Abstract
Tim54p, a component of the inner membrane TIM22 complex, does not directly mediate the import of inner membrane substrates but is required for assembly/stability of the 300-kD TIM22 complex. In addition, Δtim54 yeast exhibit a petite-negative phenotype (also observed in yeast harboring mutations in the F1Fo ATPase, the ADP/ATP carrier, mitochondrial morphology components, or the i–AAA protease, Yme1p). Interestingly, other import mutants in our strain background are not petite-negative. We report that Tim54p is not involved in maintenance of mitochondrial DNA or mitochondrial morphology. Rather, Tim54p mediates assembly of an active Yme1p complex, after Yme1p is imported via the TIM23 pathway. Defective Yme1p assembly is likely the major contributing factor for the petite-negativity in strains lacking functional Tim54p. Thus, Tim54p has two independent functions: scaffolding/stability for the TIM22 membrane complex and assembly of Yme1p into a proteolytically active complex. As such, Tim54p links protein import, assembly, and turnover pathways in the mitochondrion.
Introduction
Mitochondria have elaborate translocation machinery for the import and assembly of nuclear-coded proteins. Translocons of the outer membrane (TOM) and inner membrane (TIM) coordinate protein translocation across the outer and inner membranes (Paschen and Neupert, 2001; Truscott et al., 2003; Koehler, 2004). The TIM23 translocon mediates the import of proteins with a typical N-terminal targeting sequence. The TIM22 import pathway facilitates the TIM proteins, including the carrier family and the import components Tim22p and Tim23p. Members of the TIM22 pathway include two soluble complexes in the intermembrane space, Tim9p-Tim10p and Tim8p-Tim13p, as well as a 300-kD complex at the inner membrane, which consists of Tim12p, Tim18p, Tim22p, Tim54p, and a fraction of Tim9p and Tim10p (Koehler, 2004). The soluble complexes aid movement of the hydrophobic precursors across the aqueous intermembrane space, and the 300-kD complex mediates insertion into the inner membrane.
The essential components in the TIM22 translocon are Tim9p, Tim10p, Tim12p, and Tim22p (Sirrenberg et al., 1996, 1998; Kerscher et al., 1997; Koehler et al., 1998a,b), which all bind directly to the imported substrate as shown by chemical cross-linking. Tim18p and Tim54p, however, seem to play secondary roles in protein translocation because a direct interaction with a translocating TIM22 substrate has yet to be demonstrated. Tim18p is an accessory protein, and the 300-kD TIM22 complex is present, albeit decreased in molecular mass, in strains lacking tim18 (Kerscher et al., 2000; Koehler et al., 2000). Tim54p was identified in a two-hybrid screen using the cytosolic domain of Mmm1p, an outer membrane protein involved in the maintenance of mitochondrial morphology and mitochondrial DNA, as the bait (Kerscher et al., 1997). This interaction may be reflected by Mmm1p's role in assembly of β-barrel outer membrane proteins (Meisinger et al., 2007). Jensen and colleagues specifically showed that Tim54p partners with Tim22p in the 300-kD complex and is essential for viability (Kerscher et al., 1997), suggesting that Tim54p might assemble or stabilize the 300-kD complex. However, subsequent studies by Pfanner and Jensen showed that Tim54p is not essential under certain conditions (Kovermann et al., 2002), and raised a general question about the role of Tim54p in mitochondrial biogenesis.
Surprisingly, cells lacking tim18 have a petite-negative phenotype, which is revealed by inviability on glucose medium in the presence of ethidium bromide (Dunn and Jensen, 2003). In Saccharomyces cerevisiae, mitochondrial respiration is not essential for viability and is referred to as petite-positivity because of the ability to grow on fermentable carbon sources when the mitochondrial genome is lost (ρ0) or contains large deletions (ρ−; Ephrussi, 1953; Bulder, 1964). However, most yeast strains, including S. pombe and K. lactis, are petite-negative and require a functional mitochondrial genome for growth (Chen and Clark-Walker, 2000). Petite-positivity is achieved because a mitochondrial membrane potential (Δψ) can be maintained in the respiration-deficient state by the import of ATP (via the ADP/ATP carrier, AAC) in exchange for ADP produced by the F1-ATPase, which acts as an ATP hydrolase (Giraud and Velours, 1997).
In addition to TIM18, mutations in several nuclear genes can render S. cerevisiae petite-negative (Contamine and Picard, 2000). These nuclear genes include those for the F1 portion of the ATPase (Weber et al., 1995; Giraud and Velours, 1997), AAC (Kovác, 1967), and the mitochondrial inner membrane protease, Yme1p (Kominsky and Thorsness, 2000; Kominsky et al., 2002). AAC is required for the aforementioned exchange of ATP and ADP between the cytosol and the matrix (Palmieri et al., 1996). Yme1p may affect ATPase function by catalyzing the turnover of protein inhibitors of the ATPase (Kominsky et al., 2002) or altering F1-ATPase activity, possibly by inducing structural changes (Francis et al., 2007). The petite-negative phenotype of Δtim18 cells was suppressed by several genes coding for cytosolic proteins including translation components and chaperones (Dunn and Jensen, 2003). Jensen and colleagues suggested that this set of suppressors might compensate for a defect in protein import, allowing the mitochondrion to maintain a membrane potential (Dunn and Jensen, 2003). In addition, a mutation in tim9 results in a petite-negative phenotype that has been linked to a defect in transcription (Senapin et al., 2003), and petite-negativity for cells lacking tom70 is dependent on the strain background (Dunn and Jensen, 2003). Thus, the specific mechanism(s) resulting in the synthetic lethality between loss of the mitochondrial genome and mutations in this wide array of nuclear genes is not well understood.
In this paper, we investigate the function of Tim54p in mitochondrial biogenesis and find that the Δtim54 cells are petite-negative. In contrast, strains harboring mutations in tim9, tim10, tim12, tim18, tim22, and tim23, all import components, are petite-positive in our strain background. From a systematic analysis, we show that Tim54p is not required for maintenance of mitochondrial DNA or nucleoid morphology. Rather, Tim54p is required for the stability/assembly of the TIM22 complex and, second and specifically, for the assembly of Yme1p into a proteolytically active complex. Because Yme1p is imported through the TIM23 translocon and Tim54p is a stabilizing component of the TIM22 translocon, Yme1p import and assembly represent a novel collaborative effort between the two translocons of the inner membrane. Given the role of Yme1p in protein turnover, Tim54p effectively links pathways of import, assembly, and turnover in the mitochondrion.
Results
Loss of functional Tim54p yields a decrease in mitochondrial nucleoid number
Whereas Tim9p, Tim10p, Tim12p, and Tim22p (Sirrenberg et al., 1996, 1998; Kerscher et al., 1997; Koehler et al., 1998a,b) all bind directly to an imported TIM22 substrate, we have not been successful at identifying a direct biochemical interaction between a translocating TIM22 substrate and Tim54p (unpublished data). These observations prompted us to investigate the specific role of Tim54p in mitochondrial biogenesis.
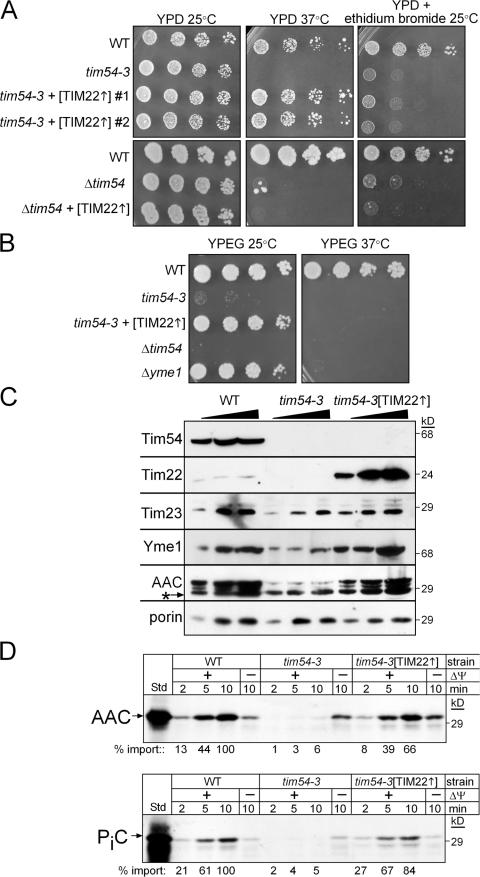
Because Jensen and colleagues originally identified Tim54p through a two-hybrid interaction with Mmm1p (Kerscher et al., 1997) and because Mmm1p has a role in mitochondrial DNA maintenance (Hobbs et al., 2001; Hanekamp et al., 2002), we probed whether Tim54p might function in mitochondrial DNA stabilization. We generated a strain deleted for TIM54 (Δtim54) using plasmid shuffling in the parental strain GA74 (genotypes for strains are listed in Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200706195/DC1) (Waldherr et al., 1993; Koehler et al., 1998a; Kovermann et al., 2002). We also generated a temperature-sensitive (ts) tim54–3 mutant using error-prone PCR in GA74 (Muhlrad et al., 1992; Leuenberger et al., 2003). Whereas the Δtim54 and tim54–3 strains grew with a doubling time of ∼3 h at 25°C on glucose media, the strains arrested growth after 6 h at 37°C. The strains also grew slowly on ethanol-glycerol media at 25°C (Fig. S1). As expected, Tim54p was not detected by immunoblot analysis in mitochondrial extracts derived from the Δtim54 strain (Fig. 1 A). Tim54p also was not detected in mitochondria purified from the tim54–3 mutant grown at 25°C; the mutant protein may be present at steady-state levels that are not detectable by the antibody, or the protein may be turned over at an increased rate compared with wild-type Tim54p.
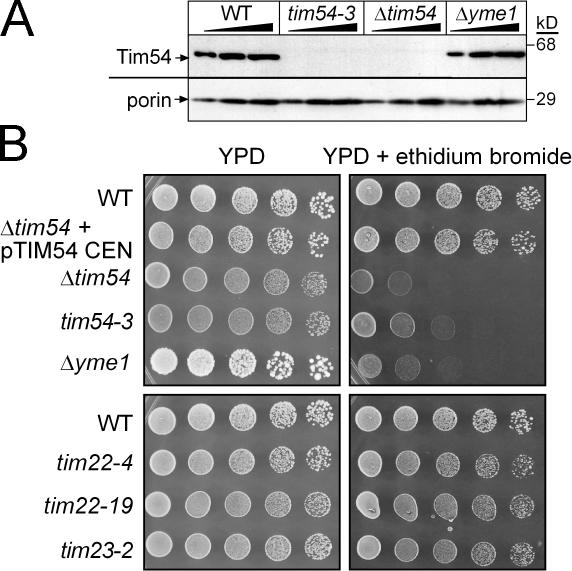
Figure 1.
Cells lacking functional Tim54p are petite-negative. (A) Increasing amounts of a mitochondrial protein lysate (50, 100, 150 μg) from a WT strain, the ts tim54-3 mutant (grown at 25°C), the strain deleted for TIM54 (Δtim54), and the Δyme1 mutant were separated by SDS-PAGE and immunoblotted with antibodies against Tim54p and porin. The strain background is GA74. (B) Yeast strains in background GA74 [WT, Δtim54 transformed with TIM54 on a centromeric plasmid (Δtim54 + pTIM54 CEN), Δtim54, tim54-3, Δyme1, and ts tim22 (tim22-4, tim22-19) and tim23 (tim23-2) mutants], were grown to mid-log phase in liquid YPD. The cultures were serially diluted by a factor of 5 and spotted on YPD plates or YPD plates containing 40 μg/ml ethidium bromide and incubated at 25°C for 4 d.
We investigated whether Tim54p might function in a pathway for the maintenance of mitochondrial DNA. We analyzed growth at 25°C by serial dilution in glucose medium supplemented with ethidium bromide, which causes cells to lose their mitochondrial DNA (referred to as ρ0; Slonimski et al., 1968) (Fig. 1 B). The Δtim54 and tim54-3 strains, like the control Δyme1 strain, were not viable on glucose media containing ethidium bromide. In contrast, ts mutants tim22-4, tim22-19, and tim23-2 (Dekker et al., 1997; Leuenberger et al., 1999) grew on media containing ethidium bromide, indicating that strains defective in protein import in the GA74 background typically do not require the mitochondrial genome for growth. Therefore, the tim54 and Δyme1 strains have a petite-negative phenotype, whereas the tim22 and tim23 mutants are petite-positive.
We checked if Tim54p was required for maintenance of mitochondrial morphology. Mitochondrial and nucleoid structure were probed in a Δtim54 strain by transforming the strain with a mitochondrial matrix-targeted GFP construct and staining with 2 μg/ml DAPI, respectively (Fig. 2 A). Compared with the parental strain with many small punctate nucleoids (25 ± 10, n = 32), Δtim54 cells contained fewer nucleoids (6 ± 3, n = 30), which is similar to that observed in the Δyme1 isogenic strain (6 ± 2, n = 34). In contrast, Δtim54 and the parental strain had a similar mitochondrial network.
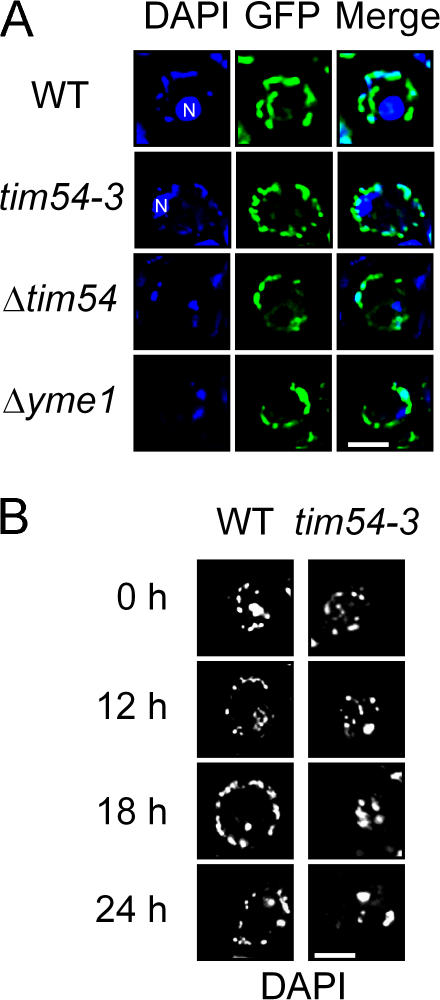
Figure 2.
Absence of Tim54p reduces nucleoid number. (A) Cells were grown to mid-log phase in YPD at 25°C, stained with 2 μg/ml DAPI, and viewed by fluorescence microscopy. The images from DAPI and GFP were superimposed (Merge). Strains analyzed were the parental strain (WT), tim54-3, Δtim54, and Δyme1 in strain background GA74. Mitochondrial morphology was visualized with a mitochondrially targeted GFP. Note: the nucleus (N) is marked in images that showed strong nuclear DAPI staining. (B) The parental strain and the tim54-3 mutant were incubated at the restrictive temperature of 37°C for 0, 12, 18, and 24 h. Nucleoid morphology was examined by fluorescence microscopy as described in A. Bar, 3 μm.
Because morphology defects can arise as a secondary defect in mitochondrial function, as has been demonstrated for mutants in mitochondrial outer membrane assembly (Stojanovski et al., 2006), we evaluated the nucleoid quantity in the tim54-3 mutant at the restrictive temperature of 37°C in a time course assay. At the initial time point and after 12 h at 37°C, the tim54-3 cells showed normal nucleoid number (20 ± 9, n = 33) (Fig. 2 B). Only after 18–24 h at 37°C were structural changes—from abundant, punctate nucleoids to fewer, coalescent nucleoids (5 ± 2, n = 20)—evident in tim54-3 cells, in contrast to the parental strain that maintained a normal nucleoid phenotype at all time points analyzed (Fig. 2 B). The aberrant nucleoid morphology thus is a secondary consequence associated with loss of functional Tim54p, indicating that Tim54p does not play a direct role in regulating mitochondrial or nucleoid structure.
Petite-negativity depends on the background of the yeast strain
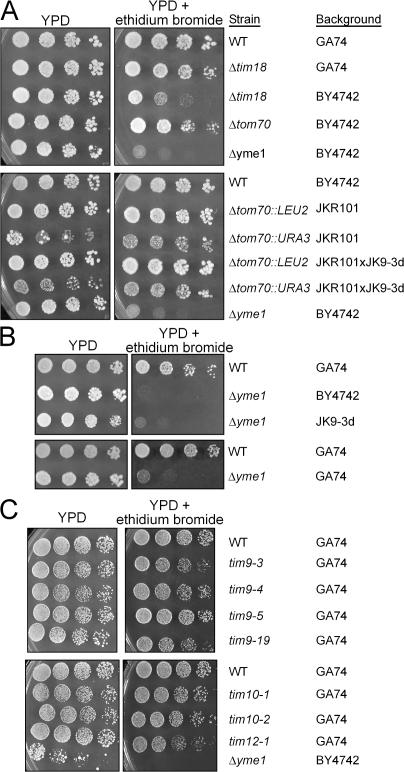
A comprehensive study by Jensen and colleagues illustrated a link between mitochondrial protein import and cytosolic protein translation and trafficking pathways (Dunn and Jensen, 2003). We assessed the petite-negativity of import components in different strain backgrounds available in our laboratory by serial dilution on glucose media supplemented with ethidium bromide (Fig. 3). Whereas the Jensen group reported previously that their Δtim18 and Δtom70 strains were petite-negative, the BY4742 strain deleted for tim18 was the only strain that showed partial sensitivity to ethidium bromide (Fig. 3 A) (Brachmann et al., 1998). Instead, our GA74 strain, as well as other backgrounds (Schmidt et al., 1987; Hines et al., 1990; Hines and Schatz, 1993), did not display petite-negativity when deleted for tim18 and tom70. In contrast, the petite-negative phenotype associated with Δyme1 was independent of strain background (Fig. 3 B) (Beilharz et al., 1998). Additionally, mutations in tim9, tim10, and tim12 did not cause petite-negativity (Fig. 3 C) (Koehler et al., 1998a,b). The studies by Jensen and colleagues show that their background choice was important for deciphering the threshold effects caused by a defect in protein import and subsequent compensatory mechanisms, whereas the GA74 strain background does not seem to display these threshold effects. Thus, petite-negativity in the GA74 strain is a unique phenotype specifically associated with mutations in tim54 and not other import components.
Figure 3.
Petite-negativity is dependent on the strain background. (A) Yeast strains (WT GA74, Δtim18 in strain GA74 and BY4742, Δtom70 in BY4742 and JKR101 and haploids derived from the cross JKR101 × JK9, and Δyme1 in BY4742) were tested for petite-negativity as described in Fig. 1 B. (B) Strains deleted for yme1 in BY4742, JK9-3d, and GA74 were tested for petite-negativity. (C) The GA74 strain expressing conditional mutants tim9-3, tim9-4, tim9-5, tim9-19, tim10-1, tim10-2, and tim12-1 and controls WT and Δyme1 in BY4742 were tested as described in Fig. 1 B. Strain background is indicated in the figure.
The TIM22 import pathway is compromised in tim54 mutants and can be restored by TIM22 overexpression
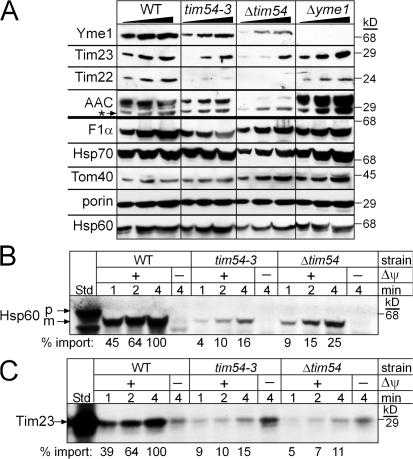
Because cells lacking Yme1p are not viable in the absence of mitochondrial DNA, we investigated the steady-state levels of Yme1p and other mitochondrial proteins in Δtim54, tim54-3, Δyme1, and wild–type (WT) mitochondria grown at 25°C (Fig. 4 A). Increasing amounts of a mitochondrial protein lysate were separated by SDS-PAGE and immunoblotted for mitochondrial proteins including Yme1p, Tim23p, Tim22p, AAC, F1α-ATPase, Hsp70, Tom40p, porin, and Hsp60. The abundance of Yme1p was decreased in the Δtim54 and tim54-3 mitochondria. The steady-state levels of Tim22p were also decreased in the tim54 mutants, but not in Δyme1 mitochondria. In addition, Tim23p and AAC levels (substrates of the TIM22 pathway) were lower in tim54 mutant mitochondria because the TIM22 import pathway is compromised in the absence of Tim54p (Kerscher et al., 1997; Kovermann et al., 2002). In contrast, the steady-state levels of porin, Tom40p, Hsp70, Hsp60, and the F1α-ATPase were similar among the tested strains. Therefore, mitochondria lacking functional Tim54p show a decreased abundance in Yme1p, as well as the previously characterized TIM22 translocon substrates, Tim23p, Tim22p, and AAC.
Figure 4.
Mitochondria lacking Tim54p display a compromised TIM22 import pathway. (A) Increasing amounts of a mitochondrial protein lysate (50, 100, 150 μg) from the parental strain (WT), the tim54-3 mutant, Δtim54, and Δyme1, were separated by SDS-PAGE and immunoblotted with antibodies against Yme1p, Tim23p, Tim22p, AAC, F1α-ATPase, Hsp70, Tom40p, porin, and Hsp60. The asterisk denotes immunoreactivity toward porin. (B) Radiolabeled Hsp60, a substrate of the TIM23 pathway, was imported into mitochondria purified from the WT strain and the Δtim54 and tim54-3 strains for 1, 2, and 4 min in the presence and absence of a membrane potential (ΔΨ). Non-imported precursor was removed by protease treatment (50 μg/ml trypsin treatment for 30 min followed by inactivation with 100 μg/ml trypsin inhibitor for 10 min). Imported proteins were separated by SDS-PAGE and visualized by fluorography. 10% of the translation reaction (Std) is included as a control. p and m indicate precursor and mature forms, respectively. (C) Radiolabeled Tim23p, a substrate of the TIM22 pathway, was imported as in B. Insertion into the inner membrane was confirmed by carbonate extraction. 10% of the translation reaction (Std) is included as a control. Import reactions were quantitated using a BioRad FX Molecular Imager and the affiliated Quantity 1 software; 100% was set as the amount of precursor imported into WT mitochondria at the endpoint in the time course.
A reason for the decreased abundance of the aforementioned proteins in the mitochondria lacking functional Tim54p might in part be caused by a general defect in protein import (Leuenberger et al., 1999). We tested the import of Hsp60 and Tim23p, which use the TIM23 and TIM22 import pathways, respectively, into mitochondria isolated from the Δtim54 and tim54-3 strains (Fig. 4, B and C). Both Hsp60 and Tim23p were imported into Δtim54 and tim54-3 strains, albeit at a slower rate than WT mitochondria. The steady-state level of Hsp60 in the tim54-3 and Δtim54 mitochondria, however, was not markedly reduced (Fig. 4 A). We have observed this scenario previously in which the in vitro import rate with isolated mitochondria is impaired compared with the steady-state levels, indicating that either import in vivo is more efficient than in vitro (Murphy, 1997; Leuenberger et al., 2003) or protein turnover is decreased to compensate for a reduced rate/abundance of import.
We investigated the function of Tim54p in the TIM22 pathway in greater detail with a combined genetic and biochemical approach. We transformed the tim54-3 and Δtim54 mutants with a high-copy plasmid (designated [TIM22]) in which Tim22p was overexpressed (Fig. 5, A and B). Overexpression of TIM22 specifically restored growth to the tim54-3 mutant at 37°C on glucose media; however, the petite-negative phenotype was not reversed (Fig. 5 A). In contrast to studies by Pfanner and Jensen and colleagues (Kovermann et al., 2002), TIM22 overexpression did not suppress the growth defect or the petite-negativity in the Δtim54 strain (Fig. 5 A), indicating that TIM22 suppression is dependent on the presence of the tim54 gene and that the tim54-3 mutant protein (albeit undetectable by immunoblot) is required for growth at 37°C. Also, TIM22 overexpression did not suppress the growth defect on rich ethanol-glycerol media at 37°C, demonstrating that tim54-3 phenocopies the Δyme1 mutant (Fig. 5 B). We also tested cold-sensitivity at 15°C on rich glucose media (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200706195/DC1). Like the Δyme1 mutant, Δtim54 and tim54-3 strains displayed cold sensitivity, which was not restored upon TIM22 overexpression (Fig. S2). In addition, the abundance of Tim22p, Tim23p, AAC, and Yme1p increased in tim54-3 mitochondria overexpressing Tim22p (Fig. 5 C), and import was restored to near-WT levels for substrates of the TIM22 pathway (Fig. 5 D; Fig. S3). This analysis indicates that overexpression of Tim22p restores defects associated with protein import and growth in the tim54-3 mutant, supporting the hypothesis that Tim54p functions as a stabilizing scaffold/assembly factor for the TIM22 complex.
Figure 5.
Overexpression of TIM22 in tim54-3 mitochondria restores viability on rich glucose media and protein import, but not petite-negativity. (A) The tim54-3 and Δtim54 mutants were transformed with a 2μ plasmid overexpressing TIM22 (designated as [TIM22]) in strain GA74. Individual transformants were serially diluted as described in Fig. 1 B onto rich glucose media in the presence and absence of ethidium bromide. Restoration of growth was tested at 37°C for the rich glucose media and 25°C for petite-negativity. (B) The WT, Δyme1, Δtim54, tim54-3, and tim54-3 + [TIM22] strains (background GA74) were plated onto rich ethanol-glyercol media and incubated at 25°C and 37°C. Plates were photographed after 3 d. (C) The steady-state levels of Tim54p, Tim22p, Tim23p, Yme1p, AAC, and porin were investigated as described in Fig. 4 A in mitochondria derived from the tim54-3 mutant and from the tim54-3 mutant overexpressing TIM22 (designated tim54-3[TIM22]). The asterisk denotes immunoreactivity toward porin. (D) Import rates for substrates of the TIM22 pathway, AAC (top) and the phosphate carrier (PiC; bottom), were analyzed as described in Fig. 4 C.
This set of experiments also suggests that Tim54p has a separate function associated with petite-negativity that TIM22 overexpression cannot suppress. Because overexpression of CCT6, identified in the screen by Jensen and colleagues was a suppressor of Δtim18 (Dunn and Jensen, 2003; Senapin et al., 2003), we investigated whether CCT6 might suppress the tim54-3 mutant (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200706195/DC1). Interestingly, CCT6 overexpression failed to restore growth of the tim54-3 mutant on ethidium bromide medium (Fig. S4). Therefore, the petite-negativity of the tim54 mutants is not seemingly caused by a defect in protein import or translation.
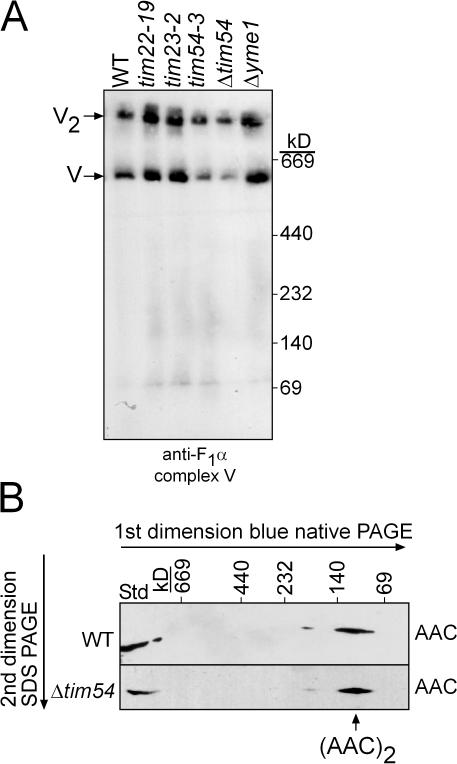
Because assembled ATPase and AAC complex are required for maintenance of a membrane potential (Giraud and Velours, 1997), we investigated the assembly state of the ATPase and AAC (Fig. 6, A and B); impaired assembly could lead to inviability in the presence of ethidium bromide. As assessed by one-dimensional blue-native PAGE, ATPase assembly was not impaired in mitochondria lacking functional Tim22p, Tim23p, Tim54p, and Yme1p (Fig. 6 A). To determine if AAC assembled normally as a dimer, mitochondrial extracts were resolved by blue-native PAGE in the first dimension and SDS-PAGE in the second dimension. AAC assembled into a dimer in Δtim54 mitochondria and WT mitochondria (Fig. 6 B) as well as the tim54-3 mutant (unpublished data). In previous studies, we have shown that AAC levels are essentially undetectable in our tim9, tim10, and tim22 mutant mitochondria (Koehler et al., 1998a; Murphy et al., 2001; Leuenberger et al., 2003). However, given that these mutants are not petite-negative as shown in Figs. 1 B and 3 C, even almost undetectable levels of AAC seem adequate to support the minimal membrane potential required for growth in the absence of respiration. Thus, the petite-negative phenotype associated with the tim54 strains is not due to a lack of assembly of the ATPase or AAC complexes.
Figure 6.
The ATPase and AAC complexes assemble correctly in tim54 mutant mitochondria. (A) Purified mitochondria from the WT, tim22-19, tim23-2, tim54-3, Δtim54, and Δyme1 strains were solubilized with 1% (wt/vol) digitonin and subjected to blue-native gel electrophoresis (5–10%). An antibody against the F1α-ATPase was used to detect complex V assembly. V and V2 indicate the assembled complex V monomer, and the assembled complex V dimer, respectively. (B) Mitochondria from WT and Δtim54 strains were solubilized in 1% digitonin and subjected first to blue-native gel electrophoresis (6–16% acrylamide) and then to SDS-PAGE (12% acrylamide). AAC was detected by immunoblotting. Note, 4× more mitochondria were loaded for the Δtim54 strain because of reduced AAC abundance. T, a sample of the total detergent-solubilized mitochondria.
Tim54p is required for Yme1p assembly into an active complex
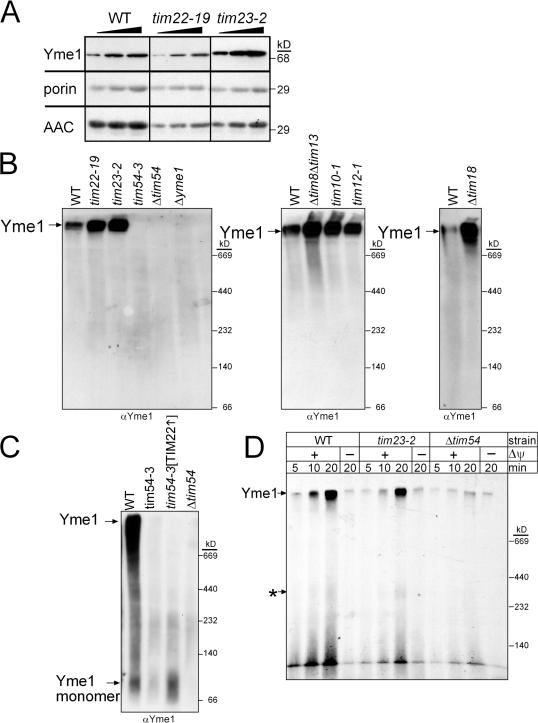
Given that the petite-negative phenotype and the respiratory-deficiency at 37°C could not be restored by overexpression of TIM22, we focused on Yme1p assembly and function. First, we evaluated Yme1p import into mitochondria defective in tim22, tim23, and tim54, as well as the tim54-3 mitochondria with overexpressed TIM22 (Fig. S5 A, available at http://www.jcb.org/cgi/content/full/jcb.200706195/DC1). Yme1p contains a typical N-terminal targeting sequence that is cleaved, presumably by the matrix processing peptidase, upon import (Klanner et al., 2001; Arnold and Langer, 2002); as such, Yme1p is predicted to use the TIM23 import pathway because of its typical presequence (Van Dyck and Langer, 1999). The Yme1p import rate was decreased in tim22-19 and tim23-2 mutant mitochondria (Fig. S5 A). Despite the decreased rate of import, however, Yme1p was present at WT levels in tim22-19 and tim23-2 mitochondria (Fig. 7 A). In addition, the rate of Yme1p into tim54 mutant mitochondria was impaired (Fig. S5 A), but Yme1 was detected in mutant tim54 mitochondria (Fig. 2 A and Fig. 5C). Finally, overexpression of Tim22p restored the rate of import (Fig. S5 A). Whereas Yme1p import is impaired, Yme1p still accumulates to levels that are detectable in WT mitochondria.
Figure 7.
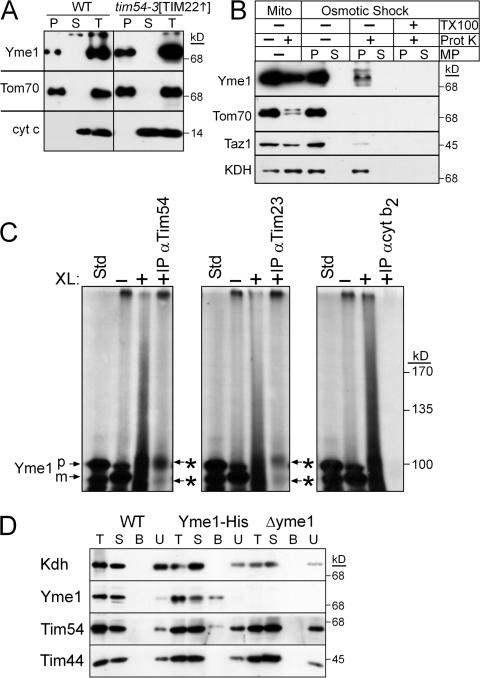
Tim54p is required for Yme1p assembly. (A) Increasing amounts of a mitochondrial protein lysate (50, 100, 150 μg) from the WT, tim22-19, and tim23-2 strains were separated by SDS-PAGE and immunoblotted with antibodies against Yme1p, porin, and AAC. (B) Mitochondria from the parent (WT), and tim22-19, tim23-2, tim54-3, Δtim54, Δyme1, Δtim8 Δtim13, tim10-1, tim12-1, and Δtim18 strains were solubilized with 1% digitonin. The lysate was separated on a 5–10% blue-native gel, and Yme1p was detected by immunoblotting. Yme1p assembles in a large complex indicated by the arrow. (C) Yme1p assembly was investigated in WT, tim54-3, Δtim54, and tim54-3[TIM22] (overexpressing Tim22p) mitochondria. Note that the gel was overexposed and migration of unassembled Yme1p can be detected. (D) Yme1p was imported as in Fig. 5 D into WT, tim23-2, and Δtim54 mitochondria. Assembly was monitored on blue-native gels. The top arrow indicates assembled Yme1p, and the bottom arrow marked by an asterisk indicates a potential assembly intermediate.
Yme1p assembles into a large complex with a predicted molecular weight of ∼1 MDa (Klanner et al., 2001). Using blue-native gel analysis, we tested whether Yme1p assembled in mitochondria defective in Tim54p function. Mitochondria were solubilized in 1.0% (wt/vol) digitonin and separated on a 5–10% blue-native gel (Fig. 7 B). In WT, tim22-19, tim23-2, Δtim8 Δtim13, tim10-1, tim12-1, and Δtim18 mitochondria, Yme1p assembled into a large complex migrating well above the highest molecular weight standard, consistent with the predicted molecular weight of 1 MDa obtained by size-exclusion chromatography (Klanner et al., 2001). Yme1p thus assembled in mutant tim22 and tim23 mitochondria. In addition, Yme1p assembly was not impaired in mitochondria lacking functional small Tim proteins or Tim18p. In contrast, Yme1p assembly was impaired in tim54-3 and Δtim54 mitochondria as well as tim54-3 mitochondria with overexpressed Tim22p (Fig. 7 C); when the gel was overexposed to film, a signal at the molecular mass of 80 kD—that of unassembled Yme1p (Yme1p monomer)—was detected (Fig. 7 C). We followed the assembly of Yme1p by coupling import assays and blue-native gels (Fig. 7 D). As expected, assembledYme1p accumulated into a high molecular weight complex in WT and mutant tim23-2 mitochondria but not in Δtim54 mitochondria, even after import for 20 min. Note that the small amount of Yme1p that is detected in the Δtim54 mitochondria accumulates in both the presence and absence of a membrane potential and might reflect a small amount of Yme1p that can assemble transiently in the import assay, but fails to accumulate in vivo (Fig. 7 C). Together, these data indicate that Tim54p is required for the assembly of Yme1p.
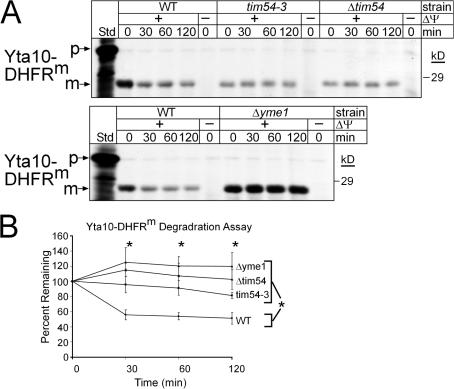
To confirm that Yme1p was not functional in mutant tim54 mitochondria, we investigated the import and degradation of a Yme1p model substrate, Yta10-DHFRm (Leonhard et al., 1999), which consists of a fusion between the N-terminal 161 amino acids of Yta10p and a loosely folded mutant of DHFR (Fig. 8, A and B). Importantly, the Yta10-DHFRm construct could be imported into WT, tim54-3, Δtim54, and Δyme1 mitochondria, although import was decreased in the tim54 mutant mitochondria (Fig. 8 A). To test if Yme1p was assembled into a proteolytically active complex, we investigated the degradation rate of imported Yta10-DHFRm in the presence of an ATP-regenerating system at 37°C (Leonhard et al., 1999). Whereas ∼50% of the Yta10-DHFRm was degraded in WT mitochondria, Yta10-DHFRm was essentially stable in mitochondria lacking functional Tim54p (even when Tim22p was overexpressed) and Yme1p (Fig. 8, A and B). Thus, Tim54p is required for assembly of a proteolytically active Yme1p complex.
Figure 8.
Tim54p mediates assembly of Yme1p into a proteolytically active complex. (A) Radiolabeled Yta10-DHFRm was imported into WT, tim54-3, Δtim54, and Δyme1 mitochondria and non-imported precursor was removed by protease treatment. The mitochondria were then incubated in the presence of an ATP- regenerating system at 37°C, and aliquots were removed at the indicated times. The samples were extracted with carbonate and the pellets were separated by SDS-PAGE and visualized by fluorography. (n = 3) (B) The rate of proteolysis from the time course in A from three independent assays was quantitated using a BioRad FX Molecular Imager and the affiliated Quantity 1 software. The amount of Yta10-DHFRm remaining at each chase time point is expressed as the percentage of the amount detected for each strain at t = 0 (set at 100%; mean ± SD, n = 3). The asterisks indicate the statistical significance for each of the mutant mitochondria relative to WT mitochondria, but not each other, at P ≤ 0.002 by multiple comparison procedures at each time point of the chase.
The observed impairment in Yme1p assembly in tim54 mitochondria may be caused by a mislocalization of Yme1p within tim54 mitochondria (Fig. 9). Initially, Yme1p localization was investigated by carbonate extraction and osmotic shock in the presence and absence of proteinase K in Δtim54 and tim54-3 mitochondria. However, Δtim54 and tim54-3 mitochondria were resistant to osmotic shock (unpublished data). We reasoned that the composition of the inner membrane might be altered, so we used the tim54-3[TIM22] mitochondria in which TIM22 was overexpressed. Indeed, these mitochondria were amenable to osmotic shock. As expected, Yme1p was an integral membrane protein because it was recovered in the pellet fraction like the integral membrane protein Tom70p after carbonate extraction (Fig. 9 A). To confirm that Yme1p was not mislocalized to the mitochondrial matrix, we used osmotic shock, which ruptures the mitochondrial outer membrane, in the presence and absence of protease to determine the location of Yme1p (Fig. 9 B; the control reaction in WT mitochondria are presented in Fig. S5 B). As expected, Yme1p and Taz1p (Claypool et al., 2006) localized to the mitochondrial intermembrane space because both were degraded by the added protease upon osmotic shock. In contrast, the matrix marker α-ketoglutarate dehydrogenase (Kdh) was protected from protease because the inner membrane remained intact and the outer membrane marker Tom70p was protease susceptible in both mitochondria and mitoplasts (MP). Yme1p thus resides in the intermembrane space in tim54-3[TIM22] mutant mitochondria.
Figure 9.
Yme1 is imported into the intermembrane space and interacts directly with Tim54p. (A) Mitochondria from the WT and tim54-3[TIM22] were subjected to alkali extraction by incubation in 100 mM Na2CO3, pH 10.9, for 30 min on ice followed by centrifugation to separate the pellet (P) from the supernatant (S). As a control, an equal volume of the lysate is included (T). (B) From the tim54-3[TIM22] strain, intact mitochondria (Mito) or mitochondria subjected to osmotic shock (mitoplasts designated MP) were incubated alone or in the presence of 10 μg/ml proteinase K (Prot K). The mitoplasts (P) were separated from the soluble intermembrane space contents (S) by centrifugation. As a control, a treatment with Triton X-100 (TX-100) has been included in the osmotic shock reaction. Equivalent amounts of each sample were resolved by SDS-PAGE and immunoblotted for Yme1p, Tom70p (outer membrane), Taz1p (intermembrane space), and Kdh (matrix). (C) Yme1p was imported into WT mitochondria for 10 min at 25°C (–XL) and an aliquot was removed. Proteins were cross-linked (+XL) by the addition of 0. 5 mM dithio-bis(succinimidyl propionate) (DSP) for 30 min followed by quenching with 0.1 M Tris-HCl. After an aliquot was removed, mitochondria were subsequently solubilized followed by immunoprecipitation with antibodies against Tim54p, Tim23p, and the negative control cytochrome b 2. Because cross-linked Yme1p migrates at a high molecular mass, cross-links were released with β-mercaptoethanol addition in the sample buffer. The asterisks indicate cross–linked Yme1p that coimmunoprecipitated with the Tim54p and Tim23p antibodies. (D) Mitochondria from a strain expressing a C-terminal hexahistidine-tagged Yme1 (Yme1-His) as well the WT and Δyme1 strains were solubilized at 2.5 mg/ml in 1% digitonin, 50 mM NaCl, 20 mM imidazole, and 10% glycerol supplemented with protease inhibitors (T). The soluble fraction obtained by centrifugation and 100 μg of the extract (S) was incubated with Ni2+-agarose beads. The beads were washed, and bound proteins (B) were eluted with SDS-PAGE sample buffer. To assess the effectiveness of binding, 100 μg of the unbound protein fraction (U) was also included. Proteins were analyzed by immunoblotting with antibodies against Kdh, Yme1, Tim54p, and Tim44p.
Because of Tim54p's role in Yme1p assembly, we predicted that Tim54p might transiently interact with Yme1p to facilitate assembly. We used a chemical cross-linking/immunoprecipitation approach coupled with the in vitro import assay (Koehler et al., 1998a) to trap an interaction between Tim54p and imported Yme1p (Fig. 9 C). Specifically, when the cross-linked partners were reduced with β-mercaptoethanol before separation by SDS-PAGE, cross-linked Yme1p was co-immunoprecipitated with antibodies against Tim54p and Tim23p. The interaction was specific because antibodies against the intermembrane space protein cytochrome b 2 failed to immunoprecipitate cross-linked Yme1p. The addition of reductant was required to detect the interaction because the cross-linked product migrated at a large molecular mass. Using another approach, we constructed a GA74 strain with a hexa-histidine tag on Yme1p (Yme1-His; Fig. 9 D); this strain grew like the WT strain, indicating that the tag did not disrupt Yme1p function. After solubilization of mitochondria, a small amount of the Tim54p, but neither Tim44p nor Kdh, interacted with tagged-Yme1p. In control reactions with WT and Δyme1 mitochondria, Tim54p did not copurify. Additional partner proteins may assist Tim54p with Yme1p assembly, because in a genetic approach, overexpression of Yme1p did not complement the petite-negativity or growth phenotype in the tim54-3 mutant (unpublished data). These studies—import/coimmunoprecipitation assays and in organello complex purification—indicate that a fraction of Tim54p interacts directly with Yme1p.
Collectively, this investigation shows that Tim54p, possibly with assistance of additional components (Dunn et al., 2005), is required for the assembly of Yme1p into a functional complex after import via the TIM23 pathway. Thus, Tim54p, a component of the TIM22 translocon, integrates functions of the two inner membrane translocases by providing an assembly activity that is independent from the translocation properties of the TIM22 translocon. As such, Tim54p serves as a link between the import, assembly, and proteolysis pathways in the mitochondrion.
Discussion
Tim54p does not play a direct role in protein import but serves as a scaffold for the TIM22 inner membrane complex
In this study, we analyzed the function of Tim54p in mitochondrial biogenesis. Previous experiments suggested that Tim54p was an essential protein of the 300-kD TIM22 translocon of the mitochondrial inner membrane. Tim22p and Tim54p coimmunoprecipitated and TIM22, expressed from a multicopy plasmid, suppressed a growth defect in a tim54-1 mutant strain (Kerscher et al., 1997). We therefore expected Tim54p to play a direct role in protein translocation. However, more recent studies suggested that Tim54p might have other functions in mitochondrial biogenesis. Specifically, TIM54 was not essential for viability and a functional translocon consisting of only Tim22p was subsequently purified (Kovermann et al., 2002). Moreover, a translocation intermediate between a TIM22 pathway substrate and Tim54p has not yet been demonstrated (Leuenberger et al., 1999). These studies suggest that, although an integral subunit of the TIM22 complex, Tim54p most likely functions in an alternative aspect of mitochondrial biogenesis.
This study confirms that Tim54p does not play a direct role in import of the known TIM22 translocon substrates. In vitro import of Tim23p and Hsp60, however, was decreased in Δtim54 and tim54-3 mitochondria. This decrease in import is potentially caused by a decrease in Tim22p and Tim23p levels (Fig. 4 A). Indeed, Tim54p is required for stabilization of Tim22p, even though a functional translocon can be assembled in the absence of Tim54p (Kerscher et al., 1997; Kovermann et al., 2002). Specifically, overexpression of TIM22 from a high-copy plasmid restored growth on rich glucose medium and the import of TIM22 substrates (Fig. 5 A). Tim54p therefore serves as a scaffold/assembly factor for the TIM22 complex, potentially by stabilizing the 300-kD TIM22 complex in the inner membrane.
Tim54p is required for Yme1p assembly and functions in a new intermembrane space biogenesis pathway
However, overexpression of TIM22 did not restore the petite-negativity of the Δtim54 and tim54-3 strains. Rather, our studies show that Tim54p plays a direct role in the assembly of a proteolytically active Yme1p complex. Two specific defects associated with mitochondria lacking functional Tim54p that are not observed with mitochondria defective in other components of the TIM22 pathway are that tim54 mutant mitochondria (1) require mitochondrial DNA for viability and (2) are defective in Yme1p assembly into a large functional complex. Thus, Tim54p seems to play a direct role in Yme1p biogenesis independent from the classical function assigned the TIM22 translocon, namely, translocation and insertion of polytopic membrane proteins into the inner membrane.
In addition to mutations in yme1, several defects in mitochondrial function contribute to petite-negativity in yeast. Other components involved in mitochondrial biogenesis are inviable when the mitochondrial genome is lost in particular genetic backgrounds (Dunn and Jensen, 2003; Senapin et al., 2003); certain strains lacking functional Tim18p, Tom70p, and Tim9p cannot tolerate loss of the mitochondrial genome. For Tom70p and Tim18p, Jensen and colleagues suggested that the decreased import efficiency in combination with a decreased membrane potential resulted in lethality when the mitochondrial genome was absent and overexpression of cytosolic proteins that improved import efficiency could suppress the defect (Dunn and Jensen, 2003); these studies suggest that the pathways to maintain a membrane potential are obviously complex. Surprisingly, overexpression of TIM22 in tim54 mutant mitochondria did not suppress the petite-negativity or the respiratory deficiency at 37°C. In addition, overexpression of CCT6 (Dunn and Jensen, 2003) suppressed petite-negativity in the Δtim18 strain, but not tim54-3. These results argue that the petite-negativity caused by a defect in tim54 is not the result of a combined impairment in import efficiency and a decrease in membrane potential. AAC function and abundance also is critical for maintaining a membrane potential in the absence of a mitochondrial genome (Contamine and Picard, 2000). Because AAC depends on the TIM22 import pathway for biogenesis and AAC levels are lower in the Δtim54 and tim54-3 mutants, defects in AAC function may contribute to petite- negativity in these mutant strains. However, because our tim22, tim9, and tim10 mutants, all with severely lowered levels of AAC, are petite-positive (Fig. 1 B and Fig. 3 C) and AAC levels are increased when TIM22 is overexpressed in the tim54-3 mutant, our data that tim54 mutants are petite-negative support the postulation that Tim54p has an alternative function in mitochondrial biogenesis, namely assembly of Yme1p.
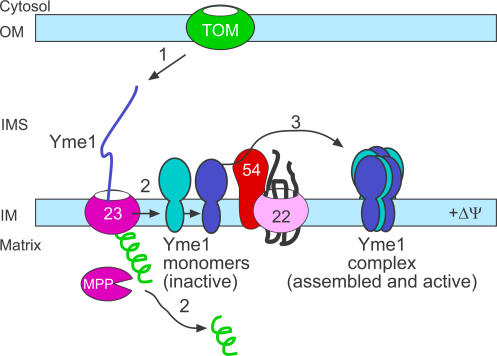
We therefore propose the following model in which Tim54p mediates Yme1p assembly (Fig. 10). In step 1, Yme1p is imported via the TIM23 translocon. After cleavage of the presequence by the matrix processing protease, the monomer is released into the inner membrane (step 2). Subsequently, Tim54p mediates assembly of the Yme1p monomers into a functional complex (step 3). Indeed, in mitochondria defective in Tim54p function, the Yme1 monomer is detected in blue-native gels (Fig. 7 C). Our cross-linking studies in which Tim54p binds to both processed and unprocessed Yme1 suggests that the TIM22 and TIM23 translocons are proximally associated in the inner membrane. In addition, Tim54p and Tim23p may be simultaneously interacting with Yme1p in transit; Tim54p might serve as a tether for the Yme1 substrate as it enters the intermembrane space, similar to a role for Yme1p in the import of PNPase (Rainey et al., 2006). Our studies suggest that the interaction between Tim54p and Yme1p is transient because freezing a stable interaction required cross-linking and only a small fraction of the Tim54p interacted with Yme1p-His. We suggest that Tim54p is a bonafide member of the TIM22 complex, because Tim54p comigrates in the 300-kD complex with Tim22p in blue-native gels and coimmunoprecipitates with Tim22p (Leuenberger et al., 1999; Kovermann et al., 2002). However, it is formally possible that Tim54p assembles in other complexes (of similar size on blue-native gels) and might partner with additional proteins to assist in the assembly of Yme1p independent of its association with the TIM22 complex. As an example, a recent study by Jensen and colleagues suggests that Mgr1p might be such a candidate because deletion of MGR1 resulted in a defect in Yme1p assembly (Dunn et al., 2005); however, Yme1p still assembled into a high molecular weight complex, in contrast to our studies in which Yme1p completely failed to assemble.
Figure 10.
A schematic showing the import and assembly pathway of Yme1p. Details are presented in the Discussion.
Our present study shows that Tim54p functions in a new pathway for assembly of Yme1p. Because Tim54p is a component of the TIM22 complex and Yme1p is imported via the TIM23 translocon, Yme1p, after import via the TIM23 pathway, requires an assembly activity provided by Tim54p of the TIM22 translocon for generating a functional complex. Thus, Tim54p functions to integrate the activities of the two major translocons of the mitochondrial inner membrane. Moreover, given that Yme1p is the i-AAA protease involved in basic quality control of the inner membrane, Tim54p functionally links pathways of protein import, assembly, and turnover within the mitochondrion. Does this pathway function in other organisms? Tim54p homologues have been identified in other fungi, but close homologues have not been identified in higher organisms including worms, fly, mouse and humans. However, the SIMAP database (Similarity of Matrix Proteins, http://boinc.bio.wzw.tum.de/boincsimap/; Rattei et al., 2006) suggests that Tim54p is similar to a mitochondrial multi-substrate lipid kinase (identity 21%, similarity 41%; Van Overloop et al., 2006), suggesting that Tim54p might have multiple functions. Additional investigations into the mammalian TIM22 pathway will be required to identify the true components.
Materials and methods
Plasmids and strains
Standard techniques were used for growth, manipulation and transformation of yeast strains (Guthrie and Fink, 1991; Jarosch et al., 1996; Gietz and Sugino, 1988). Details on the strains used in this study are listed in Table S1. A 1,830-bp fragment containing the TIM54 gene and its promoter was cloned into pYCPlac33, using the KpnI–XbaI sites, to form pCTIM54:URA3. The yeast strain deleted for TIM54 (Δtim54) was constructed as previously described by plasmid shuffling (Kovermann et al., 2002). The gene for TIM54 was replaced with the HIS3 gene flanked by the TIM54 promoter and terminator regions by homologous recombination. The haploid yeast strain containing the disruption and pCTIM54:URA3 was grown at 25°C on minimal media with 2% glucose and 5-fluoroortic acid (5-FOA) to select for loss of the plasmid. The Δyme1∷KANMX was disrupted in strain BY4742 (ResGen) and the Δyme1 strain was generated by deletion of YME1 with HIS3MX in GA74. The Yme1-His strain was generated by integrating a hexahistindine tag in frame to the C terminus of Yme1 in strain GA74. The ts strains tim22-4 has been described previously and tim22-19 was isolated from the same study (Yaffe and Schatz, 1984; Leuenberger et al., 1999). The tim23-2 strain was provided by Dr. Pfanner (University of Freiburg, Freiburg, Germany; Dekker et al., 1997), and the Yta10-DHFRm construct was provided by Dr. Langer (University of Cologne, Cologne, Germany; Leonhard et al., 1999). For in vitro transcription/translation, the DNA fragments encoding the substrates were cloned into pSP65 (Promega).
A ts tim54-3 strain was constructed using error prone PCR as described previously (Leuenberger et al., 2003). Amplified tim54 and gapped pRS314 (CEN, TRP1) were co-transformed into the Δtim54 strain. After auxotrophic selection, the mutant tim54 plasmid was selected by plasmid shuffling in the presence of 5-FOA. Resulting colonies were then screened for ts growth arrest at 37°C. The mutant tim54:TRP1 plasmid was recovered and used to reconstruct the ts mutant, confirming that the ts phenotype was plasmid dependent. For microscopy experiments, plasmids containing Su-GFP fusion were transformed into the WT, Δtim54, and tim54-3 strains.
Import of radiolabeled proteins into isolated mitochondria and manipulation of mitochondria
Mitochondria were purified from yeast cells grown in YPEG at 25°C (Glick and Pon, 1995) and assayed for protein import as previously described (Glick et al., 1992; Rospert and Schatz, 1998). Proteins were synthesized in a rabbit reticulocyte lysate in the presence of [35S]-methionine after in vitro transcription of the corresponding gene by SP6 or T7 polymerase. The reticulocyte lysate containing radiolabeled precursor was incubated at 25°C with isolated mitochondria in import buffer (1 mg/ml bovine serum albumin, 0.6 M sorbitol, 150 mM KCl, 10 mM MgCl2, 2.5 mM EDTA, 2 mM ATP, 2 mM NADH, and 20 mM Hepes-KOH, pH 7.4). Where indicated, the potential across the mitochondrial inner membrane was dissipated using 1 μM valinomycin and 25 μM FCCP. Non-imported radiolabeled precursor was removed by treatment with 100 μg/ml trypsin or 50 μg/ml proteinase K for 15–30 min on ice. Trypsin was inhibited with 400 μg/ml soybean trypsin inhibitor and proteinase K with 1 mM phenylmethylsulfonyl fluoride (PMSF).
For osmotic shock treatment to disrupt the mitochondrial outer membrane, the mitochondria were pelleted by centrifugation, suspended to 1 mg/ml in breaking buffer (0.6 M sorbitol and 20 mM Hepes–KOH, pH 7.4) and then diluted 20-fold with 20 mM Hepes-KOH, pH 7.4, and incubated on ice in the presence or absence of 10 μg/ml proteinase K. Mitoplasts were collected by centrifugation at 21,000 g for 10 min. For alkali extraction, the mitochondria were pelleted by centrifugation, suspended to 0.1 mg/ml in 0.1 M Na2CO3, and incubated for 30 min on ice (Fujiki et al., 1982). The membrane fraction was collected by centrifugation at 21,000 g for 15 min.
The degradation assay for Yta10-DHFRm was performed as described previously (Leonhard et al., 1999; Dunn et al., 2005). After import of Yta10-DHFRm into mitochondria, non-imported precursor was removed by protease treatment. Mitochondria were then incubated at 37°C in the presence of an energy regenerating system (200 μg/ml creatine phosphokinase and 10 mM creatine phosphate) and aliquots were removed over a 60-min time period. Samples were subjected to alkali extraction and separated by SDS-PAGE. Data was collected using a BioRad FX Molecular Imager and the affiliated Quantity 1 software. Data from three independent assays was pooled and statistical comparisons were performed using SigmaStat 3 software (Jandel Corp.).
Blue-native gel analysis and immunoprecipitation assays
Mitochondria (2.5 mg/ml) were solubilized in 20 mM Hepes–KOH, pH 7.4, 50 mM NaCl, 10% glycerol, 2.5 mM MgCl2, 1 mM EDTA, and 1% digitonin for 30 min on ice. Insoluble material was removed by centrifugation at 21,000 g for 15 min. Solubilized proteins were analyzed by blue-native gel electrophoresis on the linear polyacrylamide gels as indicated in the figure legends (Schägger et al., 1994). After transfer to polyvinylidene fluoride membranes, proteins were detected by immunoblotting with the indicated primary antibodies. Detection was performed either using HRP-conjugated secondary antibodies and ECL (Pierce Chemical Co.) or [125I]-protein A and autoradiography. Cross-linking reactions and immunoprecipitation assays were done as reported previously (Leuenberger et al., 1999).
Microscopy
Yeast strains were grown at their permissive temperature to mid-log phase in YPD and then incubated with 2 μg/ml DAPI for 15 min at room temperature (Hobbs et al., 2001). The cells were then washed in water, immobilized on poly-lysine/concanavalin A coated coverslips, and mounted with the Prolong AntiFade kit (Molecular Probes). The cells were visualized at 25°C immediately on a Deltavision Spectrics Applied Precision model 52–000067-002 Olympus IX71 microscope. Images were taken at 0.2-μm steps through the sample using an oil immersion 100× NA1.35 objective. The fluorochromes were DAPI (visualized with the standard DAPI filter set), and GFP (visualized with the standard GFP filter set). Images were taken with the Photometric Coolsnap HQ camera using the Deltavision Softworx version 3.3.5 program. The images were processed with constrained iterative deconvolution at 10 iterations, using the software's proprietary algorithms. Representative sections containing mitochondria were converted to TIFF format and exported to Adobe Photoshop. In a time-course assay, cells were grown at 25°C to mid-log phase and then shifted to 37°C. Aliquots were removed at t = 0, 12, 18, and 24 h after shifting to 37°C and immediately visualized. Nucleoids were counted in at least 20 separate cells. Ten focal planes were analyzed for each cell.
Online supplemental material
Table S1 lists the strains that were used in this study. The supplemental figures include additional control experiments for the results. Fig. S1 presents growth curves for the tim54 mutant strains. Fig. S2 analyzes cold sensitivity and show that tim54 mutants are cold sensitive, which was not rescued by TIM22 overexpression. Fig. S3 shows the import of Hsp60 into tim54-3 [TIM22] mitochondria. Fig. S4 presents a growth analysis and shows that CCT6 overexpression suppresses the petite-negativity of Δtim18 but not tim54-3. Fig. S5 control reactions for the import of Yme1 (import is decreased, but not absent, in tim54 mutant mitochondria.) and localization of Yme1 during osmotic shock in WT mitochondria. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200706195/DC1.
Supplementary Material
Acknowledgments
We kindly thank Dr. Peter Thorseness for antibodies against Yme1p, Dr. Thomas Langer for the Yta10–DHFRm construct, Dr. Nikolaus Pfanner for the tim23-2 mutant, and Dennis Wong, David Wong, and James Yen for excellent technical assistance.
This work was supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation (DRS18), Burroughs Wellcome Fund New Investigator in the Toxicological Sciences (1001120), the Arnold and Mabel Beckman Foundation, the American Heart Association, and the National Institutes of Health (R01 GM61721). C.M. Koehler is an American Heart Established Investigator. D.K. Hwang and H.L. Tienson are funded by the United States Public Health Service National Research Service Award (GM07185). S.M. Claypool was an American Heart Postdoctoral Fellow.
D. Leuenberger's present address is Department of Biology and Howard Hughes Medical Institute, Stanford University School of Medicine, Stanford, CA 94305-5439.
Abbreviations used in this paper: TIM, translocons of the inner membrane; TOM, translocons of the outer membrane; WT, wild type.
References
- Arnold, I., and T. Langer. 2002. Membrane protein degradation by AAA proteases in mitochondria. Biochim. Biophys. Acta. 1592:89–96. [DOI] [PubMed] [Google Scholar]
- Beilharz, T., C.K. Suzuki, and T. Lithgow. 1998. A toxic fusion protein accumulating between the mitochondrial membranes inhibits protein assembly in vivo. J. Biol. Chem. 273:35268–35272. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., A. Davies, G.J. Cost, E. Caputo, J. Li, P. Hieter, and J.D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Bulder, C.J.E.A. 1964. Induction of petite mutation and inhibition of respiratory enzymes in various yeasts. Antonie Van Leeuwenhoek. 30:1–9. [DOI] [PubMed] [Google Scholar]
- Chen, X.J., and G.D. Clark-Walker. 2000. The petite mutation in yeasts: 50 years on. Int. Rev. Cytol. 194:197–238. [DOI] [PubMed] [Google Scholar]
- Claypool, S.M., J.M. McCaffery, and C.M. Koehler. 2006. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J. Cell Biol. 174:379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine, V., and M. Picard. 2000. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64:281–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker, P.J., F. Martin, A.C. Maarse, U. Bomer, H. Muller, B. Guiard, M. Meijer, J. Rassow, and N. Pfanner. 1997. The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J. 16:5408–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, C.D., and R.E. Jensen. 2003. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics. 165:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, C.D., M.S. Lee, F.A. Spencer, and R.E. Jensen. 2005. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol. Biol. Cell. 17:213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi, B. 1953. Nucleo-Cytoplasmic Relations in Microorganisms, Their Bearing on Cell Heredity and Differentiation. Claredon Press, Oxford. 127 pp.
- Francis, B.R., K.H. White, and P.E. Thorsness. 2007. Mutations in the Atp1p and Atp3p subunits of yeast ATP synthase differentially affect respiration and fermentation in Saccharomyces cerevisiae. J. Bioenerg. Biomembr. 39:127–144. [DOI] [PubMed] [Google Scholar]
- Fujiki, Y., A.L. Hubbard, S. Fowler, and P.B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534. [DOI] [PubMed] [Google Scholar]
- Giraud, M.F., and J. Velours. 1997. The absence of the mitochondrial ATP synthase delta subunit promotes a slow growth phenotype of rho-yeast cells by a lack of assembly of the catalytic sector F1. Eur. J. Biochem. 245:813–818. [DOI] [PubMed] [Google Scholar]
- Glick, B.S., and L. Pon. 1995. Isolation of highly purified mitochondria from S. cerevisiae. Methods Enzymol. 260:213–233. [DOI] [PubMed] [Google Scholar]
- Glick, B.S., A. Brandt, K. Cunningham, S. Muller, R.L. Hallberg, and G. Schatz. 1992. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 69:809–822. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA. 933 pp.
- Hanekamp, T., M.K. Thorsness, I. Rebbapragada, E.M. Fisher, C. Seebart, M.R. Darland, J.A. Coxbill, D.L. Updike, and P.E. Thorsness. 2002. Maintenance of mitochondrial morphology is linked to maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Genetics. 162:1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines, V., and G. Schatz. 1993. Precursor binding to yeast mitochondria. A general role for the outer membrane protein Mas70p. J. Biol. Chem. 268:449–454. [PubMed] [Google Scholar]
- Hines, V., A. Brandt, G. Griffiths, H. Horstmann, H. Brutsch, and G. Schatz. 1990. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 9:3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs, A.E., M. Srinivasan, J.M. McCaffery, and R.E. Jensen. 2001. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152:401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch, E., G. Tuller, G. Daum, M. Waldherr, A. Voskova, and R.J. Schweyen. 1996. Mrs5p, an essential protein of the mitochondrial intermembrane space, affects protein import into yeast mitochondria. J. Biol. Chem. 271:17219–17225. [DOI] [PubMed] [Google Scholar]
- Kerscher, O., J. Holder, M. Srinivasan, R.S. Leung, and R.E. Jensen. 1997. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 139:1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., N.B. Sepuri, and R.E. Jensen. 2000. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell. 11:103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klanner, C., H. Prokisch, and T. Langer. 2001. MAP-1 and IAP-1, two novel AAA proteases with catalytic sites on opposite membrane surfaces in mitochondrial inner membrane of Neurospora crassa. Mol. Biol. Cell. 12:2858–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C.M. 2004. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20:309–335. [DOI] [PubMed] [Google Scholar]
- Koehler, C.M., E. Jarosch, K. Tokatlidis, K. Schmid, R.J. Schweyen, and G. Schatz. 1998. a. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 279:369–373. [DOI] [PubMed] [Google Scholar]
- Koehler, C.M., S. Merchant, W. Oppliger, K. Schmid, E. Jarosch, L. Dolfini, T. Junne, G. Schatz, and K. Tokatlidis. 1998. b. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 17:6477–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C.M., M.P. Murphy, N. Bally, D. Leuenberger, W. Oppliger, L. Dolfini, T. Junne, G. Schatz, and E. Or. 2000. Tim18p, a novel subunit of the inner membrane complex that mediates protein import into the yeast mitochondrial inner membrane. Mol. Cell. Biol. 20:1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominsky, D.J., and P.E. Thorsness. 2000. Expression of the Saccharomyces cerevisiae gene YME1 in the petite-negative yeast Schizosaccharomyces pombe converts it to petite-positive. Genetics. 154:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominsky, D.J., M.P. Brownson, D.L. Updike, and P.E. Thorsness. 2002. Genetic and biochemical basis for viability of yeast lacking mitochondrial genomes. Genetics. 162:1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovác, L. 1967. Biochemical genetics of oxidative phosphorylation. Science. 158:1564–1567. [DOI] [PubMed] [Google Scholar]
- Kovermann, P., K.N. Truscott, B. Guiard, P. Rehling, N.B. Sepuri, H. Muller, R.E. Jensen, R. Wagner, and N. Pfanner. 2002. Tim22, the essential core of the mitochondrial protein insertion complex, forms a voltage-activated and signal-gated channel. Mol. Cell. 9:363–373. [DOI] [PubMed] [Google Scholar]
- Leonhard, K., A. Stiegler, W. Neupert, and T. Langer. 1999. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature. 398:348–351. [DOI] [PubMed] [Google Scholar]
- Leuenberger, D., N.A. Bally, G. Schatz, and C.M. Koehler. 1999. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 18:4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger, D., S.P. Curran, D. Wong, and C.M. Koehler. 2003. The role of Tim9p in the assembly of the TIM22 import complexes. Traffic. 4:144–152. [DOI] [PubMed] [Google Scholar]
- Meisinger, C., S. Pfannschmidt, M. Rissler, D. Milenkovic, T. Becker, D. Stojanovski, M.J. Youngman, R.E. Jensen, A. Chacinska, B. Guiard, et al. 2007. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 26:2229–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast. 8:79–82. [DOI] [PubMed] [Google Scholar]
- Murphy, M.P. 1997. Selective targeting of bioactive compounds to mitochondria. Trends Biotechnol. 15:326–330. [DOI] [PubMed] [Google Scholar]
- Murphy, M.P., D. Leuenberger, S.P. Curran, W. Oppliger, and C.M. Koehler. 2001. The essential function of the small Tim proteins in the TIM22 import pathway does not depend on formation of the soluble 70-kilodalton complex. Mol. Cell. Biol. 21:6132–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri, F., F. Bisaccia, L. Capobianco, V. Dolce, G. Fiermonte, V. Iacobazzi, C. Indiveri, and L. Palmieri. 1996. Mitochondrial metabolite transporters. Biochim. Biophys. Acta. 1275:127–132. [DOI] [PubMed] [Google Scholar]
- Paschen, S.A., and W. Neupert. 2001. Protein import into mitochondria. IUBMB Life. 52:101–112. [DOI] [PubMed] [Google Scholar]
- Rainey, R.N., J.D. Glavin, H.W. Chen, S.W. French, M.A. Teitell, and C.M. Koehler. 2006. A new function in translocation for the mitochondrial i-AAA protease Yme1: import of polynucleotide phosphorylase into the intermembrane space. Mol. Cell. Biol. 26:8488–8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattei, T., R. Arnold, P. Tischler, D. Lindner, V. Stumpflen, and H.W. Mewes. 2006. SIMAP: the similarity matrix of proteins. Nucleic Acids Res. 34:D252–D256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospert, S., and G. Schatz. 1998. Protein translocation into mitochondria. In Cell Biology: A Laboratory Handbook. Vol. 2. J.E. Celis, editor. Academic Press, San Diego. 277–285.
- Schägger, H., W.A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two- dimensional native electrophoresis. Anal. Biochem. 217:220–230. [DOI] [PubMed] [Google Scholar]
- Schmidt, C., T. Söllner, and R.J. Schweyen. 1987. Nuclear suppression of a mitochondrial RNA splice defect: nucleotide sequence and disruption of the MRS3 gene. Mol. Gen. Genet. 210:145–152. [DOI] [PubMed] [Google Scholar]
- Senapin, S., X.J. Chen, and G.D. Clark-Walker. 2003. Transcription of TIM9, a new factor required for the petite-positive phenotype of Saccharomyces cerevisiae, is defective in spt7 mutants. Curr. Genet. 44:202–210. [DOI] [PubMed] [Google Scholar]
- Sirrenberg, C., M.F. Bauer, B. Guiard, W. Neupert, and M. Brunner. 1996. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature. 384:582–585. [DOI] [PubMed] [Google Scholar]
- Sirrenberg, C., M. Endres, H. Folsch, R.A. Stuart, W. Neupert, and M. Brunner. 1998. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 391:912–915. [DOI] [PubMed] [Google Scholar]
- Slonimski, P.P., G. Perrodin, and J.H. Croft. 1968. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal “petites”. Biochem. Biophys. Res. Commun. 30:232–239. [DOI] [PubMed] [Google Scholar]
- Stojanovski, D., M. Rissler, N. Pfanner, and C. Meisinger. 2006. Mitochondrial morphology and protein import-A tight connection? Biochim. Biophys. Acta. 1763:414–421. [DOI] [PubMed] [Google Scholar]
- Truscott, K.N., K. Brandner, and N. Pfanner. 2003. Mechanisms of protein import into mitochondria. Curr. Biol. 13:R326–R337. [DOI] [PubMed] [Google Scholar]
- Van Dyck, L., and T. Langer. 1999. ATP-dependent proteases controlling mitochondrial function in the yeast Saccharomyces cerevisiae. Cell. Mol. Life Sci. 56:825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Overloop, H., S. Gijsbers, and P.P. Van Veldhoven. 2006. Further characterization of mammalian ceramide kinase: substrate delivery and (stereo)specificity, tissue distribution, and subcellular localization studies. J. Lipid Res. 47:268–283. [DOI] [PubMed] [Google Scholar]
- Waldherr, M., A. Ragnini, B. Jank, R. Teply, G. Wiesenberger, and R.J. Schweyen. 1993. A multitude of suppressors of group II intron-splicing defects in yeast. Curr. Genet. 24:301–306. [DOI] [PubMed] [Google Scholar]
- Weber, E.R., R.S. Rooks, K.S. Shafer, J.W. Chase, and P.E. Thorseness. 1995. Mutations in the mitochondrial ATP synthase gamma subunit suppress a slow-growth phenotype of yme1 yeast lacking mitochondrial DNA. Genetics. 140:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.P., and G. Schatz. 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA. 81:4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.