yang for genotyping; Abdel Elkahloun for microarray analysis; Travis Moreland for building the database; Qian-Chun Yu and Neelima Shah for EM expertise; Arturo Incao for performing the graft surgeries; Julia Fekecs and Darryl Leja for assistance with preparing the figures; Stuart Yuspa for insightful discussions; and David Bodine, Tiffany Scharschmidt, and Paul Liu for critical evaluation of the manuscript. We also thank members of the laboratory, in particular, Satyakam Patel, Jennifer Yang, and Christina Feng for their underlying contributions.
This work was supported by the National Human Genome Research Institute intramural program.
Abbreviations used in this paper: CE, cornified envelope; ChIP, chromatin immunoprecipitation; E, embryonic day; K, cytokeratin; Klf4, Kruppel-like factor 4; SC, stratum corneum.
References
- Anderson, K.P., S.C. Crable, and J.B. Lingrel. 2000. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood. 95:1652–1655. [PubMed] [Google Scholar]
- Andl, T., K. Ahn, A. Kairo, E.Y. Chu, L. Wine-Lee, S.T. Reddy, N.J. Croft, J.A. Cebra-Thomas, D. Metzger, P. Chambon, et al. 2004. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 131:2257–2268. [DOI] [PubMed] [Google Scholar]
- Aszterbaum, M., K.R. Feingold, G.K. Menon, and M.L. Williams. 1993. Glucocorticoids accelerate fetal maturation of the epidermal permeability barrier in the rat. J. Clin. Invest. 91:2703–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain, C., W.E. Lowry, A. Geoghegan, L. Polak, and E. Fuchs. 2004. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 118:635–648. [DOI] [PubMed] [Google Scholar]
- Braff, M.H., A. Bardan, V. Nizet, and R.L. Gallo. 2005. Cutaneous defense mechanisms by antimicrobial peptides. J. Invest. Dermatol. 125:9–13. [DOI] [PubMed] [Google Scholar]
- Byrne, C., M. Tainsky, and E. Fuchs. 1994. Programming gene expression in developing epidermis. Development. 120:2369–2383. [DOI] [PubMed] [Google Scholar]
- Dai, X., and J.A. Segre. 2004. Transcriptional control of epidermal specification and differentiation. Curr. Opin. Genet. Dev. 14:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jongh, G.J., P.L. Zeeuwen, M. Kucharekova, R. Pfundt, P.G. van der Valk, W. Blokx, A. Dogan, P.S. Hiemstra, P.C. van de Kerkhof, and J. Schalkwijk. 2005. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J. Invest. Dermatol. 125:1163–1173. [DOI] [PubMed] [Google Scholar]
- Elias, P.M. 2005. Stratum corneum defensive functions: an integrated view. J. Invest. Dermatol. 125:183–200. [DOI] [PubMed] [Google Scholar]
- Furuse, M., M. Hata, K. Furuse, Y. Yoshida, A. Haratake, Y. Sugitani, T. Noda, A. Kubo, and S. Tsukita. 2002. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J. Cell Biol. 156:1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goker-Alpan, O., K.S. Hruska, E. Orvisky, P.S. Kishnani, B.K. Stubblefield, R. Schiffmann, and E. Sidransky. 2005. Divergent phenotypes in Gaucher disease implicate the role of modifiers. J. Med. Genet. 42:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin, J., and C. Lockwood. 2004. Skin tight: cell adhesion in the epidermis of Caenorhabditis elegans. Curr. Opin. Cell Biol. 16:486–492. [DOI] [PubMed] [Google Scholar]
- Hardman, M.J., P. Sisi, D.N. Banbury, and C. Byrne. 1998. Patterned acquisition of skin barrier function during development. Development. 125:1541–1552. [DOI] [PubMed] [Google Scholar]
- Holleran, W.M., E.I. Ginns, G.K. Menon, J.U. Grundmann, M. Fartasch, C.E. McKinney, P.M. Elias, and E. Sidransky. 1994. Consequences of beta-glucocerebrosidase deficiency in epidermis. Ultrastructure and permeability barrier alterations in Gaucher disease. J. Clin. Invest. 93:1756–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, M.D., R.L. Gallo, M. Boguniewicz, J.F. Jones, C. Wong, J.E. Streib, and D.Y. Leung. 2006. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 24:341–348. [DOI] [PubMed] [Google Scholar]
- Ito, M., Y. Liu, Z. Yang, J. Nguyen, F. Liang, R.J. Morris, and G. Cotsarelis. 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11:1351–1354. [DOI] [PubMed] [Google Scholar]
- Jaubert, J., J. Cheng, and J.A. Segre. 2003. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 130:2767–2777. [DOI] [PubMed] [Google Scholar]
- Kaufman, C.K., P. Zhou, H.A. Pasolli, M. Rendl, D. Bolotin, K.C. Lim, X. Dai, M.L. Alegre, and E. Fuchs. 2003. GATA-3: an unexpected regulator of cell lineage determination in skin. Genes Dev. 17:2108–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, P.J., P.A. de Viragh, E. Scharer, D. Bundman, M.A. Longley, J. Bickenbach, Y. Kawachi, Y. Suga, Z. Zhou, M. Huber, et al. 2000. Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J. Cell Biol. 151:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, K., and J.H. Rothman. 2001. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 128:2867–2880. [DOI] [PubMed] [Google Scholar]
- Koster, M.I., and D.R. Roop. 2004. p63 and epithelial appendage development. Differentiation. 72:364–370. [DOI] [PubMed] [Google Scholar]
- Kuo, C.T., and J.M. Leiden. 1999. Transcriptional regulation of T lymphocyte development and function. Annu. Rev. Immunol. 17:149–187. [DOI] [PubMed] [Google Scholar]
- Law, S., P.W. Wertz, D.C. Swartzendruber, and C.A. Squier. 1995. Regional variation in content, composition and organization of porcine epithelial barrier lipids revealed by thin-layer chromatography and transmission electron microscopy. Arch. Oral Biol. 40:1085–1091. [DOI] [PubMed] [Google Scholar]
- Lechler, T., and E. Fuchs. 2005. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 437:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer, R.I. 2005. In defense of skin. J. Invest. Dermatol. 125:viii–ix; discussion x–xi. [DOI] [PubMed] [Google Scholar]
- Levy, V., C. Lindon, B.D. Harfe, and B.A. Morgan. 2005. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev. Cell. 9:855–861. [DOI] [PubMed] [Google Scholar]
- List, K., C.C. Haudenschild, R. Szabo, W. Chen, S.M. Wahl, W. Swaim, L.H. Engelholm, N. Behrendt, and T.H. Bugge. 2002. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene. 21:3765–3779. [DOI] [PubMed] [Google Scholar]
- List, K., R. Szabo, P.W. Wertz, J. Segre, C.C. Haudenschild, S.Y. Kim, and T.H. Bugge. 2003. Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J. Cell Biol. 163:901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B., Y.J. Jiang, Y. Zhou, F.Y. Xu, G.M. Hatch, and P.C. Choy. 2005. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem. J. 385:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatta, A., T. DiColandrea, K. Groot, and F.M. Watt. 2001. Gene targeting of envoplakin, a cytoskeletal linker protein and precursor of the epidermal cornified envelope. Mol. Cell. Biol. 21:7047–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini, G., S. Lindow, H. Brismar, B. Stabi, V. Berggren, A.K. Ulfgren, S. Lonne-Rahm, B. Agerberth, and G.H. Gudmundsson. 2002. The newborn infant is protected by an innate antimicrobial barrier: peptide antibiotics are present in the skin and vernix caseosa. Br. J. Dermatol. 147:1127–1134. [DOI] [PubMed] [Google Scholar]
- Marshall, D., M.J. Hardman, K.M. Nield, and C. Byrne. 2001. Differentially expressed late constituents of the epidermal cornified envelope. Proc. Natl. Acad. Sci. USA. 98:13031–13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika, M., and S.H. Orkin. 1993. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 13:3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, R.J., Y. Liu, L. Marles, Z. Yang, C. Trempus, S. Li, J.S. Lin, J.A. Sawicki, and G. Cotsarelis. 2004. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22:411–417. [DOI] [PubMed] [Google Scholar]
- Nischt, R., D.R. Roop, T. Mehrel, S.H. Yuspa, M. Rentrop, H. Winter, and J. Schweizer. 1988. Aberrant expression during two-stage mouse skin carcinogenesis of a type I 47-kDa keratin, K13, normally associated with terminal differentiation of internal stratified epithelia. Mol. Carcinog. 1:96–108. [DOI] [PubMed] [Google Scholar]
- Page, B.D., W. Zhang, K. Steward, T. Blumenthal, and J.R. Priess. 1997. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. 11:1651–1661. [DOI] [PubMed] [Google Scholar]
- Pai, S.Y., M.L. Truitt, C.N. Ting, J.M. Leiden, L.H. Glimcher, and I.C. Ho. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 19:863–875. [DOI] [PubMed] [Google Scholar]
- Palmer, C.N., A.D. Irvine, A. Terron-Kwiatkowski, Y. Zhao, H. Liao, S.P. Lee, D.R. Goudie, A. Sandilands, L.E. Campbell, F.J. Smith, et al. 2006. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 38:441–446. [DOI] [PubMed] [Google Scholar]
- Pandolfi, P.P., M.E. Roth, A. Karis, M.W. Leonard, E. Dzierzak, F.G. Grosveld, J.D. Engel, and M.H. Lindenbaum. 1995. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat. Genet. 11:40–44. [DOI] [PubMed] [Google Scholar]
- Presland, R., J.A. Rothnagel, and O.T. Lawrence. 2006. Profilaggrin and the fused S100 family of calcium-binding proteins. In Skin Barrier. P. Elias and K. Feingold, editors. Taylor & Francis, New York, NY. 111–140.
- Proksch, E., K.R. Feingold, M.Q. Man, and P.M. Elias. 1991. Barrier function regulates epidermal DNA synthesis. J. Clin. Invest. 87:1668–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel, J.R., L.J. Garrett, D.M. Allen, T.A. Carter, L. Randolph-Moore, M.J. Gambello, F.H. Gage, A. Wynshaw-Boris, and C. Barlow. 2003. An inbred 129SvEv GFPCre transgenic mouse that deletes loxP-flanked genes in all tissues. Nucleic Acids Res. 31:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., L. Elnitski, M. Li, M. Weirauch, C. Riemer, A. Smit, E.D. Green, R.C. Hardison, and W. Miller. 2003. MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 31:3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre, J.A. 2006. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 116:1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre, J.A., C. Bauer, and E. Fuchs. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 22:356–360. [DOI] [PubMed] [Google Scholar]
- Sidransky, E., D.M. Sherer, and E.I. Ginns. 1992. Gaucher disease in the neonate: a distinct Gaucher phenotype is analogous to a mouse model created by targeted disruption of the glucocerebrosidase gene. Pediatr. Res. 32:494–498. [DOI] [PubMed] [Google Scholar]
- Tumbar, T., G. Guasch, V. Greco, C. Blanpain, W.E. Lowry, M. Rendl, and E. Fuchs. 2004. Defining the epithelial stem cell niche in skin. Science. 303:359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wani, M.A., S.E. Wert, and J.B. Lingrel. 1999. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J. Biol. Chem. 274:21180–21185. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., K. Lindblad-Toh, E. Birney, J. Rogers, J.F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature. 420:520–562. [DOI] [PubMed] [Google Scholar]
- Wertz, P.W., and B. van den Bergh. 1998. The physical, chemical and functional properties of lipids in the skin and other biological barriers. Chem Phys. Lipids. 91:85–96. [DOI] [PubMed] [Google Scholar]
- Wingender, E., X. Chen, R. Hehl, H. Karas, I. Liebich, V. Matys, T. Meinhardt, M. Pruss, I. Reuter, and F. Schacherer. 2000. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 28:316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, P., and P.A. Coulombe. 2003. Loss of keratin 6 (K6) proteins reveals a function for intermediate filaments during wound repair. J. Cell Biol. 163:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., M.M. Lu, L. Zhang, J.A. Whitsett, and E.E. Morrisey. 2002. GATA6 regulates differentiation of distal lung epithelium. Development. 129:2233–2246. [DOI] [PubMed] [Google Scholar]
- Zasloff, M. 2002. Antimicrobial peptides in health and disease. N. Engl. J. Med. 347:1199–1200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.