Abstract
The molecular role of poly (ADP-ribose) polymerase-1 in DNA repair is unclear. Here, we show that the single-strand break repair protein XRCC1 is rapidly assembled into discrete nuclear foci after oxidative DNA damage at sites of poly (ADP-ribose) synthesis. Poly (ADP-ribose) synthesis peaks during a 10 min treatment with H2O2 and the appearance of XRCC1 foci peaks shortly afterwards. Both sites of poly (ADP-ribose) and XRCC1 foci decrease to background levels during subsequent incubation in drug-free medium, consistent with the rapidity of the single-strand break repair process. The formation of XRCC1 foci at sites of poly (ADP-ribose) was greatly reduced by mutation of the XRCC1 BRCT I domain that physically interacts with PARP-1. Moreover, we failed to detect XRCC1 foci in Adprt1–/– MEFs after treatment with H2O2. These data demonstrate that PARP-1 is required for the assembly or stability of XRCC1 nuclear foci after oxidative DNA damage and suggest that the formation of these foci is mediated via interaction with poly (ADP-ribose). These results support a model in which the rapid activation of PARP-1 at sites of DNA strand breakage facilitates DNA repair by recruiting the molecular scaffold protein, XRCC1.
INTRODUCTION
Single-strand breaks (SSBs) can arise directly (e.g. by fragmentation of damaged deoxyribose) or indirectly (e.g. during the enzymatic excision of abasic sites) (1,2). The importance of repairing SSBs is suggested by the increased spontaneous and/or induced genetic instability of cell lines in which polypeptide components of this process are absent or mutated (3–7). It is likely that, if not repaired, SSBs are converted into DSBs during DNA replication (8).
A critical component of SSBR is the polypeptide XRCC1. Mutant cell lines lacking XRCC1 display hypersensitivity to a broad range of genotoxins, increased frequencies of translocations and deletions, and a reduced ability to rapidly repair chromosomal SSBs (9). Mice with targeted disruption of both Xrcc1 alleles exhibit early embryonic lethality (10). At the molecular level, XRCC1 appears to coordinate or stimulate enzymatic components of SSBR by physical interaction (2,11). XRCC1 interacts with APE1 (12), DNA polymerase β (13,14), DNA polynucleotide kinase(11), and DNA ligase IIIα (15–18). In addition, XRCC1 also interacts with poly (ADP-ribose) polymerase-1 (PARP-1) (13,19).
PARP-1 rapidly binds to DNA strand breaks (SSBs) and is thereby activated, covalently automodifying itself and, to a lesser extent, proximal acceptor proteins with poly (ADP-ribose) polymer (PAR) (20,21). XRCC1 preferentially interacts with automodified PARP-1, most likely via a PAR-binding motif present within the internal BRCT I domain of XRCC1 (19,22). Consistent with this, inhibiting PAR synthesis with 3-aminobenzamide greatly reduces or ablates the interaction of XRCC1 with PARP-1 (23,24). These data have prompted the hypothesis that one function of PARP-1 is to recruit XRCC1 to sites of SSBs. In agreement with this, the XRCC1 BRCT I domain is critical for SSBR and for cell survival mediated by XRCC1 after DNA alkylation (25). However, evidence that PARP-1 might recruit XRCC1 to SSBs is currently lacking.
Here, we have addressed the spatial and temporal relationship between PAR synthesis and the assembly of XRCC1 subnuclear foci during the repair of oxidative SSBs induced by H2O2. We show that H2O2 induces the rapid and transient formation of PAR nuclear foci, indicative of the binding and activation of PARP-1 at DNA strand breaks, followed soon after by the appearance of XRCC1 nuclear foci at the same sites. Mutation of the XRCC1 BRCT I domain that interacts with PAR reduces or prevents the appearance of XRCC1 foci, and XRCC1 foci fail to appear after H2O2 in mouse cells harbouring a targeted disruption of the Adprt1 gene encoding Parp-1. These data provide evidence that PARP-1 is required for the recruitment of XRCC1 to sites of chromosomal DNA strand breakage.
MATERIALS AND METHODS
Cells and cell culture
CHO cells were cultured as monolayers in α-MEM (Gibco, Invitrogen Corporation) supplemented with 10% fetal calf serum (Gibco). The expression constructs empty pcD2E, pcD2E-XH, pcD2E-XHC389A, or pcD2E242–533 were transfected into EM9 cells by calcium phosphate coprecipitation and pooled populations of >50 transfectants were selected in the presence of G418 (1.5 mg/ml) for 10–14 days, creating the cell lines EM9-V, EM9-XH, EM9-XHC389A and EM9-XH242–533, respectively. The cell lines EM9-XHR399Q, EM9-XHW385D, EM9-XHLI360/361DD and the wild-type control cell lines for the latter two mutants (EM9-XH5) have been described previously (25). Spontaneously immortalized Adprt1+/+ and Adprt1–/– mouse embryonic fibroblasts (26) were cultured as monolayers in DMEM (Gibco) supplemented with 10% fetal calf serum.
Expression constructs
The mammalian cell expression vector pcD2E and the derivative pcD2EXH encoding C-terminally histidine-tagged XRCC1 (XRCC1-His) have been described previously (17,27). Oligonucleotide primers (Forward; 5′-CCCGAATTCGTTGACATGCACCATCACCATCACCATGGGATCCCCAAAGGGAAGAGGAAGTTG-3′ and Reverse; 5′-CCCG AATTCTGCAGTCAATCAGGAGGCTCCTGGTGTTC-3′) were used to amplify XRCC1242–533 using PCR. The PCR product was cloned into pCR2.1 TOPO vector (Invitrogen), confirmed by sequencing, and the XRCC1242–533 ORF subcloned into the EcoR1 site of pcD2E. Derivatives of pcD2EXH harbouring mutations within the BRCT I domain were described previously (25).
Indirect immunofluorescence
Cells were grown on coverslips, rinsed in PBS, and treated with either 10 mM H2O2 in PBS at room temperature for 20 min, or with 20 or 40 µg/ml MMS for 20 min at 37°C. Rinsed coverslips were then incubated for the indicated times in drug-free medium at 37°C. Coverslips were rinsed in PBS and fixed with methanol–acetone (1:1, v/v) for 10 min at 4°C. Fixed cells were rinsed in PBS and permeabilized for 5 min at 4°C in 20 mM HEPES, pH 7.4, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton X-100. Permeabilized cells were rinsed in PBS and incubated for 20 min at 37°C with either anti-phospho H2AX monoclonal antibody (Upstate; clone JBW301, 1/800 dilution) or a mixture of anti-poly (ADP-ribose) monoclonal antibodies (10H; a gift from Gilbert de Murcia, 1:200 dilution in PBS supplemented with 2% bovine serum albumin, fraction V, Sigma) and anti-XRCC1 polyclonal antibodies (AHP428, Serotec, 1:200 dilution as above). After rinsing in PBS, coverslips were incubated in a mixture of either fluorescein isothiocyanate-conjugated anti-mouse IgG (DAKO) secondary antibody, or a mixture of fluorescein isothiocyanate-conjugated anti-rabbit IgG (DAKO) and TRITC-conjugated anti-mouse IgG (Sigma) secondary antibodies, at 1:200 dilution in PBS supplemented with 2% bovine serum albumin, fraction V for 20 min at 37°C. Nuclei were counterstained with 0.000025% 4′,6′-diamidino-2-phenylindole (DAPI). Cells were analysed and photographed with a Zeiss Axioplan-2 fluorescence microscope equipped with a model RTI/CCD-1300/Y chilled charge-coupled device digital camera (Princeton, NJ) and Metamorph imaging software. Photographs were taken at 100× magnification.
Survival curves
CHO cells or MEFs (250–1000) were plated in 10-cm dishes in duplicate and exposed to the indicated concentrations of H2O2 for 10 min in PBS, or MMS for 20 min in complete medium, at 37°C. Cells were washed with PBS (×3) and then incubated for 7–10 days in drug-free medium to form macroscopic colonies. The surviving fraction was calculated by dividing the average number of colonies on treated plates by the average number on untreated plates. For each survival curve, data are the mean (±1 S.D.) of three independent experiments.
Cell extracts and immunoblotting
Whole-cell extracts were prepared from frozen pellets of transfected EM9 cells (2 × 106) or MEFs (6 × 105) by resuspension in 0.2 ml of hot 2× Laemmli SDS–PAGE loading buffer [100 mM Tris–Cl (pH 6.8), 4% SDS, 0.2% Bromophenol Blue, 20% glycerol, 200 mM β-mercaptoethanol]. High-molecular-weight DNA was sheared by passage through a narrow-gauge needle and solubilized proteins fractionated by SDS–PAGE and transferred to Nitrocellulose. Filters were rinsed with TBST (25 mM Tris–HCl, 0.5 M NaCl, 0.1% Tween-20), blocked in TBST containing 5% non-fat dried milk, and incubated for 1 h at room temperature with anti-XRCC1 polyclonal antibody (AHP428, Serotec, 1:3000 dilution in TBST supplemented with 1% milk) or anti-Actin monoclonal antibody (clone AC-40, Sigma, 1:1000 dilution). Filters were washed in TBST (3 × 15 min) and incubated with HRP-conjugated rabbit IgG (DAKO, 1:5000 dilution) or HRP-conjugated mouse IgG (DAKO, 1:5000 dilution) for 1 h at room temperature. Filters were washed in TBST (4 × 15 min) and subject to chemiluminescence detection (Amersham Biosciences).
RESULTS
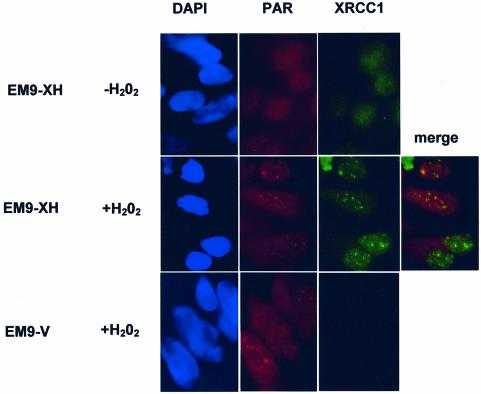
To examine whether XRCC1 is recruited to sites of poly (ADP-ribose) (PAR) synthesis at single-strand breaks induced by DNA oxidation, immunofluorescence experiments were conducted in CHO cells treated with H2O2. The CHO cells employed were XRCC1-mutant EM9 cells harbouring either empty pCD2E vector (EM9-V cells) or vector encoding human histidine-tagged XRCC1 (EM9-XH cells). The two cell lines were examined for co-localization of XRCC1 and PAR before and after a 20 min treatment with 10 mM H2O2. Immunostaining with anti-PAR Mabs revealed discrete nuclear foci in both EM9-V and EM9-XH cells after H2O2 treatment and these foci were absent from untreated cells, consistent with the rapid activation of PARP at H2O2-induced SSBs (Fig. 1, red). XRCC1 was similarly detected as nuclear immunofoci after H2O2 treatment, a large proportion of which co-localized with those of PAR (Fig. 1, green and ‘merge’). As expected, XRCC1 signal was absent from EM9-V cells harbouring empty vector (Fig. 1 bottom right panel). Next, the kinetics at which PAR and XRCC1 foci were formed during a 2-h repair-incubation after H2O2 treatment was examined. The appearance of PAR foci in EM9-XH cells was very rapid, peaking during the 20-min incubation with drug, after which the number of foci-positive cells fell to background within 1 h (Fig. 2A, filled squares). The appearance of XRCC1 foci displayed similar kinetics, though these foci peaked a little later, after ∼10 min of repair-incubation, and persisted longer (Fig. 2A, open squares). The rapid and transient formation of PAR and XRCC1 foci is consistent with the kinetics of the rapid SSBR process.
Figure 1.
Co-localization of XRCC1 foci at sites of poly (ADP-ribose) synthesis after oxidative DNA damage. EM9 CHO cells expressing either human XRCC1 (EM9-XH) or harbouring empty vector (EM9-V) were mock-treated (–H2O2) or treated with 10 mM H2O2 for 20 min and then incubated in drug-free medium for 10 min. Untreated and treated cells were then fixed with methanol/acetone and immunostained with anti-poly (ADP-ribose) (‘PAR’) monoclonal antibody 10H and anti-XRCC1 polyclonal antibody (AHP428, Serotec) for analysis by indirect immunofluorescence. Representative images (×100 magnification) were photographed and coloured using an RTI/CCD-1300/Y digital camera and Metamorph software.
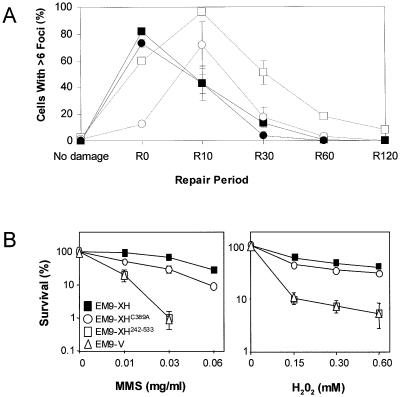
Figure 2.
Temporal assembly of PAR foci and XRCC1 foci in EM9 CHO cells expressing wild-type human XRCC1 or XRCC1C389A. (A) EM9-XH cells (squares) or EM9-XHC389A cells (circles) were mock-treated (‘no damage’) or treated with 10 mM H2O2 for 20 min and then incubated for 0, 10, 30, 60 or 120 min as indicated (‘R0-R120’) in drug-free medium to allow time for repair. Cells were then fixed and immunostained with anti-PAR antibodies (solid lines) and anti-XRCC1 antibodies (dotted lines) as described for Figure 1. For each time point, 100 cells were scored for the presence of six or more foci per cell. Data points are the mean of two or three experiments and error bars represent the data range or 1 S.D. from the mean (R10), respectively. Where not visible, error bars are smaller than the symbols. (B) 250 cells of the indicated cell lines were plated in 10 cm dishes in duplicate and either mock-treated or treated with the indicated concentrations of H2O2 for 10 min or MMS for 1 h. Cells were then rinsed in PBS and incubated in drug-free medium for 7–10 days to allow formation of macroscopic colonies. Survival was calculated by dividing the average number of colonies on treated plates by the average number on untreated plates. Data are the mean of three independent experiments for each drug and error bars represent ±1 S.D. Where not visible, error bars are smaller than the symbols.
The observation that XRCC1 foci appeared with slightly delayed kinetics when compared to those of PAR is consistent with the hypothesis that PARP-1 binds DNA strand breaks and is activated, and then recruits XRCC1 to those breaks to facilitate repair. To test whether the formation of XRCC1 foci may reflect the recruitment of XRCC1 by PAR, we examined the formation of XRCC1 foci in EM9-XHC389A cells expressing XRCC1 with a mutated BRCT I domain. The kinetics with which PAR foci appeared and disappeared in EM9-XHC389A cells was similar to that in EM9-XH cells (Fig. 2A, compare filled squares and filled circles). In contrast, the appearance of XRCC1 foci in EM9-XHC389A cells in response to H2O2 was reduced in comparison to EM9-XH cells, as measured by the number of foci-positive cells (cells with >6 foci) observed at different times during the repair incubation (Fig. 2A, compare open circles and open squares). The reduced appearance of XRCC1 foci in EM9-XHC389A cells was not explained by lower levels of XRCC1 protein because this mutant cell line exhibited slightly higher levels of XRCC1 than did EM9-XH cells (see later, Fig. 3C).
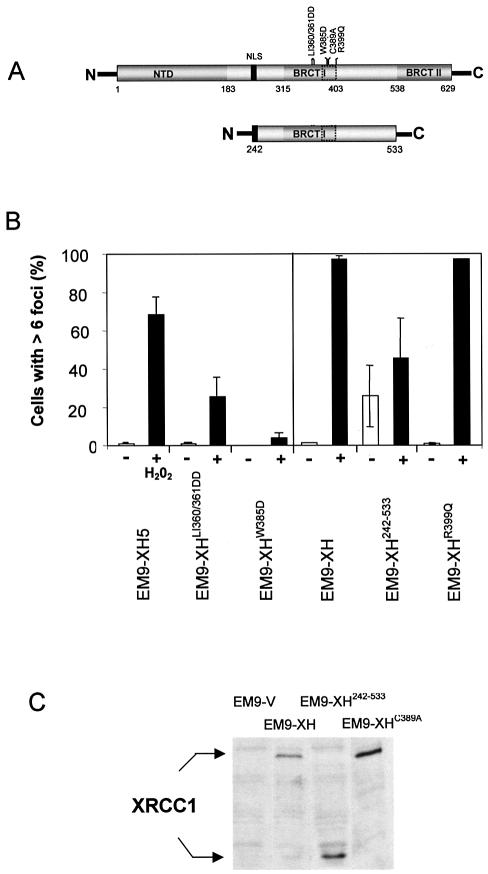
Figure 3.
The BRCT I domain is required for appearance of XRCC1 nuclear foci after oxidative DNA damage. (A) Schematic of full-length XRCC1 (top) and XRCC1242–533 (bottom) depicting the proposed PAR- binding motif (dotted box) located within BRCT I domain. The position of the nuclear localization signal (NLS) and of the mutations employed in this study are shown. (B) Mutation of the BRCT I domain inhibits assembly of XRCC1 nuclear foci after H2O2 treatment. The indicated cell lines were mock-treated (‘–’) or treated (‘+’) with H2O2 for 20 min, incubated in drug-free medium for 10 min, and then processed for anti-XRCC1 immunofluorescence as described for Figure 1. 100 cells from each cell line were scored for the presence or absence of six or more foci. Each data point represents the mean of two independent experiments and error bars reflect the data range. EM9-XH5 is a single transfectant clone and is the wild-type control for EM9-XHLI360/361DD and EM9-XHW385D, with which it expresses similar levels of recombinant human XRCC1 (25). EM9-XH is a pooled population of more than 50 transfectants and is the control cell line for EM9-XH242–533 and EM9-XHR399Q (25). (C) XRCC1 protein levels in EM9-V, EM9-XH, EM9-XH242–533 and EM9-XHC389A cells. Protein extracts from the indicated cell lines were fractionated by SDS–PAGE, transferred to nitrocellulose and immunoblotted with anti-XRCC1 polyclonal antibody.
EM9-XHC389A cells were only slightly more sensitive to MMS and H2O2 than were EM9-XH cells (Fig. 2B). This small effect on survival is in contrast to two other mutations in BRCT I domain that we have examined previously, including one mutation located only four amino acids from C389, which entirely prevents the ability of XRCC1 to maintain cell survival after MMS (25). We therefore examined the impact of these additional BRCT I mutations on assembly of XRCC1 foci. One of the mutations is a double substitution of Leu360/Ileu361 with two Asp residues and the second is a single substitution of Trp385 with Asp (Fig. 3A). Similar to C389, Trp385 lies within the PAR binding motif (22), and mutation of Leu360 has previously been reported to prevent the interaction between XRCC1 and PARP-1 (28). The appearance of XRCC1 nuclear foci was reduced in both EM9-XHLI360/361DD and EM9-XHW385D cells after H2O2 (Fig. 3B). EM9-XHW385D cells exhibited the greatest reduction, with ∼10-fold fewer XRCC1 foci observed 10 min after H2O2 treatment than was observed in wild-type EM9-XH5 cells. In contrast to the LI360/361DD and W385D mutations, a naturally occurring human polymorphism (XRCC1 399Q) located at the very end of the BRCT I domain did not measurably affect the appearance of XRCC1 foci (Fig. 3B). This is consistent with our previous observation that this polymorphism does not measurably impact on SSBR in CHO cells (25).
The BRCT I domain and the linker located between this domain and the C-terminal BRCT II domain are the most evolutionary conserved regions of XRCC1 (25). Indeed, a putative homologue in Arabidopsis thaliana is comprised of just these domains. We therefore expressed a human equivalent of the A.thaliana XRCC1, encoding residues 242–533 (see Fig. 3A and C), to examine whether these two evolutionary conserved domains are sufficient to allow assembly of human XRCC1 into nuclear foci. Interestingly, EM9-XH242–533 cells exhibited high levels of XRCC1 foci even in the absence of H2O2, though an increase in foci-positive cells was observed after H2O2 (Fig. 3B). The BRCT I domain and the linker region located downstream of this domain are thus sufficient for assembly of XRCC1 in subnuclear foci, though other domains of XRCC1 may be important to regulate this process. Perhaps consistent with this, XRCC1242–533 failed to correct the sensitivity of EM9 cells to either H2O2 or MMS (Fig. 2B, open squares).
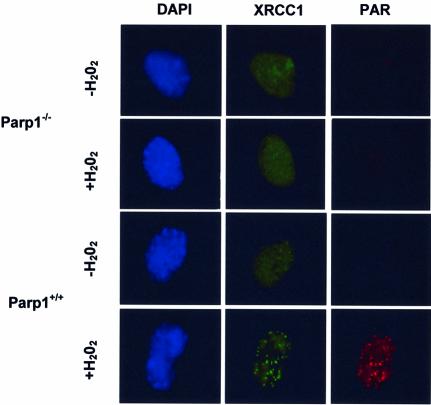
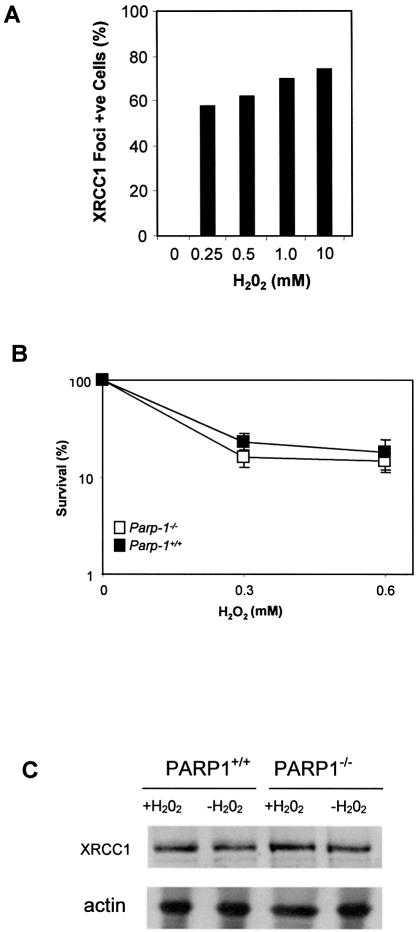
If PARP-1 activity is required to recruit XRCC1 to sites of SSBs, then XRCC1 should fail to assemble into foci in cells lacking PARP-1. We therefore examined the formation of Xrcc1 foci in MEFs harbouring targeted disruption of exon 1 of the Adprt-1 gene (from here on termed Parp1 for clarity) that encodes Parp-1 (26). As expected, immunostaining with anti-PAR antibodies revealed nuclear foci in Parp1+/+ MEFs after H2O2, but few or no foci in Parp1+/+ MEFs before H2O2 or in Parp1–/– MEFs after H2O2 (Fig. 4, red). Similarly, Xrcc1 foci formed in Parp1+/+ MEFs after H2O2, and many of these co-localized with PAR (Fig. 4, green). Typically, we observed between 10 and 30 Xrcc1/PAR foci per cell. Strikingly, however, Xrcc1 nuclear foci were not detected in Parp1–/– cells after H2O2, demonstrating that PARP-1 is required for the appearance of Xrcc1 foci after exposure to this drug. Although the dose of H2O2 employed in these experiments (10 mM) is lethal, similar numbers of Xrcc1 foci-positive cells were observed at lower, more physiological, doses of H2O2 in Parp1+/+ cells (Fig. 5A and B). Once again, between 10 and 30 foci were typically observed per cell, and these foci were both similar in appearance to those observed after 10 mM H2O2 and were similarly co-localized with sites of PAR (data not shown). The absence of Xrcc1 foci did not reflect an impact of PARP-1 on Xrcc1 protein levels because Xrcc1 was present at similar levels in Parp1+/+ and Parp1–/– cell extracts, both before and after H2O2 treatment (Fig. 5C).
Figure 4.
PARP-1 is required for appearance of XRCC1 nuclear foci at sites of oxidative DNA damage. Parp1+/+ or Parp1–/– MEFs were mock-treated or treated for 20 min with 10 mM H2O2 and then incubated in drug-free medium for 10 min. Cells were then fixed in methanol/acetone and processed for indirect immunofluorescence using anti-PAR monoclonal antibody and anti-XRCC1 polyclonal antibody and counterstained with DAPI. Representative cells are shown (×100 magnification).
Figure 5.
XRCC1 protein levels, nuclear foci and survival to H2O2 in Parp1+/+ or Parp1–/– MEFs. (A) Parp1+/+ MEFs were treated for 20 min with the indicated concentrations of H2O2 and then incubated in drug-free medium for 10 min. Cells were immunostained for Xrcc1 and PAR as described in Figure 4 and the fraction of cells with >6 Xrcc1 foci was determined. The absence of visible bars for untreated cells indicates the absence of detectable foci-positive cells. Data are the mean of duplicate samples from a single experiment, and the data range for each duplicate was less than five. (B) The clonogenic survival of Parp1+/+ or Parp1–/– MEFs treated with the indicated concentrations of H2O2 was determined as described in Figure 2B. (C) Protein extracts from mock-treated or H2O2-treated Parp1+/+ or Parp1–/– MEFs were fractionated by SDS–PAGE, transferred to nitrocellulose and immunoblotted with anti-XRCC1 polyclonal, or anti-actin monoclonal, antibody.
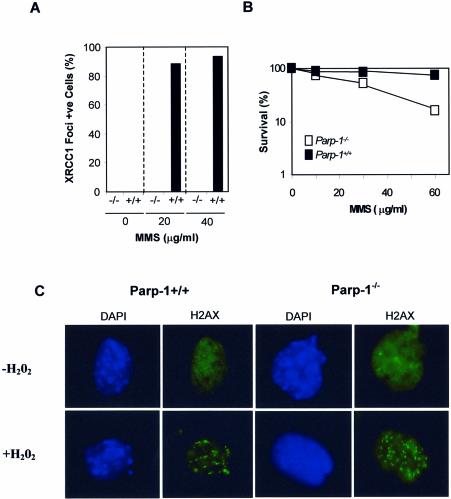
Similar results were observed when the response of Parp1+/+ and Parp1–/– cells to the alkylating agent methyl methanesulphonate (MMS) was examined. Whereas most Parp1+/+ cells exhibited Xrcc1 and PAR foci that co-localized extensively after exposure to physiological doses of MMS, Parp1–/– cells did not (Fig. 6A and B). Finally, to rule out the possibility that the Parp1–/– MEFs employed here were unable to mount any response to DNA damage under the conditions employed, we compared these cells with Parp1+/+ MEFs for the appearance of phospho H2AX foci, an early event triggered in response to DNA strand breaks. In contrast to XRCC1 foci, the appearance of H2AX foci was observed in both Parp1+/+ and Parp1–/– cells, after exposure to 10 mM H2O2 (Fig. 6C).
Figure 6.
XRCC1 and H2AX nuclear foci in Parp1+/+ and Parp1–/– MEFs following exposure to MMS and H2O2. (A) Parp1+/+ (‘+/+’) and Parp1–/– (‘–/–’) MEFs were treated for 20 min with the indicated concentrations of MMS and then incubated in drug-free medium for 10 min. Cells were immunostained for Xrcc1 and PAR as described in Figure 4 and the fraction of cells with >6 Xrcc1 foci was determined. The absence of visible bars for untreated cells and for treated Parp1–/– MEFs indicates the absence of detectable foci-positive cells. Data are the mean of duplicate samples from a single experiment, and the data range for each duplicate was less than five. (B) The clonogenic survival of Parp1+/+ or Parp1–/– MEFs treated with the indicated concentrations of MMS for 1 h was determined as described in Figure 2B. Where not visible, error bars are smaller than the symbols. (C) Parp1+/+ or Parp1–/– MEFs were mock-treated or treated for 20 min with 10 mM H2O2 as indicated and then incubated in drug-free medium for 10 min. Cells were then fixed in methanol/acetone and processed for indirect immunofluorescence using anti-phospho H2AX monoclonal antibody (Upstate; clone JBW301) and counterstained with DAPI. Representative cells are shown (×100 magnification).
DISCUSSION
The phenotype of cells in which PARP-1 is inhibited or absent is consistent with a role in DNA repair (21,29,30), but the nature of this role is unclear. A direct physical link between PARP-1 and the DNA strand break repair machinery emerged with the discovery that PARP-1 interacts with the BRCT I domain of XRCC1 (13,19). XRCC1 appears to operate as a molecular scaffold protein that interacts with and stimulates enzymatic components of single strand break repair (2,31). In this study, we have shown that PARP-1 is required for the rapid appearance of XRCC1 nuclear foci after oxidative damage and that these foci co-localize with sites of PAR synthesis. PAR synthesis was the product of PARP-1 activity because it was absent from MEFs in which Parp-1 was disrupted. The appearance of XRCC1 foci was dependent upon the central BRCT I domain in this polypeptide, which has been shown previously to interact with PAR polymer and to be critical for the role of XRCC1 in SSBR and for cell survival (22,25).
What are the nature of the breaks at which PARP-1 and XRCC1 foci assemble? Given the extensive involvement of XRCC1 in SSBR (2,31), it seems likely that SSBs are responsible for the foci observed after H2O2, though an involvement of DSBs is possible (32). It is likely that the XRCC1/PAR foci reported here are physiologically relevant sites of DNA strand break repair because they rapidly disappeared during a repair incubation in drug-free medium. These foci could reflect the steady state number of DNA strand breaks undergoing repair at the time of fixation, or alternatively could reflect a discrete number of ‘repair factories’ into which DNA strand breaks are recruited.
XRCC1 is required for at least two types of SSBR process, a rapid pathway that occurs throughout the cell cycle and a second pathway that is specific for S/G2 phase (33). In addition, there exists at least one additional SSBR pathway that is XRCC1 independent. Of the two identified XRCC1-dependent processes, the S/G2 pathway appears to be the most important for cell survival. Indeed, the S/G2 process can largely compensate for absence of the rapid pathway, possibly by removing unrepaired SSBs when they are encountered by the replication fork (33,34). It is unclear whether the XRCC1/PAR foci observed here are associated with one or both of these processes, though the rapid appearance of PAR foci and XRCC1 foci correlates best with the rapid pathway.
The requirement for PARP-1 for assembly of XRCC1 foci could be direct, via the ability of these polypeptides to interact. Alternatively, it could reflect an indirect impact of PARP-1 on the expression of other repair genes or on chromatin architecture (35–37). However, we favour the model that PARP-1 directly recruits XRCC1 via the binding of PAR polymer to XRCC1, for several reasons. First, XRCC1 interacts preferentially with ribosylated PARP-1 and XRCC1 binds purified PAR polymer in vitro (19,22–24). This strongly suggests that PARP-1 recruits XRCC1 by physical interaction once PAR synthesis is triggered at a DNA strand break. Second, the interaction of XRCC1 with PAR polymer occurs via a motif within the BRCT I domain, mutations within which reduced the formation of XRCC1 foci after oxidative DNA damage. Also, a truncated XRCC1 polypeptide encoding just the BRCT I domain and the linker domain between BRCT I and BRCT II was able to form XRCC1 foci, albeit in an apparently deregulated fashion that may not be responsive to oxidative DNA damage. The difference observed in the impact of the three mutations on foci formation may be due to leakiness, since there is a high degree of redundancy amongst PAR-binding motifs (22). The LI360/361DD and W385D mutations had the greatest effect on foci formation, reducing their appearance by ∼3- and 10-fold, respectively. These mutations appear to impact specifically on functions of the BRCT I domain because we have shown previously that they do not prevent protein–protein interactions mediated by flanking domains (25). Together, these data best support a model in which PARP-1 mediates the assembly of XRCC1 foci after oxidative DNA damage by direct interaction of the XRCC1 BRCT I domain with PAR polymer, thereby recruiting XRCC1 to sites of DNA strand breakage.
A requirement for PARP-1 for recruiting XRCC1 to strand breaks during DNA repair reactions in vitro has not been reported. This may reflect that the active recruitment of XRCC1 to breaks is not required for repair in simple cell-free reactions, because the high concentration of DNA breaks and soluble repair factors in such experiments prevent the detection of DNA breaks from being a rate limiting step. However, active recruitment of XRCC1 and its protein partners may be important to accelerate the detection and repair of SSBs within the context of the nuclear genome, in which DNA breaks are relatively rare and distributed amongst many kilobases of undamaged DNA. It is also possible that the recruitment of XRCC1 is important for repair of chromatin, rather than naked plasmid DNA or oligonucleotide duplex.
We have suggested that PARP-1 may primarily be required to recruit XRCC1 to direct DNA breaks, which are ‘un-scheduled’ and require rapid detection (31). It is feasible that PARP-1 also recruits XRCC1 to SSBs arising during BER, since both PAR and XRCC1 foci were absent from Parp1–/– MEFs after exposure to the alkylating agent, MMS. However, it is more difficult to envisage a need for PARP to ‘detect’ breaks during early stages of BER, since current models suggest that breaks created by APE1 can be passed directly to DNA polymerase β (38–40) which in turn may itself then recruit XRCC1/DNA ligase IIIα for the completion of DNA repair (13,14,31). Perhaps PAR synthesis after base damage reflects binding by PARP-1 to SSBs at a later stage of BER, since PARP-1 may influence the choice of whether short patch or long patch repair is employed during DNA gap filling (23,35,41,42). Alternatively, PAR synthesis after base damage could reflect a fraction of abasic sites that become ‘uncoupled’ from APE1 after cleavage or that are cleaved by an AP lyase activity, either of which might be expected to result in ‘exposed’ SSBs that require rapid detection by PARP-1.
In summary, these data show that PARP-1 is required for assembly or stability of XRCC1 nuclear foci after oxidative DNA damage, consistent with a model in which poly (ADP-ribose) synthesis serves to recruit XRCC1 to sites of DNA strand breakage.
Acknowledgments
ACKNOWLEDGEMENTS
S.E.K. is funded by an MRC Programme Grant (G0001259) to K.W.C. We thank Gilbert de Murcia for the gift of anti-PAR antibodies.
REFERENCES
- 1.Xu Y.J., Kim,E.Y. and Demple,B. (1998) Excision of C-4′-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase beta. J. Biol. Chem., 273, 28837–28844. [DOI] [PubMed] [Google Scholar]
- 2.Caldecott K.W. (2001) Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays, 23, 447–455. [DOI] [PubMed] [Google Scholar]
- 3.Ochs K., Sobol,R.W., Wilson,S.H. and Kaina,B. (1999) Cells deficient in DNA polymerase beta are hypersensitive to alkylating agent-induced apoptosis and chromosomal breakage. Cancer Res., 59, 1544–1551. [PubMed] [Google Scholar]
- 4.Thompson L.H., Brookman,K.W., Dillehay,L.E., Carrano,A.V., Mazrimas,J.A., Mooney,C.L. and Minkler,J.L. (1982) A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat. Res., 95, 427–440. [DOI] [PubMed] [Google Scholar]
- 5.Dominguez I., Daza,P., Natarajan,A.T. and Cortes,F. (1998) A high yield of translocations parallels the high yield of sister chromatid exchanges in the CHO mutant EM9. Mutat. Res., 398, 67–73. [DOI] [PubMed] [Google Scholar]
- 6.Oph V., Zdzienicka,M.Z., Vrieling,H., Lohman,P.H. and van Zeeland,A.A. (1994) Molecular analysis of ethyl methanesulfonate-induced mutations at the hprt gene in the ethyl methanesulfonate-sensitive Chinese hamster cell line EM-C11 and its parental line CHO9. Cancer Res., 54, 3001–3006. [PubMed] [Google Scholar]
- 7.Zdzienicka M.Z., Vanderschans,G.P., Natarajan,A.T., Thompson,L.H., Neuteboom,I. and Simons,J.W.I.M. (1992) A Chinese-hamster ovary cell mutant (Em-c-11) with sensitivity to simple alkylating-agents and a very high-level of sister chromatid exchanges. Mutagenesis, 7, 265–269. [DOI] [PubMed] [Google Scholar]
- 8.Kuzminov A. (2001) Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl Acad. Sci. USA, 98, 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson L.H. and West,M.G. (2000) XRCC1 keeps DNA from getting stranded. Mutat. Res., 459, 1–18. [DOI] [PubMed] [Google Scholar]
- 10.Tebbs R.S., Flannery,M.L., Meneses,J.J., Hartmann,A., Tucker,J.D., Thompson,L.H., Cleaver,J.E. and Pedersen,R.A. (1999) Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev. Biol., 208, 513–529. [DOI] [PubMed] [Google Scholar]
- 11.Whitehouse C.J., Taylor,R.M., Thistlethwaite,A., Zhang,H., Karimi-Busheri,F., Lasko,D.D., Weinfeld,M. and Caldecott,K.W. (2001) XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell, 104, 107–117. [DOI] [PubMed] [Google Scholar]
- 12.Vidal A.E., Boiteux,S., Hickson,I.D. and Radicella,J.P. (2001) XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein–protein interactions. EMBO J., 20, 6530–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D.E. and Lindahl,T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 15.Mackey Z.B., Ramos,W., Levin,D.S., Walter,C.A., Mccarrey,J.R. and Tomkinson,A.E. (1997) An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol., 17, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nash R.A., Caldecott,K.W., Barnes,D.E. and Lindahl,T. (1997) XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry, 36, 5207–5211. [DOI] [PubMed] [Google Scholar]
- 17.Caldecott K.W., Tucker,J.D., Stanker,L.H. and Thompson,L.H. (1995) Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res., 23, 4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldecott K.W., Mckeown,C.K., Tucker,J.D., Ljungquist,S. and Thompson,L.H. (1994) An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol., 14, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masson M., Niedergang,C., Schreiber,V., Muller,S., Menissier-de Murcia,J. and de Murcia,G. (1998) XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol., 18, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkle A. (2001) Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays, 23, 795–806. [DOI] [PubMed] [Google Scholar]
- 21.Shall S. and De Murcia,G. (2000) Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res., 460, 1–15. [DOI] [PubMed] [Google Scholar]
- 22.Pleschke J.M., Kleczkowska,H.E., Strohm,M. and Althaus,F.R. (2000) Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem., 275, 40974–40980. [DOI] [PubMed] [Google Scholar]
- 23.Dantzer F., de La,R.G., Menissier-de Murcia,J., Hostomsky,Z., De Murcia,G. and Schreiber,V. (2000) Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry, 39, 7559–7569. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber V., Ame,J.C., Dolle,P., Schultz,I., Rinaldi,B., Fraulob,V., Menissier-de Murcia,J. and De Murcia,G. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem., 277, 23028–23036. [DOI] [PubMed] [Google Scholar]
- 25.Taylor R.M., Thistlethwaite,A. and Caldecott,K.W. (2002) Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol. Cell. Biol., 22, 2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masutani M., Suzuki,H., Kamada,N., Watanabe,M., Ueda,O., Nozaki,T., Jishage,K., Watanabe,T., Sugimoto,T., Nakagama,H. et al. (1999) Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc. Natl Acad. Sci. USA, 96, 2301–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadkhodayan S., Salazar,E.P., Lamerdin,J.E. and Weber,C.A. (1996) Construction of a functional cDNA clone of the hamster ERCC2 DNA repair and transcription gene. Somat. Cell Mol. Genet., 22, 453–460. [DOI] [PubMed] [Google Scholar]
- 28.Kubota Y. and Horiuchi,S. (2003) Independent roles of XRCC1’s two BRCT motifs in recovery from methylation damage. DNA Repair (Amst.), 2, 407–415. [DOI] [PubMed] [Google Scholar]
- 29.Masutani M., Nozaki,T., Nakamoto,K., Nakagama,H., Suzuki,H., Kusuoka,O., Tsutsumi,M. and Sugimura,T. (2000) The response of Parp knockout mice against DNA damaging agents. Mutat. Res., 462, 159–166. [DOI] [PubMed] [Google Scholar]
- 30.Herceg Z. and Wang,Z.Q. (2001) Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res., 477, 97–110. [DOI] [PubMed] [Google Scholar]
- 31.Caldecott K.W. (2003) DNA single-strand break repair and spinocerebellar ataxia. Cell, 112, 7–10. [DOI] [PubMed] [Google Scholar]
- 32.Nocentini S. (1999) Rejoining kinetics of DNA single- and double-strand breaks in normal and DNA ligase-deficient cells after exposure to ultraviolet C and gamma radiation: An evaluation of ligating activities involved in different DNA repair processes. Radiat. Res., 151, 423–432. [PubMed] [Google Scholar]
- 33.Taylor R.M., Moore,D.J., Whitehouse,J., Johnson,P. and Caldecott,K.W. (2000) A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol. Cell. Biol., 20, 735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore D.J., Taylor,R.M., Clements,P. and Caldecott,K.W. (2000) Mutation of a BRCT domain selectively disrupts DNA single-strand break repair in noncycling Chinese hamster ovary cells. Proc. Natl Acad. Sci. USA, 97, 13649–13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanderson R.J. and Lindahl,T. (2002) Down-regulation of DNA repair synthesis at DNA single-strand interruptions in poly(ADP-ribose) polymerase-1 deficient murine cell extracts. DNA Repair (Amst.), 1, 547–558. [DOI] [PubMed] [Google Scholar]
- 36.Simbulan-Rosenthal C.M., Ly,D.H., Rosenthal,D.S., Konopka,G., Luo,R., Wang,Z.Q., Schultz,P.G. and Smulson,M.E. (2000) Misregulation of gene expression in primary fibroblasts lacking poly(ADP-ribose) polymerase. Proc. Natl Acad. Sci. USA, 97, 11274–11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tulin A. and Spradling,A. (2003) Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science, 299, 560–562. [DOI] [PubMed] [Google Scholar]
- 38.Bennett R.A., Wilson,D.M.,III, Wong,D. and Demple,B. (1997) Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl Acad. Sci. USA, 94, 7166–7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson S.H. and Kunkel,T.A. (2000) Passing the baton in base excision repair. Nat. Struct. Biol., 7, 176–178. [DOI] [PubMed] [Google Scholar]
- 40.Mol C.D., Izumi,T., Mitra,S. and Tainer,J.A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature, 403, 451–456. [DOI] [PubMed] [Google Scholar]
- 41.Prasad R., Lavrik,O.I., Kim,S.J., Kedar,P., Yang,X.P., Vande Berg,B.J. and Wilson,S.H. (2001) DNA polymerase beta-mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem., 276, 32411–32414. [DOI] [PubMed] [Google Scholar]
- 42.Le Page F., Schreiber,V., Dherin,C., De Murcia,G. and Boiteux,S. (2003) Poly(ADP-ribose) polymerase-1(PARP-1) is required in murine cell lines for base excision repair of oxidative DNA damage in absence of DNA polymerase beta. J. Biol. Chem., 278, 18471–18477. [DOI] [PubMed] [Google Scholar]