Abstract
Embryonic stem (ES) cells are able to grow indefinitely (self-renewal) and have the potential to differentiate into all adult cell types (pluripotency). The regulatory network that controls pluripotency is well characterized, whereas the molecular basis for the transition from self-renewal to the differentiation of ES cells is much less understood, although dynamic epigenetic gene silencing and chromatin compaction are clearly implicated. In this study, we report that UTF1 (undifferentiated embryonic cell transcription factor 1) is involved in ES cell differentiation. Knockdown of UTF1 in ES and carcinoma cells resulted in a substantial delay or block in differentiation. Further analysis using fluorescence recovery after photobleaching assays, subnuclear fractionations, and reporter assays revealed that UTF1 is a stably chromatin-associated transcriptional repressor protein with a dynamic behavior similar to core histones. An N-terminal Myb/SANT domain and a C-terminal domain containing a putative leucine zipper are required for these properties of UTF1. These data demonstrate that UTF1 is a strongly chromatin-associated protein involved in the initiation of ES cell differentiation.
Introduction
Mouse embryonic stem (ES) cells derived from the inner cell mass of blastocyst embryos have the ability to self-renew and are pluripotent. ES cell pluripotency is maintained via the LIF-gp130-STAT3, bone morphogenetic protein (BMP)–Smad-Id, and probably Wnt and mTOR signaling cascades (Smith et al., 1988; Williams et al., 1988; Niwa et al., 1998; Matsuda et al., 1999; Ying et al., 2003; Gangloff et al., 2004; Murakami et al., 2004; Sato et al., 2004). Intracellular regulators of ES cell self-renewal include Oct4, Sox2, Nanog, and the recently implicated transcription factors Sall4, Esrrb, Tbx3, and Tcl1 (Yuan et al., 1995; Nichols et al., 1998; Niwa et al., 2002; Chambers et al., 2003; Mitsui et al., 2003; Ivanova et al., 2006; Zhang et al., 2006).
Using chromatin immunoprecipitation on chip analyses to map Oct4, Sox2, and Nanog target genes, a large group of genes was identified that is coregulated by these factors in different combinations, although the majority of genes was cooccupied by Oct4, Sox2, and Nanog (Boyer et al., 2005; Loh et al., 2006). Interestingly, many of these target genes are not expressed in ES cells.
Recent reports showed that in ES cells, many differentiation genes are silenced by Polycomb group (PcG) complexes, indicating that the epigenetic regulation of gene expression is essential for maintaining ES cell pluripotency (Azuara et al., 2006; Bernstein et al., 2006; Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006; Loh et al., 2006). Interestingly, many of the repressed Nanog, Oct4, and Sox2 target genes were cooccupied by PcG complexes, suggesting that ES cells are poised to enter differentiation programs but are held in check by PcG-mediated chromatin modifications. The suggestion that epigenetic regulation is an important instrument to control ES cell pluripotency versus their capacity to differentiate is further supported by the findings that the PcG protein Suz12 is required for ES cell differentiation (Pasini et al., 2007) and that a functional NuRD (nucleosome remodeling and disruption) complex, which is involved in nucleosome remodeling, is required for the lineage commitment of ES cells (Kaji et al., 2006).
Apart from Oct4, Sox2, and Nanog, other genes are also highly and almost exclusively expressed during early embryogenesis (Mitsui et al., 2003; Ivanova et al., 2006). One of these genes, the UTF1 (undifferentiated embryonic cell transcription factor 1) gene, is specifically expressed in the inner cell mass and primitive ectoderm and is down-regulated at early primitive streak stages (Okuda et al., 1998). Expression is maintained in the primordial germ cells in developing embryos and in the gonads in adult animals (Chuva de Sousa Lopes et al., 2005). Promoter analysis indicated that the murine UTF1 gene is transcriptionally regulated by Oct4 and Sox2 (Nishimoto et al., 1999). The UTF1 protein was shown to repress transcription (Fukushima et al., 1999), to activate reporter genes in an ATF2-dependent manner, and to interact with the basal transcription factor TFIID (Fukushima et al., 1998; Okuda et al., 1998). A recent study suggested a role for UTF1 in the proliferation rate and teratoma-forming capacity of ES cells (Nishimoto et al., 2005).
The purpose of this study was to determine the requirement of UTF1 for ES cell self-renewal and/or differentiation and to gain insight into its mechanistic properties. Using knockdown (KD) strategies, we determined that UTF1 is involved in ES cell differentiation. UTF1 KD perturbed ES and embryonic carcinoma (EC) cell differentiation, whereas their ability to self-renew was unaffected. UTF1 displays transcriptional repressor activity, and a combination of localization experiments, FRAP protocols, and subcellular fractionation assays indicated that UTF1 is stably chromatin associated with dynamics and biochemical properties similar to core histones.
Results
UTF1 is required for EC cell differentiation
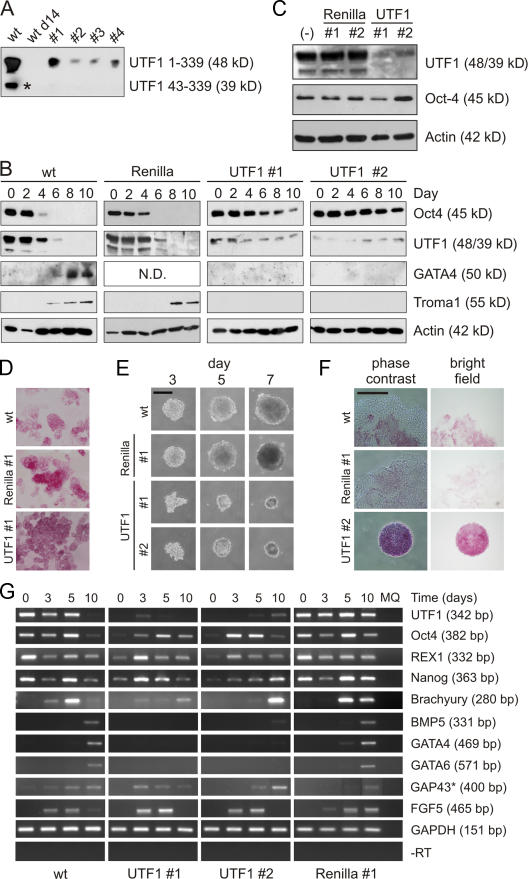
To study the potential role of mouse UTF1 (mUTF1; hereafter UTF1) in ES and EC cell differentiation, we stably expressed UTF1 and Renilla luciferase (hereafter Renilla) siRNAs in P19CL6 EC cells. UTF1 expression levels were substantially decreased in all clones tested (Fig. 1 A), whereas expression levels of the pluripotency marker Oct4 were not affected (Fig. 1 B). Next, DMSO-induced differentiation of wild-type (wt), Renilla, and UTF1 KD cells was analyzed (Fig. 1 B). wt and Renilla KD cells differentiated normally, which was reflected by a drastic reduction in Oct4 levels around day 4, decreased UTF1 levels between days 4 and 6, and detectable GATA4 (not determined for Renilla) and Troma1 expression by day 8. Actin was used as a protein loading control. In UTF1 KD lines, the differentiation-induced down-regulation of Oct4 was either delayed (#1) or minor (#2), and both GATA4 and Troma1 were not detected. Residual UTF1 protein levels were not further down-regulated, most likely as a consequence of high Oct4 levels, a transcriptional activator of the UTF1 gene.
Figure 1.
UTF1 is involved in the differentiation of EC and ES cells. (A) UTF1 expression in P19CL6 EC cells (wt), 14-d DMSO-differentiated EC cells (wt d14), and four independent UTF1 EC KD clones (UTF1 #1–#4). The asterisk indicates a shorter variant of mUTF1 (aa 43–339) generated by transcription from an alternative start site (Nishimoto et al., 2001). (B) DMSO-induced differentiation of wt, Renilla luciferase KD (Renilla), and UTF1 KD (UTF1 #1 and #2) EC cells. Cell lysates were analyzed with antibodies against Oct4, UTF1, GATA4, and Troma1. Actin staining was performed as a loading control. (C) Western analysis of wt, Renilla luciferase KD (Renilla #1 and #2), and UTF1 KD (UTF1 #1 and #2) IB10 ES cells. Cell lysates were analyzed with antibodies against UTF1 and Oct4. Actin levels were determined to correct for gel loading. (D) Brightfield images of wt, Renilla luciferase KD (Renilla #1), and UTF1 KD (UTF1 #1) ES cells stained for AP activity. (E) Phase-contrast images of EBs from wt, Renilla luciferase KD (Renilla #1), and UTF1 KD (UTF1 #1 and #2) ES cells after 3, 5, and 7 d. (F) Phase-contrast and brightfield images of day 8 EBs from wt, Renilla luciferase KD (Renilla #1), and UTF1 KD (UTF1 #2) ES cells stained for AP activity. (G) Expression levels of markers for ES cells (UTF1, Oct4, REX1, and Nanog), ectoderm (FGF5 and GAP43), mesoderm (Brachyury and BMP5), and endoderm (GATA4 and GATA6) were measured by semiquantitative RT-PCR in undifferentiated ES cells and EBs cultured for 3, 5, and 10 d. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used as a control. In the −RT lanes, reverse transcriptase was omitted from the reverse transcriptase reactions to control for genomic DNA contamination and amplified using glyceraldehyde-3-phosphate dehydrogenase primers. A representative experiment is depicted. The asterisk indicates that the 5- and 10-d GAP43 RT-PCR products were not loaded in adjacent lanes. Bars, 250 μm.
As UTF1 was previously reported to be involved in ES cell proliferation (Nishimoto et al., 2005), we determined the doubling times of wt, Renilla KD, and UTF1 KD EC cells. UTF1 KD cells showed a 24% and 17% increase in doubling time (8.9 ± 0.3 h) compared with wt EC (7.2 ± 0.1 h) and Renilla KD (7.6 ± 0.3 h) cells, respectively. Next, the differentiation of wt and UTF1 KD EC cells was performed with different cell numbers to rule out potential cell density effects on differentiation (0.5 and 2 times the number of cells: 1.8 × 105 and 7.3 × 105 cells, respectively). Irrespective of the initial number of cells, the differentiation of UTF1 KD cells was always delayed or blocked, whereas wt cells differentiated normally (unpublished data). Summarizing, these data indicate that in EC cells, UTF1 KD results in an abrogated differentiation capacity and persistent Oct4 expression under differentiation-inducing conditions.
UTF1 is involved in ES cell differentiation
To extend these findings to a nontransformed mouse cell line, we tested the effect of UTF1 KD on IB10 ES cell differentiation. Renilla KD clones expressed normal levels of UTF1 and Oct4, whereas in UTF1 KD cell lines, UTF1 levels were reduced, but Oct4 expression was not affected (Fig. 1 C). In addition, UTF1 and Renilla KD ES cells are positive for AP, confirming their ES cell phenotype (Fig. 1 D). To determine whether UTF1 down-regulation also affected the differentiation potential of these cells, embryoid bodies (EBs) were generated. Where wt and Renilla KD cells formed normal EBs with high efficiency, UTF1 KD–derived EBs were irregularly shaped, much smaller in size, formed with low efficiency, and compaction was not observed (Fig. 1 E).
In agreement with observations by Nishimoto et al. (2005), UTF1 KD affected (although less dramatically) the doubling time of ES cells: UTF1 KD ES cells have a doubling time of 11.8 ± 0.7 h compared with 9.6 ± 0.7 h and 10.2 ± 0.1 h for wt (23% increase) and Renilla (16% increase) ES cells, respectively. Because UTF1 KD abrogated the ability of EC cells to differentiate, we tested whether EBs from UTF1 KD ES cells also failed to differentiate. AP staining showed that day 8 wt and Renilla EBs are largely AP negative, whereas UTF1 KD EBs still displayed substantial AP activity, suggesting that UTF1 is involved in ES cell differentiation (Fig. 1 F). To further validate this observation, the expression pattern of several germ layer–specific marker genes during EB development was determined by RT-PCR (Fig. 1 G). Both wt and Renilla ES cells show a clear up-regulation of various lineage markers. At days 3–5, Brachyury (early mesoderm) was detected, and at day 10, BMP5 (dorsal mesoderm) was detected. Endoderm markers GATA4 and GATA6 were detected at day 10, and ectoderm markers GAP43 and FGF5 were both detected at days 3–10. In contrast, both UTF1 KD cell lines showed either an absence (GATA6), minor (GATA4 and BMP5), or delayed (Brachyury) expression of these markers. However, ectoderm markers FGF5 and GAP43 were detected from day 3. Pluripotency markers like Oct4, REX1, and Nanog were detected at all time points in the various EBs, most likely as a result of the incomplete differentiation of a subset of cells.
The increased doubling time of UTF1 KD ES cells could (partially) be responsible for the observed differentiation defect. However, already after 48 h, when differences in doubling times have not yet resulted in substantial differences in cell numbers, we observed that UTF1 KD cells often failed to form aggregates (unpublished data). This strongly suggests that the observed effects on EB formation and differentiation cannot solely be explained by the increased doubling time of UTF1 KD cells. Collectively, these data show that UTF1 KD in ES cells results in perturbed EB formation and a severely reduced differentiation potential in the endodermal and mesodermal lineages.
UTF1 is a chromatin-associated transcriptional repressor
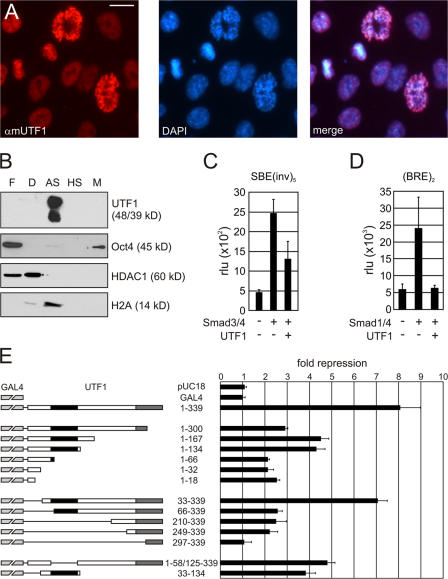
To understand the mechanistic properties of UTF1 that underlie its involvement in EC and ES cell differentiation, a series of experiments were performed to molecularly characterize the protein. First, we determined the subcellular localization of UTF1 in EC cells (Fig. 2 A). UTF1 was clearly localized to the nucleus and excluded from the nucleoli. In addition, we found UTF1 to localize to the chromosomes at different stages during cell division. To further characterize this potential interaction between UTF1 and the DNA/chromatin, we performed subnuclear fractionations of EC cells separating free-diffusing proteins (cytosolic and nuclear), weak/strong DNA-associated proteins, and nuclear matrix (associated) proteins. UTF1 is observed exclusively in the ammonium sulfate fraction known to contain strongly DNA-associated proteins, like core histone H2A (Fig. 2 B). In contrast, Oct4 primarily localized to the free-diffusing fraction and, to some extent, to the nuclear matrix fraction, indicating that Oct4 and UTF1 have distinct chromatin-binding characteristics. To compare the observed behavior of UTF1 to that of chromatin-modifying proteins, we determined the fractions containing histone deacetylase 1 (HDAC1; Fig. 2 B). Unlike UTF1, HDAC1 is found in the fractions containing free-diffusing and weak DNA-associated proteins. Collectively, these data suggest that UTF1 is a protein with a high affinity for chromatin, similar to that of core histones and different from chromatin-modifying proteins like HDAC1.
Figure 2.
Characterization of localization, fractionation, and reporter activity of UTF1. (A) Immunofluorescence analysis of endogenous UTF1 in EC cells using an antibody directed against UTF1. (B) Subnuclear fractionation of EC cells: F, free-diffusing protein fraction; D, DNaseI fraction; AS, ammonium sulfate fraction; HS, high salt fraction; M, nuclear matrix fraction. Fractions were immunostained with antibodies recognizing UTF1, Oct4, HDAC1, and histone H2A. (C and D) Reporter analysis of HepG2 cells transfected with the SBE(inv)5 reporter, (BRE)2 reporter, Smad 3/4, Smad 1/4, and UTF1 as indicated. In all transfections, a LacZ expression plasmid, pDM2-LacZ, was included as an internal standard, and relative luciferase units (rlu) are depicted as the mean with SD (error bars). In all samples, equal amounts of expression plasmids were present by the addition of empty pcDNA3 plasmid when required. (E) Mapping the UTF1 repressor domains. A schematic representation of GAL4-UTF1 constructs used in this experiment; the Myb/SANT domain and CD2 are represented by black boxes and gray boxes, respectively. Different GAL4-UTF1 constructs and a constitutive active TK-luciferase reporter containing five GAL4-binding sites (UAS-TK-Luc) were transfected into HepG2 cells. The inhibitory effect of UTF1 on reporter activity is depicted as fold repression compared with GAL4 alone. In all transfections, a LacZ expression plasmid, pDM2-LacZ, was included as an internal standard, and normalized luciferase activity is depicted as the mean with SD. Bar, 15 μm.
As a chromatin-associated protein, UTF1 is likely to be involved in gene expression regulation. To determine the effect of UTF1 on promoter activity, reporter assays were performed using constructs containing multiple copies of either the Smad-binding element (SBE) or BMP-responsive element (BRE). These reporters were used because we previously identified UTF1 as an SBE-interacting protein in a yeast 1 hybrid screen. However, more detailed analysis showed that UTF1 is not specifically involved in Smad signaling. The reporters are activated by the cotransfection of either Smad3 and 4 (SBE) or Smad1 and 4 (BRE; Fig. 2, C and D). Cotransfection of UTF1 reduced the activity of Smad-stimulated SBE and BRE reporters by approximately twofold and fourfold, respectively. These data indicate that UTF1 is a transcriptional repressor.
Mapping of UTF1 repressor domains
UTF1 contains two conserved domains (CDs): CD1 (aa 55–124), which shares high homology with Myb/SANT DNA-binding domains, and CD2 (aa 271–334), which contains a putative leucine zipper. To identify its repressor domains, the effect of a series of GAL4-UTF1 (deletion) constructs was tested on a thymidine kinase (TK) luciferase reporter containing five copies of the GAL4 target sequence (UAS-TK-Luc; Fig. 2 E). As expected, UTF1 repressed UAS-TK-Luc reporter activity (eightfold reduction compared with GAL4). Deletion of the very C-terminal 39 aa resulted in an almost 2.8-fold reduction in repressor activity. Further C-terminal deletions only marginally affected repressor activity, but when the Myb/SANT domain (aa 55–124) was deleted, an additional drop in repressor activity compared with the 1–167 and 1–134 constructs was observed. The finding that both the C terminus and Myb/SANT domain are involved in transcriptional repression was confirmed using a series of progressive N-terminal deletions. Deletion of aa 1–65 resulted in a 3.1-fold reduction of repressor activity (compare 1–339 with 66–339), and further N-terminal deletions did not affect UTF1 repressor activity except for the deletion construct (297–339) that misses part of the CD2 domain, which completely lacked repressor activity. To address the importance of the Myb/SANT domain, we generated a mutant lacking this region, which reduced repressor activity by 1.7-fold, indicating that it is important for transcriptional repression by UTF1. In addition, the Myb/SANT domain alone (33–134) also displayed considerable repressor activity (3.9-fold repression). Collectively, these experiments indicate that both the Myb/SANT domain and the extreme C terminus of UTF1 are important for transcriptional repression by UTF1.
Live cell distribution of UTF1
To study its localization in living cells, UTF1 was fused to enhanced GFP (eGFP), creating eGFP-HA-UTF1 (hereafter GFP-UTF1), and was stably expressed in EC cells. To prevent localization artifacts, we used a clone that underexpressed GFP-UTF1 compared with the endogenous protein (Fig. 3 A). Subnuclear fractionation showed that GFP-UTF1, like endogenous UTF1, is almost exclusively found in the strongly DNA-associated fraction (Fig. 3 B). Reporter (UAS-TK-Luc) assays in HepG2 cells showed that GFP-UTF1 acted as a transcriptional repressor as well (see Fig. 5 A). These data indicate that fusing GFP to the N terminus of UTF1 does not interfere with the function of the protein.
Figure 3.
Cellular localization and subnuclear fractionation of GFP-UTF1 in EC and ES cells. (A) Western blot analysis of wild type (wt) EC cells and a clone stably expressing GFP-UTF1 (#1) using an antibody directed against UTF1. (B) Subnuclear fractionation of EC and ES cells, both expressing GFP-UTF1. Immunoblot analysis was performed with an antibody directed against UTF1 and the HA tag of the fusion protein. F, free-diffusing protein fraction; D, DNaseI fraction; AS, ammonium sulfate fraction; HS, high salt fraction; M, nuclear matrix fraction. (C) Confocal and transmission image of living EC cells expressing GFP-UTF1. Nucleoli are indicated by arrows. (D) Time-lapse imaging of a GFP-UTF1–expressing EC cell going through mitosis. (E) Confocal images of a mitotic GFP-UTF1–expressing cell treated with Hoechst. (F) Confocal and transmission images of GFP-UTF1–expressing ES cells grown on an STO cell feeder layer. The arrow indicates a nucleolus, and the arrowhead points to mitoticchromosomes. (G) Transmission image of AP staining of GFP-UTF1 ES cells. The underlying STO feeder cells are negative for AP activity. Bars, 15 μm.
Figure 5.
Analysis of subcellular localization and mobility of wt and mutated GFP-UTF1. (A) A schematic representation and repressor activity of various GFP-UTF1 mutants. The Myb/SANT domain (aa 55–124) and conserved domain 2 (CD2) are indicated by black boxes and gray boxes, respectively. The W63G and E67K point mutations are indicated by asterisks. Repressor activity of the GFP-UTF1 fusion proteins was measured on a constitutively active UAS-TK-Luc reporter in transiently transfected HepG2 cells. Negative controls include pUC18 and a peGFP-C1 plasmid. Error bars represent SD. (B) Confocal images of living cells expressing GFP-UTF1 (1–339), GFP-UTF1 W63G E67K (W63G E67K), GFP-UTF1 1–300 (1–300), and GFP-UTF1 W63G E67K 1–300 (W63G E67K 1–300). (C) FRAP analysis of EC cells expressing either GFP (green line), GFP-UTF1 (1–339; blue line), or GFP-UTF1 W63G E67K (W63G E67K; red line). (D) FRAP experiment of EC cells expressing either GFP (green line), GFP-UTF1 (1–339; blue line), or GFP-UTF1 1–300 (1–300; red line). (E) FRAP experiment of EC cells expressing either GFP (green line), GFP-UTF1 (1–339; blue line), or GFP-UTF1 W63G E67K 1–300 (W63G E67K 1–300; red line). (F) Subnuclear fractionations of stable cell lines expressing GFP-UTF1 (1–339), GFP-UTF1 W63G E67K (W63G E67K), GFP-UTF1 1–300 (1–300), and GFP-UTF1 W63G E67K 1–300 (W63G E67K 1–300). Blots were developed with an antibody against HA. F, free-diffusing protein fraction; D, DNaseI fraction; AS, ammonium sulfate fraction; HS, high salt fraction; M, nuclear matrix fraction. Bar, 15 μM.
Confocal microscopy of living cells showed that GFP-UTF1 localized to the nucleus with an inhomogeneous distribution in a similar fashion as the endogenous protein (Figs. 2 A and 3 C). GFP-UTF1 is excluded from the nucleoli (Fig. 3 C, arrows). The punctate localization is more intense around the nucleoli and in the nuclear periphery. Time-lapse imaging of a cell counterstained with Hoechst showed the chromosomal localization of GFP-UTF1 during metaphase, anaphase, and telophase (Fig. 3, D and E).
In ES cells, a similar GFP-UTF1 distribution was observed: localized to the nucleus, excluded from the nucleoli (Fig. 3 C, arrows), and chromosome associated during mitosis (Fig. 3 F, arrowhead). GFP-UTF1 ES cells were AP positive (Fig. 3 G) and expressed Oct4 (not depicted). Fractionation of GFP-UTF1 ES cells showed that both endogenous UTF1 (αmUTF1) and GFP-UTF1 (αHA) localized to the fraction containing strongly DNA-associated proteins (Fig. 3 B).
Mobility of UTF1 in living cells
To study the observation that UTF1 is a stably chromatin-associated protein in a more physiological context, we analyzed the dynamic properties of UTF1 in living cells using a FRAP protocol. In EC cells, GFP-UTF1 molecules were bleached in a small strip spanning the nucleus, and subsequent fluorescent recovery in the strip was measured at 20-ms intervals (Hoogstraten et al., 2002; van den Boom et al., 2004). The mean fluorescence intensity in the strip of several cells was plotted against time relative to the prebleach level. GFP-expressing cells showed a fast recovery of fluorescence in the strip (Fig. 4 A, green line), indicating a highly mobile protein. Fluorescence in the strip did not recover to prebleach levels as a result of the permanent bleaching of a fraction of the molecules. In contrast to GFP, GFP-UTF1 (Fig. 4 A, blue line) showed only little recovery after bleaching, indicating that the vast majority is long-term immobilized, at least for the duration of the FRAP experiments. Because mobility measurements of GFP and GFP-UTF1 in ES cells produced identical results (Fig. 4 B, green line and blue line, respectively), EC cells were used for all subsequent mobility measurements.
Figure 4.
Strip-FRAP analysis of GFP, GFP-UTF1, H2B-GFP, and Oct4-GFP. (A) FRAP analysis of EC cells expressing GFP (green line) or GFP-UTF1 (blue line). The graph shows the relative fluorescent recovery directly after bleaching. The prebleach level is normalized to 1. GFP displays a quick fluorescent recovery in the bleached region, whereas GFP-UTF1 shows only little recovery directly after photobleaching. (B) FRAP analysis of ES cells expressing GFP (green line) or GFP-UTF1 (blue line). GFP shows a quick recovery after bleaching, whereas GFP-UTF1 displays only a marginal recovery. (C) FRAP experiment of EC cells expressing either GFP-UTF1 (blue line) or histone H2B-GFP (red line). GFP-UTF1 and H2B-GFP both show only a small recovery directly after photobleaching. (D) FRAP experiment of EC cells expressing GFP (green line), GFP-UTF1 (blue line), or Oct4-GFP (red line). Oct4-GFP shows a quick recovery after fluorescence, although slower than GFP.
In terms of localization (Figs. 2 A and 3 C) and subnuclear fractionation behavior (Fig. 2 B), UTF1 greatly resembles core histones (Kanda et al., 1998). To further substantiate this observation, the mobilities of UTF1 and core histone H2B (Kimura and Cook, 2001) were compared (Fig. 4 C). FRAP curves for GFP-UTF1 (Fig. 4 C, blue line) and H2B-GFP (Fig. 4 C, red line) were virtually identical, indicating the similar molecular kinetics of these proteins. Computer simulations of the FRAP procedure were used to fit the experimental data, yielding diffusion constants, immobile fractions, and residence times of all proteins tested (Table I and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200702058/DC1). Both the population of GFP-UTF1 and H2B-GFP molecules displayed an immobile fraction of ∼90% (Table I and Fig. S1, A and B). The duration of immobilization was much longer than the 20-s time scale of our experiments and, therefore, could only be determined with limited accuracy. For both GFP-UTF1 and H2B-GFP, a residence time in the order of minutes to hours was determined, which is in agreement with the findings of Kimura and Cook (2001).
Table I.
Diffusion constants, immobile fractions, and residence times of tested constructs derived from FRAP data fitting
| Protein | Diffusion constant |
Immobile fraction |
Residence time |
|---|---|---|---|
| μm2/s | % | s | |
| GFP | 14 ± 1.8 | 0 | 0 |
| Oct4-GFP | 3 ± 1.30 | 10 ± 0.1 | 0.1 ± 0.2 |
| H2B-GFP | 1 ± 0.38 | 90 ± 0.01 | 1,024 ± 105 |
| GFP-UTF1 | 0.6 ± 2.74 | 90 ± 0.02 | 512 ± 35 |
| GFP-UTF1 W63G E67K | 0.6 ± 0.5 | 60 ± 0.04 | 1,024 ± 95 |
| GFP-UTF11-300 | 14 ± 2.74 | 85 ± 0.01 | 0.25 ± 0.04 |
| GFP-UTF1 W63G E67K 1–300 |
14 ± 2.40 | 25 ± 0.04 | 0.25 ± 0.03 |
Subsequently, we compared the dynamic behavior of GFP-UTF1 and Oct4-GFP (Fig. 4 D). In contrast to GFP-UTF1 (Fig. 4 D, blue line), Oct4-GFP (Fig. 4 D, red line) is largely mobile. Note that Oct4-GFP fluorescence recovery is much slower than that of GFP (Fig. 4 D, green line). Computer simulations indicated that 10% of the Oct4-GFP molecules are immobile with a residence time in the order of 0.1 s (Table I and Fig. S1 C). In addition, the diffusion rate of Oct4-GFP (3 μm2/s) suggested that the protein resides in a high molecular weight complex. The highly dynamic behavior of Oct4-GFP molecules is similar to what is found for several other DNA transacting factors like the transcription/repair factor TFIIH, the homologous recombination protein Rad54, and TFIIB during interphase (McNally et al., 2000; Phair and Misteli, 2000; Chen et al., 2002; Essers et al., 2002; Hoogstraten et al., 2002; Phair et al., 2004; van den Boom et al., 2004; Houtsmuller, 2005). These data indicate that the dynamic behavior of UTF1 is similar to that of core histones but not to that of transcription factors like Oct4.
Localization and mobility of GFP-UTF1 mutants
Using GAL4-UTF1 fusions, we identified the putative Myb/SANT domain and the C terminus of UTF1 as repressor domains (Fig. 2 E). To investigate the requirement of these domains in UTF1 localization and mobility, a series of GFP-UTF1 mutants was generated. First, the repressor activity of wt and mutant GFP-UTF1 proteins was determined in reporter assays. Mutation of aa 63 (W→G) and 67 (E→K; GFP-UTF1 W63G E67K; Fig. 5 A), two amino acids highly conserved in Myb/SANT domains, and/or deletion of the C-terminal 39 aa (GFP-UTF1 W63G E67K 1–300; GFP-UTF1 1–300) resulted in a complete loss of UTF1 repressor activity (Fig. 5 A).
In terms of localization, GFP-UTF1 and GFP-UTF1 W63G E67K display a similar distribution. Deletion of the entire Myb/SANT domain resulted in an almost completely cytoplasmic localized fusion protein (unpublished data). GFP-UTF1 1–300 also interacted with mitotic chromosomes, whereas during interphase, the protein seemed to be more dispersed (Fig. 5 B). GFP-UTF1 W63G E67K 1–300 showed a completely homogenous nuclear distribution in combination with nucleolar exclusion. Furthermore, association with mitotic chromosomes was never observed (Fig. 5 B). These data indicate that both the Myb/SANT domain and C terminus of UTF1 are required for proper localization of the protein during interphase as well as mitosis.
To determine the role of the Myb/SANT domain and C terminus in UTF1 mobility, FRAP analyses were performed. GFP-UTF1 W63G E67K–expressing cells showed an increased recovery of fluorescence in the strip (Fig. 5 C, red line) compared with GFP-UTF1 (Fig. 5 C, blue line), indicating a reduced binding efficiency. The rate of fluorescence recovery after the initial influx resembled that of GFP-UTF1, implying that the residence time of individual molecules was not affected. Computer simulations showed that the residence time of GFP-UTF1 W63G E67K molecules is similar to GFP-UTF1 molecules (in the order of minutes to hours) but that the mean immobile fraction was smaller (∼60%; Table I and Fig. S1 D).
GFP-UTF1 1–300–expressing cells showed a complete recovery after bleaching (Fig. 5 D, red line), demonstrating that the C terminus is required for the long-term immobilization of GFP-UTF1. However, the initial fluorescence recovery in the strip was substantially slower than that of GFP (Fig. 5 D), suggesting that UTF1 resides in a high molecular weight complex and/or is still capable of transiently interacting with sites of affinity. Simulations showed that 85% of the GFP-UTF1 1–300 molecules are immobilized with a residence time in the order of 0.25 s (Table I and Fig. S1 E). These data indicate that the C terminus of UTF1 is required for the long-term stabilization of interactions with sites of affinity, most likely chromatin.
Remarkably, GFP-UTF W63G E67K 1–300 showed a much faster recovery of fluorescence than GFP-UTF1 1–300 and an only slightly slower recovery than GFP (Fig. 5 E), indicating that this mutant is freely mobile. This was further supported by computer simulations that predicted that 25% of the GFP-UTF1 W63G E67K 1–300 molecules was immobilized with a short residence time of 0.25 s (Table I and Fig. S1 F). The UTF1 mutants lacking their C-terminal 39 aa (GFP-UTF1 1–300 and GFP-UTF1 W63G E67K 1–300) displayed a marked increase in their diffusion constants compared with GFP-UTF1 and GFP-UTF1 W63G E67K (14 vs. 0.6 μm2/s; Table I). However, because the model used for fitting the data only included one pair of binding constants (immobile fraction and residence time) and the stable binding of GFP-UTF1 and GFP-UTF1 W63G E67K is dominant in the FRAP curve, the observed low mobility of 0.6 μm2/s is most likely the result of additional transient interactions similar to those of the C-terminal mutants.
To investigate whether the differential mobilities of the mutant proteins are reflected by altered distribution over subnuclear fractions, cell lines stably expressing mutant GFP-UTF1 proteins were analyzed (Fig. 5 F). As shown before (Fig. 3 B), GFP-UTF1 localized to the strongly DNA-associated protein fraction. The majority of the GFP-UTF1 W63G E67K proteins was also strongly DNA associated but was detected in the free-diffusing protein fraction as well, indicating the presence of an increased portion of mobile molecules, which is in agreement with the FRAP data. Both GFP-UTF1 1–300 and GFP-UTF1 W63G E67K 1–300 were found in the free-diffusing protein fraction, indicating that both mutants are fully mobile. Note that GFP-UTF1 1–300 was still capable of binding to mitotic chromosomes and had a punctate nuclear localization, suggesting that this protein is capable of transient interactions with sites of affinity. Throughout these experiments, endogenous UTF1 was always detected in the fraction containing strong DNA-associated proteins (unpublished data).
Discussion
These results show that UTF1 is required for the proper differentiation of EC and ES cells. KD of UTF1 expression in EC and ES cells resulted in blocked or delayed differentiation but did not affect the self-renewal capacity of these cells, indicating that UTF1 is not required for ES cell self-renewal. In addition, reporter assays, subnuclear fractionations, and FRAP analyses showed that UTF1 is a stably chromatin-associated transcriptional repressor with histone-like properties like long-term DNA association and a majority of immobilized molecules. The UTF1–chromatin interaction is dependent on two separate interaction domains: the Myb/SANT domain and the extreme C terminus. The concerted action of both interaction domains causes ∼90% of the molecules to bind to sites of affinity for times similar to those of H2B (Table I; Kimura and Cook, 2001). Summarizing, these data indicate that UTF1 is strongly associated with chromatin in EC and ES cells and most likely also during the early stages of embryogenesis. UTF1 may establish a chromatin state that renders an ES cell susceptible to the activation of differentiation programs in response to appropriate stimuli. Despite being prone to differentiation, ES cells are kept in a self-renewing state by the combined action of self-renewal regulators like Nanog, Oct4, Sox2, and Sall4 and the recently identified Esrrb, Tbx3, and Tcl1 proteins that interfere with differentiation to epiblast-derived lineages (Yuan et al., 1995; Nichols et al., 1998; Chambers et al., 2003; Mitsui et al., 2003; Ivanova et al., 2006; Zhang et al., 2006). Although the expression patterns of UTF1 and these genes are identical (Mitsui et al., 2003; Ivanova et al., 2006), the function of UTF1 seems opposite; where UTF1 is not required for ES cell self-renewal, it is involved in ES cell differentiation. The fact that UTF1 expression is down-regulated during ES cell differentiation is probably a consequence of the inactivation of the Oct4 gene, a transcriptional activator of the UTF1 gene (Nishimoto et al., 1999).
Molecularly, UTF1 may be necessary for signaling to these self-renewal factors to allow differentiation to commence, explaining why UTF1 is dispensable for ES cell self-renewal and that its expression is down-regulated upon the initiation of differentiation. More likely, in view of the histone-like properties of UTF1, its function could be the maintenance of a specific epigenetic profile required for differentiation either by attracting chromatin-modifying proteins or by chromatin compaction. This hypothesis is supported by the observation that UTF1 has transcriptional repressor activity, an observation also made by Fukushima et al. (1999), who showed that UTF1 in the absence of ATF2 could repress the activity of various reporter genes.
Recent reports have emphasized the role of the epigenetic regulation of gene expression in ES cell self-renewal and differentiation (Azuara et al., 2006; Bernstein et al., 2006; Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006). The PcG proteins were found to silence a large set of developmental differentiation genes in ES cells. Many of these PcG targets in ES cells are cooccupied by Oct4, Sox2, and Nanog (Boyer et al., 2005; Loh et al., 2006), suggesting that to maintain the self- renewing state of ES cells, stem cell self-renewal regulators may directly regulate the targeting and/or activity of chromatin remodeling complexes.
The phenotype of UTF1 KD ES cells is similar to that of ES cells lacking the PcG protein Suz12 (Pasini et al., 2007). Pasini et al. (2007) show that Suz12−/− ES cells fail to differentiate, underlining the important role of PcG-mediated silencing in lineage specification. Similarly, UTF1 may either directly or indirectly influence the epigenetic state of ES cells, thereby allowing the initiation of lineage-specific differentiation. Down-regulation of UTF1 may allow ES cells to establish a new, more somatic type of chromatin, which is analogous to observations made by Meshorer et al. (2006), who showed that architectural chromatin proteins bind loosely to chromatin in ES cells and become immobilized upon differentiation.
In this study, we show for the first time that the ES cell protein UTF1 is a stably chromatin-associated protein that is involved in initiation of the differentiation program of ES cells. We propose that with UTF1, we have identified a principal component of the complex regulatory gene network underlying initiation of the lineage-committed differentiation of ES cells.
Materials and methods
Constructs
The mUTF1 cDNA was provided by H. Stunnenberg (Nijmegen Centre for Molecular Life Sciences, Nijmegen, Netherlands). BamHI (5′) and EcoRI (3′) sites were added by PCR, and this fragment was BamHI–EcoRI ligated into pcDNA3-HA, resulting in pcDNA3-HA-mUTF1. For PCR, the primers mUTF1 forward (5′-ATATGATATCGGATCCATGCTGCTTCGTCCCCGGAG-3′) and mUTF1 reverse (5′-ATATGAATTCTTATTGGCGCAAGTCCCCAAG-3′) were used.
pSG424-UTF1 constructs.
pSG424mUTF1 1–300 was generated by BamHI–SalI digestion of pcDNA3-HA-mUTF1 and ligation of the fragment into pSG424. pSGS424mUTF1 1–339 was generated by NheI–SacI digestion of pcDNA3-HA-mUTF1 and ligating the fragment into pSG424mUTF1 1–300 digested with NheI–SacI. pSG424-mUTF1 1–167 was cloned by SacII (T4 DNA polymerase) and SalI (Klenow) digestion of pSG424mUTF1 and subsequent self-ligation. pSG424-mUTF1 1–134 was generated by BspEI–XbaI (Klenow) digestion of pSG424mUTF1 and self-ligation. pSG424-mUTF1 1–66 was cloned by the digestion of pSG424mUTF1 with BsmBI–XbaI (Klenow) and self-ligation. pSG424-mUTF1 1–32 was generated by AspI–XbaI digestion (Klenow) of pSG424mUTF1 and self-ligation. pSG424-mUTF1 1–18 was generated by NheI–SalI digestion (Klenow) of pSG424mUTF1 and self-ligation. pSG424mUTF1 33–339 was cloned by ligating the AspI (Klenow)–SacII fragment from pSG424mUTF1 1–339 into pSG424mUTF1 1–339 digested with BamHI (Klenow)–SacII. pSG424mUTF1 66–339 was generated by ligation of a BamHI–SacII- digested PCR fragment (forward: 5′-ATATGGATCCTTCGAGAGACGGAGCTACTTC-3′; reverse: 5′-ATATGAATTCTTATTGGCGCAAGTCCCCAAG-3′) into pSG424mUTF 1–339 digested with BamHI–SacII. pSG424mUTF1 210–339 was generated by digesting pSG424mUTF1 with BamHI–AspI (Klenow) and self-ligation. pSG424mUTF1 249–339 was cloned by BamHI–DraIII digestion (Klenow) of pSG424mUTF1 and self-ligation. pSG424mUTF1 297–339 was cloned by EcoRI digestion of a PCR fragment (forward: 5′-ATGAATTCCAGCTGTCGACCCTGAAC-3′; reverse: 5′-ATATGAATTCTTATTGGCGCAAGTCCCCAAG-3′) and subsequent ligation in pSG424 digested with EcoRI. To delete the Myb/SANT domain, a PCR fragment (forward: 5′-ATATGATATCAGATCTATGCTGCTTCTGCCCCGGAG-3′; reverse: 5′-AGGGTCCGGACGGCTGGCCCTGGGAGTCTCGGAGCGCCGAGTCCGGGGACAC-3′) was BglII–BspEI digested and BamHI–BspEI ligated into pcDNA3-HA-mUTF1. To generate pSG424mUTF1 1–55/125–339, the NheI–SacII fragment was isolated from pcDNA3-HA-mUTF1 1–55/125–339 and ligated into pSG424mUTF1 1–339 digested with NheI–SacII. pSG424mUTF1 33–134 was generated by BspE1–XbaI digestion (Klenow) of pSG424mUTF 33–339 and subsequent self-ligation.
peGFP-UTF1 constructs.
peGFP-HA-mUTF1 was generated by HindIII and EcoRI digestion of pcDNA3-HA-UTF1 and HindIII–EcoRI ligation into peGFP-C1 (CLONTECH Laboratories, Inc.). peGFP-HA-mUTF1 1–300 was cloned by SalI digestion of peGFP-HA-mUTF1 and ligating the resulting 941-bp fragment into the peGFP-HA-mUTF1 backbone followed by orientation check. peGFP-HA-mUTF1 W63G E67K was generated by HindIII–EcoRI digestion of pcDNA3-HA-mUTF1 W63G E67K and ligation into peGFP-C1 digested with HindIII–EcoRI. pcDNA3-HA-mUTF1 W63G E67K was generated by fusion PCR (forward: 5′-TATAGGATCCATGCTGCTTCTGCCCCGGA-3′; reverse: 5′-GTCTTTCGGGCACTCCCGGGCG-3′; and forward: 5′-GCCCGGGAGTTGCCCGAAAGACG-3′; reverse: 5′-ATATGAATTCTTATTGGCGCAAGTCCCCAA-3′) and cloned into pcDNA3-HA using BamHI and EcoRI. peGFP-HA-mUTF1 W63G E67K 1–300 was cloned by SalI digestion of peGFP-HA-mUTF1 W63G E67K and ligation of the 941-bp fragment into the SalI-digested peGFP-HA-mUTF1 W63G E67K backbone followed by orientation check.
siRNA constructs.
For stable UTF1 KD, a specific short hairpin repeat of a 19-nucleotide sequence directly downstream of the stop codon (AGCTTTGTTATCAGTCCTC) was cloned into pSuper (Brummelkamp et al., 2002) after digestion with BglII and HindIII. Similarly, a sequence targeting Renilla luciferase mRNA (AAACATGCAGAAAATGCTG) was cloned into pSuper. pBos-H2B-GFP was obtained from BD Biosciences.
Cell culture and transfections
P19CL6 EC cells (Habara-Ohkubo, 1996) were grown in α-MEM (Invitrogen) supplemented with antibiotics and 10% FBS (Hyclone) at 37°C and 5% CO2. HepG2 cells were maintained in DME with antibiotics and 10% FBS. For differentiation of P19CL6 cells, 365,000 cells were seeded in 6-cm ø plates in culture medium supplemented with 1% DMSO (Sigma-Aldrich). Embryonic day 14 ES cells (subclone IB10) were grown on gelatin-coated dishes in buffalo rat liver cell–conditioned medium supplemented with 1,000 U/ml leukemia inhibitory factor (Chemicon), nonessential amino acids, and 0.1 mM 2-mercaptoethanol. For confocal laser-scanning imaging, IB10 cells were seeded on a layer of STO feeder cells on gelatinized glass coverslips. To generate stably transfected cell lines, 107 cells were electroporated with 13.5 μg plasmid DNA and 1.5 μg pPGK-Hyg plasmid. Selection was performed using 200 μg/ml hygromycin, clones were picked, and cell lysates were analyzed. For analyses of AP activity, an AP detection kit (Chemicon) was used. To generate stably transfected P19CL6 cell lines, cells were transfected with FuGENE 6 (Roche) and selected with 600 μl/ml G418 or 600 μg/ml hygromycin, and clones were picked. For DNA staining, cells were cultured for 2 h in the presence of 10 μg/ml Hoechst 33258. For transient transfections, 250,000 HepG2 cells were seeded per 3.5-cm ø well. Transfections were performed using calcium phosphate coprecipitation. After 48 h, cells were harvested (reporter lysis buffer; Promega), and luciferase activity was measured (LucLite; Packard). To normalize luciferase activities, a β-galactosidase expression plasmid (pDM2LacZ) was cotransfected. β-Galactosidase activity was determined in 100 mM Na2HPO4/NaH2PO4, 1 mM MgCl2, 100 mM 2-mercaptoethanol, and 0.67 mg/ml O-nitrophenylgalactopyranoside.
EB formation
For EB formation, ES cells were suspended from the lids of 10-cm ø dishes in 20-μl drops (5 × 104 cells/ml). After 48 h, EBs were transferred to bacterial grade Petri dishes. On day 7, EBs were transferred to gelatinized 3.5-cm ø six-well plates. On days 3, 5, and 7, pictures were taken, and total RNA was isolated on days 3, 5, and 10.
RNA isolation and RT-PCR analyses
Total RNA was extracted with TRIzol (Invitrogen), treated with DNaseI (Fermentas), and reverse transcribed (RevertAid M-MuLV Reverse Transcriptase; Fermentas). Details of primer sets, cycle numbers, and annealing temperatures used in subsequent PCR reactions can be found in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200702058/DC1). PCR products were analyzed on 2% agarose gels.
Western blot analysis and subnuclear fractionation
Cells were washed with cold PBS and incubated in lysis buffer (400 mM NaCl, 20 mM Tris-HCl, pH 7.8, 1% NP-40, 0.5% sodium deoxycholate, 2 mM EDTA, 2 mM DTT, and protease inhibitors) for 30 min on ice. Next, cell lysates were collected by scraping, subsequently sonicated, and cleared by centrifugation at 4°C and 14,000 rpm for 10 min. For western analysis, the following primary antibodies were used: mUTF1 (rabbit polyclonal raised by Eurogentec), Oct-4 (H-134; Santa Cruz Biotechnology, Inc.), HDAC1 (H-51; Santa Cruz Biotechnology, Inc.), histone H2A (acidic patch; Upstate Biotechnology), GATA4 (C20; Santa Cruz Biotechnology, Inc.), actin (C4; MP Biomedicals), and HA (3F10; Roche). Secondary immunodetection was performed using donkey anti–rabbit IgG-HRP (GE Healthcare), rabbit anti–rat IgG-HRP (DakoCytomation), goat anti–mouse IgG-HRP (Santa Cruz Biotechnology, Inc.), and donkey anti–goat IgG-HRP (Santa Cruz Biotechnology, Inc.). Subnuclear fractionation was performed as previously described (Citterio et al., 2004).
Microscopy and image analysis
For immunofluorescence analysis, P19CL6 cells were cultured on poly-d- lysine–coated glass coverslips and fixed in 2% PFA in PBS for 10 min at RT. After fixing, cells were permeabilized with 0.1% Triton X-100 in PBS. Endogenous UTF1 was detected using our UTF1 antibody followed by a goat anti–rabbit tetramethylrhodamine IgG (H+L)-conjugated secondary antibody (Invitrogen). Fluorescent images were made using a microscope (Axiophot; Carl Zeiss MicroImaging, Inc.) with a plan-NEOFLUAR 40× NA 0.70 lens. Confocal laser-scanning microscopy images of live cells were recorded with a microscope (LSM 510; Carl Zeiss MicroImaging, Inc.). GFP signal was detected using a 488-nm argon laser line and a bandpass 500–550-nm filter. Hoechst signal was monitored by excitation with a Titanium Sapphire 810-nm dual-photon laser and a bandpass 390–465-nm filter.
FRAP
For FRAP experiments, a confocal laser-scanning microscope (LSM 510; Carl Zeiss MicroImaging, Inc.) was used. To measure FRAP, a 10-μm-wide strip spanning the nucleus was bleached for 120 ms at the highest intensity of the 488-nm line of a 30-mW argon laser focused by a plan Apochromat 63× NA 1.4 oil differential interference contrast lens (Carl Zeiss MicroImaging, Inc.). Recovery of fluorescence in the strip was monitored at 20-ms intervals at 0.5% of the laser intensity used for bleaching. For emission detection, a bandpass 500–550-nm filter was used.
Computer simulations
For analysis of FRAP data, FRAP curves were normalized to prebleach values, and the best fitting curve (least squares) was picked from a large set of computer-simulated FRAP curves in which three parameters representing mobility properties were varied: diffusion rate (ranging from 0.04 to 25 μm2/s), immobile fraction (0, 10, 20, 30, 40, and 50%), and time spent in the immobile state (2, 4, 8, 16, 32, 64, 128, and ∞ s). Monte Carlo computer simulations used to generate FRAP curves were based on a model of random diffusion in an ellipsoid volume representing the cell nucleus and simple binding kinetics representing binding to immobile elements in the cell nucleus. Simulations were performed at unit time steps corresponding to the experimental sample rate of 21 ms.
Diffusion was simulated by each step, deriving novel positions M(x + dx, y + dy, and z + dz) for all mobile molecules M(x, y, and z), where dx = G(r1), dy = G(r2), dz = G(r3), ri is a random number (0 ≤ ri ≤ 1) chosen from a uniform distribution, and G(ri) is an inversed cumulative Gaussian distribution with μ = 0 and σ2 = 6Dt, where D is the diffusion coefficient and t is time measured in unit time steps.
Immobilization was based on simple binding kinetics described by kon/koff = Fimm/(1 − Fimm), where Fimm is the relative number of immobile molecules. The chance for each particle to become immobilized (representing chromatin binding) was defined as Pimmobilize = kon = koff Fimm/(1 − Fimm), where koff = 1/timm and timm is the mean time spent in immobile complexes measured in unit time steps; the chance to release was Pmobilize = koff = 1/timm. In simulations of two immobile fractions with different kinetics, two immobilization/mobilization chances were evaluated for each unit time step.
The FRAP procedure was simulated on the basis of an experimentally derived 3D laser intensity profile, providing a chance based on 3D position for each molecule to get bleached or to be sent to a temporary dark state (blinking) during simulation of the bleach pulse. The profile was derived from confocal images (z stacks) of chemically fixed nuclei containing GFP that were exposed to a stationary laser beam at various intensities and varying exposure times.
For each set of parameters, a FRAP curve was generated based on 106 molecules per nucleus (which yields similar results as averaging 10 cells containing 105 molecules or 100 cells containing 104 molecules). This number was empirically determined to produce a curve with a limited fluctuation of fluorescence (as a result of diffusion) after complete recovery.
Online supplemental material
Table S1 provides the primer sequences used for the PCR reactions displayed in Fig. 1 G and their product sizes, annealing temperatures, and number of PCR cycles. Fig. S1 shows experimental FRAP curves and computer-simulated curves for the constructs GFP-UTF1, H2B-GFP, Oct4-GFP, GFP-UTF1 W63D E67K, GFP-UTF1 1–300, and GFP-UTF1 1–300 W63D E67K. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200702058/DC1.
Supplementary Material
Acknowledgments
We thank Henk Stunnenberg for supplying the mUTF1 cDNA, Duanqing Pei for sending the Oct4-GFP construct, and Peter ten Dijke for providing the (BRE)2-luc reporter.
This work was supported by the Groningen Biomolecular Sciences and Biotechnology Institute.
V. van den Boom and S.M. Kooistra contributed equally to this paper.
V. van den Boom's present address is Department of Cell Biology, Section of Stem Cell Biology, University Medical Centre Groningen, 9700 RB Groningen, Netherlands.
Abbreviations used in this paper: BMP, bone morphogenetic protein; BRE, BMP-responsive element; CD, conserved domain; EB, embryoid body; EC, embryonic carcinoma; eGFP, enhanced GFP; ES, embryonic stem; HDAC1, histone deacetylase 1; KD, knockdown; mUTF1, mouse UTF1; PcG, Polycomb group; SBE, Smad-binding element; TK, thymidine kinase; UTF1, undifferentiated embryonic cell transcription factor 1; wt, wild type.
References
- Azuara, V., P. Perry, S. Sauer, M. Spivakov, H.F. Jorgensen, R.M. John, M. Gouti, M. Casanova, G. Warnes, M. Merkenschlager, and A.G. Fisher. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8:532–538. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.E., T.S. Mikkelsen, X. Xie, M. Kamal, D.J. Huebert, J. Cuff, B. Fry, A. Meissner, M. Wernig, K. Plath, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125:315–326. [DOI] [PubMed] [Google Scholar]
- Boyer, L.A., T.I. Lee, M.F. Cole, S.E. Johnstone, S.S. Levine, J.P. Zucker, M.G. Guenther, R.M. Kumar, H.L. Murray, R.G. Jenner, et al. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, L.A., K. Plath, J. Zeitlinger, T. Brambrink, L.A. Medeiros, T.I. Lee, S.S. Levine, M. Wernig, A. Tajonar, M.K. Ray, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 441:349–353. [DOI] [PubMed] [Google Scholar]
- Bracken, A.P., N. Dietrich, D. Pasini, K.H. Hansen, and K. Helin. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20:1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553. [DOI] [PubMed] [Google Scholar]
- Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 113:643–655. [DOI] [PubMed] [Google Scholar]
- Chen, D., C.S. Hinkley, R.W. Henry, and S. Huang. 2002. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell. 13:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuva de Sousa Lopes, S.M., S. van den Driesche, R.L. Carvalho, J. Larsson, B. Eggen, M.A. Surani, and C.L. Mummery. 2005. Altered primordial germ cell migration in the absence of transforming growth factor beta signaling via ALK5. Dev. Biol. 284:194–203. [DOI] [PubMed] [Google Scholar]
- Citterio, E., R. Papait, F. Nicassio, M. Vecchi, P. Gomiero, R. Mantovani, P.P. Di Fiore, and I.M. Bonapace. 2004. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol. Cell. Biol. 24:2526–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers, J., A.B. Houtsmuller, L. van Veelen, C. Paulusma, A.L. Nigg, A. Pastink, W. Vermeulen, J.H. Hoeijmakers, and R. Kanaar. 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21:2030–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima, A., A. Okuda, M. Nishimoto, N. Seki, T.A. Hori, and M. Muramatsu. 1998. Characterization of functional domains of an embryonic stem cell coactivator UTF1 which are conserved and essential for potentiation of ATF-2 activity. J. Biol. Chem. 273:25840–25849. [DOI] [PubMed] [Google Scholar]
- Fukushima, A., M. Nishimoto, A. Okuda, and M. Muramatsu. 1999. Carboxy-terminally truncated form of a coactivator UTF1 stimulates transcription from a variety of gene promoters through the TATA Box. Biochem. Biophys. Res. Commun. 258:519–523. [DOI] [PubMed] [Google Scholar]
- Gangloff, Y.G., M. Mueller, S.G. Dann, P. Svoboda, M. Sticker, J.F. Spetz, S.H. Um, E.J. Brown, S. Cereghini, G. Thomas, and S.C. Kozma. 2004. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol. Cell. Biol. 24:9508–9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habara-Ohkubo, A. 1996. Differentiation of beating cardiac muscle cells from a derivative of P19 embryonal carcinoma cells. Cell Struct. Funct. 21:101–110. [DOI] [PubMed] [Google Scholar]
- Hoogstraten, D., A.L. Nigg, H. Heath, L.H. Mullenders, R. van Driel, J.H. Hoeijmakers, W. Vermeulen, and A.B. Houtsmuller. 2002. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell. 10:1163–1174. [DOI] [PubMed] [Google Scholar]
- Houtsmuller, A.B. 2005. Fluorescence recovery after photobleaching: application to nuclear proteins. Adv. Biochem. Eng. Biotechnol. 95:177–199. [DOI] [PubMed] [Google Scholar]
- Ivanova, N., R. Dobrin, R. Lu, I. Kotenko, J. Levorse, C. DeCoste, X. Schafer, Y. Lun, and I.R. Lemischka. 2006. Dissecting self-renewal in stem cells with RNA interference. Nature. 442:533–538. [DOI] [PubMed] [Google Scholar]
- Kaji, K., I.M. Caballero, R. MacLeod, J. Nichols, V.A. Wilson, and B. Hendrich. 2006. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 8:285–292. [DOI] [PubMed] [Google Scholar]
- Kanda, T., K.F. Sullivan, and G.M. Wahl. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385. [DOI] [PubMed] [Google Scholar]
- Kimura, H., and P.R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.I., R.G. Jenner, L.A. Boyer, M.G. Guenther, S.S. Levine, R.M. Kumar, B. Chevalier, S.E. Johnstone, M.F. Cole, K. Isono, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 125:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, Y.H., Q. Wu, J.L. Chew, V.B. Vega, W. Zhang, X. Chen, G. Bourque, J. George, B. Leong, J. Liu, et al. 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38:431–440. [DOI] [PubMed] [Google Scholar]
- Matsuda, T., T. Nakamura, K. Nakao, T. Arai, M. Katsuki, T. Heike, and T. Yokota. 1999. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 18:4261–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, J.G., W.G. Muller, D. Walker, R. Wolford, and G.L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 287:1262–1265. [DOI] [PubMed] [Google Scholar]
- Meshorer, E., D. Yellajoshula, E. George, P.J. Scambler, D.T. Brown, and T. Misteli. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 10:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 113:631–642. [DOI] [PubMed] [Google Scholar]
- Murakami, M., T. Ichisaka, M. Maeda, N. Oshiro, K. Hara, F. Edenhofer, H. Kiyama, K. Yonezawa, and S. Yamanaka. 2004. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 24:6710–6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 95:379–391. [DOI] [PubMed] [Google Scholar]
- Nishimoto, M., A. Fukushima, A. Okuda, and M. Muramatsu. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19:5453–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto, M., A. Fukushima, S. Miyagi, Y. Suzuki, S. Sugano, Y. Matsuda, T. Hori, M. Muramatsu, and A. Okuda. 2001. Structural analyses of the UTF1 gene encoding a transcriptional coactivator expressed in pluripotent embryonic stem cells. Biochem. Biophys. Res. Commun. 285:945–953. [DOI] [PubMed] [Google Scholar]
- Nishimoto, M., S. Miyagi, T. Yamagishi, T. Sakaguchi, H. Niwa, M. Muramatsu, and A. Okuda. 2005. Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Mol. Cell. Biol. 25:5084–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, H., T. Burdon, I. Chambers, and A. Smith. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12:2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, H., S. Masui, I. Chambers, A.G. Smith, and J. Miyazaki. 2002. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol. Cell. Biol. 22:1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, A., A. Fukushima, M. Nishimoto, A. Orimo, T. Yamagishi, Y. Nabeshima, M. Kuro-o, Y. Nabeshima, K. Boon, M. Keaveney, et al. 1998. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 17:2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini, D., A.P. Bracken, J.B. Hansen, M. Capillo, and K. Helin. 2007. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 27:3769–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair, R.D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D.T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, N., L. Meijer, L. Skaltsounis, P. Greengard, and A.H. Brivanlou. 2004. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10:55–63. [DOI] [PubMed] [Google Scholar]
- Smith, A.G., J.K. Heath, D.D. Donaldson, G.G. Wong, J. Moreau, M. Stahl, and D. Rogers. 1988. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 336:688–690. [DOI] [PubMed] [Google Scholar]
- van den Boom, V., E. Citterio, D. Hoogstraten, A. Zotter, J.M. Egly, W.A. van Cappellen, J.H. Hoeijmakers, A.B. Houtsmuller, and W. Vermeulen. 2004. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J. Cell Biol. 166:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R.L., D.J. Hilton, S. Pease, T.A. Willson, C.L. Stewart, D.P. Gearing, E.F. Wagner, D. Metcalf, N.A. Nicola, and N.M. Gough. 1988. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 336:684–687. [DOI] [PubMed] [Google Scholar]
- Ying, Q.L., J. Nichols, I. Chambers, and A. Smith. 2003. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 115:281–292. [DOI] [PubMed] [Google Scholar]
- Yuan, H., N. Corbi, C. Basilico, and L. Dailey. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9:2635–2645. [DOI] [PubMed] [Google Scholar]
- Zhang, J., W.L. Tam, G.Q. Tong, Q. Wu, H.Y. Chan, B.S. Soh, Y. Lou, J. Yang, Y. Ma, L. Chai, et al. 2006. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 8:1114–1123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.