Abstract
While molecular beacons are primarily known as biosensors for the detection of nucleic acids, it has proven possible to adapt other nucleic acid binding species (aptamers) to function in a manner similar to molecular beacons, yielding fluorescent signals only in the presence of a cognate ligand. Unfortunately, engineering aptamer beacons requires a detailed knowledge of aptamer sequence and structure. In order to develop a general method for the direct selection of aptamer beacons we have first developed a selection method for molecular beacons. A pool of random sequence DNA molecules were immobilized via a capture oligonucleotide on an affinity column, and those variants that could be released from the column by a target oligonucleotide were amplified. After nine rounds of selection and amplification the elution characteristics of the population were greatly improved. A fluorescent reporter in the selected beacons was located adjacent to a DABCYL moiety in the capture oligonucleotide; addition of the target oligonucleotide led to release of the capture oligonucleotide and up to a 17-fold increase in fluorescence. Signaling was specific for the target oligonucleotide, and occurred via a novel mechanism, relative to designed molecular beacons. When the target oligonucleotide is bound it can form a stacked helical junction with an intramolecular hairpin in the selected beacon; formation of the intramolecular hairpin in turn leads to release of the capture oligonucleotide. The ability to select molecular beacons may prove useful for identifying available sites on complex targets, such as mRNAs, while the method for selection can be easily generalized to other, non-nucleic acid target classes.
INTRODUCTION
Molecular beacons are oligonucleotide probes that assume a hairpin structure in which the single-stranded loop can pair with complementary sequences and the paired stem contains fluorescent reporters (or a fluorophore and a quencher) that interact with one another (1). Hybridization of a complementary target sequence leads to the formation of a long duplex region, destabilization of the hairpin, and a spatial separation between the two dyes. Ultimately, interaction with target oligonucleotides leads to either the loss of fluorescence resonance energy transfer (FRET) or to dequenching of a fluorophore, optical signals that can be readily detected. An alternate form of molecular beacons called tripartite molecular beacons have been engineered by Nutiu and Li (2). The hairpin–stem of the tripartite molecular beacon does not have the fluorophore and quencher directly attached to it, but rather is extended by two universal single-stranded arms that bind single-stranded oligonucleotides having fluorophore or quencher attached to them. The simplicity of the transduction mechanism and the corresponding ease with which molecular beacons can be designed has led to their adoption in a variety of applications, including identifying single nucleotide polymorphisms (3–5), detecting pathogens (6,7), monitoring the amplification of nucleic acids during real-time PCR (8,9), and detecting DNA–RNA hybridization in real-time in living cells (10,11). Immobilized molecular beacons have also been used to detect DNA–RNA hybridization (12,13), including in DNA arrays (14). The oligonucleotide-dependent conformational changes that characterize molecular beacons have also been engineered into deoxyribozymes, yielding oligonucleotide-dependent changes in catalytic activity, or so-called catalytic molecular beacons (15,16).
However, while molecular beacons can detect nucleic acid targets with high specificity and with single mismatch discrimination, their ability to function as biosensors for the detection of analytes other than nucleic acids has so far been relatively limited. Molecular beacons have been used to probe the interactions of known nucleic acid binding proteins with single-stranded DNA; for example, single-stranded DNA binding proteins can open a molecular beacon as well or better than a complementary target (17,18). However, these approaches have not proven to be generalizable. Sensors similar to molecular beacons have also been adapted to the detection of proteins that interact with double-stranded DNA targets (19). In this instance, the DNA binding protein assembles two sub-fragments that contain different dyes, leading to a FRET signal.
In comparison, it has proven possible to select nucleic acid binding species (aptamers) that can bind to a wide variety of targets, from small organic molecules to large supramolecular structures (20–22). Aptamers can potentially be adapted to function as nucleic acid biosensors in a variety of ways (reviewed in 23,24). For example, aptamers frequently undergo small ligand-induced conformational changes. When fluorescent labels were introduced into conformationally labile positions in anti-adenosine aptamers, the resultant ‘signaling aptamers’ showed ATP-dependent increases in fluorescence intensity and could track the concentration of free ATP in solution (25). Larger conformational changes can also be exploited. An anti-thrombin DNA aptamer was known to assume an equilibrium between random coil and quadruplex structures. By labeling the aptamer with either a fluorophore and a quencher or two fluorophores (26), addition of thrombin shifted the equilibrium to the quadraplex conformer and resulted in fluorescence quenching or FRET.
In addition to exploiting the inherent conformational changes that aptamers undergo, aptamers that are similar to molecular beacons have been generated by engineering the aptamer such that the addition of an analyte resulted in a large conformational change and concomitant diminution or increase in a fluorescent signal. In a strategy similar to that described above for the thrombin biosensor, an anti-cocaine aptamer was mutated and its secondary structure destabilized; the addition of cocaine resulted in stem formation and fluorescence quenching (27). In contrast, the addition of sequences to the anti-thrombin DNA quadruplex forced the adoption of an alternate, hairpin conformation, similar to the hairpin or ‘closed’ conformation of a molecular beacon. Upon addition of thrombin, the aptamer resumed its quadruplex conformation, splitting apart an appended fluorophore and quencher and yielding a fluorescent signal (28). Finally, even quaternary structural changes have been exploited to create aptamer biosensors. An anti-Tat aptamer was split into two pieces, one of the pieces was converted into a molecular beacon, and the Tat-dependent reassembly of the aptamer resulted in the opening of the beacon and the generation of a fluorescent signal (29). Anti-cocaine and anti-rATP aptamers have also been converted into fluorescent sensors for their respective analytes using a similar strategy of target-mediated assembly, although in this instance analyte binding leads to hairpin stem formation and hence to fluorescence quenching (30). Recently, Nutiu and Li employed a similar strategy to convert anti-ATP and anti-thrombin aptamers into signaling aptamers. In this case, an antisense oligonucleotide bound to, denatured, and quenched a fluorescently labeled aptamer. Target binding stabilized the native conformation of the aptamer and resulted in fluorescence dequenching (31).
However, all of these methods ultimately rely upon engineering known aptamers to generate signals and frequently require a prior knowledge of the detailed secondary or tertiary structure of the aptamer. Even when these details are known, multiple, different constructs must frequently be generated in order to identify those that signal (or that signal most efficiently). If possible, it would be much simpler to directly couple selection with signaling. We have previously employed selected aptamers from pools that randomly incorporated fluorescent nucleotide derivatives, and that therefore pre-positioned fluorophores into functional nucleic acid structures (32). However, since the selected pool could still only be selected for binding, rather than for signaling, the selected aptamers had to be individually screened to identify those that could not only bind but signal. Not surprisingly, not all high-affinity binding species were also good at signal transduction; only one family of aptamers was found to have significant signaling abilities.
In order to generalize the utility of molecular beacons to analyte classes other than nucleic acids, there was a continuing need for a method for the direct selection of signaling aptamers. To this end, we developed a method that directly couples selection for ligand binding to a nucleic acid conformational change that in turn leads to the production of a fluorescent signal. As a proof-of-principle, we have directly selected molecular beacons that are responsive to oligonucleotide effectors. The selected molecular beacons have properties similar to designed molecular beacons, but utilize a quite novel mechanism for signaling. The success of the method should allow its application to a variety of ligand classes, while the novel molecular beacons may find new applications in their own right.
MATERIALS AND METHODS
Synthetic DNA
All oligonucleotides were either made in our laboratory on an Expedite 8909 DNA synthesizer (PE Biosystems, Foster City, CA) using synthesis reagents purchased from Glen Research (Sterling, VA), or were ordered from Integrated DNA Technologies (Coralville, IA).
A single-stranded DNA pool containing 20 randomized positions (N20: 5′-GTCACTGTCTTCATAGGTTG-N20-GAATCAGTGAGACATCCC-3′) was synthesized using previously reported methodology (33), and was used as a starting point for in vitro selection. The pool was amplified using primers 20n.20 (5′-GTCACTGTCTTCATAGGTTG-3′) and 38.20 (5′-TTCTAATACGACTCACTATAGGGATGTCT CACTGATTC-3′), where the underlined residues indicate the non-transcribed portions of a T7 RNA polymerase promoter. A primer that contained biotin at its 5′ end, 18.20 (5′-Biotin-GGGATGTCTCACTGATTC-3′), was used instead of 38.20 during later rounds of selection.
In experiments designed to optimize the length of the capture oligonucleotide, four different biotinylated oligonucleotides that were complementary to the N20 5′ constant region were utilized: 7.oa20 (5′-AGTGACT-Biotin-3′), 13.oa20 (5′-GAAGACAGTGACT-Biotin-3′), 15.oa20 (5′-TATGAAGACAGTGAC-Biotin-3′) and 19.oa20 (5′-CAACCTATGAAGACAGTGA-Biotin-3′). Oligonucleotides 7.oa20 and 13.oa20 had an additional residue at their 3′ ends to reduce steric interference between biotin and streptavidin.
For selection experiments, a fluorescein-dT residue (Glen Research) was incorporated at the 11th position of 20n.20 (20.11f: 5′-GTCACTGTCTTCATAGGTTG-3′), where the underlined residue corresponds to the site of insertion. DABCYL (Glen Research) was incorporated into the capture oligonucleotide at its 5′ end (q13.20: 5′-DABCYL-GAAGACAGTGACT-Biotin-3′). Two 16mer oligonucleotides, ot1.20 (5′-ATGCGATCTAGTCTGC-3′) and ot2.20 (5′-TAGCACGTCTGATCTC-3′) were used as selection targets. In those instances where negative selections were applied, five different oligonucleotides were used: RO1.18 (5′-GTAG TGCTCCGTGGATTG-3′), RO2.20 (5′-TCGAGGGAGAGC CATACCTG-3′), RO3.20 (5′-TGCATGAGGATGCAGG ATGC-3′), RO4.20 (5′-ATTGATGAGTCTGACTGCCT-3′) and RO5.19 (5′-GCGACTGGACATCACGAGA-3′).
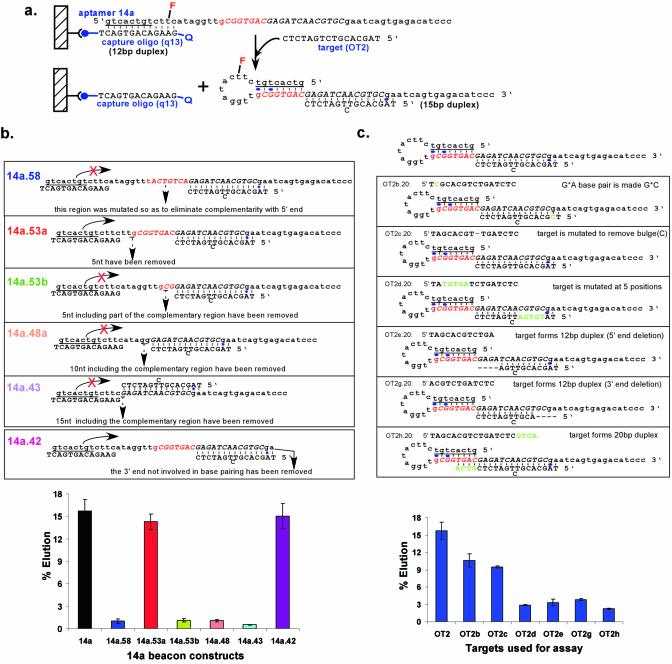
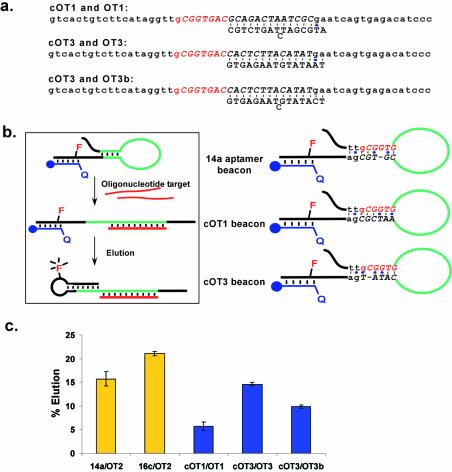
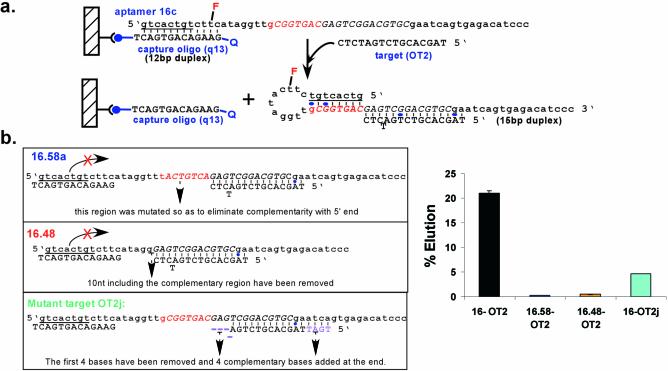
Variants of selected beacons 14a and 16c were designed and synthesized to test the mechanism of oligonucleotide-dependent elution. For beacon 14a, these included the oligonucleotides 14a.58, 14a.53a, 14a.53b, 14a.48a, 14a.43 and 14a.42 and for beacon 16c, the oligonucleotides 16.58 and 16.48. In addition, variant target oligonucleotides were assayed, including OT2b.20, OT2c.20, OT2d.20, OT2e.20, OT2g.20, OT2h.20 and OT2j.20. In order to test the generality of the signaling mechanism, a beacon cOT1 was designed that was complementary to oligonucleotide target OT1, and a beacon cOT3 was designed that was complementary to an unrelated 16mer oligonucleotide target, OT3 or OT3b. The sequences of these various oligonucleotides have been listed in Figures 4b and c, 5a and 6b.
Figure 4.
Mechanism of elution for beacon 14a. (a) Proposed mechanism of elution. Representations are as in Figure 3c. Hybridization of the oligonucleotide target OT2 stabilizes the formation of a hairpin stem and disrupts interactions with the capture oligonucleotide. (b) Assaying the mechanism of elution with beacon variants. Beacons designed to assess interactions with OT2 or the formation of the intramolecular hairpin (underlined) are shown. The elution characteristics of these constructs with target OT2 were assessed as in Figure 3a. (c) Assaying the mechanism of elution with target oligonucleotide variants. Targets designed to assess interactions between OT2 and beacon 14a are shown. The elution characteristics of beacon 14a with the target variants were assessed as in Figure 3a.
Figure 5.
Designed molecular beacons. (a) Based on the proposed elution mechanism for beacon 14a, two molecular beacons (cOT1 and cOT3) and three target oligonucleotides (OT1, OT3 and OT3b), were designed. (b) Proposed role of a hypothesized, immobilized secondary structure in the mechanism of elution. The hypothesized secondary structures of the selected and designed beacons are shown. (c) The elution characteristics of beacons cOT1 and cOT3 with targets OT1, OT3 and OT3b were assessed as in Figure 3a. The representations are as in Figure 3c.
Figure 6.
Mechanism of elution for beacon 16c. (a) Proposed mechanism of elution. Representations are as in Figure 3c. (b) Assaying the mechanism of elution. Beacons and target oligonucleotides designed to assess the mechanism of elution are shown. The elution characteristics of beacon 16c were assessed as in Figure 3a.
The wavelength-shifting molecular beacon constructs were 14mb.58 (5′-DABCYL-GTCACTGTCTTCATAGGTTGCG GTGACGAGATCAACGTGCGAATCAGTGAGACATCC C-3′) and 16mb.58 (5′-DABCYL-GTCACTGTCTTCATAG GTTGCGGTGACGAGTCGGACGTGCGAATCAGTGAG ACATCCC-3′). In addition to the 5′ DABCYL, these constructs contained a fluorescein-dT at position 11 and an amino modified-dC (Glen Research) at the 27th position, as indicated by underlines.
N20 pool construction
A single-stranded DNA pool was generated by a combination of chemical synthesis, PCR amplification, in vitro transcription and reverse transcription. Following chemical synthesis, the N20 DNA pool was purified on an 8% denaturing polyacrylamide gel. The gel-purified pool (32 µg) was amplified in a 25 ml PCR using the non-fluoresceinated primers 20n.20 and 38.20. Only 15% of the initial pool could be extended by Taq DNA polymerase. However, since the theoretical pool size was relatively small (420 = 1.1 × 1012 molecules), there should have been an average 135 copies of each species in the pool. The PCR yielded over 2000 pool equivalents. Primer 38.20 contained a T7 RNA polymerase promoter, and 65 pool equivalents were transcribed using the Ampliscribe T7 In Vitro Transcription Kit (Epicenter Technologies, Madison, WI). The resultant RNA was gel purified on an 8% denaturing polyacrylamide gel. Greater than 2000 RNA pool equivalents were reverse transcribed with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) using the fluoresceinated 5′ primer, 20.11f, in a 500 µl reverse transcription reaction. The reverse transcription reaction was performed according to the protocol provided with the SuperScript II reverse transcriptase enzyme using 5 µg of RNA template per 20 µl reverse transcription reaction. The cDNA:RNA duplexes were digested with RNase A (Ambion, Austin, TX) and Ribonuclease H (Ambion) (37°C, 25 min; 80°C, 1 min; 37°C, 30 min) to remove template RNA, and the remaining fluoresceinated cDNA was purified on an 8% denaturing polyacrylamide gel.
Optimization of capture oligonucleotide length
The nascent, single-stranded DNA pool was 5′ end-labeled with T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP (2.0 mCi, 7000 Ci/mmol; ICN Biomedicals, Costa Mesa, CA). Following gel purification, 50 pmol of the labeled pool were annealed with 100 pmol of the biotinylated capture oligonucleotide 7oa.20 in a 20 µl reaction volume. The annealing reaction was heated at 94°C for 30 s and 45°C for 90 s, and was then cooled to room temperature. The annealing reaction was diluted to 500 µl using binding buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2), and the bound pool was captured on streptavidin–agarose (Sigma-Aldrich, St Louis, MO) over a period of 25 min. The streptavidin–agarose was transferred to a column (Bio-Rad, Hercules, CA), and the amount of radioactivity in the eluant was determined using a scintillation counter. The column was washed 10 times with 1 ml of binding buffer, fractions were collected, and the amount of radioactivity in the fractions was determined. Similar assays were carried out with the other capture oligonucleotides, 13.oa20, 15.oa20 and 19.oa20. The total amount of pool that could be recovered from the capture oligonucleotide columns was determined by carrying out similar experiments in parallel, except that the columns were eluted with denaturing buffer (7 M urea, 0.1 M sodium citrate, 3 mM EDTA, pH 5).
In vitro selection
To initiate the selection, the fluoresceinated, single-stranded N20 DNA pool (100 pool equivalents) was annealed with a 2-fold molar excess of the biotinylated capture oligonucleotide q13.20 in 100 µl of water. The annealing reaction was heated to 94°C for 30 s and 45°C for 90 s, and was then cooled to room temperature. The capture oligonucleotide and bound pool were immobilized on streptavidin–agarose (Sigma-Aldrich) and transferred to a column. The column was equilibrated with selection buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2) by repeated washing. An equimolar mixture of the two oligonucleotide targets in selection buffer was heat denatured, cooled to room temperature, added to the column containing the immobilized pool, and allowed to react for 10 min at room temperature with occasional mixing. The column was then drained and washed three times with 200 µl aliquots of binding buffer. All the eluates were collected and the eluted DNA was precipitated with ethanol. The eluted DNA was amplified by the PCR, transcribed to generate RNA, and reverse transcribed. The cDNA was purified by RNase digestion and gel purification, as before, and was used for the next round of selection.
Seven rounds of selection and amplification were performed as described above, except that the number of washes that occurred prior to elution was successively increased. The pool to target ratio was also increased from 1:1 in the first three rounds to 2:1 in the next four rounds. The increased competition of pool molecules for targets should have increased the stringency of the selection. During the eighth and ninth rounds of selection, a negative selection step was introduced. The immobilized pool was first incubated with a mixture of five different oligonucleotides (RO1.18, RO2.20, RO3.20, RO4.20 and RO5.19) that bore no sequence similarity to the targets. Any pool members that eluted with these random oligonucleotides were discarded. The remaining pool was then eluted with the two target oligonucleotides, except that the pool to target ratio was again increased, to 5:1 in the eighth round and to 10:1 in the ninth round. Also, instead of amplifying selected molecules via transcription and reverse transcription, DNA species from Rounds 7 and 8 were PCR-amplified with the biotinylated primer 18.20 and the fluoresceinated primer 20.11f, the double-stranded PCR products were captured on streptavidin–agarose, and fluoresceinated, single-stranded DNA molecules were eluted with 0.2 N NaOH. These eluates were immediately neutralized by adding 3 M NaOAc (pH 5.2) and precipitated with ethanol. While this latter, faster method for single-strand DNA preparation could have been used in earlier rounds as well, the reverse transcription method was adopted because it gave consistently better yields of single-stranded DNA (often >70% of input) and therefore helped to maximize the recovery of the amplified single-strand DNA pool during the early rounds of selection.
Ultimately, the Round 9 selected pool was cloned (TA Cloning Kit; Invitrogen) and sequenced using the Dye Terminator Cycle Sequencing Kit (Beckman Coulter, Fullerton, CA) and a CEQ 2000 XL DNA sequencer (Beckman Coulter) (34).
Binding assays
Following Rounds 5, 7 and 9, the amplified, single-stranded DNA pools were 5′ end-labeled using T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP (2.0 mCi, 7000 Ci/mmol; ICN Biomedicals). Binding assays were performed in a manner similar to the selection experiments themselves, except that fractions were collected for scintillation counting. In short, 50 pmol of gel-purified, labeled single-stranded DNA pool were annealed with 100 pmol of the capture oligonucleotide q13 in a 50 µl reaction, as described above. The radiolabeled pool was immobilized on 60 µl of streptavidin–agarose (Sigma-Aldrich) and the unbound fraction was collected. The column was washed three times with 300 µl of selection buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2) and the washes were again collected. A mixture of the two oligonucleotide targets in binding buffer was prepared such that each target would be at the same final concentration as the pool. The oligonucleotide targets were heat denatured and added to the immobilized pool in a total volume of 200 µl. As with the selections, binding reactions were incubated for 10 min prior to washing the column two times with 10× column volumes of binding buffer. All the eluants and the remaining solid resins were preserved, radioactivity was quantitated using a scintillation counter, and the proportions of the pools that were specifically eluted by target oligonucleotides were determined.
Binding assays with individual, selected beacons and designed variants were also performed as described above. The designed variants 14a.58, 14a.53a, 14a.53b, 14a.48a, 14a.43, 14a.42, 16.58 and 16.48 were all assayed for their ability to be eluted by target OT2. Beacon 14a was assayed for its ability to be eluted by targets OT2b.20, OT2c.20, OT2d.20, OT2e.20, OT2g.20 and OT2h.20. Beacon 16c was assayed for its ability to be eluted by target OT2j.20. Similarly, the designed beacons cOT1 and cOT3 were assayed for their ability to be eluted by their respective targets, OT1 and OT3 (or OT3b).
Construction of doubly labeled molecular beacons
The two amine-modified beacon constructs, 14mb.58 and 16mb.58, were conjugated to the succinimydyl ester derivative of Texas Red-X (Molecular Probes, Eugene, OR). The coupling protocol provided with the dye was used, with the exceptions that the oligonucleotide solution was heat denatured at 75°C for 3 min prior to setting up the reaction, and the coupling reaction was carried out at a 4:1 dye:oligonucleotide ratio. After incubation for 12–16 h at room temperature, the labeled oligonucleotides were ethanol precipitated two times and then gel purified on a 12% denaturing polyacrylamide gel.
Beacon preparation for fluorescence measurements
Beacons were generated by reverse-transcribing RNA with Superscript II reverse transcriptase at 42°C for 55 min. Template RNA was removed by ribonucleolytic degradation with several different ribonucleases. RNase A and Ribonuclease H were first added to the reverse transcription reaction and the mixture was incubated at 37°C for 30 min, followed by 85°C for 90 s and 37°C for 2 min. Next, RNase I (Ambion) and Riboshredder (Epicenter Technologies) were added and the sample was further incubated at 37°C for 1 h. The single-stranded DNA was extracted with a mixture of phenol–chloroform, then chloroform alone, gel purified on a 10% denaturing polyacrylamide gel, eluted overnight and ethanol precipitated.
Fluorescence measurements
All fluorescence measurements were made on a PTI Quantamaster QM-4/2003SE spectrofluorimeter (Photon Technology International, Ontario, Canada). The beacons (50 nM final concentrations) were annealed with capture oligonucleotide (100 nM) in selection buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 10 mM MgCl2) by heating to 94°C for 30 s and 45°C for 90 s, and cooling to room temperature over 10 min. Background fluorescence was first measured by adding 480 µl of selection buffer to a fluorimeter cell. The beacon–capture oligonucleotide complexes (10 µl) were then added and fluorescence was monitored over time. Once a steady fluorescent signal had been achieved, a heat-denatured solution of oligonucleotide target (10 µl) was added. The fluorescence response was monitored over 15 min by exciting the samples at 494 nm (the λex for fluorescein) and measuring the fluorescence intensity at 518 nm (the λem for fluorescein). The signal-to-background ratio was calculated as:
I = (Fopen – Fbuffer) / (Fclosed – Fbuffer)
where Fopen was the fluorescence of the beacon–capture oligonucleotide complex (14a or 16c hybridized to q13) in the presence of target, Fclosed was the fluorescence of the beacon–capture oligonucleotide complex in the absence of target, and Fbuffer was the background fluorescence of the buffer solution alone.
The limits-of-detection for the selected molecular beacons were calculated by plotting I versus concentration at 15 min in the linear response range (0–100 nM target oligonucleotide). Best-fit lines were calculated, and the limits-of-detection were taken to be the concentration value at which I = 3, a value previously used by Liu and Tan (12) and Zhang et al. (35).
For the doubly labeled molecular beacons, the real time fluorescence response was monitored for both fluorescein and Texas Red (λex = 595 nm and λem = 615 nm) at each concentration of target oligonucleotide. The fluorescence response of each fluorophore was measured separately by exciting the beacon–capture oligonucleotide complex in the presence of various concentrations of target oligonucleotide at the excitation maxima of the fluorophore and recording the emission at the emission maxima. The value of I was evaluated after 15 min and the percent change in fluorescence was normalized according to the equation:
I = (I – Imin) / (Imax – Imin)
where I was the value at any particular target concentration, Imin was the minimum value of I (i.e. the emission of fluorescein at 0 nM target and of Texas Red at 1000 nM target), and Imax was the maximum value of I (i.e. the emission of fluorescein at 1000 nM target and of Texas Red at 0 nM target). The data set for fluorescein was fit to the equation:
Y = AX / (X + B)
where Y was the percent change in the fluorescence of fluorescein at a given target concentration, X was I, A was the increase in the fluorescence of fluorescein at saturating target concentrations, and B was the apparent dissociation constant value. Conversely, the data set for Texas Red was fit to the equation:
Y = 100 + [AX / (X + B)]
where Y was the percent change in the fluorescence of Texas Red at a given target concentration, X was I, A was the decrease in the fluorescence of Texas Red at saturating target concentrations, and B was the apparent dissociation constant value.
RESULTS AND DISCUSSION
Design and optimization of molecular beacon selection
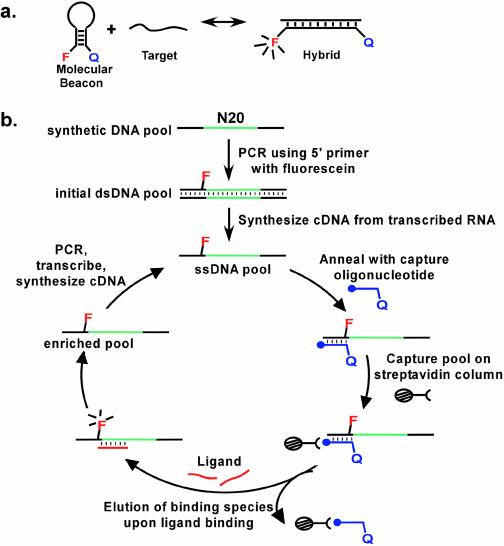
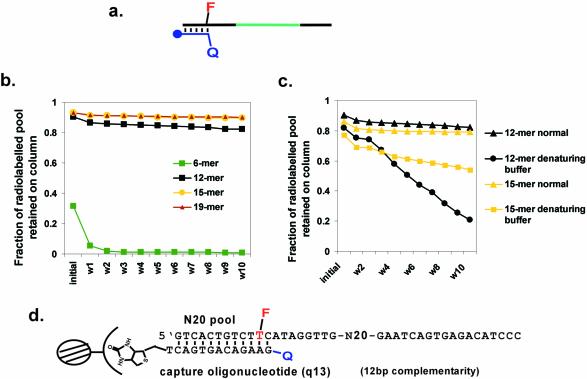
Most molecular beacons rely upon the hybridization of an oligonucleotide to the hairpin loop of a stem–loop structure, which in turn results in a conformational change that separates a fluorescent reporter from an adjacent quencher (Fig. 1a). Molecular beacons have primarily been designed to recognize oligonucleotides, although it has also been proven possible to engineer protein-dependent conformational changes (17–19). The generality of extending molecular beacon design to other types of analytes is currently unclear. Therefore, we have attempted to develop a method for the direct selection of molecular beacons from a random sequence population (Fig. 1b). To this end, we wanted to select for the analyte-dependent separation of an oligonucleotide from its complement. An oligonucleotide was immobilized on a column, and a DNA pool was hybridized to the oligonucleotide via one of its constant sequence regions (Fig. 1b). It was anticipated that upon the addition of an analyte that some species would undergo an analyte-dependent conformational change that would in turn disrupt the complementary interaction and result in the release of the species from the oligonucleotide affinity column. Those species that were released could be collected, amplified and carried into additional rounds of selection.
Figure 1.
Selection scheme for molecular beacons. (a) Conformational changes in designed molecular beacons. F represents an embedded fluorophore, Q a quencher. (b) In vitro selection of molecular beacons. The closed circle at the termini of the capture oligonucleotide represents biotin. The selection protocol is described in greater detail in Materials and Methods, and Results and Discussion.
The strength of the interaction between the oligonucleotide affinity column and the pool should in large measure determine whether and what kind of analyte-dependent conformational change could be selected. Therefore, we first attempted to poise the complementary interaction so that the DNA pool might be readily released from the affinity column following interactions with an analyte. Oligonucleotide affinity columns of different lengths were constructed, and their abilities to capture and release DNA pools containing complementary constant regions were determined (Fig. 2a). The capture oligonucleotides were complementary to 6, 12, 15 or 19 residues in the constant region of the pool. The shortest hybridization interaction, 6 bp, was insufficient to hold the pool on the column at room temperature. However, capture oligonucleotides that formed 12, 15 or 19 bp with the pool were all effective in immobilization (Fig. 2b). The elution profiles of the 12 and 15 residue capture oligonucleotides with appended DABCYL were further investigated by eluting the captured pools with denaturing buffer to determine the total amount of pool that can be recovered (Fig. 2c). Since the 12 bp interaction could be more easily disrupted, the corresponding capture oligonucleotide was chosen for selection experiments (Fig. 2d).
Figure 2.
The effect of oligonucleotide length on affinity column retention. (a) The capture oligonucleotide was designed to be complementary to the 5′ end of the pool. Symbols are as in Figure 1. (b) Comparison of four different capture oligonucleotides. W1–W10 indicate fractions obtained after washing the column with one column volume of selection buffer. (c) Residual retention of the pool on the column. To determine the extent of elution with the 12mer and 15mer capture oligonucleotides the experiments described in (b) were repeated, except that denaturing buffer containing 7 M urea was used. Fractions are the same as in (b). (d) The sequences adopted for selection experiments. F indicates fluorescein, while Q indicates a pendant DABCYL.
Selection of beacons that elute with oligonucleotide targets
A single-stranded DNA pool containing 20 randomized positions (N20 pool) was used as a starting point for selection. A relatively small random sequence region was employed in order to make the eventual analysis of any selected molecular beacon mechanisms more straightforward. A fluorescent reporter was introduced into the pool via a 5′ primer that contained a fluorescent thymidine residue at position 11 (T11), in which fluorescein was conjugated to position 5 of the nucleobase. In each round of selection, the fluorescent, single-stranded DNA pool was annealed to the 12 residue capture oligonucleotide and the duplex was in turn immobilized on streptavidin–agarose. The capture oligonucleotide also had a fluorescence quencher (DABCYL) at its 5′ end. Upon hybridization, the DABCYL was in proximity to the fluorescein on the pool (Fig. 2d).
The column containing the immobilized pool was washed several times with buffer to remove any unbound or poorly bound species. The pool was then eluted from the column with two, different 16mer target oligonucleotides, OT1 and OT2. Neither oligonucleotide had any sequence similarity with the 5′ and 3′ constant regions of the pool and any eluted products would therefore presumably have to undergo a conformational change that would lead to release from the capture oligonucleotide. The species eluted by the target oligonucleotides were collected, amplified and carried into the next round of selection.
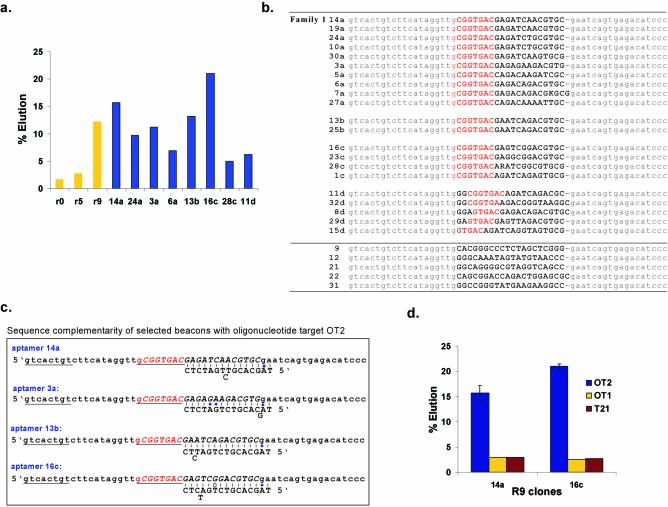
After seven rounds of selection and amplification, only a small improvement was observed in target-dependent elution. An additional negative selection step was therefore introduced into the eighth and ninth rounds of selection. The pool was first incubated with a set of five different oligonucleotides, all 18–20 nt long and bearing no resemblance to the two target oligonucleotides used for elution. It was hypothesized that pre-elution with these non-target oligonucleotides would improve the eventual sequence selectivity of any selected beacons. The selection was then carried out as before with the two specific oligonucleotide targets, except that the ratio of the targets to the single-stranded DNA pool was progressively decreased so as to increase the stringency of the selection (beacons would have to compete more fiercely for targets). As expected, the Round 9 pool showed significant target-specific elution from the column (Fig. 3a).
Figure 3.
Progress of the selection. (a) Binding assays with Round 9 beacons. The horizontal axis indicates the percents of the pools or beacons that were specifically eluted with target from an oligonucleotide affinity column. Yellow bars show elution with a mixture of oligonucleotide targets OT1 and OT2, while the blue bars show elution with oligonucleotide target OT2 alone. (b) Sequences of Round 9 beacons. The constant primer binding regions are shown in lower-case gray while the random region has been shown in upper-case bold. The common octamer motif is shown in bold red. (c) Beacon sequence complementarity. Predicted base pairings to OT2 are shown. A stem that is predicted to form between the constant region and the common octamer (red) is shown underlined. The constant regions have been shown in lower-case and the random region has been shown in upper-case and italicized. (d) Specificity of elution. The total amount of beacons 14a and 16c that were eluted from an oligonucleotide affinity column with an equimolar amount of the oligonucleotide targets, OT2, OT1 and T21 is shown.
Twenty six individual beacons from Round 9 were cloned and sequenced (Fig. 3b). Most (21) of the selected beacons contained from five to seven common residues at or near the 5′ end of the random region (Family 1). Some beacons differed from one another by only one residue, and may have been derived from a common ancestor (e.g. compare clones 16c and 23c). Other beacons shared a core of sequence similarities, but otherwise differed at several positions (e.g. compare clones 19a, 24a and 3a). A number of outlier sequences were also still present in the population.
Selected sequences that contained the heptamer motif were also generally complementary to OT2 but not to OT1 (Fig. 3c), and in fact the beacons were specifically eluted with OT2, as opposed to OT1 or another unrelated oligonucleotide target, T21 (Fig. 3d). While we could not have predicted this particular behavior in advance, this was why two different oligonucleotide targets were originally used during the course of the selection: to allow the evolution of either specificity or a lack of specificity for oligonucleotide targets, and to determine whether some oligonucleotide targets were more amenable to selecting beacons than others. The two beacons, 14a and 16c, which showed the greatest elution with the oligonucleotide target OT2 were chosen for further analysis.
Mechanism of oligonucleotide-dependent elution
The selected heptamer motif and a single residue from the constant region comprised an octamer motif that could potentially form a stem–loop structure with the constant region. The predicted stem–loop (Fig. 3c) should in turn disrupt hybridization to the capture oligonucleotide. A possible mechanism for target-dependent elution might therefore be binding by the target oligonucleotide and concomitant re-folding of the 5′ constant region to form a stacked helical junction (Fig. 4a).
To further investigate the mechanism of elution, beacon variants were designed based on the beacons 14a (Fig. 4a and b) and 16c (Fig. 5a and b). When the octamer motif within beacon 14a (5′-GCGGTGAC) was mutated to eliminate potential complementarity with the 5′ constant region (construct 14a.58), the beacon could no longer be eluted by the target oligonucleotide (Fig. 4b). Similarly, deletion constructs (14a.53b, 14a.48a and 14a.43) that partially or completely removed the octamer motif could no longer be eluted by the target oligonucleotide. In contrast, when five residues outside the octamer motif were deleted (14a.53a), the elution characteristics of the beacon remained almost unchanged.
Mutant target oligonucleotides were also assayed for their ability to elute beacons (Fig. 4c). When the predicted complementarity of the target oligonucleotide was mutated either by changing five residues in tandem (OT2d.20) or by deleting residues (OT2e.20 and OTg.20) the extent of elution was decreased, further confirming that helix formation between the target oligonucleotide and the beacon was important for elution. In addition, when the predicted complementarity of the target oligonucleotide was extended into the region required for the formation of the predicted hairpin stem (OT2h.20), the extent of elution was decreased. These latter results were consistent with the postulated model, since any new base pairs that were formed with the target oligonucleotide would have interfered with the formation of the stem–loop and stacked helical junction, and thus with the dissociation of the selected beacon from the capture oligonucleotide.
It is interesting to note that ∼3% of the wild-type beacon was eluted with mutant target oligonucleotides, while much lower levels of mutant beacons were eluted with the wild-type target oligonucleotide. These results are consistent with the finding that the wild-type beacon has a background elution rate of 3% (data not shown). Mutant beacons are less able to undergo the selected conformational change, and thus should show a lower intrinsic elution rate, irrespective of the nature of the target oligonucleotide.
While almost all results were consistent with the proposed structural hypothesis, a few substitutions could not be completely rationalized. Target oligonucleotide OT2b.20 contained an A to C substitution that should have replaced a postulated G:A base pair in beacon 14a with a more stable G:C base pair (Fig. 4c). Similarly, target oligonucleotide OT2c.20 was mutated to delete a predicted bulged base (Fig. 4c). In both instances, the fractions of eluted beacons unexpectedly decreased by slight amounts. These results indicate that a secondary or tertiary structure more complex than a simple Watson–Crick paired duplex may assist in the formation of the stacked helical junction.
It is also possible that the secondary structure of the aptamer beacons immobilized on the capture oligonucleotide may contribute to the mechanism of elution. For example, when the beacon is annealed to the capture oligonucleotide, the remaining non-paired portion of the beacon might fold to form a stem–loop structure that presents the oligonucleotide binding site (much as a regular molecular beacon folds to present its oligonucleotide binding site). If so, oligonucleotide binding and thus elution might be facilitated. For the selected beacon 14a, it was possible that the sequences ttgCGGTG and CGTCGga (that encompass the 5′ and 3′ ends of the random region, respectively) might pair with one another.
To test this hypothesis and the generality of our proposed mechanism, we designed two additional aptamer beacons, one that was complementary to target OT1 (designated cOT1) and the other to a new and unrelated 16mer sequence OT3 (designated cOT3; Fig. 5a). Beacon cOT1 was designed to form a stronger hypothesized stem structure; beacon cOT3 was designed to not form a stem structure (or to form an extremely weak structure, see Fig. 5b). Both beacons showed oligonucleotide-specific elution (>4-fold above background elution), although they eluted less well than the original, selected beacons (Fig. 5c). A variant of target oligonucleotide OT3 (designated OT3b) was also assayed, and eluted cOT3 less well than target oligonucleotide OT3. Similarly, a number of the originally selected molecular beacons should have formed the same hypothesized stem structure, but showed very different elution characteristics with the same oligonucleotide target, OT2 (e.g. compare 16c and 24a). These results seem to indicate that the way in which an oligonucleotide binding site was presented was much less important for elution than the sequence of the oligonucleotide target and its binding site. However, these results further emphasize the generality and utility of our method. In addition, the selection appears to have led to the optimization of sequence and structural contributions to the elution mechanism. For example, the fact that the selected beacon that bound OT2 was eluted better than an equivalent designed beacon that bound OT1 may explain why only OT2-dependent beacons were derived from the original selection.
Assays were also performed with beacon 16c to determine its mechanism of elution (Fig. 6a and b). When the octamer motif (5′-GCGGTGAC) was again mutated (16.58a) or deleted (16.48) to eliminate complementarity the beacon no longer eluted in the presence of the target oligonucleotide (Fig. 6b), as previously observed with beacon 14a.58. In addition, when the target oligonucleotide was mutated to interfere with the formation of the stacked helical junction (OT2j.20), elution again went down markedly, further corroborating the suggested mechanism.
If the basic mechanism for beacon elution had been established by these experiments, then it should prove possible to more significantly modify the beacons and perhaps to design new beacons. To this end, a minimal version of beacon 14a was designed in which the 3′ portion of the molecule, beyond the target binding domain, was removed (14a.42; Fig. 4b). As expected, the beacon continued to show robust target-dependent elution.
Selected beacons show oligonucleotide-dependent increases in fluorescence
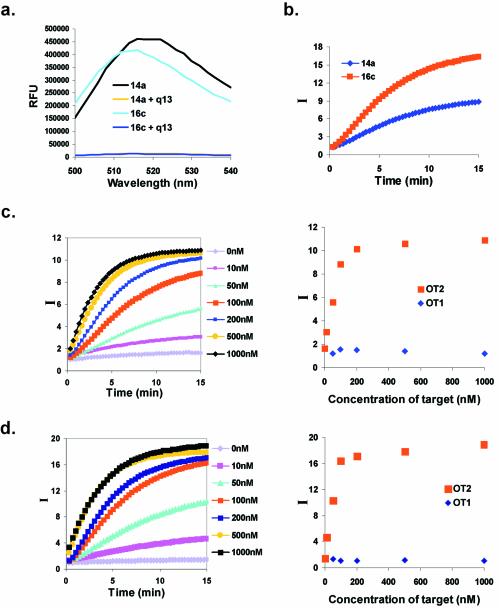
In order to determine if the eluted species could function like molecular beacons, changes in fluorescence upon the addition of target oligonucleotides were measured. Selected beacons bearing a fluorescein on T11 within the 5′ constant region were hybridized with a capture oligonucleotide containing a DABCYL moiety at its 5′ end. These positions were chosen to juxtapose the fluorescent reporter and the quencher, so that the fluorescent signal would be low in the absence of the target oligonucleotide. In fact, the hybridization of the two oligonucleotides resulted in up to 30-fold quenching for both beacons 14a and 16c (Fig. 7a).
Figure 7.
Fluorescence responsivities of selected beacons. (a) Fluorescence quenching in the presence of the capture oligonucleotide. The capture oligonucleotide q13 was present in a 2:1 (100:50 nM) molar excess to beacons 14a or 16c. (b) Target-dependent increase in fluorescence. Complexes with the capture oligonucleotide were formed as in (a), the target oligonucleotide OT2 was added in 2-fold excess, and the time-dependent development of the signal was monitored. (c) Concentration-dependent response of beacon 14a to target oligonucleotide OT2. I is the signal-to-background ratio, as defined in Materials and Methods. These data are also shown as a function of concentration, rather than time, for the t = 15 min time point. (d) Concentration-dependent response of beacon 16c to target oligonucleotide OT2. These data are also shown as a function of concentration for the t = 15 min time point.
Addition of the oligonucleotide target (OT2) was predicted to facilitate the same conformational change that led to release from the column, and thus should displace the capture oligonucleotide bearing the quencher and also result in an increase in fluorescence intensity. Beacon 14a showed a 9.5-fold increase in fluorescence in the presence of a 2-fold molar excess of OT2, whereas beacon 16c showed a 16.5-fold increase in fluorescence (Fig. 7b). Both beacons 14a (Fig. 7c) and 16c (Fig. 7d) exhibited target concentration-dependent increases in fluorescence. The apparent Kd of beacon 14a for OT2 was 37 ± 11 nM, and for beacon 16c a similar value was obtained, 34 ± 8 nM. The limit of target detection was ∼14 nM for beacon 14a (Fig. 7c) and 3.6 nM for beacon 16c (Fig. 7d), values which are similar to the limits previously demonstrated for at least some designed molecular beacons (12,14,18). The rate of fluorescence response increased with both target concentration (Fig. 7c and d) and beacon concentration (data not shown), and appeared to follow second-order kinetics as seen with other designed molecular beacons (1). As expected based on the elution data, the selected beacons showed no fluorescence response in the presence of the non-hybridizing target oligonucleotide OT1 (Fig. 7c and d).
The responsivities and kinetics of the selected molecular beacons were comparable with those of many designed molecular beacons (1). While an optimally designed molecular beacon can show a 200-fold increase in fluorescence intensity upon interaction with its target sequence, more commonly reported increases in fluorescence range from 10- to 60-fold (1,10,29,36–38). The increases in fluorescence intensity that we observed with our selected molecular beacons fell within this range. The fluorescence responses of the selected molecular beacons were observable at room temperature and reached a stable level within 10–15 min, values that were again similar to those seen for designed molecular beacons.
However, the selected molecular beacons did not exhibit complete restoration of fluorescence intensity, even upon addition of a 20-fold excess of the complementary OT2 target. The selected molecular beacon 14a exhibited only 34% of its maximum possible fluorescence response, while beacon 16c exhibited an 85% response. In contrast, when an oligonucleotide target binds to a designed molecular beacon an almost complete dequenching of a fluorescent reporter is frequently observed (8), although this is not always the case (35). In this latter instance, Tan et al. observed that the fluorescence intensity of the reporter fluorophore coumarin increased further upon DNase I digestion of a complex of a molecular beacon with its complementary oligonucleotide target (35). This incomplete response may have been due to a significant overlap between the emission spectra of coumarin and DABCYL. The selected molecular beacons utilized fluorescein as a reporter, would not have been subject to this limitation, and a different rationale for partial responsivity must therefore be hypothesized.
The relatively low responsivity of beacon 14a may be a consequence of the pool size used for selection. Designed molecular beacons (39) and other sequence sensors (40) form structures that can be readily disrupted by target oligonucleotides. The selected molecular beacons that we have described disrupt a 12 bp, perfectly paired duplex, and instead are predicted to form an 8 bp stem–loop that contains two non-Watson–Crick pairings, and a 15 bp duplex that contains at least one non-Watson–Crick pairing and frequently bulge residues as well (Fig. 3c). The 23 bp in the two stacked helices completely span the 20 nt random sequence region and extend into the constant regions. It is possible that if a longer random sequence region was used, a more stable, target-dependent conformer would have been selected, and the fluorescence response obtained would have been greater. Nevertheless, even with the short pool that was employed at least one of the selected beacons (16c) was able to obtain efficiencies similar to those seen for designed beacons. It is not immediately apparent why beacon 16c is much more efficient than beacon 14a, although again it is possible that structures more complex than a Watson–Crick duplex may ultimately be responsible for the function of selected beacons.
An additional caveat is that the selected beacons were generated by reverse transcription followed by RNase digestion of template RNA, and optimal fluorescence response was found to be dependent on the purity of the samples. While the selected beacons always demonstrated a target-dependent increase in fluorescence, irrespective of how they were prepared, the magnitude of the fluorescence response decreased if the RNA template was incompletely degraded and removed. In order to optimize the responsivities of selected beacons, the cDNA produced by reverse transcrip tion had to be sequentially treated with several different ribonucleases.
Chemical synthesis of selected beacons was an obvious solution to the problems posed by ribonuclease digestion and cDNA purification. However, chemically synthesized beacons showed a smaller (4–5-fold, as opposed to 10–20-fold) target-dependent increase in fluorescence. This may be due to the accumulation of additional chemical lesions in the longer synthetic DNAs, as opposed to the shorter primers for cDNA synthesis. It has previously been shown that as the length of synthetic DNA goes up, chemical lesions accumulate and its overall integrity and quality decreases (41). In this regard, it is interesting to note that the impact of chemical synthesis on the functionality of designed beacons is largely unknown, since designed beacons have almost always been generated by chemical synthesis. When we generated the same beacon by chemical versus enzymatic methods, extension with reverse transcriptase indicated that only 35% of the chemically synthesized DNA did not contain lesions, as opposed to over 90% of the enzymatically synthesized DNA (data not shown). Thus, one of the unanticipated insights of our studies is that the enzymatic rather than chemical preparation of molecular beacons may lead to more robust performance.
An alternative possibility for the observed differences between enzymatically and chemically synthesized beacons is that digested RNA fragments might act to potentiate the target-dependent conformational change; for example, by providing additional stacking interactions for the initial formation and stabilization of the OT2–beacon complex. This possibility is deemed unlikely for a variety of reasons. First, the cDNAs were gel purified, and would have likely contained only a very small amount of contaminating, smaller RNA molecules. In addition, cDNAs that were purified following multiple treatments with ribonucleases gave uniformly better fluorescence responses. Similarly, cDNAs purified following extensive alkaline hydrolysis of RNA yielded increases in target-dependent fluorescence that were identical to those observed following thorough ribonuclease digestion. In contrast, residual RNA contamination was found to interfere with the elution of molecular beacons from affinity columns.
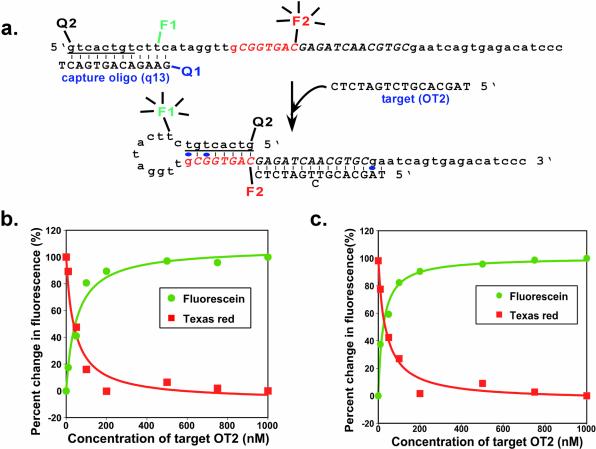
Wavelength-shifting molecular beacons
Two separate conformational changes occur during the target-dependent activation of the selected beacons: loss of the capture oligonucleotide and formation of a hairpin stem. In order to exploit both conformational changes and expand the potential utility of the selected beacons, a second quencher (DABCYL) was introduced at the 5′ end of beacon constructs during chemical synthesis, and a second fluorescent reporter (Texas Red) was appended to a cytidine residue in the octamer motif (position 27) via post-synthetic chemical coupling (shown for beacon 14a in Fig. 8a). In the absence of target, fluorescein should be quenched by the adjacent DABCYL on the capture oligonucleotide, and Texas Red should fluoresce. In the presence of target, the conformational changes will bring the 5′ DABCYL into apposition with Texas Red, and fluorescein should instead fluoresce (Fig. 8a). While these doubly fluorescent selected molecular beacons are similar in conception to the wavelength-shifting molecular beacons designed by Tyagi et al. (42), the selected color-switching beacons undergo a complete color change in which only one dye at a time is active, while the designed color-switching beacons rely upon FRET between two dyes to yield a target-dependent change in color.
Figure 8.
Wavelength-shifting beacons. (a) Sequence and predicted structure of a wavelength-shifting beacon based on beacon 14a. Representations are as in Figure 3c. The previously introduced fluorescein-dT is now labeled F1, while a second fluorophore (Texas Red) is F2. There are two DABCYL moieties, at positions Q1 (previously introduced) and Q2. (b) Fluorescence response at different wavelengths of beacon 14a to target OT2. The normalized signal-to-background ratio (percent change in fluorescence) is shown, as described in Materials and Methods. (c) Fluorescence response at different wavelengths of beacon 16c to target OT2.
The relative increase and decrease in these two fluorescent signals as a function of target concentration is shown for derivatives of beacon 14a (Fig. 8b) and beacon 16c (Fig. 8c). The complementary responsivities of the two dyes, while predicted and expected, nonetheless strongly confirm the models we have put forth for the selected beacons (Fig. 4a). While it is possible that FRET can take place between fluorescein and Texas Red, the color switching that was observed is more consistent with a dequenching of fluorescein followed by a subsequent quenching of Texas Red than with changes in FRET between the dyes themselves. The apparent Kd of the OT2–14a beacon complex was calculated based on fluorescein response to be 52 ± 12 nM and on Texas Red response to be 43 ± 11 nM. These values were slightly higher than those observed for the selected beacons with a single fluorophore, possibly due to the interference of the second dye or quencher with OT2 binding or the conformational transition. The response times for the color-switching beacons were in the order of 10 min, as previously observed for the selected beacons with a single fluorophore.
CONCLUSIONS
The selected molecular beacons have performance characteristics comparable with those of designed molecular beacons. While it is of course relatively simple to design molecular beacons, the ability to select beacons should prove useful. For example, a complex target, such as a mRNA molecule, will have regions that are more or less accessible to a designed molecular beacon, due to the formation of secondary and even tertiary structures and the binding of accessory proteins. Using the described method, molecular beacons could be selected de novo and would likely interact with those portions of a mRNA that were most intrinsically accessible. Nonetheless, designed molecular beacons are capable of mismatch discrimination and can be readily used in applications such as real-time PCR. Further research will be required to determine to what extent the selected molecular beacons or the novel mechanism for molecular beacon signaling can also be adapted to these requirements and applications.
The identification of a novel mechanism for signal transduction by molecular beacons may also yield applications. For example, to the extent that the structure-forming beacons we have described require interactions with the free 3′ end of a target molecule, these beacons could be used to specifically detect the 3′ ends of particular RNA molecules, including during RNA processing events. The more complex conformational changes that occur in selected molecular beacons, relative to designed molecular beacons, lend themselves more easily to more complex signaling modalities, such as the true two-color beacons that we have designed and proofed. Finally, the property that was originally selected for, release from immobilization, could also be employed in the design of oligonucleotide-specific actuators for nanoscale devices or nucleic acid-based machines (43–45). In contrast, it is difficult to imagine how designed molecular beacons could be engineered to undergo target-dependent release from a capture oligonucleotide.
Ultimately, though, the importance of these experiments is that they potentially provide a general method for the selection of aptamer beacons. The ligands used for elution of species from the affinity column need not be restricted to oligonucleotides, but could be almost any molecule, from small ions to peptides to proteins to supramolecular complexes. Since any ligand-dependent release from a column will of necessity also lead to a separation of a fluorescent reporter from a quencher, selection for binding and elution will select for signaling.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge the Foundation for Research for support of this project and Matthew Levy for the synthesis of many of the oligonucleotides used in these studies.
REFERENCES
- 1.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 2.Nutiu R. and Li,Y. (2002) Tripartite molecular beacons. Nucleic Acids Res., 30, E94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marras S.A., Kramer,F.R. and Tyagi,S. (1999) Multiplex detection of single-nucleotide variations using molecular beacons. Genet. Anal., 14, 151–156. [DOI] [PubMed] [Google Scholar]
- 4.Marras S.A., Kramer,F.R. and Tyagi,S. (2003) Genotyping SNPs with molecular beacons. Methods Mol. Biol., 212, 111–128. [DOI] [PubMed] [Google Scholar]
- 5.Kostrikis L.G., Tyagi,S., Mhlanga,M.M., Ho,D.D. and Kramer,F.R. (1998) Spectral genotyping of human alleles. Science, 279, 1228–1229. [DOI] [PubMed] [Google Scholar]
- 6.Vet J.A., Majithia,A.R., Marras,S.A., Tyagi,S., Dube,S., Poiesz,B.J. and Kramer,F.R. (1999) Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl Acad. Sci. USA, 96, 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piatek A.S., Tyagi,S., Pol,A.C., Telenti,A., Miller,L.P., Kramer,F.R. and Alland,D. (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol., 16, 359–363. [DOI] [PubMed] [Google Scholar]
- 8.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 9.Leone G., van Schijndel,H., van Gemen,B., Kramer,F.R. and Schoen,C.D. (1998) Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res., 26, 2150–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sokol D.L., Zhang,X., Lu,P. and Gewirtz,A.M. (1998) Real time detection of DNA.RNA hybridization in living cells. Proc. Natl Acad. Sci. USA, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perlette J. and Tan,W. (2001) Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal. Chem., 73, 5544–5550. [DOI] [PubMed] [Google Scholar]
- 12.Liu X. and Tan,W. (1999) A fiber-optic evanescent wave DNA biosensor based on novel molecular beacons. Anal. Chem., 71, 5054–5059. [DOI] [PubMed] [Google Scholar]
- 13.Liu X., Farmerie,W., Schuster,S. and Tan,W. (2000) Molecular beacons for DNA biosensors with micrometer to submicrometer dimensions. Anal. Biochem., 283, 56–63. [DOI] [PubMed] [Google Scholar]
- 14.Steemers F.J., Ferguson,J.A. and Walt,D.R. (2000) Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. Nat. Biotechnol., 18, 91–94. [DOI] [PubMed] [Google Scholar]
- 15.Stojanovic M.N., de Prada,P. and Landry,D.W. (2001) Catalytic molecular beacons. Chembiochem, 2, 411–415. [DOI] [PubMed] [Google Scholar]
- 16.Stojanovic M.N., Mitchell,T.E. and Stefanovic,D. (2002) Deoxyribozyme-based logic gates. J. Am. Chem. Soc., 124, 3555–3561. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Fang,X., Schuster,S., Liu,X. and Tan,W. (2000) Molecular beacons: a novel approach to detect protein–DNA interactions. Angew. Chem. Int. Ed. Engl., 39, 1049–1052. [DOI] [PubMed] [Google Scholar]
- 18.Fang X., Li,J.J. and Tan,W. (2000) Using molecular beacons to probe molecular interactions between lactate dehydrogenase and single-stranded DNA. Anal. Chem., 72, 3280–3285. [DOI] [PubMed] [Google Scholar]
- 19.Heyduk T. and Heyduk,E. (2002) Molecular beacons for detecting DNA binding proteins. Nat. Biotechnol., 20, 171–176. [DOI] [PubMed] [Google Scholar]
- 20.Hesselberth J., Robertson,M.P., Jhaveri,S. and Ellington,A.D. (2000) In vitro selection of nucleic acids for diagnostic applications. Rev. Mol. Biotechnol., 74, 15–25. [DOI] [PubMed] [Google Scholar]
- 21.Jayasena S.D. (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem., 45, 1628–1650. [PubMed] [Google Scholar]
- 22.Wilson D.S. and Szostak,J.W. (1999) In vitro selection of functional nucleic acids. Annu. Rev. Biochem., 68, 611–647. [DOI] [PubMed] [Google Scholar]
- 23.Rajendran M. and Ellington,A.D. (2002) Nucleic acids for reagentless biosensors. In Ligler,F.S. and Rowe Taitt,C.A. (eds), Optical Biosensors: Present and Future. Elsevier Science BV, Amsterdam, pp. 369–396. [Google Scholar]
- 24.Rajendran M. and Ellington,A.D. (2002) Selecting nucleic acids for biosensor applications. Comb. Chem. High Throughput Screen., 5, 263–270. [DOI] [PubMed] [Google Scholar]
- 25.Jhaveri S., Kirby,R., Conrad,R., Maglott,E.J., Bowser,M., Kennedy,R.T., Glick,G. and Ellington,A.D. (2000) Designed signaling aptamers that transduce molecular recognition to changes in fluorescence intensity. J. Am. Chem. Soc., 122, 2469–2473. [Google Scholar]
- 26.Li J.J., Fang,X. and Tan,W. (2002) Molecular aptamer beacons for real-time protein recognition. Biochem. Biophys. Res. Commun., 292, 31–40. [DOI] [PubMed] [Google Scholar]
- 27.Stojanovic M.N., de Prada,P. and Landry,D.W. (2001) Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc., 123, 4928–4931. [DOI] [PubMed] [Google Scholar]
- 28.Hamaguchi N., Ellington,A. and Stanton,M. (2001) Aptamer beacons for the direct detection of proteins. Anal. Biochem., 294, 126–131. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto R., Baba,T. and Kumar,P.K. (2000) Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells, 5, 389–396. [DOI] [PubMed] [Google Scholar]
- 30.Stojanovic M.N., de Prada,P. and Landry,D.W. (2000) Fluorescent sensors based on aptamer self-assembly. J. Am. Chem. Soc., 122, 11547–11548. [DOI] [PubMed] [Google Scholar]
- 31.Nutiu R. and Li,Y. (2003) Structure-switching signaling aptamers. J. Am. Chem. Soc., 125, 4771–4778. [DOI] [PubMed] [Google Scholar]
- 32.Jhaveri S., Rajendran,M. and Ellington,A.D. (2000) In vitro selection of signaling aptamers. Nat. Biotechnol., 18, 1293–1297. [DOI] [PubMed] [Google Scholar]
- 33.Robertson M.P. and Ellington,A.D. (1999) In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol., 17, 62–66. [DOI] [PubMed] [Google Scholar]
- 34.Cox J.C., Hayhurst,A., Hesselberth,J., Bayer,T.S., Georgiou,G. and Ellington,A.D. (2002) Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res., 30, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P., Beck,T. and Tan,W. (2001) Design of a molecular beacon DNA probe with two fluorophores. Angew. Chem. Int. Ed. Engl., 40, 402–405. [DOI] [PubMed] [Google Scholar]
- 36.Nutiu R. and Li,Y. (2002) Tripartite molecular beacons. Nucleic Acids Res., 30, e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan W., Fang,X., Li,J. and Liu,X. (2000) Molecular beacons: a novel DNA probe for nucleic acid and protein studies. Chemistry, 6, 1107–1111. [DOI] [PubMed] [Google Scholar]
- 38.Poddar S.K. (1999) Detection of adenovirus using PCR and molecular beacon. J. Virol. Methods, 82, 19–26. [DOI] [PubMed] [Google Scholar]
- 39.Tsourkas A., Behlke,M.A., Rose,S.D. and Bao,G. (2003) Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res., 31, 1319–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Q., Luan,G., Guo,Q. and Liang,J. (2002) A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res., 30, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green R., Ellington,A.D., Bartel,D.P. and Szostak,J.W. (1991) In vitro genetic analysis: selection and amplification of rare functional nucleic acids. Methods, 2, 75–86. [Google Scholar]
- 42.Tyagi S., Marras,S.A. and Kramer,F.R. (2000) Wavelength-shifting molecular beacons. Nat. Biotechnol., 18, 1191–1196. [DOI] [PubMed] [Google Scholar]
- 43.Seeman N.C. (2003) DNA in a material world. Nature, 421, 427–431. [DOI] [PubMed] [Google Scholar]
- 44.Yurke B., Turberfield,A.J., Mills,A.P.,Jr, Simmel,F.C. and Neumann,J.L. (2000) A DNA-fuelled molecular machine made of DNA. Nature, 406, 605–608. [DOI] [PubMed] [Google Scholar]
- 45.Alberti P. and Mergny,J.L. (2003) DNA duplex–quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA, 100, 1569–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]