Abstract
Multiple commercial microarrays for measuring genome-wide gene expression levels are currently available, including oligonucleotide and cDNA, single- and two-channel formats. This study reports on the results of gene expression measurements generated from identical RNA preparations that were obtained using three commercially available microarray platforms. RNA was collected from PANC-1 cells grown in serum-rich medium and at 24 h following the removal of serum. Three biological replicates were prepared for each condition, and three experimental replicates were produced for the first biological replicate. RNA was labeled and hybridized to microarrays from three major suppliers according to manufacturers’ protocols, and gene expression measurements were obtained using each platform’s standard software. For each platform, gene targets from a subset of 2009 common genes were compared. Correlations in gene expression levels and comparisons for significant gene expression changes in this subset were calculated, and showed considerable divergence across the different platforms, suggesting the need for establishing industrial manufacturing standards, and further independent and thorough validation of the technology.
INTRODUCTION
A powerful application of microarray technology is in discovery-based biomedical research. Under the discovery based approach, DNA microarrays are used as screening tools to identify genes associated with biological processes of interest. Using microarrays, a genome-wide assay can be conducted and researchers can mine the resulting experimental data to screen a large subset of the genome to discover sets of genes associated with the biological phenomena of interest (1). Once target genes are identified, additional laboratory resources may be invested to validate this list and to further characterize the relationship of their biological functions to the process under study (2). The efficiency of knowledge discovery using this high-throughput experimental process depends upon the reliability of the microarray technology used in the initial screening experiments. Researchers planning to utilize microarray experiments for discovery-based research must evaluate available commercial technologies when allocating laboratory resources for prospective experiments.
Several formats of microarrays for measuring genome-wide gene expression levels are currently available (3). Important factors for selecting an appropriate microarray platform would include sensitivity, specificity and both inter- and intra-assay reproducibility. Also important is knowledge of the degree of cross-platform agreement, as interchangeability amongst various microarray formats would allow for the utility of gene expression data without regard to platform. Having such a property would allow researchers from independent laboratories to make direct comparisons on data produced from different types of available platforms, and would reduce the need to replicate experiments (4). Such cross-platform comparisons ideally require that corresponding RNA expression measurements be concordant. Previous comparisons of microarray formats suggested that expression data on the NCI60 cell lines from spotted cDNA mircroarrays could not be directly combined with data from synthesized oligonucleotide arrays (5). This finding was determined using identical originating cell lines; however, cell culturing, mRNA preparation and hybridization of targets were all performed separately. In this study we analyzed identical RNA preparations using three commercially available high-density microarray platforms. This experimental design allowed us to compare the microarray formats while controlling for variation that may have arisen from independent cell culturing, RNA isolation and purification.
Three major commercial microarray platforms were evaluated by using standardized input RNA sample, and ensuring that all microarray experiments were carried out by technologists specialized in each particular microarray labeling and hybridization protocol. In addition, the analysis of data from a number of biological and experimental replicates allowed us to implement robust statistical methods to select differentially expressed genes from each platform. Interestingly, during the course of data analysis, we discovered that there was substantial variation in the data generated from the individual platforms. Hence, we attempted to determine the extent to which discovery-based research using microarrays from different commercial vendors would produce either overlapping or divergent target gene sets.
MATERIALS AND METHODS
Gene expression data
PANC-1 cells were grown in serum-rich medium, trypsinized and collected immediately and at 24 h following transfer of these cells to serum-free medium. RNA was promptly isolated using Trizol reagent (Invitrogen) and RNeasy (Qiagen), and their quality checked using the Bioanalyzer (Agilent). Sufficient RNA for each biological replicate was extracted to run many microarray experiments, and stored in ethanol at –70°C until the time of the assay, when it was solubilized in RNAse-free water. Thus, each one of the microarray platforms utilized a common sample pool of RNA from control PANC-1 cells which have a pancreatic ductal cell phenotype or from an early stage of their differentiation to a pancreatic islet phenotype (Hardikar et al., manuscript submitted). RNA was labeled and hybridized to microarrays from Affymetrix (U95Av.2 GeneChips, multiple 25mer oligonucleotide probe sets), Agilent (Human 1, cDNA probes) and Amersham (Codelink UniSet Human I Bioarrays, 30mer oligonucleotide probes) according to manufacturers’ guidelines. For the single-channel-type arrays (Affymetrix, Amersham), a total of 10 microarrays were used to hybridize RNA collected from cells from the two time points. For each time point, three arrays were hybridized with RNA derived from one of the PANC-1 cell cultures (technical replicates); the remaining two microarrays were hybridized with RNA from two independent cell cultures (biological replicates), thus generating five data points for each probe at each time point. For the cDNA arrays (two-channel type) RNA from the two time points were cohybridized on a single array. Each of the five Agilent slides contained two sets of coordinate arrays, where samples labeled with the opposite dye (Cy5 and Cy3) configurations were hybridized. Because of the dye-swap replication used in the Agilent system, 10 data values were generated for each time point. For this dataset, dye-swap replicates produced by repeated measurement using the green and red fluorochromes were averaged resulting in five data values for each time point. Probe (Agilent and Amersham) or probe set (Affymetrix) signals were obtained using manufacturers’ standard software and normalization procedures. For the Agilent cDNA arrays, the default settings of the Agilent G2566AA Feature Extraction Software (v.A.5.1.1) were used, which selects the LOWESS (locally weighted linear regression curve fit) normalization method (6). For the Amersham Codelink Array, the BioDiscovery ImaGene (v.5) software was used, and for Affymetrix GeneChips, the Microarray Suite software (MAS 5.0) was used, both of which utilize global (linear) normalization procedures. Data flagged as being poor quality by the Agilent and Amersham data extraction software were removed from the analysis. Each microarray platform reported the GenBank ID of the sequence interrogated by each of the probes or probe sets on the array. These GenBank IDs were compared across platforms to identify a group of 2009 common genes present on all three of the platforms. Signal values were averaged in cases where multiple probes for a given GenBank ID were present on the array. The method of matching probes by GenBank IDs was chosen over matching by Unigene ID, since this could have introduced additional confounding factors such platform-dependent probes for different splice variants (7) across the arrays. Despite the increased number of common genes (4012) when analyzing measurements matched by the Unigene IDs, we did find results similar to those presented in this paper (see supplemental Tables 1, 2 and 3 available as Supplementary Material at NAR Online).
Table 1. Pearson’s product-moment and Spearman’s rank-order correlation coefficients of gene expression measurements from three commercial microarray technologies.
| Comparison | Platform A | Platform B | Pearson’s | P-value | Spearman’s | P-value | n |
|---|---|---|---|---|---|---|---|
| 1 | Amersham | Agilent | 0.47767 | <0.0001 | 0.47760 | <0.0001 | 4018 |
| 2 | Amersham | Affymetrix | 0.59594 | <0.0001 | 0.59127 | <0.0001 | 4018 |
| 3 | Agilent | Affymetrix | 0.50498 | <0.0001 | 0.50322 | <0.0001 | 4018 |
P-values of the hypothesis of no correlation are also reported.
Table 2. Pearson’s product-moment and Spearman’s rank-order correlation coefficients of the log ratio of times 0 and 24 h measurements.
| Comparison | Platform A | Platform B | Pearson’s | P-value | Spearman’s | P-value | n |
|---|---|---|---|---|---|---|---|
| 1 | Amersham | Agilent | 0.59171 | <0.0001 | 0.52132 | <0.0001 | 2009 |
| 2 | Amersham | Affymetrix | 0.52159 | <0.0001 | 0.50853 | <0.0001 | 2009 |
| 3 | Agilent | Affymetrix | 0.53443 | <0.0001 | 0.53262 | <0.0001 | 2009 |
P-values of the hypothesis of no correlation are also reported.
Table 3. Summary of cross platform concordance levels.
| Platform A | Platform B | Bonferroni alpha = 0.05 | Alpha = 0.001 | Alpha = 0.001 and fold change > 2 | Alpha = 0.01 and fold change > 2 |
|---|---|---|---|---|---|
| Affymetrix | Agilent | 9 | 4 | 19 | |
| A 34 (26%) | A 22 (18%) | A 84 (23%) | |||
| B 67 (13%)a | B 36 (11%)a | B 66 (29%)a | |||
| Affymetrix | Amersham | 5 | 2 | 22 | |
| A 34 (15%) | A 22 (9%) | A 84 (26%) | |||
| B 117 (4%) | B 56 (4%) | B 153 (14%)a | |||
| Agilent | Amersham | 1 | 23 | 16 | 34 |
| A 12 (8%) | A 67 (34%) | A 36 (44%) | A 66 (52%) | ||
| B 19 (5%) | B 117 (20%)a | B 56 (29%)a | B 153 (22%)a |
Column titles indicate the cut-off criterion for determination of differential gene expression. In each cell, the first row represents the number of genes found common to the technologies; the second and third rows report the total number found by technologies A and B. The percent overlap is also reported for each technology.
aSignificant non-random association (see supplemental Tables 6, 7 and 8).
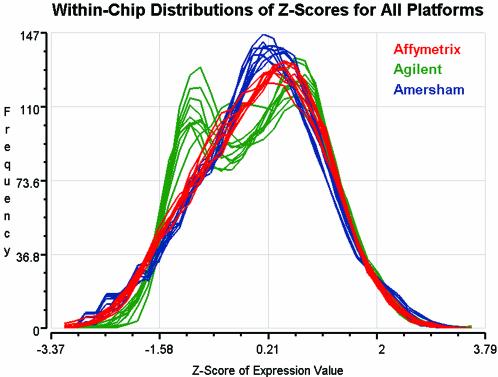
In a manner similar to the ANOVA analysis performed by Kerr et al. (8) and Wolfinger et al. (9), in this study we analyzed base 2 logarithms of the original fluorescent signals when modeling differential expression with an ANOVA model and computing correlations of signal across platforms (see below). However, since gene expression measurements were reported in units unique to each platform, to directly compare data between platforms (Figs 1 and 3) required that these measurements be converted to a single common scale. To accomplish this, we applied a Z-transformation so that the mean and variance (mean = 0 and standard deviation = 1) of the signals for the 2009 common GenBank IDs were equivalent across microarray chips, and platforms (Figs 1 and 3). Thus, analysis of the Z-scores permitted direct comparisons of signal distributions and error levels across technologies.
Figure 1.
A comparison of distributions of log signal intensity values from repeated experiments on three different commercial microarray technologies. Heterogeneous intensity scales across platforms were rescaled using a Z-transformation with mean = 0 and standard deviation = 1.
Figure 3.
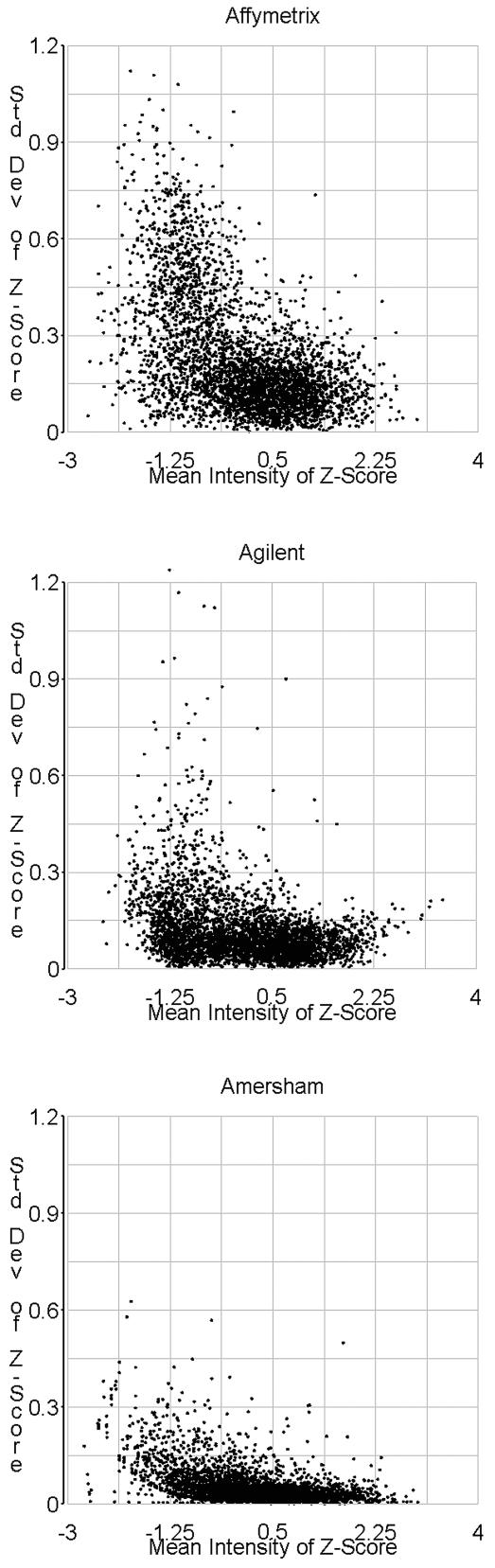
Experimental error plots of Z-scores as a function of mean intensity. Data from experimental replicates were used for standard deviation calculations.
Correlation computations
We computed Pearson linear correlation coefficients and Spearman rank-order correlation coefficients between the first and second technical replicates and the first and second biological replicates for each platform to assess the intra-platform correlations. To assess the cross-platform correlation of gene-expression measurements we computed three Pearson linear correlation coefficients and Spearman rank-order correlation coefficients for sequence-matched gene measurements for each platform pair. The expression measure of a gene for a given platform was computed as the mean of the biological replicates (where the average of the experimental replicates was used for the first biological replicate). Since data from two time points were available, we computed correlation coefficients across 4018 matched measurement pairs. To examine the comparability of intra-assay relationships found within each of the microarray runs, we also calculated correlation coefficients for the log ratio calculated as the base 2 logarithm of the ratio of averaged signal of the biological replicates (where the average of the experimental replicates was used for the first biological replicate) from time point 0 h and time point 24 h (n = 2009). With three sets of microarray measurements, three two-way comparisons of these measurements can be performed. Statistical significance of each correlation coefficient was determined using a Bonferroni corrected alpha of 0.05/3 or ∼0.017. The analysis was performed using SAS statistical software.
Analysis of variance
As an exercise to examine whether data from each of the commercial microarray technologies would lead to similar results in a knowledge discovery experiment, we analyzed the overlap of the target gene lists produced by data from the different platforms. Presence or absence of differential gene expression between time points 0 and 24 h was used as the criterion for classification of the genes as associated or not associated with the biological phenomenon of phenotypic differentiation. To determine significant differences in gene expression levels between the two time points, gene expression measurements for each gene on each array were modeled using a two-factor mixed-model nested ANOVA. For each gene in our set of 2009 common genes the expression measurements were modeled as follows:
ygij = µ + Ti + (TD)ij + εgij
Where ygij is the base 2 logarithm measurement from gene g(g = 1,…,2009), treatment i(I = 1,2) and dish j(j = 1,2,3). Estimates of the parameters of this model were computed using type III sums of squares for each of the 2009 genes and each microarray platform. Statistics from this model were used to produce dichotomous classifications of the genes. Genes were considered to have exhibited differential expression (‘yes’ or ‘no’) if the treatment effect showed statistical significance tested at various alpha levels. Statistical tests using an uncorrected (lenient) P-value criterion of 0.001 and Bonferroni corrected (stringent) criterion of 0.05/2009 or 2.489 E-05 were performed. Two-way contingency tables for each of the three possible platform pairs were created using both the lenient and stringent classifications. Fisher’s exact test and McNemar’s test were performed on these tables using a Bonferroni corrected alpha of 0.05/3 or ∼0.017. The ANOVA and the various post-hoc analyses were performed using SAS statistical software.
RESULTS
Gene expression data
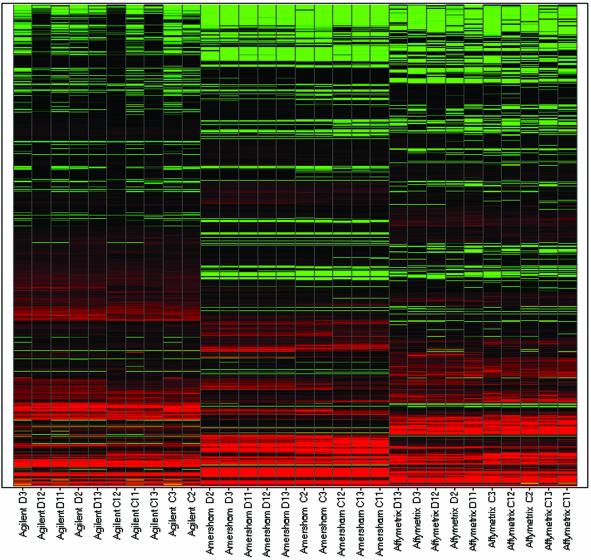
To assess the overall similarity of the gene expression measurements produced by each platform we examined the distributions of the Z-scores of expression measurements (Fig. 1). As indicated in Figure 1, the distribution of intensity values from the Affymetrix GeneChip and Amersham Codelink technologies appear approximately bell-shaped, whereas the distribution for the Agilent cDNA array appears bimodal. To see whether an unsupervised clustering method would group technologies, we subjected the data to hierarchical clustering after global normalization of the 2009 data points from each group to one of the Affymetrix arrays (Fig. 2). The dendogram shows that the three platform groups form distinct clusters at the top level indicating that between-platform variability is greater than within-platform variability.
Figure 2.
Heat map of gene expression measurements normalized to a single array showing genes in rows and samples in columns.
To compare the error profiles for the three platforms, scatterplots of the standard deviation of gene expression measurements for the technical replicates of each gene versus the mean signal of these replicates were compared (Fig. 3). Previous studies have shown that gene expression measurements of low-abundance transcripts are more variable than high-abundance transcripts (10,11). As indicated in Figure 3, this trend is exhibited by the Affymetrix GeneChip data and to a lesser degree by data produced by the Agilent technology. Variability of the Amersham Codelink expression data was low overall but still somewhat signal dependent.
Correlation coefficients
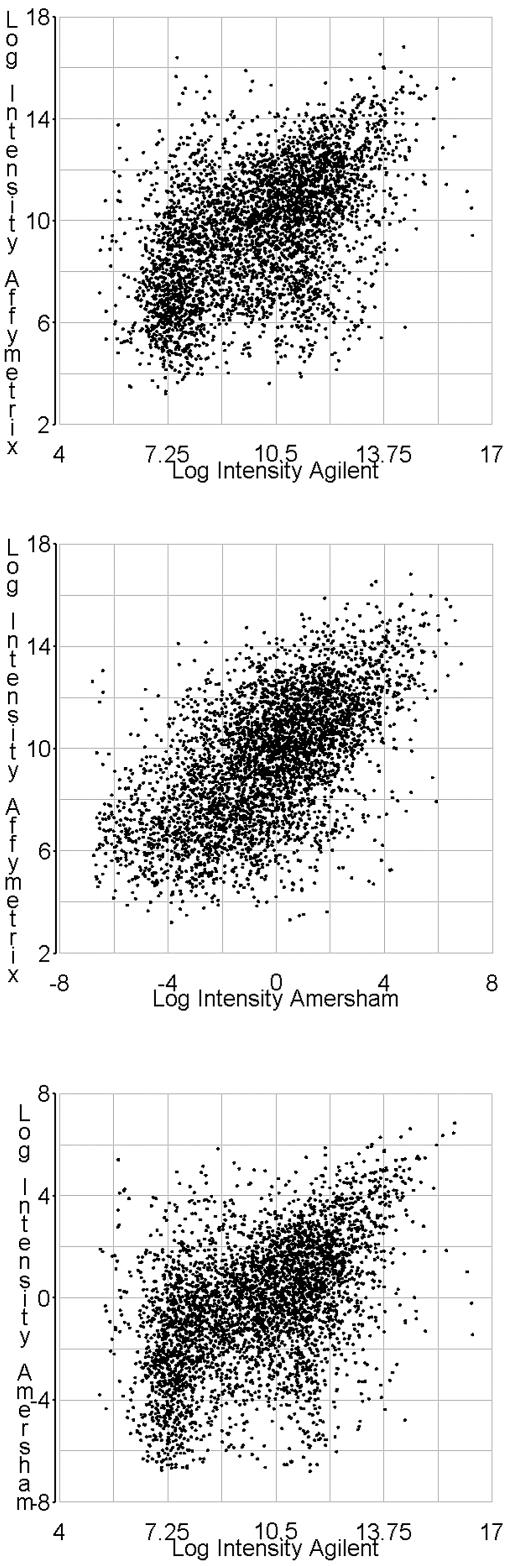
We observed intra-platform correlation coefficients >0.9 for both technical and biological replicates on all platforms (supplemental Table 4, supplemental Figs 1 and 2, available at NAR Online). In comparison, we found the correlation of matched gene measurements from different microarray technologies to be modest, with an average Pearson’s correlation coefficient of 0.53. The Pearson’s correlation coefficients were similar across any combination of platforms. The oligonucleotide arrays (Affymetrix and Amersham) showed modestly higher correlation at 0.59; whereas the Pearson’s correlation coefficient for the Agilent and Amersham data was 0.48, and between the Agilent and Affymetrix data was 0.50 (Table 1 and Fig. 4). Each of these correlations were signficantly positive using a Bonferroni corrected alpha for multiple comparisons (P < 0.017). The level and significance of correlation was not substantially altered by computation of Spearman rank-order coefficients (Table 1). Linear correlation of log ratio measures showed similar results (Table 2). Hypothesis test of no correlation of log ratio measurements could also be rejected at a Bonferroni corrected alpha of 0.017. Additionally, the greatest average distance of log ratio observed was ∼0.15 (supplemental Table 5). Previous research has found that signal strength can affect the level of cross-platform correlation, suggesting that discordance occurred mostly with expression data in the low signal range (5). We were interested in assessing the presence of this property in our data, however we were unable to make any definitive inferences regarding this issue since creating subsets of our data by signal strength would lead to the confounding problem of restriction of range, making any potential inferences about the effect of signal strength upon the level of correlation ambiguous. Nevertheless, from gross inspection of the correlation plots, a distinctly ‘rocket-shaped’ appearance, which would suggest the presence of this trend, was not apparent.
Table 4. Classifications based upon highest 5, 10, 25, 50 and 100 F statistics in each direction (up- and down-regulation).
| Platform A | Platform B | Highest 5 | Highest 10 | Highest 25 | Highest 50 | Highest 100 |
|---|---|---|---|---|---|---|
| Agilent | Affymetrix | 1/10 10% | 2/20 10% | 12/50 24% | 27/100 27% | 70/200 35% |
| Amersham | Affymetrix | 1/10 10% | 1/20 5% | 9/50 18% | 17/100 17% | 52/200 26% |
| Amersham | Agilent | 2/10 20% | 3/20 15% | 9/50 18% | 24/100 24% | 64/200 32% |
The number of genes found common to the technologies in either the up- or down-regulated sets over the total number in both sets found by each technology and the percentage this represents is reported.
Figure 4.
Correlations of the gene-matched mRNA measurements. Scatter plots are of the mean of log intensity values. Pearson’s correlation coefficients of these means and 95% confidence intervals are reported in Table 1.
Table 5. Classifications based upon highest 5, 10, 25, 50 and 100 log2 ratio in each direction (up- and down-regulation).
| Platform A | Platform B | Highest 5 | Highest 10 | Highest 25 | Highest 50 | Highest 100 |
|---|---|---|---|---|---|---|
| Agilent | Affymetrix | 2/10 20% | 2/20 10% | 11/50 22% | 25/100 25% | 57/200 29% |
| Amersham | Affymetrix | 2/10 20% | 3/20 15% | 10/50 20% | 25/100 25% | 60/200 30% |
| Amersham | Agilent | 4/10 40% | 8/20 40% | 17/50 34% | 35/100 35% | 78/200 39% |
The number of genes found common to the technologies in either the up- or down-regulated sets over the total number found in both sets by each technology and the percentage this represents is reported.
Analysis of variance
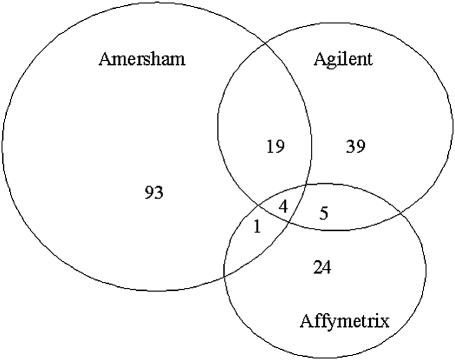
Using a Bonferroni correction for multiple tests (P < 2.489 E-05) resulted in one overlap in the lists of genes differentially expressed across three technologies. Therefore, we analyzed the ANOVA model using a more relaxed alpha to examine a higher power and less conservative approach often taken in knowledge discovery experimentation where false negatives may be more undesirable than false positives (12). Using an alpha of 0.001, we did observe larger overlaps between the gene lists, which are summarized in Table 3 (see also supplemental Table 6). Thus, the unions of differential gene sets that included the Agilent gene list showed the highest overall level of agreement (23 and 9) when compared to the union of the two oligomer probe based platforms, Affymetrix and Amersham; 5, (Fig. 5). The highest percent overlap observed (34%) was the percent of differentially expressed genes detected by Agilent that were also detected by the Amersham platform. Further analysis of the gene lists showed that there were no instances of conflicting results, i.e. genes that were determined to be up-regulated by one technology and down-regulated by another. Similar results were observed using an additional 2-fold change (in both directions) minimum criterion upon any genes found significant at a 0.001 alpha level and a reduced alpha cut-off of 0.01 with a 2-fold minimum criterion (Table 3, see also supplemental Tables 7 and 8).
Table 6. GO biological process categories represented in the lists of down-regulated genes detected by each platform (top 200 ranked by F statistics).
| Multiple comparison adjusted P-values | List hits | |||||
|---|---|---|---|---|---|---|
| GO Biological Process | Amersham | Agilent | Affymetrix | Amersham | Agilent | Affymetrix |
| Cell cycle | <0.01 | <0.01 | <0.01 | 86 | 48 | 35 |
| Nucleotide metabolism | <0.01 | <0.01 | 0.14 | 13 | 5 | 3 |
| DNA metabolism | 0.03 | <0.01 | 0.02 | 47 | 34 | 21 |
| Ribosome biogenesis | 0.03 | <0.01 | 1.00 | 6 | 5 | 2 |
| RNA metabolism | 0.19 | <0.01 | 1.00 | 29 | 13 | 8 |
| Transcription from Pol I promoter | 0.28 | 0.02 | 1.00 | 7 | 6 | 3 |
Figure 5.
Venn diagram of genes classified as differentially expressed by each platform using a mixed-model nested ANOVA and an alpha cut-off of 0.001.
Additionally, we applied a ‘sliding scale’ analysis to our data to explore the effect of using an alternative to pre-set alpha and fold change cut-offs. First, a directional F statistic was produced by giving the F statistic computed with the ANOVA model a negative sign if the calculated log ratio of the gene was negative. Genes were then classified as differentially expressed if this directional F statistic ranked among the top 100 (up-regulated) or among the bottom 100 (down-regulated) directional F statistics for each platform, altogether encompassing ∼10% of the common gene list and corresponding to a maximum P-value of 0.035. This analysis was repeated using the top 5, 10, 25, 50 and 100 ranked directional F statistics in each direction. The highest rate of agreement produced by this ‘sliding scale’ approach was 35% (Table 4), which is similar to the best rate of overlap to the method of using pre-set alpha and fold change cut-offs. When applying this sliding scale analysis to log ratio instead of the F statistic (Table 5), the highest rate of agreement was 40%; the lowest log ratio observed was 0.7 or ∼62% percent change.
Biological process themes
In addition to measuring cross-platform agreement on a gene-by-gene level, we also explored the level of concordance of the biological themes represented in the data across platforms. Determination of the biological processes over-represented in the differentially expressed gene lists of 200 of the top-ranking (by F statistics) down- and up-regulated genes was performed using the Expression Analysis Systematic Explorer (EASE) software system (13). We selected Gene Ontology (GO) biological process terms that had a multiple comparison adjusted EASE score <0.05 for each of the platforms. The multiple correction adjusted EASE P-value score was determined using a bootstrap method that calculates the probability of biological themes occurring by chance alone by permuting 100 randomly generated lists of genes of equal size to the original list of genes (13). We then condensed the biological process terms to the most common parent without going higher than the fourth GO level below biological process. Biological themes represented in the down-regulated gene lists showed some concordance; the highest rate of concordance observed in this analysis (4/6, 67%) was the set of biological process themes found by the Amersham and Agilent platforms, which included cell cycle, nucleotide metabolism, DNA metabolism and ribosome biogenesis (Table 6). The pattern of down-regulation of genes related to the cell cycle evident from data on all three technologies is consistent with additional experimental cell cycle analysis which indicated that ∼40% of PANC-1 cells growing in serum-containing medium were in the DNA synthesis phase while ∼15% of cells were in this phase after 24 h in serum-free medium (14). Among the up-regulated gene lists, only two predominating themes (carboxylic acid metabolism, organic acid metabolism) were detected with the Agilent platform, whereas Amersham and Affymeterix platforms showed no significant predominating biological theme. Similar results were obtained if up- or down-regulated genes were selected at an alpha cut-off of <0.05.
DISCUSSION
Presently, biomedical researchers using commercially available microarray assays can choose among a wide variety of products based upon different adaptations of the technology. Consistency of gene expression measurements across the different platforms would allow researchers to directly compare these measurements. Results of our unsupervised clustering and PCA analysis (see Supplementary Material), however, suggest that the largest variation in measurements from these commercial microarrays is attributable to the differences contributed by the platforms themselves. Additionally, our results indicate that the Pearson’s linear correlation for gene expression measurements across platforms was modest, ranging from 0.48 to 0.60. It is difficult to comment on which platforms might show greater similarity, since the interdependence in the correlational analysis precludes us from making a direct comparison across the platform pairs. For example: in Table 1, each of the measurements used in the correlation calculation in row 1 were used once in the calculations made in rows 2 and 3. This dependence between the datasets would confound any inferences we could make about differences in correlations Therefore, although we found slight variation in the linear correlations between data, determination whether differences in correlation were statistically significant could not be made.
While the study was mainly focused on evaluating the performance of individual microarray platforms, we unexpectedly found from our exploratory analysis of the datasets that substantial differences existed across platforms. After dichotomous classification of the genes for presence or absence of differential expression using a Bonferroni-corrected alpha, we observed virtually no intersection of the target gene sets. However, by using a less conservative alpha of 0.001, we were able to observe a non-random association between the classified data in comparisons between the cDNA(Agilent) and oligonucleotide technologies (Amersham and Affymetrix). Nevertheless, despite the greater similarities in the hardware and protocols utilized by the two oligonucleotide platforms, there was no significant agreement between classifications produced by Affymetrix and Amersham. Overall, using a variety of call criteria, our results show that the best level of agreement between the target gene sets was only 21% (between Amersham and Agilent, using a 2-fold and P < 0.001 criteria), when calculated as the intersection of two platforms as a percentage of total number of differentially expressed genes detected by both platforms. Thus, although the gene sets overlapped to some extent across platforms, the majority of genes identified as differentially expressed by each technology were uniquely identified by that technology. This result confounds interpretation of a target gene list found by any one of the microarray platforms. We feel that the low level of overlap between the gene lists indicates that the prognosis for the interchangeability of microarray platforms in this type of experiment is currently poor. However, when looking at a higher level of comparison such as biological themes on the 200 highest ranking down-regulated genes, it appeared that there was better concordance between the Agilent and Amersham platforms, suggesting that enough genes within distinct GO categories were detected by each platform to arrive at a common biological theme.
Previous cross-platform microarray studies also found considerable discordance between gene expression measurements (5,15). Kuo et al. observed little reproducibility of the hierarchical clusters of the NCI60 cell lines using gene expression measurements using oligonucleotide and cDNA microarray measurements. In contrast to this analysis of the NCI60 data, our current experimental design enabled us to examine the comparability of repeated measurements of a single RNA preparation where the only factor distinguishing these measurements was the type of microarray assay employed. With the experimental design presented here, we feel that the source of the discordance observed in our results may likely be attributable to differences inherent in each technology. Additionally, because of the greater level of experimental control within this study design, we expected a higher level of comparibility of our results than that observed in the previous analysis of the NCI60 data. Despite the removal of potential sources of variability, we arrive at a similar conclusion that the gene expression results depend to a large degree upon the type of microarrays used in the experiment.
Since there are alternative algorithms for normalization and probe level analysis of Affymetrix GeneChips, we investigated the possibility that use of these algorithms could improve the level of agreement between Affymetrix and other platforms. We reanalyzed the raw cel files from Affymetrix using two algorithms, selected because of their wide acceptance among Affymetrix GeneChip users: dChip (DNA-Chip Analyzer, PM-MM model) which uses model-based expression indexes (11) and RMA (robust multi-array analysis, default parameters) (16). Using the same mixed-model nested ANOVA method (P < 0.001), gene expression measurements arising from both algorithms slightly improved the cross-comparability of differential gene expression classifications for the Affymetrix platform when compared against the Amersham platform, but not against the Agilent platform (see supplemental Tables 9 and 10). Furthermore, the maximum agreement rate reached was not >11%. Interestingly, the results across three different algorithms within the Affymetrix platform showed a level of discordance similar to that observed for cross-platform comparisons (supplemental Table 11). Hence, the possible contribution of an algorithmic component of the Affymetrix platform towards the discordant gene expression measurements cannot be ruled out. However, it appears that the single probe platforms (Agilent and Amersham), despite having more straightforward analytical methods, were not more concordant.
Overall, the results of this study suggest that cross-platform differences arise from the intrinsic properties of the microarrays themselves, and/or the processing and analytical steps of these microarrays. Possible causes for platform-dependent differential gene expression results may include: probe sequence differences, variations in labeling and hybridization conditions and ultimately factors that derive from an overall lack of industrial standards across multiple technologies. In addition, the notion that the lack of concordance might in part be attributed to the detection of distinct types or sets of alternately spliced transcript variants among the technologies is one we are actively pursuing. Although we are continuing to study the contribution of probe sequence differences to the platform-dependent differential gene expression results, at present we only have full access to the Affymetrix sequences, and are negotiating full access to Codelink probe sequences. In addition, there is limited sequence information (last 100 bp on the 3′-end) for the cDNA probes from the Agilent platform, which would preclude us from making a more comprehensive sequence comparison. We believe that continued refinement of these technologies is necessary before measurements from various commercial technologies can be directly transformed to a universal gene expression index. Our study underscores the importance of follow-up verification of results from exploratory microarray experiments. Previous exploratory studies have found agreement between genes screened with microarray data and subsequent northern blot or real-time PCR verification of expression measurements of screened genes (17–19). In a similar manner, we are planning to use real-time PCR as an independent method for resolving the conflicting gene expression results.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
NOTE ADDED IN PROOF
While revising our manuscript, Barczak et al. (20) reported a study of gene expression measurements of identical RNA preparations using Affymetrix and long oligonucleotide arrays that found a high level of correlation between relative gene expression measurements made on each technology. However, a direct comparison of these two studies may not be appropriate since the results presented here do not include a long oligo format. Preliminary analysis of the probe sequences used in that study suggested a high degree of overlap, which we have yet to test with the platforms in this study.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Scott Vacha and Janet Herbert from Agilent Tehcnologies and Kari Wanat and Vipin Adhlakha from Amersham Biosciences for providing arrays and technical expertise, and to Dr Robert Nadon for helpful comments.
REFERENCES
- 1.Yang X., Pratley,R.E., Tokraks,S., Bogardus,C. and Permana,P.A. (2002) Microarray profiling of skeletal muscle tissues from equally obese, non-diabetic insulin-sensitive and insulin-resistant Pima Indians. Diabetologia, 45, 1584–1593. [DOI] [PubMed] [Google Scholar]
- 2.Chuaqui R.F., Bonner,R.F., Best,C.J., Gillespie,J.W., Flaig,M.J., Hewitt,S.M., Phillips,J.L., Krizman,D.B., Tangrea,M.A., Ahram,M., Linehan,W.M., Knezevic,V. and Emmert-Buck,M.R. (2002) Post-analysis follow-up and validation of microarray experiments. Nature Genet., 32, 509–514. [DOI] [PubMed] [Google Scholar]
- 3.Holloway A.J., Van Laar,R.K., Tothil,R.W. and Bowtell,D.D.L. (2002) Options available—from start to finish—for obtaining data from DNA microarrays II. Nature Genet., 32, 481–489. [DOI] [PubMed] [Google Scholar]
- 4.Edgar R., Domrachev,M. and Lash,A.E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuo W.P., Jenssen,T.K., Butte,A.J., Ohno-Machado,L. and Kohane,I.S. (2002) Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics, 18, 405–412. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y.H., Dudoit,S., Luu,P., Lin,D.M., Peng,V., Ngai,J. and Speed,T.P. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res., 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouck J., Yu,W., Gibbs,R. and Worley,K. (1999) Comparison of gene indexing databases. Trends Genet., 15, 159–162. [DOI] [PubMed] [Google Scholar]
- 8.Kerr M.K., Martin,M. and Churchill,G.A. (2000) Analysis of variance for gene expression microarray data. J. Comput. Biol., 7, 819–837. [DOI] [PubMed] [Google Scholar]
- 9.Wolfinger R.D., Gibson,G., Wolfinger,E.D., Bennett,L., Hamadeh,H., Bushel,P., Afshari,C. and Paules R.S. (2001) Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol., 8, 625–637. [DOI] [PubMed] [Google Scholar]
- 10.Churchill G.A. and Oliver B. (2001) Sex, flies and microarrays. Nature Genet., 29, 355–356. [DOI] [PubMed] [Google Scholar]
- 11.Cheng L. and Wong,W.H. (2001a) Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection, Proc. Natl Acad. Sci. USA, 98, 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadon R. and Shoemaker,J. (2002) Statistical issues with microarrays: processing and analysis. Trends Genet., 18, 265–271. [DOI] [PubMed] [Google Scholar]
- 13.Hosack D.A., Dennis,G., Sherman,B.T., Lane,H.C. and Lempicki,R.A. (2003) Identifying biological themes within lists of genes with EASE. Genome Biol., 4, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardikar A., Raaka,B., Geras-Raaka,E. and Gershengorn,M. (2003) Human pancreatic precursor cells can be induced to differentiate into beta cells, de-differentiate, proliferate and differentiate again. 63rd Scientific Sessions, American Diabetes Association, Diabetes, 52, A43. [Google Scholar]
- 15.Kothapalli R., Yoder,S.J., Mane,S. and Loughran,T.P. (2002) Microarray results: how accurate are they? Bioinformatics, 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irizarry R.A., Bolstad,B.M., Collin,F., Cope,L.M., Hobbs,B. and Speed,T.P. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res., 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka T.S., Jaradat,S.A., Lim,M.K., Kargul,G.J., Wang,X., Grahovac,M.J., Pantano,S., Sano,Y., Piao,Y., Nagaraja,R., Doi,H., Wood,W.H.,III, Becker,K.G. and Ko,M.S. (2000) Genome-wide expression profiling of mid-gestation placenta and embryo using a 15 000 mouse developmental cDNA microarray. Proc. Natl Acad. Sci. USA, 97, 9127–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniguchi M., Miura,K., Iwao,H. and Yamanaka,S. (2001) Quantitative assessment of DNA microarrays—comparison with Northern blot analyses. Genomics, 71, 34–39. [DOI] [PubMed] [Google Scholar]
- 19.Al Moustafa A.E., Alaoui-Jamali,M.A., Batist,G., Hernandez-Perez,M., Serruya,C., Alpert,L., Black,M.J., Sladek,R. and Foulkes,W.D. (2002) Identification of genes associated with head and neck carcinogenesis by cDNA microarray comparison between matched primary normal epithelial and squamous carcinoma cells. Oncogene, 21, 2634–2640. [DOI] [PubMed] [Google Scholar]
- 20.Barczak A., Rodriguez,M.W., Hanspers,K., Koth,L.L., Tai,Y.C., Bolstad,B.M., Speed,T.P. and Erle,D.J. (2003) Spotted long oligonucleotide arrays for human gene expression analysis. Genome Res., 13, 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.